Abstract
Background
Sugarcane (Saccharum spp.) is predominantly an autopolyploid plant with a variable ploidy level, frequent aneuploidy and a large genome that hampers investigation of its organization. Genetic architecture studies are important for identifying genomic regions associated with traits of interest. However, due to the genetic complexity of sugarcane, the practical applications of genomic tools have been notably delayed in this crop, in contrast to other crops that have already advanced to marker-assisted selection (MAS) and genomic selection. High-throughput next-generation sequencing (NGS) technologies have opened new opportunities for discovering molecular markers, especially single nucleotide polymorphisms (SNPs) and insertion-deletion (indels), at the genome-wide level. The objectives of this study were to (i) establish a pipeline for identifying variants from genotyping-by-sequencing (GBS) data in sugarcane, (ii) construct an integrated genetic map with GBS-based markers plus target region amplification polymorphisms and microsatellites, (iii) detect QTLs related to yield component traits, and (iv) perform annotation of the sequences that originated the associated markers with mapped QTLs to search putative candidate genes.
Results
We used four pseudo-references to align the GBS reads. Depending on the reference, from 3,433 to 15,906 high-quality markers were discovered, and half of them segregated as single-dose markers (SDMs) on average. In addition to 7,049 non-redundant SDMs from GBS, 629 gel-based markers were used in a subsequent linkage analysis. Of 7,678 SDMs, 993 were mapped. These markers were distributed throughout 223 linkage groups, which were clustered in 18 homo(eo)logous groups (HGs), with a cumulative map length of 3,682.04 cM and an average marker density of 3.70 cM. We performed QTL mapping of four traits and found seven QTLs. Our results suggest the presence of a stable QTL across locations. Furthermore, QTLs to soluble solid content (BRIX) and fiber content (FIB) traits had markers linked to putative candidate genes.
Conclusions
This study is the first to report the use of GBS for large-scale variant discovery and genotyping of a mapping population in sugarcane, providing several insights regarding the use of NGS data in a polyploid, non-model species. The use of GBS generated a large number of markers and still enabled ploidy and allelic dosage estimation. Moreover, we were able to identify seven QTLs, two of which had great potential for validation and future use for molecular breeding in sugarcane.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-016-3383-x) contains supplementary material, which is available to authorized users.
Keywords: Saccharum spp, Polyploidy, SNPs, Molecular markers, Allelic dosage, Quantitative traits
Background
Sugarcane (Saccharum spp.) has a complex genome because of its variable ploidy level, frequent aneuploidy and large genome size of approximately 10 Gb [1–5]. This crop is a member of the Poaceae family and the Andropogoneae tribe, which includes maize and sorghum [6, 7]. Modern sugarcane cultivars are the result of interspecific crosses between the domesticated species Saccharum officinarum L. (2n = 80) and the wild species S. spontaneum L. (2n = 40–120), followed by several backcrosses with S. officinarum [6, 8]. These cultivars have chromosome numbers ranging from 100 to 130, are vegetatively propagated, and result from the selection of populations derived from outcrossing heterozygous parents [1, 9]. Sugarcane has a very high photosynthetic efficiency and is a crop with major economic importance in many tropical and subtropical countries primarily because of its use in the production of sugar and bioethanol [10–12].
Polyploidy, an important driver of plant evolution in natural populations, has played a crucial role in the domestication of crops such as wheat, sugarcane, cotton and potato [13–16]. Sugarcane is predominantly an autopolyploid plant, and the understanding of its genome organization is limited [4, 7]. One possible way to increase knowledge of the genome organization of this species is by using genetic maps. High-resolution genetic linkage mapping may be used for quantitative trait loci (QTL) studies in mapping poulations and also such as a first step toward potential marker-assisted selection (MAS) in plants [17–22]. Several genetic linkage maps of sugarcane have been generated since a methodology based on single-dose markers (SDMs) was proposed by Wu et al. [23]. SDMs that segregate 1:1 and 3:1 in full-sib progenies (F1 populations) [24] or 3:1 in populations created by selfing an individual are commonly used for constructing genetic maps in sugarcane [4, 25–35]. An integrated map of sugarcane with different types of molecular markers, such as microsatellites or single sequence repeats (SSRs) and target region amplification polymorphism (TRAP), extended the characterization of polymorphic variation throughout the entire genome [36–38]. However, in outcrossing heterozygous species such as sugarcane, for each segregating loci, different numbers of segregating alleles can exist, and a relative large number of markers is required to guarantee reasonable coverage of its genome [37, 39, 40].
Currently, high-throughput next-generation sequencing (NGS) technologies have provided new opportunities for discovering molecular markers, especially single nucleotide polymorphisms (SNPs), at the genome-wide level [41–43]. Some of these techniques, e.g., restriction-site associated DNA sequencing (RAD-seq) [44] and genotyping-by-sequencing (GBS) [42, 45, 46], employ a reduced genome representation that is achieved through restriction enzyme digestion, which could be particularly helpful for a complex genome such as that of a polyploid [47]. Moreover, these strategies can be used to species without reference sequence [48]. The GBS protocol has been widely used in a range of genetic studies in several species such as apple, barley, lettuce, switchgrass, maize, rice, wheat, and soybean [42, 45, 46, 49–55]. SNP datasets generated from GBS can be analyzed to detect associations between genotypes and phenotypes, perform diversity analyses, and construct genetic maps, among other applications [39, 56–58].
QTL mapping in sugarcane is a promising tool for characterizing the genetic architecture of several yield component traits of interest, such as sucrose yield, cane yield, stalk diameter, stalk height, stalk number, and stalk weight, as well as resistance to diseases, pests and abiotic stresses [10, 59–62]. Sugarcane is a semi-perennial crop with repeated measures data obtained for several harvests and locations, and QTL mapping studies are usually performed in two steps. First, adjusted phenotypic means are obtained; second, these means are searched for associations with molecular markers and/or along genetic maps [31, 32, 63]. Gazaffi et al. [62] proposed a method that considers an integrated genetic map in which QTL mapping is performed based on the advantages of the composite interval mapping (CIM) approach [64]. Briefly, a mapping model with three genetic effects is considered for genome scanning [62]. It is assumed that a QTL may also segregate in different patterns in progeny as a function of its genetic effects and of the linkage phase between markers and QTL alleles.
This study is the first to report on the development and application of GBS for mapping studies in sugarcane. Our objectives were to (i) establish a pipeline for identifying SNPs and insertion-deletion (indels) from GBS data in a sugarcane F1 population, (ii) construct the first GBS-based integrated genetic map with additional SSR and TRAP markers in this bi-parental mapping population, (iii) identify QTLs related to yield component traits based on the integrated genetic map, and (iv) perform annotation of the sequences that originated the associated markers with mapped QTLs to search putative candidate genes that may be involved in yield traits in sugarcane. We discuss these results in the context of where GBS is likely to be most useful in sugarcane crop development.
Methods
Mapping population and DNA extraction
The mapping population consisted of 151 full sibs derived from a commercial cross between the SP80-3280 (female parent) and RB835486 (male parent) sugarcane cultivars. The parents are broadly cultivated throughout Brazil because of their high biomass and sugar yields. SP80-3280 (SP71-1088 × H57-5028) was one of the cultivars with transcriptome sequencing performed previously by SUCEST [65] and RNA-seq [66] projects; its genome is currently being completely sequenced by the Brazilian initiative [67]. This cultivar is resistant to brown rust (Puccinia melanocephala), whereas RB835486 (L60-14 × ?) is susceptible to fungal disease. The parents have been used in studies of evolutionary relations in putative tandem gene duplication [68] and retrotransposon-based insertion polymorphisms [69]. Total genomic DNA samples from parents and progeny were extracted from the 1+ internode (leaf primordia) as proposed by Al-Janabi et al. [70], with modifications.
GBS-based markers
GBS was performed by the Institute for Genomic Diversity (Cornell University, Ithaca, NY, USA) according to the protocol described in detail by Elshire et al. [45]. Samples from both parents of the population were replicated three times for sequencing. Each individual within a library was part of a 96-plex reaction (including one blank sample each). To provide a higher sequence depth, libraries were obtained by digestion with PstI, a partially methyl-sensitive six-base-pair site restriction enzyme. Additionally, the 96-plex libraries were run in two distinct lanes each on a HiSeq™ 2000 platform (Illumina® Inc., San Diego, CA, USA).
To discover polymorphisms, we initially used the Tassel-GBS pipeline [71], which was implemented in Tassel software (v. 4.3.8). Because this pipeline requires a reference genome and because the complete sugarcane genome sequencing is in progress [67], we proposed the use of four alternative pseudo-references: (i) a methyl-filtered sugarcane genome (~674 Mb arranged in 1,109,444 scaffolds) [72], (ii) the Sorghum bicolor genome (v. 2.1; ~726 Mb arranged in 10 chromosomes and 1,600 unassembled scaffolds) [73], (iii) an RNA-seq sugarcane transcriptome (~780 Mb arranged in 119,768 transcripts) [66], and (iv) sequences from the SUCEST project (~152 Mb of a total of 237,954 sequences) [65]. The Bowtie2 (v. 2.2.1) algorithm was used to map the 64-bp-long tags against each reference with default parameters and the very sensitive-local argument. The exact reference and alternative sequence depths (read counts) were recorded in variant call format (VCF) files. To perform this task, we modified the GBS-Tassel pipeline to record a maximum value of 32,767 counts for each allele.
Allelic dosage estimation and marker curation
The GBS technique generated allele-specific read count data in the form D = {(x 1, y 1), (x 2, y 2), …, (x n, y n)} for each biallelic locus from individuals i = 1, 2, …, n. Data D from each locus were analyzed in SuperMASSA software [74]. As a prior quality control, markers with more than 25% missing data were filtered out. We also excluded GBS loci data with fewer than 50 read counts for the reference allele on average. In addition, individual data points with the radial coordinate smaller than (0.10) × max(r 1, r 2, …, r n) were removed.
All even-numbered ploidy levels ranging from 2 to 20 were tested [39]. The ploidy that returned the highest likelihood was selected after fitting a subjacent F1 segregation model into SuperMASSA. The replicated parental data provided additional constraints during estimation. Following the recommendation reported in Serang et al. [74] to find the maximum a posteriori (MAP) solution for the estimates, the SuperMASSA naive posterior report threshold was set to zero. Afterward, the values of individual posterior probability given the selected ploidy (6 through 14) were also calculated; these values indicated the maximum threshold that would allow individual assignment to a certain dosage cluster. Only ploidies ranging from 6 to 14 were selected because they were more likely to appear in the sugarcane genome and exhibited a greater number of SDMs [39].
For posterior quality control, we only selected SDMs for which the median of all individual posterior probabilities was higher than 0.80. The SuperMASSA dosage outputs were recoded for mapping purposes in R software by substituting the respective reference and alternative codominant alleles for a and b. Redundant loci within each reference and between references were inspected and excluded based on the recoded genotype calls. Here, only non-redundant loci were used for linkage mapping analysis. A circular plot was used to summarize the duplicate loci within and between references using the R circlize package [75].
Gel-based SSR and TRAP markers
A total of 120 SSR markers were genotyped in the 151 full-sib progeny and in the two parents. SSR markers were derived from both ESTs and genomic sequences. There were 98 EST-SSRs named SCA [76, 77], SCB [76], SCC [76, 78] and IISR [79], and 16 genomic SSRs named SMC [80] and CIR [81]. In addition, there were six SSR markers (named SB, Xtxp, CNL and SvPEPCAA [82–84]) from a genic sorghum library. PCRs were performed in a final volume of 20 μL as described by Oliveira et al. [76].
For TRAP markers four fixed and three arbitrary primers (named ARB1, ARB2 and ARB3) were used. The arbitrary primers were adapted of Li and Quiros [85]. Two fixed primers were designed from sucrose phosphate synthase (SuPS) [86, 87], and one primer each was designed from caffeic acid 3-O-methyltransferase (COMT) and cinnamoyl-CoA reductase (CCR) [88] gene sequences. PCRs were performed in a final volume of 20 μL [89].
Amplicons of SSR and TRAP markers were denatured at 90 °C for 3 min in an equal volume of loading buffer (formamide containing 0.8 mM EDTA and traces of bromophenol blue and xylene cyanol), snap-cooled on ice, and electrophoresed in 6% denaturing polyacrylamide gels in 1X TBE buffer. The samples were loaded on a dual vertical electrophoresis system (CBS Scientific) and were run at 75 W for 1 to 3 h depending on the fragment sizes to be separated. A 10-bp ladder was used as a standard size. The bands were visualized by silver staining according to Creste et al. [90].
