Abstract
Background
Plants respond to abiotic stress on physiological, biochemical and molecular levels. This includes a global change in their cellular proteome achieved by changes in the pattern of their protein synthesis and degradation. The ubiquitin-proteasome system (UPS) is a key player in protein degradation in eukaryotes. Proteins are marked for degradation by the proteasome by coupling short chains of ubiquitin polypeptides in a three-step pathway. The last and regulatory stage is catalyzed by a member of a large family of substrate-specific ubiquitin ligases.
Results
We have identified AtPUB46 and AtPUB48—two paralogous genes that encode ubiquitin ligases (E3s)—to have a role in the plant environmental response. The AtPUB46, −47, and −48 appear as tandem gene copies on chromosome 5, and we present a phylogenetic analysis that traces their evolution from an ancestral PUB-ARM gene. Single homozygous T-DNA insertion mutants of AtPUB46 and AtPUB48 displayed hypersensitivity to water stress; this was not observed for similar mutants of AtPUB47. Although the three genes show a similar spatial expression pattern, the steady state levels of their transcripts are differentially affected by abiotic stresses and plant hormones.
Conclusions
AtPUB46 and AtPUB48 encode plant U-Box E3s and are involved in the response to water stress. Our data suggest that despite encoding highly homologous proteins, AtPUB46 and AtPUB48 biological activity does not fully overlap.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0963-5) contains supplementary material, which is available to authorized users.
Background
Plants respond to abiotic stress with major physiological, biochemical and molecular changes that lead to a new homeostasis. These changes include a global alteration of the plant transcriptome, proteome, and metabolome that result from a new balance between the rates of cellular biosynthesis and degradation activities. Enhanced protein degradation in stress conditions leads to a reduced steady state level of proteins whose optimal levels are much lower in stress conditions than in non-stress conditions. Furthermore, abiotic stress conditions induce the production of reactive oxygen species (ROS) [1] that can also result in irreversibly oxidized proteins and other biologically active polymers that are targeted for degradation [2–4].
The ubiquitin-proteasome system (UPS) is a central eukaryotic system for regulated protein degradation [5, 6]. The proteolytic activity resides in the 26S proteasome present in both the cytoplasm and the nucleus. Proteins are targeted for degradation by the 26S proteasome by covalent attachment of a short chain of ubiquitin molecules [5] performed by a sequence of three enzymes, a ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2), and a ubiquitin ligase (E3) that recognizes the substrate [7]. Thus, it is the E3 that determines whether a given protein will be sent for degradation by the 26S proteasome. Although protein ubiquitylation is mostly associated with degradation, ubiquitylation also plays a role in signaling and modification of protein activities [6] giving the E3s a critical role in cell function.
Over 5% of Arabidopsis genes encode proteins of the UPS with the majority of UPS-related genes (ca 1,700) encoding E3s. E3s can be divided into subfamilies based on their structure and primary amino acid sequence. These include multimeric E3s such as cullin-based E3s, and monomeric E3s such as the RING and U-box protein families [5, 6]. Many UPS genes are induced in response to abiotic stress: these include genes that encode ubiquitin, E2s, E3s and proteasome subunits (reviewed by [8–11]).
Plant U-box (PUB) proteins are a small family of proteins with the U-box motif [10, 12, 13]. The U-box comprises ca. 70 amino acids and resembles a modified RING finger that forms a similar structure stabilized by salt-bridges and hydrogen bonds [14, 15]. PUBs have E3 activity [10, 12, 14]. Of the more than 1000 E3 genes found in Arabidopsis and rice, the PUBs comprise a small family of 64 and 77 genes, respectively [12, 13, 16]. Although PUBs are just a small fraction of the plant E3s, they are far more abundant than in other organisms such as yeast and animals—each of these encodes fewer than ten U-box E3s [10, 12, 13]. PUBs are monomeric: the largest PUB subgroup comprises proteins with a N-terminal U-box followed by Armadillo (ARM) repeats. The ARM motif is a ca. 40 residue long that often appears in tandem non-identical copies in a single PUB and functions in protein-protein interactions [17, 18].
PUB E3s are involved in diverse biological processes such as development, self-incompatibility, and response to hormones. They are widely connected with the plant stress response [8–11, 19, 20]. PUBs play an essential role in drought [21–24], salt stress [21, 25, 26], temperature stress [21, 27], oxidative stress [28], and in the response to phosphate starvation [29].