Linkage map construction and homo(eo)logous group assignment
Linkage mapping analysis was performed over non-duplicated SDMs using the Onemap (v. 2.0-4) R package [91]. This analysis allowed simultaneous estimation of linkage and linkage phases between markers [92] and marker ordering using multipoint likelihood through hidden Markov models [93, 94] from a mixed set of different marker segregation patterns. The markers were coded according to the notation proposed by Wu et al. [92]. In brief, the codominant alleles were coded as a and b, while the null alleles were coded as o and treated as recessive alleles. GBS-based codominant markers were used to assess segregation with the following three cross types: ‘B3.7’ (ab × ab), ‘D1.10’ (ab × aa) and ‘D2.15’ (aa × ab). In addition, SSR and TRAP gel-based dominant markers were used to assess three more cross types that are traditionally used in integrated sugarcane maps: ‘C.8’ (ao × ao), ‘D1.13’ (ao × oo) and ‘D2.18’ (oo × ao). ‘D1’ and ‘D2’ stand for crosses in which the marker locus is heterozygous (and hence informative) only to SP80-3280 or to RB835486, respectively; they are both expected to segregate in a 1:1 ratio. ‘B3’ and ‘C’ stand for crosses in which the marker locus is heterozygous and symmetric in both parents; the former is expected to segregate in a 1:2:1 ratio, whereas the latter will segregate in a 3:1 ratio. Because SuperMASSA is able to predict parental genotypes using the population data even when parental data are missing, all B3-type markers could be recovered. However, D1- and D2-type markers were only recovered when read counts for at least one parental were available; with no parental data, these markers were discarded.
For gel-based markers, chi-square tests were conducted in R software according to the expected segregation ratios inferred through parental genotypes, and then p-values were corrected using false discovery rate control for non-dependent tests as implemented in the ‘p.adjusted’ R function. For GBS-based markers, segregation had already been considered during dosage estimation in SuperMASSA software according to the F1 model [74].
To obtain the genetic map, we first performed a two-point test to identify linkage groups (LGs). Any pairwise markers that showed a LOD Score > 9.0 and a recombination fraction < 0.10 were considered linked. Afterward, we applied ordering algorithms to each group. For the groups with less than six markers, the best order was obtained by performing an exhaustive search with the ‘compare’ function. For those groups with more than six markers, the ‘order.seq’ command was used, i.e., an initial set of the five most informative markers (preferentially B3- and C-type markers) was sampled for an exhaustive search. The best order was used as a frame for the consecutive inclusion of new markers. Once these groups were obtained, we used the ‘try.seq’ function to verify markers that were considered unlinked according to the initial procedure, and it was possible to integrate the pre-ordered groups. In this step, the following other markers were also tested: (i) markers at the ends of the LGs more than 20 centiMorgans (cM) far from the closest marker; and (ii) markers belonging to very small LGs (with sizes less than 1 cM or containing only two loci). As a final step, the LGs with more than five markers were refined using the ‘ripple’ algorithm within a sliding window of five markers. The ordered group heatmap plots were inspected visually, and manual correction was performed when needed throughout the map building process. The LGs were drawn in MapChart software [95]. The homo(eo)logous groups (HGs) were defined according to the sugarcane reference scaffolds shared by the GBS-based markers. Gel-based markers were also checked because they can produce different alleles that share the same primer pair.
Phenotypic data
The mapping population was planted in 2010 at two locations (Araras, located at 22°21′25″ S, 47°23′03″ W, and Ipaussu, located at 23°08′44″ S, 49°23′23″ W; both in the State of Sao Paulo, Brazil) and evaluated during three harvest years for several yield component traits, including sucrose content of cane (POL%C, in %), soluble solid content (BRIX, in °Brix), stalk diameter (SD, in mm) and fiber (FIB, in %). At each location, the experimental design consisted of an augmented randomized incomplete block design, which was fully replicated three times. For each trait, a multiple-harvest-location trial was considered under a mixed linear model approach for each yield component [10].
QTL mapping
The joint adjusted phenotypic means by location for each trait were used for QTL mapping. The QTL mapping methodology applied in this work was presented by Gazaffi et al. [62], and it expands the CIM method [64] to full-sib families. In brief, the model has three genetic effects, with two for additive effects (one for each parent) and one dominance effect (intra-loci interaction). To infer the conditional probabilities of QTL genotypes, multipoint probabilities were obtained using hidden Markov models at each 1 cM from the genetic map.
The mapping strategy was based on three steps. First, an interval mapping (IM) [96] search was carried out in order to select marker cofactors. The peaks with a LOD Score greater than 2 were sampled for inclusion in the QTL detection procedure. If the peak was not coincident with a marker, the closest one was considered as cofactor. Second, the QTL search was performed along the genome and considered the cofactors located outside the linkage group under analysis. To declare a QTL, the threshold for each search was obtained from 1,000 permutations with a significance level of 0.95 [97]. Finally, the peaks above the permutation threshold were fully characterized, i.e., the significance of each genetic effect was tested along with the linkage phase between markers and QTLs and the QTL segregation pattern. The proportions of phenotypic variance (R 2) as explained by each detected QTL were obtained for all the effects simultaneously. All the analyses were performed in R software [98].
Sequence annotation
Functional annotation of the regions of adjacent markers from the mapped QTLs was performed using sequence information from the scaffolds of the methyl-filtered sugarcane genome, sugarcane transcriptome from RNA-seq assembly, sequences from the SUCEST project and sequences with 400 nucleotides in length at both sides of the SNP/indel position for mapped markers from the sorghum genome. These scaffolds and sequences were annotated using Blast2GO software version 3.1 [99] on the non-redundant NCBI database with E values ≤ 1 × 10−3, and the Phytozome website [100] was used to align the data against the Viridiplantae protein databases.
Results
GBS-based marker polymorphism discovery
Short-read sequences were obtained from the mapping population and triple-replicated parents after double sequencing the 96-plex PstI libraries. Of the 330 million good barcoded reads, more than 3.1 million resulting tags were obtained for alignment against four different pseudo-references. Three of four pseudo-references originated from sugarcane DNA or RNA libraries. The methyl-filtered sugarcane genome resulted in the highest alignment rate, with 87.94% (2,729,457) aligned tags. Regarding RNA-based references, 38.53% (1,195,723) and 23.89% (741,537) tags were aligned with the RNA-seq transcriptome and SUCEST project sequences, respectively; however, their rates of non-unique alignment differed greatly (Additional file 1: Table S1). Finally, the reference for the close-relative genome of sorghum had 42.29% (1,312,661) aligned tags.
From 39,058 to 151,755 biallelic variants were identified, depending on the reference. Furthermore, for all the references, SNPs were identified more often than indels, at a 2.6:1 ratio on average (Table 1). With respect to SNPs, transitions (purine-purine or pyrimidine-pyrimidine interchanges) were identified 1.4 times more often than transversions (purine-pyrimidine interchanges). Approximately 12% of the markers with more than 25% missing data were filtered out per reference (Table 1) because current missing data imputation methods are not able to handle the complexity of the sugarcane genome. Low-coverage or ambiguous loci were also broadly present. At this stage of analysis, ~64% underrepresented loci were also excluded by considering a minimum of 50 read counts on average for the reference alleles. Although a large number of loci were removed, from 8,885 to 38,378 high-coverage loci, missing-filtered polymorphic loci were subjected to SuperMASSA quantitative genotyping analyses. The remaining redundancy was investigated only after these analyses.
Table 1.
Number of markers generated after GBS-Tassel pipeline analyses to map the GBS sugarcane population data
| GBS-Tassel pipeline pseudo-references | SNPs | Indels | Total | Excluded data | Filtered polymorphic sites | |
|---|---|---|---|---|---|---|
| Missing dataa | Low coverage locib | |||||
| Methyl-filtered sugarcane genome | 110,261 | 41,494 | 151,755 | 16,815 (11.1%) | 96,562 (63.6%) | 38,378 (25.3%) |
| Sorghum bicolor genome (v. 2.1) | 84,757 | 35,447 | 120,204 | 13,773 (11.5%) | 78,914 (65.6%) | 27,517 (22.9%) |
| RNA-seq sugarcane transcriptome | 73,275 | 26,778 | 100,053 | 11,809 (11.8%) | 63,658 (63.6%) | 24,586 (24.6%) |
| SUCEST project sequences | 29,238 | 9,820 | 39,058 | 4,878 (12.5%) | 25,295 (64.8%) | 8,885 (22.7%) |
Notes
a More than 25% of the population is missing data
b Less than 50 reads on average for the reference alleles
Ploidy and allelic dosage estimation
Ploidy levels ranging from 2 to 20 were evaluated with SuperMASSA software. Once the more likely ploidy was acknowledged, the software provided the individual posterior probability for each individual that was allocated in one of the expected dosage clusters. For the ploidy levels considered in these analyses, the number of loci varied within each ploidy class (Fig. 1), and an average of 10.7% loci were classified as having ploidies of 2 or 4, 60.3% as ploidies 6 through 14, and 29.0% as ploidies 16 through 20. Here, we used the same ad hoc criteria to classify each locus into one quality category based on their posterior probabilities for each ploidy; categories A and B included loci with either the highest or the sum of the two highest posterior probabilities that were greater than or equal to 0.80, respectively, and category C included all other cases [39]. Categories A, B and C represented 60.4%, 26.6% and 12.9% of the loci on average for all the ploidies, respectively (Fig. 1).
Fig. 1.
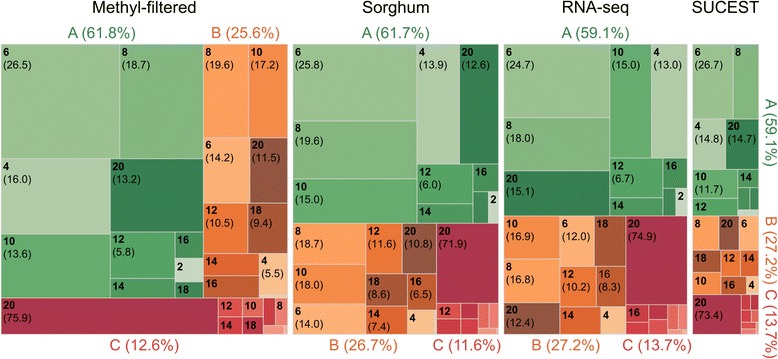
Mosaic plot showing the ploidy levels that produced the highest posterior probabilities for the mapping of GBS sugarcane population data considering the following four pseudo-references: the methyl-filtered sugarcane genome, Sorghum bicolor genome, RNA-seq sugarcane transcriptome and sequences from the SUCEST project. The areas of the rectangles are proportional to the number of loci that have the same ploidy level, as indicated within each rectangle in parentheses. According to the posterior probabilities calculated for each even-numbered ploidy level within a range from 2 to 20, each locus was classified into one category using the following ad hoc criteria: Category A (green), when the highest posterior probability was greater than or equal to 0.80; Category B (yellow), when no single value of the posterior probability was higher than 0.80 but the sum of the two highest ones was greater than or equal to 0.80; and Category C (red), which included all other cases. In parentheses: the number of loci as a percentage within the given ploidy level and category
For the linkage map construction, we selected the loci that were classified into category A and the ploidies ranging from 6 to 14, which represented 40.7% of loci on average from the total input in SuperMASSA or from 3,433 to 15,906 markers depending on the reference (Table 2). In addition, we characterized these remaining good-quality loci according to dosage. SDMs and multi-dose markers (MDMs) were equally represented by GBS, with approximately 50% on average for each one. As a final quality control analysis of the GBS data, we selected the loci with the median of all individual posterior probabilities greater than 0.80. This ad hoc criterion aimed to ensure that the loci used in genetic mapping had at least 50% of the individuals with a high probability (superior to 0.80) of being in their given clusters for the selected ploidy. Depending on the number of clusters, the SDMs were classified as segregating in a 1:2:1 ratio (three clusters) or in a 1:1 ratio (two clusters). On average, the percentages that represented each segregation class were 16.4% and 83.6%, respectively (Table 2).
Table 2.
Selected loci with good quality (category A) and ploidy (6 through 14) were classified as single-dose markers (SDMs) or multi-dose markers (MDMs). High-quality SDMs (median of all individual a posteriori probabilities > 0.80) were also characterized according to their segregation pattern in the sugarcane mapping population
| Reference | Total | Dosage | High-quality SDM | Segregation pattern | ||
|---|---|---|---|---|---|---|
| MDM | SDM | 1:2:1 | 1:1 | |||
| Methyl-filtered sugarcane genome | 15,906 | 7,014 (44.1%) | 8,892 (55.9%) | 5,266 | 912 (17.3%) | 4,354 (82.7%) |
| Sorghum bicolor genome (v. 2.1) | 11,789 | 5,784 (49.1%) | 6,005 (50.9%) | 3,433 | 605 (17.6%) | 2,828 (82.4%) |
| RNA-seq sugarcane transcriptome | 9,808 | 4,959 (50.6%) | 4,849 (49.4%) | 2,869 | 469 (16.4%) | 2,400 (83.6%) |
| SUCEST project sequences | 3,433 | 1,736 (50.6%) | 1,697 (49.4%) | 983 | 141 (14.3%) | 842 (85.7%) |
The redundancy of the SDM was inspected after quality and ploidy filtration with the alleles recoded as a or b. All the references showed very similar levels of redundancy within and between them (Fig. 2). For instance, the same overall level of 22.7% for within-redundancy was found. Only 84 SNPs markers were attributed equally to all four references and represented approximately 3.8% of each reference. Interestingly, each reference provided 39.6% new loci on average. By keeping only one call for each ambiguous marker, we obtained 7,049 loci in total for mapping. Of these loci, 5,757 (81.67%) and 1,292 (18.33%) segregated 1:1 and 1:2:1, respectively (Table 3).
Fig. 2.
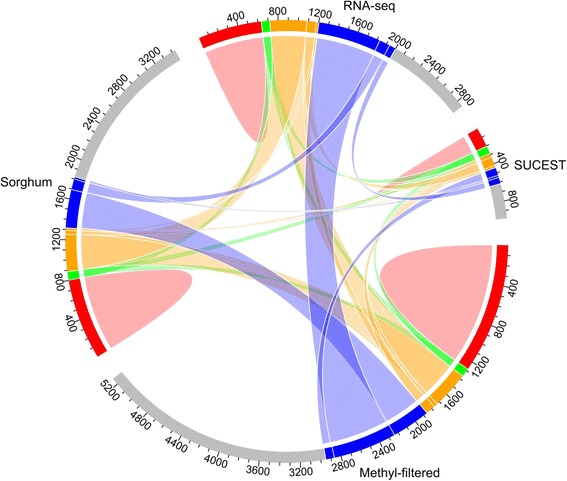
Circular plot showing the redundancy between single-dose markers from four pseudo-references (methyl-filtered sugarcane genome, Sorghum bicolor genome, RNA-seq sugarcane transcriptome and SUCEST project sequences) that were used to align the GBS sugarcane tags. The red regions represent redundancy within each pseudo-reference, whereas the green, orange and blue regions represent redundancy between four, three and two pseudo-references, respectively. The remaining grey regions represent loci that are unique to each pseudo-reference
Table 3.
Overall single-dose gel-based and GBS-based markers screened for the progeny of the cross between sugarcane cultivars SP80-3280 and RB835486
| Markers | Gel-based markers | GBS-based markers | |||
|---|---|---|---|---|---|
| Genomic SSR | Genic SSR | TRAP | Total | ||
| Number of SDMs evaluated (gel-based and GBS-based markers) | 109 | 842 | 80 | 7,049 | 8,080 |
| SDMs with 1:1 segregation | 66 | 456 | 23 | 5,757 | 6,302 |
| SDMs with 1:2:1 segregation (GBS-based markers) | - | - | - | 1,292 | 1,292 |
| Double SDMs (gel-based markers) with 3:1 segregation | 43 | 386 | 57 | - | 486 |
| Number of markers with distorted segregation | 25 | 336 | 41 | 0 | 402 |
| Total number (1:1, 1:2:1 and 3:1) feasible for linkage analysis | 84 | 506 | 39 | 7,049 | 7,678 |
Gel-based marker genotyping
A total of 120 SSRs and four combinations of TRAP markers (COMT + ARB2, SuPS + ARB1, CCR + ARB1, and CCR + ARB3) produced 1,031 polymorphic bands. Of these 1,031 bands, 545 (52.86%) were tested for 1:1 segregation, and 486 (47.14%) were tested for 3:1 segregation. The number of SDMs available for linkage analysis was 629 (61%), of which 506, 84 and 39 originated from genic SSR, genomic SSR and TRAP markers, respectively (Table 3).
Genetic map
An integrated genetic map was constructed using 151 full sibs generated from a cross between SP80-3280 and RB835486 (Additional file 1: Figure S1). Of the 8,080 SDMs that were scored (Table 3), 7,678 were used for linkage analysis (7,049 GBS-based markers and 629 gel-based markers), and 993 (12.93%) were placed in the linkage map (Tables 3 and 4). The mapped markers included 934 GBS-based markers and 59 SSRs. The distribution of the segregation patterns of mapped markers were 254 B3-type markers (1:2:1), 15 C-type markers (3:1), 518 D1-type markers or that were informative only for SP80-3280 (500 GBS-based markers and 18 gel-based markers) and 206 D2-type markers or that were informative only for RB835486 (180 GBS-based markers and 26 gel-based markers) (Table 4). The markers were distributed throughout the 223 LGs, with a cumulative map length of 3,682.04 cM and an average marker density of 3.70 cM (Table 5). The length of LGs ranged from 1.06 cM (LG 70) to 235.67 cM (LG46), with an average of 16.51 cM; 56 LGs displayed lengths shorter than 2 cM, 95 LGs exhibited lengths greater or equal to 2 cM and smaller than 10 cM, and the other 72 LGs had lengths greater or equal to 10 cM.
Table 4.
Distribution of the different marker types as mapped according to their cross type
| Cross type | Number of markers | ||||
|---|---|---|---|---|---|
| Gel-based markers | |||||
| Genomic SSR | Genic SSR | TRAP | GBS-based markers | Total | |
| D1.10 (ab x aa) | - | - | - | 500 | 500 |
| D1.13 (ao x oo) | 4 | 14 | 0 | - | 18 |
| D2.15 (aa x ab) | - | - | - | 180 | 180 |
| D2.18 (oo x ao) | 4 | 22 | 0 | - | 26 |
| B3.7 (ab x ab) | - | - | - | 254 | 254 |
| C.8 (ao x ao) | 2 | 13 | 0 | - | 15 |
| Total | 10 | 49 | 0 | 993 | 993 |
Table 5.
Number of each type of mapped marker within each homo(eo)logous group (HG), number of linkage groups (LGs) within each HG, the length of each HG in centimorgans (cM) and the marker density in cM of each HG for the genetic map construct from a progeny of a cross between sugarcane cultivars SP80-3280 and RB835486
| HG | No. LGs | No. SSR | No. GBS-based markers | No. mapped markers | Length of HG (cM) | Marker density (cM) |
|---|---|---|---|---|---|---|
| 1 | 2 | 0 | 11 | 11 | 48.90 | 4.44 |
| 2 | 2 | 0 | 8 | 8 | 61.72 | 7.71 |
| 3 | 3 | 5 | 21 | 26 | 157.14 | 6.04 |
| 4 | 2 | 0 | 9 | 9 | 59.09 | 6.56 |
| 5 | 4 | 5 | 17 | 22 | 100.45 | 4.56 |
| 6 | 2 | 2 | 14 | 16 | 144.24 | 9.01 |
| 7 | 2 | 1 | 9 | 10 | 21.45 | 2.14 |
| 8 | 2 | 0 | 8 | 8 | 37.84 | 4.73 |
| 9 | 2 | 4 | 5 | 9 | 53.72 | 5.96 |
| 10 | 3 | 0 | 19 | 19 | 120.08 | 6.32 |
| 11 | 5 | 13 | 31 | 44 | 273.66 | 6.22 |
| 12 | 3 | 5 | 7 | 13 | 40.64 | 3.12 |
| 13 | 2 | 0 | 7 | 7 | 2.91 | 0.41 |
| 14 | 2 | 0 | 9 | 9 | 60.21 | 6.69 |
| 15 | 3 | 0 | 13 | 13 | 69.55 | 5.35 |
| 16 | 4 | 10 | 8 | 18 | 120.73 | 6.70 |
| 17 | 3 | 0 | 14 | 14 | 15.35 | 1.09 |
| 18 | 2 | 4 | 3 | 7 | 37.26 | 5.32 |
| Unassigned in HG | 175 | 10 | 721 | 730 | 2,257.10 | 3.09 |
| Total | 223 | 59 | 934 | 993 | 3,682.04 | 3.70 |
A total of 18 HGs were formed based on the common genomic origins of mapped loci from different LGs, which were provided by SSR and GBS-based markers. The number of LGs allocated into HGs ranged from two (HG1, HG2, HG4, HG6, HG7, HG8, HG9, HG13, HG14 and HG18) to five (HG11). The coverage within the HGs varied from 2.91 cM (HG13) to 273.66 cM (HG11). A total of 175 LGs with 730 markers remained unassigned to any HG (Table 5 and Additional file 1: Figure S1).
QTL mapping
QTL mapping was performed for POL%C, BRIX, SD and FIB traits [10] by applying a CIM model [62] to the integrated genetic map. Considering all traits, 24 cofactors were found for each location, Araras and Ipaussu. The trait with more cofactors was SD, with eight cofactors identified for each location (Additional file 1: Table S2).
To declare the significant QTLs, a permutation test was performed for each phenotype [96]. The values of the LOD Score threshold for BRIX, POL%C, SD and FIB at Araras and Ipaussu were 3.79 and 3.77, 3.80 and 3.86, 4.20 and 4.28, and 4.45 and 4.14, respectively. In this case, we were able to declare seven QTLs. For the respective locations, Araras and Ipaussu, BRIX had two and one QTLs, POL%C had one and one QTL, SD had zero and one QTL, and FIB had one and zero QTLs.
The global LOD Score values ranged from 4.17 to 6.02, and the R 2 values ranged from 2.71% to 9.19%. The highest LOD Score for the additive effect was 5.22 for parental SP80-3280 for a QTL associated with BRIX at Araras (B1 at LG4). The highest LOD Score for the dominance effect was 3.93 for a QTL associated with FIB at Araras (FIB1 at LG46). The segregation patterns of the QTLs were as follows: 1:1 (42.85%), 1:2:1 (14.30%) and 3:1 (42.85%). The results of the QTL mapping are summarized in Table 6 and Fig. 3.
Table 6.
QTLs mapped for BRIX, POL%C, SD and FIB traits by applying a CIM model in Araras (Location 1) and Ipaussu (Location 2)
| Location | QTL | Trait | LGa | Position (cM)a | Flanking Markersb | Global LODb | R2 c | Additive effect SP80-3280d | LODe | Additive effect RB835486d | LODe | Dominance effectd | LODe | Segregf |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | B1 | BRIX | 4 | 43.32 | mf60753_2013 (QTL) | 5.75 | 9.19 | −0.31 | 5.22 | 0.08 | 0.25 | 0.04 | 0.09 | 1:1 |
| 1 | B2 | BRIX | 47 | 40.00 | SCSFAM1074E10_287 - QTL - mf16592_3766 | 4.79 | 4.78 | −0.23 | 2.22 | 0.28 | 3.77 | −0.17 | 1.26 | 3:1 |
| 2 | B3 | BRIX | 4 | 43.32 | mf60753_2013 (QTL) | 4.24 | 7.65 | −0.23 | 3.84 | 0.06 | 0.16 | 0.01 | 0.01 | 1:1 |
| 1 | P1 | POL%C | 4 | 43.32 | mf60753_2013 (QTL) | 4.18 | 8.10 | −0.23 | 4.14 | 0.14 | 0.84 | 0.08 | 0.27 | 1:2:1 |
| 2 | P2 | POL%C | 4 | 43.32 | mf60753_2013 (QTL) | 4.17 | 8.09 | −0.23 | 4.13 | 0.14 | 0.83 | 0.08 | 0.27 | 1:1 |
| 2 | SD1 | SD | 29 | 2.00 | sb2_61882838 - QTL - sb2_61882853 | 6.02 | 5.38 | 0.36 | 1.66 | 0.64 | 4.57 | −0.54 | 3.32 | 3:1 |
| 1 | FIB1 | FIB | 46 | 171.00 | sb1_29954417 - QTL - mf125302_409 | 4.77 | 2.71 | −0.47 | 3.11 | −0.75 | 3.95 | 0.38 | 3.93 | 3:1 |
aLG: Linkage groups; Position (cM): QTL position on LG; bAdjacent markers for QTLs and associated LODs; cExplained phenotypic variation; dAdditive effects of parents and dominance effects; eLODs of the additive and dominance effects; fEstimation of the segregation pattern of the QTLs
Fig. 3.
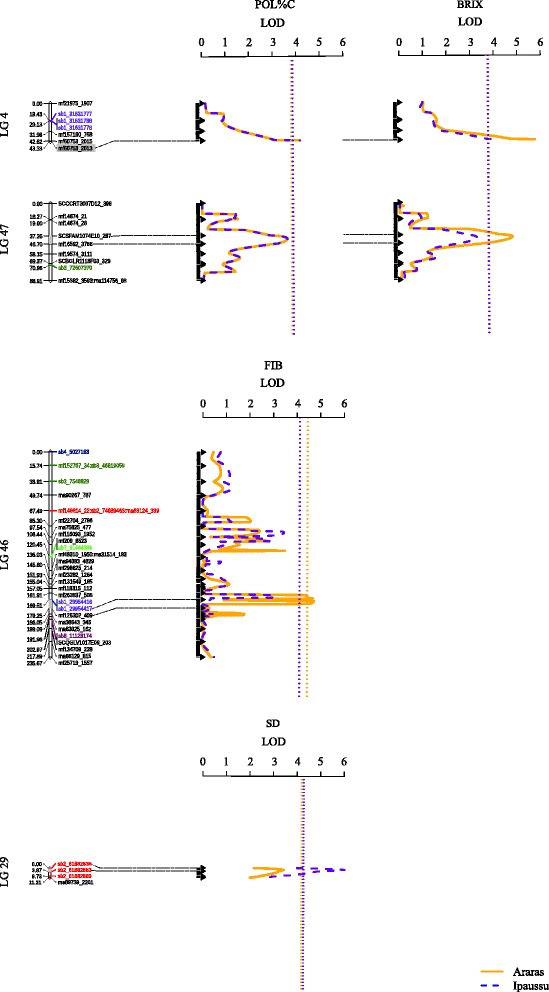
Composite interval mapping (CIM) for soluble solid content (BRIX, in °Brix), sucrose content of cane (POL%C, in %), stalk diameter (SD, in mm) and fiber content (FIB, in %) from the SP80-3280 and RB835486 F1 population. Blue and yellow dotted lines indicate the LOD thresholds for Ipaussu-SP and Araras-SP, respectively, obtained after permutation tests. The portions highlighted in gray in the linkage groups show the positions of the QTLs
For BRIX, two QTLs (B1 and B2) explained 10.54% of the phenotypic variation in Araras. The QTL identified in Ipaussu (B3 at LG4) was also part of a set found in Araras, i.e., it could be near in the LG at both locations and showed similar effects; this QTL had a significant additive effect for parental SP80-3280 and a segregation pattern of 1:1. In addition, the mapping analysis showed that the region of QTLs B1 and B3 at LG4 was also associated with POL%C (P1 and P2). QTLs for SD and FIB (SD1 and FIB1) showed larger dominance effects that were negative for SD and positive for FIB (Table 6 and Fig. 3).
Sequence annotation
Sequence similarity was found for six out of seven adjacent markers of the mapped QTLs, with homologies for S. bicolor, Solanum tuberosum and Zea mays. A functional description of the sequences showed possible candidate genes for BRIX, SD and FIB traits, whereas the sequence from the marker associated with the QTL found for POL%C did not show similarity or a characterized protein. Of the total mapped QTLs, three presented adjacent markers and were located in LG47 (B2), LG29 (SD1) and LG46 (FIB1) for BRIX, SD and FIB traits, respectively (Tables 6 and 7).
Table 7.
Functional description of the sequences that gave rise to adjacent markers of the mapped QTLs for the traits BRIX, POL%C, SD and FIB, and references regarding their functions in plants
| Marker | QTLs | LG | Locations | Traits | Description | e-value | Reference |
|---|---|---|---|---|---|---|---|
| mf60753_2013 | B1, B3, P1, P2 | 4 | 1 and 2 | BRIX and POL%C | No homology found | - | - |
| SCSFAM1074E10_287 | B2 | 47 | 1 | BRIX | Extended synaptotagmin-1-like [Zea mays] | 2.6e-26 | [137–142] |
| mf16592_3766 | B2 | 47 | 1 | BRIX | Hypothetical protein SORBIDRAFT_03 g038130 [Sorghum bicolor] | 1.2e-18 | Unknown function |
| sb2_61882838 | SD1 | 29 | 2 | SD | Hypothetical protein SORBIDRAFT_02 g026690 [Sorghum bicolor] | 4.0e-13 | Unknown function |
| sb2_61882853 | SD1 | 29 | 2 | SD | Hypothetical protein SORBIDRAFT_02 g026690 [Sorghum bicolor] | 4.0e-13 | Unknown function |
| mf125302_409 | FIB1 | 46 | 1 | FIB | Zinc finger protein CONSTANS-LIKE 15 [Solanum tuberosum] | 0.0 | [143–148] |
| sb1_29954417 | FIB1 | 46 | 1 | FIB | Transposon mutator sub-class [Sorghum bicolor] | 2.0e-177 | [149] |
For BRIX, the QTL B2 had two adjacent markers that were each identified in a different reference. The region of the GBS-based marker SCSFAM1074E10_287, which originated from SUCEST sequences, showed similarity with extended synaptotagmin-1-like, which is a member of a family of membrane-trafficking proteins. The second adjacent GBS-based marker of this QTL, mf16592_3766, which originated from the methyl-filtered sugarcane genome, showed similarity with a hypothetical protein in S. bicolor (Tables 6 and 7).
For SD, the QTL SD1 had two adjacent GBS-based markers, which were both identified from the sorghum genome. The markers sb2_61882838 and sb2_61882853 have a small physical distance in the sorghum genome and share almost the same sequence determine. These two markers showed similarity with a hypothetical protein in S. bicolor (Tables 6 and 7).
For FIB, the QTL FIB1 had two adjacent GBS-based markers from two distinct references. Moreover, each of the two adjacent markers showed different similarity by descent. The region of the sb1_29954417, which originated from sequences of the sorghum genome, had similarity with transposon mutator sub-class, and the second adjacent marker, mf125302_409, which originated from the methyl-filtered sugarcane genome, had similarity with zinc finger protein CONSTANS-LIKE 15 (Tables 6 and 7).
Discussion
The simultaneous identification and genotyping of SNPs and indels is possible because of important recent advances in sequencing [41–50]. GBS is the preferred high-throughput genotyping method for plants with some level of genetic complexity; this method involves complexity reduction and multiplex sequencing to produce high-quality polymorphism data at a relatively low cost per sample [101]. Using four pseudo-references to discover GBS-based markers, we obtained more markers suitable for linkage analysis (Table 3) than any other previously published study on sugarcane mapping. The strategies adopted for the discovery of GBS-based markers allowed us to relate the sugarcane markers to sorghum chromosomes [73] and to potential genetic regions sampled from the methyl-filtered sugarcane genome [72], RNA-seq sugarcane transcriptome [66] and SUCEST project sequences [65].
The highest alignment (87.94%) of the 3.1 million resulting tags against the methyl-filtered sugarcane genome also contains most of the high-quality SDMs (Table 2). This great alignment rate may be related to the greater amount of scaffolds for this reference compared with the other three references and to the fact that the PstI enzyme used for library formation is sensitive to DNA methylation [102]; thus, more polymorphic sites are expected from the methyl-filtered genome. Additionally, GBS-based markers had more markers mapped to parent SP80-3280 than to parent RB835486 (Table 4). This result can be explained by some factors: a) the possible presence of more PstI enzyme restriction sites in cultivar SP80-3280 than in RB835486, leading to more polymorphic sites in the first cultivar, as shown for barley cultivars by Liu et al. [54]; b) also could be due to more similarity between the genome of SP80-3280 with the reference; or c) due to different methylation effects between the two cultivars. Furthermore, inconsistencies in the number of sites sequenced per sample [102] and in the number of reads per site [103, 104], in addition to the filtering steps applied to the GBS libraries to obtain the markers, can influence the observed result. Other factors that can influence these results are the quality and quantity of the biological replicates used for GBS-based marker calling. Because the sequencing of the samples can present failures that will be included in the downstream process, better dosage and ploidy level estimates for each marker in the SuperMASSA software can be hampered.
The analysis of the loci with high coverage that were filtered for missing data after analysis using SuperMASSA software showed that for the ploidy levels under consideration, the number of loci varied within each ploidy class (Fig. 1), suggesting that the number of chromosomes within the HGs is not constant in sugarcane, as reported previously [39, 105]. As stated by Garcia et al. [39], technique artifacts yielding either strong bias or too much noise should explain marker misclassification, i.e., loci not included in the 6-14 expected ploidy range. In fact, the graphical Bayesian model used in the analyses benefits smaller ploidies due to parsimony when the skewed clusters are confounded or, conversely, favors a higher number of clusters by attempting to explain a diffuse scatterplot [106]. In addition, Garcia et al. [39] hypothesized that poor-quality data can also be generated by biological events such as copy number variations or paralogous regions. We used the same ad hoc criteria as Garcia et al. [39] to classify each locus into one quality category based on the a posteriori probabilities for each ploidy category A as represented by 60.4% of loci of all ploidies on average (Fig. 1), which is smaller than the 77.6% of Sequenom-based data that were previously studied [39]. The GBS read count data worked slightly poorer because of their broad genome coverage and eventual technique artifacts. Despite this reduction, a large part of the loci could be further exploited for mapping purposes.
The presence of repeat elements, paralogs, and incomplete or inaccurate reference genome sequences can create ambiguities in GBS-based marker calling [107]. After we selected the loci that were classified into category A and ploidies ranging from 6 to 14 (Table 2), we continued on to the final steps of quality control and redundancy analyses that showed a low redundancy considering simultaneously all four references. Aitken et al. [35] presented the first sugarcane genetic map with DArT markers and did not remove any redundant markers. Sugarcane has a large and complex genome, and a low level of redundancy is important for showing the true coverage of the genome. Heslot et al. [52] showed that DArT markers were significantly more redundant than GBS markers, and they suggested that GBS markers were significantly more evenly distributed across the wheat genome. These authors also concluded that GBS is the marker platform of choice for further diversity analyses and genomic selection.
The integrated genetic map of sugarcane obtained in this paper presents improvements in comparison with previous works. Here, the number of markers used for linkage analysis is more than twice the number of markers used for the development of the largest map [35]. Furthermore, this genetic map of sugarcane is the first to use a high-throughput approach for genotyping. Co-dominant biallelic markers can segregate in a 1:2:1 fashion (‘B3’ cross type), which is even more informative for map integration purposes. The previously published genetic maps had molecular markers treated as dominant even when are potentially co-dominant, that is, all clusters that have at least one copy of the allele will collapse into a single cluster [37]. The mapping population was formed by a cross between polyploid heterozygous parents, and for each segregating loci, there could be different numbers of segregating alleles and different dosages that are potentially expressed. Thus, accessing the dosage information of the SDMs with a segregation pattern of 1:2:1 was important; the double SDMs with a segregation pattern of 3:1 (‘C8’ type) were also important. This information was used to construct an integrated genetic map for sugarcane that increased the genome coverage.
The 223 LGs obtained here had a cumulative map length of 3,682.04 cM and an average marker density of 3.70. The number of LGs exceeds the number of chromosomes of modern sugarcane cultivars, which can range from 100 to 130 [1, 9], and 56 LGs showed lengths shorter than 2 cM. This result indicates the presence of gaps and that the map is not yet well saturated. In 2007, Oliveira and collaborators [38] claimed that because there is a constraint to discarding markers in multiples doses, i.e., duplexes of monoparental origin, triplex or higher multiplex markers, gaps are evidently expected; the same discussion should be applied to this study. The number of unlinked markers (87.07%) is higher than that obtained in other sugarcane maps [4, 31, 35, 37, 38, 81, 108–110] and reflects the highly stringent criteria used to construct an integrated genetic map of sugarcane that is reliable for performing QTL mapping analysis.
To increase the understanding of the genetic architecture of sugarcane, a necessary requirement is the availability of good genetics maps, i.e., maps with a high density of markers and with high coverage of the genome [111–113]. The complexity of the sugarcane genome, the cost of generating a large number of markers, and the absence of a statistical genetic model that could consider other segregation ratios beyond 1:1, 1:2:1 and 3:1 have limited the development of high-density genetic maps. These limitations have delayed practical applications of genomic tools in sugarcane, in contrast to other crops that have already advanced to MAS and genomic selection. Sugarcane still does not have its genome completely sequenced, and sorghum genome is widely recognized as a reference genome for comparative analysis with sugarcane [67]. The origin of modern sugarcane cultivars raises issues that are related to not only the extent and nature of the divergence of the sugarcane and sorghum genomes but also the relations (in terms of meiosis and dosage) among homo(eo)logous loci [68]. Differences in chromosome structures between the ancestor species and pairing behavior in modern cultivars suggest that the hybrid monoploid number is likely to be greater than 10 in sugarcane hybrids [38, 114]. Probably because of aneuploidy, an unequal number of chromosomes in each HG is likely to occur; this inequality was reflected in the 18 HGs with differences in genome coverage. Moreover, translocation events may have occurred in sugarcane between regions equivalent to sorghum, as discussed previously [27, 28, 115–118]. Although these comparative studies proposed hypotheses about the evolutionary aspects of sugarcane and sorghum, the results showed variations that were primarily derived from the low resolution of the genetic maps used and to the coverage of the sugarcane genome. In addition, it is important to highlight that advances in the assembly of polyploid genomes will enable the use of the full sugarcane genome as a reference in the future [119].
The genetic maps and field data obtained through designed experiments are required for QTL mapping studies. For sugarcane, multiple harvest-location trials may be used to infer the genetic architecture of quantitative traits. However, this inference makes the data analysis more complex and challenging because of the interactions that it generates, e.g., genotype by environment interactions. To solve this problem, a mixed model approach has been used to obtain highly accurate genetic estimates [10, 31, 120], and for segregating populations, these results will be the input for QTL mapping.
In this study, QTL mapping was performed by applying the statistical model proposed by Gazaffi et al. [62], which extends the CIM [64] for a full-sib progeny. The primary advantage of CIM is that it is more precise and effective at mapping QTLs in comparison with single-marker analysis and IM, especially when QTLs are present outside the mapping window [64]. The results obtained from the CIM method are usually comparable to those obtained from multi-QTL analysis if a high-density genetic map is employed to better represent the number of loci underlying the quantitative traits [79]. Several QTL mapping studies in sugarcane have been published [31, 34, 61, 81, 121–136]. The comparison between the results may be biased by several issues, such as the different rates of polymorphisms in parents, the number of progeny, the evaluation methodologies for phenotypic traits, the methodologies used for QTL detection, genetic map contruction, and experimental design, among others. For example, Pastina et al. [31] worked with a population of 100 individuals from a cross between cultivars SP80-180 and SP80-4966 to construct an integrated genetic map that was 2,468.14 cM in length. These researchers used IM to test presence of putative QTLs and a multi-QTL model to declare QTLs and found 46 QTLs. There were 13 mapped QTLs for the tonnes of cane per hectare (TCH), 14 for sugar content in tonnes of sucrose per hectare (TSH), 11 for FIB and eight for POL%C. Singh et al. [34] studied the progeny of 207 individuals derived from a cross between cultivars Co86011 and CoH70, and they constructed two separate genetic maps, one for each parent. Through the CIM model, these researchers found 31 QTLs, with seven for BRIX and four for stalk number (SN). Thus, specific objectives must be taken into consideration for QTL mapping in sugarcane.
For QTL mapping in this study, we performed a permutation test to obtain the threshold for declaring significant QTLs [97]. The CIM model was a useful tool once it was able to identify regions with next QTL considering Araras and Ipaussu over the harvests (three years of evaluations). Seven QTLs were identified, being that a region located in LG4 at 43.32 cM showed QTLs for BRIX (B1-B3) and POL%C (P1-P2). The marker associated with the QTLs was identical for both traits, and the region that gave rise to this marker could be evaluated for future applications by sugarcane breeding programs. POL%C and BRIX are correlated traits [10], and although the commercial cultivars used as parents of the mapping population presented a small contrast in terms of phenotypic averages, especially for sucrose content, these results show that a combination of different alleles in each parent segregates and contributes to the observed variation in progeny. Furthermore, this result was expected because the parents of the mapping population are cultivars that were improved primarily to increase the sucrose content.
The percentage of phenotypic variation explained by each QTL ranged from 2.71% (FIB1) to 9.19% (B1) for Araras and from 5.38% (SD1) to 8.09% (P1) for Ipaussu. The sampling of the genome in single doses requires that QTL also segregate as single doses. Furthermore, the use of improved parents that have close phenotypic averages and that have some level of fixed alleles for traits of interest could decrease the chances of detecting QTLs with high rates of explained phenotypic variation. Nevertheless, the QTLs described here can be considered reliable because they have all taken into account the phenotypic average of three harvests and because it was identified QTLs in same position into LG4 for both locations.
The common QTLs between locations may be regarded as potential regions to search genes that are involved in controlling quantitative traits. In addition, a strong marker-QTL association detected in full-sib progenies also could be an impact on crop improvement via clonal propagation because probability of crossover between the marker and the QTL is low [34]. Therefore, the similarity analysis and annotation of sequences that originated the markers with mapped QTLs are important for identified putative candidate genes in sugarcane, although the QTLs regions are relatively large and an uncertain number of genes may be involved with the evaluated traits. Sugarcane has high genetic complexity and its genome still does not was completely sequenced [67], whereas inferences about putative candidate genes could be contribute for new insights and open new fronts of research to mining and validation of genes of interest.
For the BRIX trait, we can highlight the similarity of the marker SCSFAM1074E10_287, located in QTL B2, with extended synaptotagmin-1-like, which is a member of a membrane-trafficking protein family that is characterized by an N-terminal transmembrane region, a linker of variable size, and two C-terminal C2 domains in tandem [137]. C2 domains, identified as a conserved sequence motif in protein kinase C [138], are autonomously folded protein modules that generally act as calcium (Ca2+) and phospholipid-binding domains and that were shown to represent autonomously folded Ca2+-binding domains in synaptotagmins [139]. In addition, Ca2+ acts as a second messenger in the signal transduction pathways of hormones and environmental stimuli (touch, wind, chilling, light, and elicitors) [140], and several proteins that are involved in photosynthesis depend on Ca2+ [141]. In sugarcane, Papini-Terzi et al. [142] identified differentially expressed genes in genotypes contrasting for sucrose content and showed that among these genes, nine were associated with calcium signaling and a calcium-dependent protein kinase (ScCDPK-27). Further researches are needed to expand the inference found in this study for pathways and regulatory networks of sugarcane in order to relate with sucrose biosynthesis and, consequentely, with BRIX trait.
For the FIB trait, a QTL (FIB1) showed different similarities for each of the two adjacent markers and we can highlight the similarity of the marker mf125302_409 with zinc finger protein CONSTANS-LIKE 15. The CONSTANS (CO) protein is a zinc finger transcription factor that contain two conserved domains (i.e., a B-box zinc finger domain and a CCT [CO, CO-like, TOC1] domain), located in the region near the amino- and carboxy-terminus, respectively [143, 144]. The CO proteins play a central role in the photoperiod pathway of Arabidopsis by mediating the circadian clock and floral integrators via positive regulation of FLOWERING LOCUS T expression [145–147]. In cotton (Gossypium spp.), which is the most important natural source of fiber for the textile industry, the CO5 protein (CONSTANS-LIKE 5) was up-regulated for cell wall modification and developing fibers in MD52ne, a near-isogenic line. Furthermore, using an F2 population derived from a cross between MD52ne and MD90ne, stable QTLs for bundle fiber strength and fiber length were found, and CO5 was present on the QTL region for fiber length [148]. Although there is an indicative of relation between the candidate gene and FIB trait, advanced studies should be conducted to validate and prove the real function and effects into phenotypic expression in sugarcane.
Conclusions
Our understanding of the genetic architecture of triats of interest in sugarcane is increasing with the development of new analytical methods. The estimation of ploidy and allelic dosage through markers generated by GBS as well as the inclusion of these markers in an integrated genetic map of sugarcane were first observed in this study, and these markers showed great potential for QTL mapping. The CIM approach that provided additive and dominance effects and estimated the segregation patterns for all mapped QTLs was efficient for detecting possible stable QTLs among the evaluated locations. The verification of possible candidate genes for mapped QTLs as a preliminary analysis showed importance for new insights into the comprehensive relations between phenotypes and genotypes. It is still necessary to develop statistical approaches to enable the inclusion of markers at multiple doses to enhance the coverage by linking the SDMs that are pulverized by the genome. Moreover, QTL mapping with markers in multiple doses must be considered a major step in the understanding of regions that control quantitative traits in polyploid organisms and perhaps permit the verification of the allelic expression of phenotypic traits in the future.
Acknowledgements
We gratefully acknowledge Marcelo Mollinari for sharing his experience working with SuperMASSA software.
Funding
This work was supported by grants from the FINEP (Finaciadora de Estudos e Projetos), FAPESP (Fundação de Amparo à Pesquisa de São Paulo, 08/52197-4) and INCT-Bioetanol (Instituto Nacional de Ciência e Tecnologia do Bioetanol, FAPESP 08/57908-6 and CNPq, Conselho Nacional de Desenvolvimento Científico e Tecnológico, 574002/2008-1). TWAB, GSP, MCM, EAC and CBCS received doctoral fellowships from FAPESP (10/50091-4, 12/25236-4, 10/50549-0, 10/50031-1 and 12/11109-0, respectively). COA received a doctoral fellowship from the CNPq. FZB received a master’s fellowship from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior). APS and AAFG received research fellowships from CNPq.
Availability of data and materials
The GBS sequence data files supporting the conclusions of this article are in the process of submission to the NCBI Sequence Read Archive (SRA), http://www.ncbi.nlm.nih.gov/sra, with accession number SRP092493.
Authors’ contributions
MSC and HPH conceived and designed the field experiments. TWAB, MCM, GSP and COA performed the field experiment assessments and the phenotypic data analysis. TWAB and FZB performed the DNA extraction and SSR and TRAP marker data generation. MSC, AAFG, GRAM, FZB and TWAB designed the GBS experiments. GSP, AAFG and GRAM performed the GBS data analysis. APS, CBCS and EAC provided the RNA-seq data. TWAB, GSP, AAFG and GRAM performed the genetic map analysis. RG and AAFG performed the QTL mapping analysis. MSC, TWAB, GSP, AAFG, GRAM and APS participated in the study design and integrated the analysis of results. TWAB and GSP wrote the manuscript draft, and MSC, AAFG, and APS edited and revised the manuscript. GRAM and RG critically read the manuscript. All the authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- BRIX
Soluble solid content
- CCR
Cinnamoyl-CoA reductase
- CIM
Composite interval mapping
- COMT
Caffeic acid 3-O-methyltransferase
- DNA
Deoxyribonucleic acid
- FIB
Fiber content
- GBS
Genotyping-by-sequencing
- HGs
Homo(eo)logous groups
- IM
Interval mapping
- LGs
Linkage groups
- LOD
Logarithm of the odds
- MAP
Maximum a posteriori
- MAS
Marker-assisted selection
- MDM
Multiple dose marker
- NGS
Next-generation sequencing
- PCR
Polymerase chain reaction
- POL%C
Sucrose content of cane
- QTLs
Quantitative trait loci
- RNA
Ribonucleic acid
- SD
Stalk diameter
- SDMs
Single-dose markers
- SNPs
Single nucleotide polymorphisms
- SSRs
Single sequence repeats, another term for microsatellites
- SuPS
Sucrose phosphate synthase
- TRAP
Target region amplification polymorphism
- VCF
Variant call format
Additional file
Bowtie2 alignment results of 3,103,708 million GBS tags in absolute and relative (in parentheses) values. Table S2. Markers selected as cofactors for mapping through the composite interval mapping (CIM) model for the soluble solid content (BRIX, in °Brix), sucrose content of cane (POL%C, in %), fiber content (FIB, in %) and stalk diameter (SD, in mm) traits for the locations Araras-SP and Ipaussu-SP. Figure S1. Genetic map of sugarcane that was generated with a population of 151 full sibs that originated from a commercial cross between the cultivars SP80-3280 and RB835486. The linkage groups (LGs) were separated into 18 homo(eo)logous groups (HGs). The numbers on the left of the LGs are the cumulative genetic distances in Kosambi centimorgans. The marker names are shown on the right. Markers corresponding to each sorghum chromosome are highlighted in different colors. (DOCX 2545 kb)
Contributor Information
Thiago Willian Almeida Balsalobre, Email: thiagobalsalobre@gmail.com.
Guilherme da Silva Pereira, Email: guispb@gmail.com.
Gabriel Rodrigues Alves Margarido, Email: gramarga@usp.br.
Rodrigo Gazaffi, Email: rodrigo@cca.ufscar.br.
Fernanda Zatti Barreto, Email: fernandazbarreto@gmail.com.
Carina Oliveira Anoni, Email: carina.anoni@gmail.com.
Cláudio Benício Cardoso-Silva, Email: benicio.genetic@gmail.com.
Estela Araújo Costa, Email: estela06d@gmail.com.
Melina Cristina Mancini, Email: mel.cmancini@gmail.com.
Hermann Paulo Hoffmann, Email: hermann@cca.ufscar.br.
Anete Pereira de Souza, Email: anete@unicamp.br.
Antonio Augusto Franco Garcia, Email: augusto.garcia@usp.br.
Monalisa Sampaio Carneiro, Email: monalisa@cca.ufscar.br.
References
- 1.D’Hont A, Glaszmann JC. Sugarcane genome analysis with molecular markers, a first decade of research. Proc Int Soc Sugar Cane Technol. 2001;24:556–9. [Google Scholar]
- 2.D’Hont A, Ison D, Alix K, Roux C, Glaszmann JC. Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome. 1998;41:221–5. doi: 10.1139/gen-41-2-221. [DOI] [Google Scholar]
- 3.D’Hont A. Unraveling the genome structure of polyploids using FISH and GISH; examples of sugarcane and banana. Cytogenet Genome Res. 2005;109:27–33. doi: 10.1159/000082378. [DOI] [PubMed] [Google Scholar]
- 4.Palhares AC, Rodrigues-Morais TB, Van Sluys MA, Domingues DS, Maccheroni W, Jordão H, et al. A novel linkage map of sugarcane with evidence for clustering of retrotransposon-based markers. BMC Genet. 2012;13:51. doi: 10.1186/1471-2156-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piperidis G, Piperidis N, D’Hont A. Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol Genet Genomics. 2010;284:65–73. doi: 10.1007/s00438-010-0546-3. [DOI] [PubMed] [Google Scholar]
- 6.Ha S, Moore PH, Heinz D, Kato S, Ohmido N, Fukui K. Quantitative chromosome map of the polyploid Saccharum spontaneum by multicolor fluorescence in situ hybridization and imaging methods. Plant Mol Biol. 1999;39:1165–73. doi: 10.1023/A:1006133804170. [DOI] [PubMed] [Google Scholar]
- 7.Daniels J, Roach BT. Taxonomy and evolution. In: Heinz DJ, editor. Sugarcane improvement through breeding. Amsterdam: Elsevier Press; 1987. pp. 7–84. [Google Scholar]
- 8.Irvine JE. Saccharum species as horticultural classes. Theor Appl Genet. 1999;98:186–94. doi: 10.1007/s001220051057. [DOI] [Google Scholar]
- 9.Roach BT. Cytological studies in Saccharum. Chromosome transmission in interspecific and intergeneric crosses. Proc Int Soc Sugar Cane Technol. 1969;13:901–20. [Google Scholar]
- 10.Balsalobre T, Mancini MC, Pereira GS, Anoni CO, Barreto FZ, Hoffmann HP, et al. A mixed-model approach for analysis of yield components and brown rust resistance in full-sib families of sugarcane. Agron J. 2016;108:1–14. doi: 10.2134/agronj2015.0430. [DOI] [Google Scholar]
- 11.Eksteen A, Singels A, Ngxaliwe S. Water relations of two contrasting sugarcane genotypes. Field Crops Res. 2014;168:86–100. doi: 10.1016/j.fcr.2014.08.008. [DOI] [Google Scholar]
- 12.Waclawovsky AJ, Sato PM, Lembke CG, Moore PH, Souza GM. Sugarcane for bioenergy production: an assessment of yield and regulation of sucrose content. Plant Biotechnol J. 2010;8:263–76. doi: 10.1111/j.1467-7652.2009.00491.x. [DOI] [PubMed] [Google Scholar]
- 13.Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–41. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–6. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesová M, et al. The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot. 2012;109:19–45. doi: 10.1093/aob/mcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madlung A. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity. 2013;110:99–104. doi: 10.1038/hdy.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yagi M, Yamamoto T, Isobe S, Hirakawa H, Tabata S, Tanase K, et al. Construction of a reference genetic linkage map for carnation (dianthus caryophyllus L.) BMC Genomics. 2013;14:734. doi: 10.1186/1471-2164-14-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartholomé J, Mandrou E, Mabiala A, Jenkins J, Nabihoudine I, Klopp C, et al. High-resolution genetic maps of eucalyptus improve eucalyptus grandis genome assembly. New Phytol. 2015;206:1283–96. doi: 10.1111/nph.13150. [DOI] [PubMed] [Google Scholar]
- 19.Deokar AA, Ramsay L, Sharpe AG, Diapari M, Sindhu A, Bett K, et al. Genome wide SNP identification in chickpea for use in development of a high density genetic map and improvement of chickpea reference genome assembly. BMC Genomics. 2014;15:708. doi: 10.1186/1471-2164-15-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Portis E, Mauromicale G, Mauro R, Acquadro A, Scaglione D, Lanteri S. Construction of a reference molecular linkage map of globe artichoke (Cynara cardunculus var. scolymus) Theor Appl Genet. 2009;120:59–70. doi: 10.1007/s00122-009-1159-2. [DOI] [PubMed] [Google Scholar]
- 21.Hudson CJ, Freeman JS, Kullan AR, Petroli CD, Sansaloni CP, Kilian A, et al. A reference linkage map for eucalyptus. BMC Genomics. 2012;13:240. doi: 10.1186/1471-2164-13-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong Y, Chen X, Liang X, Liu H, Zhou G, Li S, et al. A SSR-based composite genetic linkage map for the cultivated peanut (Arachis hypogaea L.) genome. BMC Plant Biol. 2010;10:17. doi: 10.1186/1471-2229-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu KK, Burnquist W, Sorrells ME, Tew TL, Moore PH, Tanksley SD. The detection and estimation of linkage in polyploids using single-dose restriction fragments. Theor Appl Genet. 1992;83:294–300. doi: 10.1007/BF00224274. [DOI] [PubMed] [Google Scholar]
- 24.Grattapaglia D, Sederoff R. Genetic linkage maps of eucalyptus grandis and eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genet. 1994;137:1121–37. doi: 10.1093/genetics/137.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grivet L, D'Hont A, Dufour P, Hamon P, Roques D, Glaszmann JC. Comparative genome mapping of sugar cane with other species within the Andropogoneae tribe. Hered. 1994;73:500–8. doi: 10.1038/hdy.1994.148. [DOI] [Google Scholar]
- 26.Da Silva J, Honeycutt RJ, Burnquist W, Al-Janabi SM, Sorrells ME, Tanksley SD, et al. Saccharum spontaneum L. ‘SES 208’ genetic linkage map combining RFLP and PCR based markers. Mol Breeding. 1995;1:165–79. doi: 10.1007/BF01249701. [DOI] [Google Scholar]
- 27.Dufour P, Deu M, Grivet L, D’Hont A, Paulet F, Bouet A, et al. Construction of a composite sorghum genome map and comparison with sugarcane, a related complex polyploid. Theor Appl Genet. 1997;94:409–18. doi: 10.1007/s001220050430. [DOI] [Google Scholar]
- 28.Ming R, Liu SC, Lin YR, da Silva J, Wilson W, Braga D, et al. Detailed alignment of Saccharum and Sorghum chromosomes: comparative organization of closely related diploid and polyploid genomes. Genetics. 1998;150:1663–82. doi: 10.1093/genetics/150.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asnaghi C, Paulet F, Kaye C, Grivet L, Deu M, Glaszmann JC, et al. Application of synteny across Poaceae to determine the map location of a sugarcane rust resistance gene. Theor Appl Genet. 2000;101:962–9. doi: 10.1007/s001220051568. [DOI] [Google Scholar]
- 30.Edmé SJ, Glynn NG, Comstock JC. Genetic segregation of microsatellite markers in Saccharum officinarum and S Spontaneum. Heredity. 2006;97:366–75. doi: 10.1038/sj.hdy.6800888. [DOI] [PubMed] [Google Scholar]
- 31.Pastina MM, Malosetti M, Gazaffi R, Mollinari M, Margarido GR, Oliveira KM, et al. A mixed model QTL analysis for sugarcane multiple-harvest-location trial data. Theor Appl Genet. 2012;124:835–49. doi: 10.1007/s00122-011-1748-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pastina MM, Pinto LR, Oliveira KM, Souza AP, Garcia AAF. Molecular mapping of complex traits. In: Henry RJ, Kole C, editors. Genetics, genomics and breeding of sugarcane. Boca Raton: CRC Press; 2010. [Google Scholar]
- 33.Andru S, Pan Y-B, Thongthawee S, Burner DM, Kimbeng CA. Genetic analysis of the sugarcane (Saccharum spp.) cultivar ‘LCP 85–384′. I. Linkage mapping using AFLP, SSR, and TRAP markers. Theor Appl Genet. 2011;123:77–93. doi: 10.1007/s00122-011-1568-x. [DOI] [PubMed] [Google Scholar]
- 34.Singh RK, Singh SP, Tiwari DK, Srivastava S, Singh SB, Sharma ML, et al. Genetic mapping and QTL analysis for sugar yield-related traits in sugarcane. Euphytica. 2013;191:333–53. doi: 10.1007/s10681-012-0841-7. [DOI] [Google Scholar]
- 35.Aitken KS, McNeil MD, Hermann S, Bundock PC, Kilian A, Heller-Uszynska K, et al. A comprehensive genetic map of sugarcane that provides enhanced map coverage and integrates high-throughput diversity array technology (DArT) markers. BMC Genomics. 2014;15:152. doi: 10.1186/1471-2164-15-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu P, Liao CY, Hu B, Yi KK, Jin WZ, Ni JJ, et al. QTLs and epistasis for aluminum tolerance in rice (Oryza sativa L.) at different seedling stages. Theor Appl Genet. 2000;100:1295–303. doi: 10.1007/s001220051438. [DOI] [Google Scholar]
- 37.Garcia AA, Kido EA, Meza AN, Souza HM, Pinto LR, Pastina MM, et al. Development of an integrated genetic map of a sugarcane (Saccharum spp.) commercial cross, based on a maximum-likelihood approach for estimation of linkage and linkage phases. Theor Appl Genet. 2006;112:298–314. doi: 10.1007/s00122-005-0129-6. [DOI] [PubMed] [Google Scholar]
- 38.Oliveira KM, Pinto LR, Marconi TG, Margarido GRA, Pastina MM, Teixeira LHM, et al. Functional integrated genetic linkage map based on EST-markers for a sugarcane (Saccharum spp.) commercial cross. Mol Breed. 2007;20:189–208. doi: 10.1007/s11032-007-9082-1. [DOI] [Google Scholar]
- 39.Garcia AA, Mollinari M, Marconi TG, Serang OR, Silva RR, Vieira ML, et al. SNP genotyping allows an in-depth characterisation of the genome of sugarcane and other complex autopolyploids. Sci Rep. 2013;3:3399. doi: 10.1038/srep03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Roe B, Macmil S, Yu Q, Murray JE, Tang H, et al. Microcollinearity between autopolyploid sugarcane and diploid sorghum genomes. BMC Genomics. 2010;11:261. doi: 10.1186/1471-2164-11-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- 42.He J, Zhao X, Laroche A, Lu ZX, Liu H, Li Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci. 2014;5:484. doi: 10.3389/fpls.2014.00484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SR, Ramos J, Ashikari M, Virk PS, Torres EA, Nissila E, et al. Development and validation of allele-specific SNP/indel markers for eight yield-enhancing genes using whole-genome sequencing strategy to increase yield potential of rice, Oryza sativa L. Rice. 2016;9:12. doi: 10.1186/s12284-016-0084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller MR, Dunham JP, Amores A, Cresko WA, Johnson EA. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 2007;17:240–8. doi: 10.1101/gr.5681207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang X, Feng Q, Qian Q, Zhao Q, Wang L, Wang A, Guan J, Fan D, Weng Q, Huang T, Dong G, Sang T, Han B. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009;19:1068–1076. doi: 10.1101/gr.089516.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y-F, Poland JA, Wight CP, Jackson EW, Tinker NA. Using genotyping-by-sequencing (GBS) for genomic discovery in cultivated oat. PLoS One. 2014;9:e102448. doi: 10.1371/journal.pone.0102448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barabaschi D, Tondelli A, Desiderio F, Volante A, Vaccino P, Valè G, et al. Next generation breeding. Plant Sci. 2016;242:3–13. doi: 10.1016/j.plantsci.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 49.Byrne S, Czaban A, Studer B, Panitz F, Bendixen C, Asp T. Genome wide allele frequency fingerprints (GWAFFs) of populations via genotyping by sequencing. PLoS One. 2013;8:e57438. doi: 10.1371/journal.pone.0057438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sonah H, Bastien M, Iquira E, Tardivel A, Légaré G, Boyle B, et al. An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLoS One. 2013;8:e54603. doi: 10.1371/journal.pone.0054603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crossa J, Beyene Y, Kassa S, Pérez P, Hickey JM, Chen C, et al. Genomic prediction in maize breeding populations with genotyping-by-sequencing. G3 (Bethesda) 2013;3:1903–26. doi: 10.1534/g3.113.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heslot N, Rutkoski J, Poland J, Jannink J-L, Sorrells ME. Impact of marker ascertainment bias on genomic selection accuracy and estimates of genetic diversity. PLoS One. 2013;8:e74612. doi: 10.1371/journal.pone.0074612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spindel J, Wright M, Chen C, Cobb J, Gage J, Harrington S, et al. Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor Appl Genet. 2013;126:2699–716. doi: 10.1007/s00122-013-2166-x. [DOI] [PubMed] [Google Scholar]
- 54.Liu H, Bayer M, Druka A, Russell JR, Hackett CA, Poland J, et al. An evaluation of genotyping by sequencing (GBS) to map the Breviaristatum-e (ari-e) locus in cultivated barley. BMC Genomics. 2014;15:104. doi: 10.1186/1471-2164-15-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verma S, Gupta S, Bandhiwal N, Kumar T, Bharadwaj C, Bhatia S. High-density linkage map construction and mapping of seed trait QTLs in chickpea (Cicer arietinum L.) using genotyping-by-sequencing (GBS) Sci Rep. 2015;5:17512. doi: 10.1038/srep17512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rafalski A. Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol. 2002;5:94–100. doi: 10.1016/S1369-5266(02)00240-6. [DOI] [PubMed] [Google Scholar]
- 57.Hackett CA, McLean K, Bryan GJ. Linkage analysis and QTL mapping using SNP dosage data in a tetraploid potato mapping population. PLoS One. 2013;8:e63939. doi: 10.1371/journal.pone.0063939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J, Izzah NK, Jayakodi M, Perumal S, Joh HJ, Lee HJ, et al. Genome-wide SNP identification and QTL mapping for black rot resistance in cabbage. BMC Plant Biol. 2015;15:32. doi: 10.1186/s12870-015-0424-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welham SJ, Gogel BJ, Smith AB, Thompson R, Cullis BR. A comparison of analysis methods for late-stage variety evaluation trials. Aust NZ J Stat. 2010;52:125–49. doi: 10.1111/j.1467-842X.2010.00570.x. [DOI] [Google Scholar]
- 60.Aitken KS, Hermann S, Karno K, Bonnett GD, McIntyre LC, Jackson PA. Genetic control of yield related stalk traits in sugarcane. Theor Appl Genet. 2008;117:1191–203. doi: 10.1007/s00122-008-0856-6. [DOI] [PubMed] [Google Scholar]
- 61.Margarido GRA, Pastina MM, Souza AP, Garcia AAF. Multi-trait multi-environment quantitative trait loci mapping for a sugarcane commercial cross provides insights on the inheritance of important traits. Mol Breeding. 2015;35:175. doi: 10.1007/s11032-015-0366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gazaffi R, Margarido GRA, Pastina MM, Mollinari M, Garcia AAF. A model for quantitative trait loci mapping, linkage phase, and segregation pattern estimation for a full-sib progeny. Tree Genet Genomes. 2014;10:791–801. doi: 10.1007/s11295-013-0664-2. [DOI] [Google Scholar]
- 63.Souza LM, Gazaffi R, Mantello CC, Silva CC, Garcia D, Le Guen V, et al. QTL mapping of growth-related traits in a full-sib family of rubber tree (hevea brasiliensis) evaluated in a sub-tropical climate. PLoS One. 2013;8:e61238. doi: 10.1371/journal.pone.0061238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeng ZB. Precision mapping of quantitative trait loci. Genet. 1994;136:1457–68. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vettore AL, da Silva FR, Kemper EL, Souza GM, da Silva AM, Ferro MI, et al. Analysis and functional annotation of an expressed sequence tag collection for tropical crop sugarcane. Genome Res. 2003;13:2725–35. doi: 10.1101/gr.1532103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cardoso-Silva CB, Costa EA, Mancini MC, Balsalobre TW, Canesin LE, Pinto LR, et al. De novo assembly and transcriptome analysis of contrasting sugarcane varieties. PLoS One. 2014;9:e88462. doi: 10.1371/journal.pone.0088462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Souza GM, Berges H, Bocs S, Casu R, D’Hont A, Ferreira JE, et al. The sugarcane genome challenge: strategies for sequencing a highly complex genome. Trop Plant Biol. 2011;4:145–56. doi: 10.1007/s12042-011-9079-0. [DOI] [Google Scholar]
- 68.de Setta N, Monteiro-Vitorello CB, Metcalfe CJ, Cruz GM, Del Bem LE, Vicentini R, et al. Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics. 2014;15:540. doi: 10.1186/1471-2164-15-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Metcalfe CJ, Oliveira SG, Gaiarsa JW, Aitken KS, Carneiro MS, Zatti F, et al. Using quantitative PCR with retrotransposon-based insertion polymorphisms as markers in sugarcane. J Exp Bot. 2015;66:4239–50. doi: 10.1093/jxb/erv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Al-Janabi SM, Forget L, Dookun A. An improved and rapid protocol for the isolation of polysaccharide-and polyphenol-free sugarcane DNA. Plant Mol Biol Rep. 1999;17:1–8. doi: 10.1023/A:1017213630972. [DOI] [Google Scholar]
- 71.Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, et al. Tassel-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One. 2014;9:e90346. doi: 10.1371/journal.pone.0090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grativol C, Regulski M, Bertalan M, McCombie WR, da Silva FR, Zerlotini Neto A, et al. Sugarcane genome sequencing by methylation filtration provides tools for genomic research in the genus Saccharum. Plant J. 2014;79:162–72. doi: 10.1111/tpj.12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, et al. The sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–6. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 74.Serang O, Mollinari M, Garcia AA. Efficient exact maximum a posteriori computation for Bayesian SNP genotyping in polyploids. PLoS One. 2012;7:e30906. doi: 10.1371/journal.pone.0030906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gu Z, Gu L, Eils R, Schlesner M, Brors B. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 76.Oliveira KM, Pinto LR, Marconi TG, Mollinari M, Ulian EC, Chabregas SM, et al. Characterization of new polymorphic functional markers for sugarcane. Genome. 2009;52:191–209. doi: 10.1139/G08-105. [DOI] [PubMed] [Google Scholar]
- 77.Pinto LR, Oliveira KM, Marconi T, Garcia AAF, Ulian EC, de Souza AP. Characterization of novel sugarcane expressed sequence tag microsatellites and their comparison with genomic SSRs. Plant Breed. 2006;125:378–84. doi: 10.1111/j.1439-0523.2006.01227.x. [DOI] [Google Scholar]
- 78.Marconi TG, Costa EA, Miranda HR, Mancini MC, Cardoso-Silva CB, Oliveira KM, et al. Functional markers for gene mapping and genetic diversity studies in sugarcane. BMC Res Notes. 2011;4:264. doi: 10.1186/1756-0500-4-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singh RK, Jena SN, Khan S, Yadav S, Banarjee N, Raghuvanshi S, et al. Development, cross-species/genera transferability of novel EST-SSR markers and their utility in revealing population structure and genetic diversity in sugarcane. Gene. 2013;524:309–29. doi: 10.1016/j.gene.2013.03.125. [DOI] [PubMed] [Google Scholar]
- 80.Cordeiro GM, Taylor GO, Henry RJ. Characterisation of microsatellite markers from sugarcane (Saccharum spp.), a highly polyploid species. Plant Sci. 2000;155:161–8. doi: 10.1016/S0168-9452(00)00208-9. [DOI] [PubMed] [Google Scholar]
- 81.Raboin L-M, Oliveira KM, Lecunff L, Telismart H, Roques D, Butterfield M, et al. Genetic mapping in sugarcane, a high polyploid, using bi-parental progeny: identification of a gene controlling stalk colour and a new rust resistance gene. Theor Appl Genet. 2006;112:1382–91. doi: 10.1007/s00122-006-0240-3. [DOI] [PubMed] [Google Scholar]
- 82.Brown SM, Hopkins MS, Mitchell SE, Senior ML, Wang TY, Duncan RR, et al. Multiple methods for the identification of polymorphic simple sequence repeats (SSRs) in sorghum [sorghum bicolor (L.) Moench] Theor Appl Genet. 1996;93:190–8. doi: 10.1007/BF00225745. [DOI] [PubMed] [Google Scholar]
- 83.Kong L, Dong J, Hart GE. Characteristics, linkage-map positions, and allelic differentiation of sorghum bicolor (L.) Moench DNA simple-sequence repeats (SSRs) Theor Appl Genet. 2000;101:438–48. doi: 10.1007/s001220051501. [DOI] [Google Scholar]
- 84.Wang ML, Wang ML, Barkley NA, Yu J, Dean RE, Newman ML, et al. Transfer of simple sequence repeat (SSR) markers from major cereal crops to minor grass species for germplasm characterization and evaluation. Plant Genet Resour Newsl. 2005;3:45–57. doi: 10.1079/PGR200461. [DOI] [Google Scholar]
- 85.Li G, Quiros CF. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction : its application to mapping and gene tagging in Brassica. Theor Appl Genet. 2001;103:455–61. doi: 10.1007/s001220100570. [DOI] [Google Scholar]
- 86.Alwala S, Suman A, Arro JA, Veremis JC, Kimbeng CA. Target region amplification polymorphism (TRAP) for assessing genetic diversity in sugarcane germplasm collections. Crop Sci. 2006;46:448–55. doi: 10.2135/cropsci2005.0274. [DOI] [Google Scholar]
- 87.Creste S, Sansoli DM, Tardiani ACS, Silva DN, Gonçalves FK, Fávero TM, et al. Comparison of AFLP, TRAP and SSRs in the estimation of genetic relationships in sugarcane. Sugar Tech. 2010;12:150–54. doi: 10.1007/s12355-010-0029-1. [DOI] [Google Scholar]
- 88.Suman A, Ali K, Arro J, Parco AS, Kimbeng CA, Baisakh N. Molecular diversity among members of the Saccharum complex assessed using TRAP Markers based on lignin-related genes. Bioenerg Res. 2012;5:197–205. doi: 10.1007/s12155-011-9123-9. [DOI] [Google Scholar]
- 89.Hu J, Vick BA. Target region amplification polymorphism: a novel marker technique for plant genotyping. Plant Mol Biol Rep. 2003;21:289–94. doi: 10.1007/BF02772804. [DOI] [Google Scholar]
- 90.Creste S, Neto AT, Figueira A. Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Mol Biol Rep. 2001;19:299–306. doi: 10.1007/BF02772828. [DOI] [Google Scholar]
- 91.Margarido GR, Souza AP, Garcia AA. OneMap: software for genetic mapping in outcrossing species. Hereditas. 2007;144:78–9. doi: 10.1111/j.2007.0018-0661.02000.x. [DOI] [PubMed] [Google Scholar]
- 92.Wu R, Ma C-X, Painter I, Zeng Z-B. Simultaneous maximum likelihood estimation of linkage and linkage phases in outcrossing species. Theor Popul Biol. 2002;61:349–63. doi: 10.1006/tpbi.2002.1577. [DOI] [PubMed] [Google Scholar]
- 93.Wu R, Ma C-X, Wu SS, Zeng Z-B. Linkage mapping of sex-specific differences. Genet Res. 2002;79:85–96. doi: 10.1017/S0016672301005389. [DOI] [PubMed] [Google Scholar]
- 94.Jiang C, Zeng Z-B. Mapping quantitative trait loci with dominant and missing markers in various crosses from two inbred lines. Genetica. 1997;101:47–58. doi: 10.1023/A:1018394410659. [DOI] [PubMed] [Google Scholar]
- 95.Voorrips RE. Computer Note MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 1994;93:77–8. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 96.Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen L, Storey JD. Relaxed significance criteria for linkage analysis. Genetics. 2006;173:2371–81. doi: 10.1534/genetics.105.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.R Development Core Team . R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 99.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 100.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–86. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li H, Vikram P, Singh RP, Kilian A, Carling J, Song J, et al. A high density GBS map of bread wheat and its application for dissecting complex disease resistance traits. BMC Genomics. 2015;16:216. doi: 10.1186/s12864-015-1424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heffelfinger C, Fragoso CA, Moreno MA, Overton JD, Mottinger JP, Zhao H, et al. Flexible and scalable genotyping-by-sequencing strategies for population studies. BMC Genomics. 2014;15:979. doi: 10.1186/1471-2164-15-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Beissinger TM, Hirsch CN, Sekhon RS, Foerster JM, Johnson JM, Muttoni G, et al. Marker density and read depth for genotyping populations using genotyping-by-sequencing. Genetics. 2013;193:1073–81. doi: 10.1534/genetics.112.147710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang Z, Wang H, Michal JJ, Zhou X, Liu B, Woods LC, et al. Genome wide sampling sequencing for SNP genotyping: methods, challenges and future development. Int J Biol Sci. 2016;12:100–8. doi: 10.7150/ijbs.13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grivet L, Arruda P. Sugarcane genomics: depicting the complex genome of an important tropical crop. Curr Opin Plant Biol. 2002;5:122–7. doi: 10.1016/S1369-5266(02)00234-0. [DOI] [PubMed] [Google Scholar]
- 106.Mollinari M, Serang O. Quantitative SNP genotyping of polyploids with MassARRAY and other platforms. Methods Mol Biol. 2015;1245:215–41. doi: 10.1007/978-1-4939-1966-6_17. [DOI] [PubMed] [Google Scholar]
- 107.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2012;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reffay N, Jackson PA, Aitken KS, Hoarau JY, D’hont A, Besse P, et al. Characterisation of genome regions incorporated from an important wild relative into Australian sugarcane. Mol Breed. 2005;15:367–81. doi: 10.1007/s11032-004-7981-y. [DOI] [Google Scholar]
- 109.Aitken KS, Jackson PA, McIntyre CL. A combination of AFLP and SSR markers provides extensive map coverage and identification of homo(eo)logous linkage groups in a sugarcane cultivar. Theor Appl Genet. 2005;110:789–801. doi: 10.1007/s00122-004-1813-7. [DOI] [PubMed] [Google Scholar]
- 110.Aitken KS, Jackson PA, McIntyre CL. Construction of a genetic linkage map for Saccharum officinarum incorporating both simplex and duplex markers to increase genome coverage. Genome. 2007;50:742–56. doi: 10.1139/G07-056. [DOI] [PubMed] [Google Scholar]
- 111.Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci U S A. 2013;110:8057–62. doi: 10.1073/pnas.1217133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Poland JA, Brown PJ, Sorrells ME, Jannink JL. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One. 2012;7:e32253. doi: 10.1371/journal.pone.0032253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang N, Li F, Chen B, Xu K, Yan G, Qiao J, et al. Genome-wide investigation of genetic changes during modern breeding of Brassica napus. Theor Appl Genet. 2014;127:1817–29. doi: 10.1007/s00122-014-2343-6. [DOI] [PubMed] [Google Scholar]
- 114.Butterfield MK, D’Hont A, Berding N. The sugarcane genome: a synthesis of current understanding, and lessons for breeding and biotechnology. Proc South African Sugar Technology Assoc. 2001;75:1–5. [Google Scholar]
- 115.Aitken KS, McNeil MD, Berkman PJ, Hermann S, Kilian A, Bundock PC, et al. Comparative mapping in the Poaceae family reveals translocations in the complex polyploid genome of sugarcane. BMC Plant Biol. 2014;14:190. doi: 10.1186/s12870-014-0190-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Glaszmann JC, Dufour P, Grivet L, D'Hont A, Deu M, Paulet F, et al. Comparative genome analysis between several tropical grasses. Euphytica. 1997;96:13–21. doi: 10.1023/A:1002987620250. [DOI] [Google Scholar]
- 117.Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008;320:486–8. doi: 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- 118.Salse J, Abrouk M, Murat F, Quraishi UM, Feuillet C. Improved criteria and comparative genomics tool provide new insights into grass paleogenomics. Brief Bioinform. 2009;10:619–30. doi: 10.1093/bib/bbp037. [DOI] [PubMed] [Google Scholar]
- 119.Margarido GRA, Heckermann D. ConPADE: Genome Assembly Ploidy Estimation from next-generation sequencing data. PLoS Comput Biol. 2015;11(4):e1004229. doi: 10.1371/journal.pcbi.1004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Malosetti M, Ribaut J-M, van Eeuwijk FA. The statistical analysis of multi-environment data: modeling genotype-by-environment interaction and its genetic basis. Front Physiol. 2013;4:44. doi: 10.3389/fphys.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Al-Janabi SM, Parmessur Y, Kross H, Dhayan S, Saumtally S, Ramdoyal K, et al. Identification of a major quantitative trait locus (QTL) for yellow spot (Mycovellosiella koepkei) disease resistance in sugarcane. Mol Breed. 2007;19:1–14. doi: 10.1007/s11032-006-9008-3. [DOI] [Google Scholar]
- 122.Sills GR, Bridges W, Al-Janabi SM, Sobral BWS. Genetic analysis of agronomic traits in a cross between sugarcane (Saccharum officinarum L.) and its presumed progenitor. (S robustum Brandes & Jesw. Ex Grassl) Mol Breed. 1995;1:355–63. doi: 10.1007/BF01248413. [DOI] [Google Scholar]
- 123.Daugrois JH, Grivet L, Roques D, Hoarau JY, Lombard H, Glaszmann JC, et al. A putative major gene for rust resistance linked with a RFLP marker in sugarcane cultivar ‘R570. Theor Appl Genet. 1996;92:1059–64. doi: 10.1007/BF00224049. [DOI] [PubMed] [Google Scholar]
- 124.Guimarães CT, Sills GR, Sobral BW. Comparativemapping of Andropogoneae: Saccharum L. (sugarcane) and its relation to sorghum and maize. Proc Natl Acad Sci U S A. 1997;94:14261–6. doi: 10.1073/pnas.94.26.14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Asnaghi C, D’hont A, Glaszmann JC, Rott P. Resistance of sugarcane cultivar R570 to Puccinia melanocephala from different geographic locations. Plant Dis. 2001;85:282–6. doi: 10.1094/PDIS.2001.85.3.282. [DOI] [PubMed] [Google Scholar]
- 126.Ming R, Liu SC, Moore PH, Irvine JE, Paterson AH. QTL analysis in a complex autopolyploid: genetic control of sugar content in sugarcane. Genome Res. 2001;11:2075–84. doi: 10.1101/gr.198801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ming R, Del Monte TA, Hernandez E, Moore PH, Irvine JE, Paterson AH. Comparative analysis of QTLs affecting plant height and flowering among closely-related diploid and polyploid genomes. Genome. 2002;45:794–803. doi: 10.1139/g02-042. [DOI] [PubMed] [Google Scholar]
- 128.Ming R, Wang W, Draye X, Moore H, Irvine E, Paterson H. Molecular dissection of complex traits in autopolyploids: mapping QTLs affecting sugar yield and related traits in sugarcane. Theor Appl Genet. 2002;105:332–45. doi: 10.1007/s00122-001-0861-5. [DOI] [PubMed] [Google Scholar]
- 129.Hoarau JY, Grivet L, Offmann B, Raboin LM, Diorflar JP, Payet J, et al. Genetic dissection of a modern sugarcane cultivar (Saccharum spp.).II. Detection of QTLs for yield components. Theor Appl Genet. 2002;105:1027–37. doi: 10.1007/s00122-002-1047-5. [DOI] [PubMed] [Google Scholar]
- 130.Silva JA, Bressiani JA. Sucrose synthase molecular marker associated with sugar content in elite sugarcane progeny. Genet Mol Biol. 2005;28:294–8. doi: 10.1590/S1415-47572005000200020. [DOI] [Google Scholar]
- 131.Aitken KS, Jackson PA, McIntyre CL. Quantitative trait loci identified for sugar related traits in a sugarcane (Saccharum spp.) cultivar x Saccharum officinarum population. Theor Appl Genet. 2006;112:1306–17. doi: 10.1007/s00122-006-0233-2. [DOI] [PubMed] [Google Scholar]
- 132.Raboin LM, Pauquet J, Butterfield M, D’hont A, Glaszmann JC. Analysis of genome-wide linkage disequilibriumin the highly polyploid sugarcane. Theor Appl Genet. 2008;116:701–14. doi: 10.1007/s00122-007-0703-1. [DOI] [PubMed] [Google Scholar]
- 133.Wei X, Jackson PA, McIntyre CL, Aitken KS, Croft B. Associations between DNA markers and resistance to diseases in sugarcane and effects of population substructure. Theor Appl Genet. 2006;114:155–64. doi: 10.1007/s00122-006-0418-8. [DOI] [PubMed] [Google Scholar]
- 134.Piperidis N, Jackson PA, D’hont A, Besse P, Hoarau J, Courtois B, et al. Comparative genetics in sugarcane enables structured map enhancement and validation of marker-trait associations. Mol Breed. 2008;21:233–47. doi: 10.1007/s11032-007-9124-8. [DOI] [Google Scholar]
- 135.Pinto LR, Garcia AAF, Pastina MM, Teixeira LHM, Bressiani JA, Ulian EC, et al. Analysis of genomic and functional RFLP derived markers associated with sucrose content, fiber and yield QTLs in a sugarcane (Saccharum spp.) commercial cross. Euphytica. 2010;172:313–27. doi: 10.1007/s10681-009-9988-2. [DOI] [Google Scholar]
- 136.Jordan DR, Casu RE, Besse P, Carroll BC, Berding N, McIntyre CL. Markers associated with stalk number and suckering in sugarcane colocate with tillering and rhizomatousness QTLs in sorghum. Genome. 2004;47:988–93. doi: 10.1139/g04-040. [DOI] [PubMed] [Google Scholar]
- 137.Craxton M. Synaptotagmin gene content of the sequencedgenomes. BMC Genomics. 2004;5:43. doi: 10.1186/1471-2164-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Coussens L, Parker PJ, Rhee L, Yang-Feng TL, Chen E, Waterfield MD, et al. Multiple, distinct forms of bovine and human protein kinase C suggest diversity in cellular signaling pathways. Science. 1986;233:859–66. doi: 10.1126/science.3755548. [DOI] [PubMed] [Google Scholar]
- 139.Schapire AL, Voigt B, Jasik J, Rosado A, Lopez-Cobollo R, Menzel D, et al. Arabidopsis Synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell. 2008;20:3374–88. doi: 10.1105/tpc.108.063859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shin DH, Choi M-G, Lee HK, Cho M, Choi S-B, Choi G, et al. Calcium dependent sucrose uptake links sugar signaling to anthocyanin biosynthesis in Arabidopsis. Biochem Biophys Res Commun. 2013;430:634–9. doi: 10.1016/j.bbrc.2012.11.100. [DOI] [PubMed] [Google Scholar]
- 141.Hochmal AK, Schulze S, Trompelt K, Hippler M. Calcium-dependent regulation of photosynthesis. Biochim Biophys Acta. 1847;2015:993–1003. doi: 10.1016/j.bbabio.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 142.Papini-Terzi FS, Rocha FR, Vêncio RZN, Felix JM, Branco DS, Waclawovsky AJ, Bem LEVD, Lembke CG, Costa MDL, Junior MYN, Vicentini R, Vincentz MGA, Ulian EC, Menossi M, Souza GM. Sugarcane genes associated with sucrose contente. BMC Genomics. 2009;10:120. doi: 10.1186/1471-2164-10-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Robson F, Costa MM, Hepworth SR, Vizir I, Piñeiro M, Reeves PH, et al. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001;28:619–31. doi: 10.1046/j.1365-313x.2001.01163.x. [DOI] [PubMed] [Google Scholar]
- 144.Chou ML, Shih MC, Chan MT, Liao SY, Hsu CT, Haung YT, et al. Global transcriptome analysis and identification of a CONSTANS-like gene family in the orchid Erycina pusilla. Planta. 2013;237:1425–41. doi: 10.1007/s00425-013-1850-z. [DOI] [PubMed] [Google Scholar]
- 145.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–94. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- 146.Kovi MR, Sablok G, Bai X, Wendell M, Rognli OA, Yu H, et al. Expression patterns of photoperiod and temperature regulated heading date genes in Oryza sativa. Comput Biol Chem. 2013;45:36–41. doi: 10.1016/j.compbiolchem.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 147.Liu T, Zhu S, Tang Q, Tang S. Identification of a CONSTANS homologous gene with distinct diurnal expression patterns in varied photoperiods in ramie (Boehmeria nivea L Gaud) Gene. 2015;560:63–70. doi: 10.1016/j.gene.2015.01.045. [DOI] [PubMed] [Google Scholar]
- 148.Islam MS, Fang DD, Thyssen GN, Delhom CD, Liu Y, Kim HJ. Comparative fiber property and transcriptome analyses reveal key genes potentially related to high fiber strength in cotton (Gossypium hirsutum L.) line MD52ne. BMC Plant Biol. 2016;16:36. doi: 10.1186/s12870-016-0727-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Manetti ME, Rossi M, Cruz GMQ, Saccaro NL, Nakabashi M, Altebarmakian V, et al. Mutator system derivatives isolated from sugarcane genome sequence. Trop Plant Biol. 2012;5:233–43. doi: 10.1007/s12042-012-9104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The GBS sequence data files supporting the conclusions of this article are in the process of submission to the NCBI Sequence Read Archive (SRA), http://www.ncbi.nlm.nih.gov/sra, with accession number SRP092493.