Given the central role of E3s in selecting specific proteins for degradation, the identification of E3s that are active in the response to a defined stress is an important step towards elucidating the pathways that regulate this response. We therefore initiated a screen of homozygous Arabidopsis E3-T-DNA insertion mutant plants for their response to water stress. The AtPUB46- T-DNA insertion mutants were found to be hypersensitive to water stress compared with WT plants. The study was then expanded to the adjacent AtPUB47 and AtPUB48 genes, which encode highly homologous proteins. As found for AtPUB46 mutants, T-DNA insertion mutants of AtPUB48 displayed increased sensitivity to water stress. On the other hand, sensitivity of AtPUB47 T-DNA insertion mutants for water stress was not affected. Cell and tissue expression patterns of the AtPUB46-48 genes are similar; however, we found that they differ in their response to hormones and abiotic stress cues. All three genes encode active E3s as shown using recombinant AtPUB46-48 proteins produced in bacteria. Thus, our results suggest that AtPUB46 and AtPUB48 play a role in establishing a new protein homeostasis via the UPS in response to drought.
Methods
Plant material
All experiments were carried out with Arabidopsis thaliana ecotype Columbia.
T-DNA insertion-mutants
T-DNA insertion lines prepared by the Salk Institute Genomic Analysis Laboratory [30] were obtained from the Arabidopsis Resource Center, Columbus Ohio. The lines were: Atpub46-1, SALK_096071, T-DNA insert in exon 1, 36 bp from the translation start codon; Atpub46-2, SALK_109233, 129 bp upstream of the translation start codon; Atpub47-1, SALK_018208, 103 bp into exon 2; Atpub47-2, SALK_056774, in exon 1, 64 bp downstream of translation start codon; Atpub48-1, SALK_057909, 5’ UTR, 97 bp upstream of the translation start codon; Atpub48-2, SALK_086659, exon 1, 285 bp downstream of the translation start codon. All lines were homozygous for T-DNA insertion. Homozygosity was confirmed by PCR analysis.
Plant transformation and selection of transgenic plants
Recombinant plasmids were introduced into Agrobacterium GV-3101, and the transformed bacteria were used for genetic transformation of Arabidopsis by the floral dip method [31]. Transgenic plants were selected on plates containing 30 μg/ml hygromycin. All experiments were performed on T3 generation homozygous plants containing single-site T-DNA inserts. At least three independent-transformant lines were used for each assay.
Construct design
Promoter::GUS constructs
DNA sequences of the respective PUB genes were isolated by PCR using Arabidopsis genomic DNA and promoter-specific DNA primer pairs (Additional file 1: Table S1), and subcloned into the pCAMBIA 1391Z vector upstream of the sequence encoding GUS. Histochemical GUS staining was performed as described [32].
Constructs for expression of AtPUBs::eGFP fusion proteins
cDNA amplified DNA fragments were fused to the N-terminus of EGFP in the pSAT4-EGFP-N1 plasmid [33] downstream of the constitutive CaMV 35S promoter. The CaMV 35S:AtPUBs::EGFP fusion cassette was ligated into pCAMBIA 1302 replacing the CaMV 35S:6xHis-GFP sequence originally found in this vector.
Constructs for expressing recombinant proteins in E. coli
The DNA sequences encoding full-length Arabidopsis proteins were prepared by PCR using cDNA from Arabidopsis seedlings as a template, and primer sets described in Additional file 1: Table S1. The resulting protein-encoding sequences were sub-cloned, in-frame, into the indicated bacterial expression vectors. The following constructs for the expression of recombinant Arabidopsis proteins in E. coli were made: UBE8 and UBE10 in pHIS-Parallel2 and Arabidopsis PUB46, PUP47, and PUB48 in pGST-Parallel2 [34]. Plasmid Ube1/PET21d, expressing 6xHis-UBE1 was purchased from Addgene (http://www.addgene.org/34965/). All constructs were sequenced to verify that they are in frame and that there are no mutations in the amplified sequences.
Plant growth and Stress application
Seeds were surface sterilized and cold treated before sowing as described [32]. Plants were grown in Petri dishes containing half strength Murashige and Skoog (0.5 x MS) nutrient solution mix [35] supplemented with 0.5% sucrose and 0.6% agarose, or in pots containing planting mix at 22–25 °C and 50% humidity in a 12 h light/12 h dark regime. Where indicated, plates also contained hormones, antibiotics, or abiotic-stress agents. Application of stress and hormones to two-week old seedlings was performed by the transfer of plate-grown seedlings to Whatman No 1 filter paper soaked in 0.5 x MS and with the indicated concentration of the hormone/stress-inducing chemical.
Seed germination and cotyledon greening assay
Surface-sterilized cold-treated seeds were sown on Petri plates containing 0.5 x MS, 0.7% agar, and when applied, the indicated MV, NaCl, or mannitol. Plates were incubated at 22 °C in a 16 h light/8 h dark regime. Green seedlings were scored 5 days later.
Drought tolerance
Plants were grown for 3 weeks in pots containing equal amounts of potting mix under non-stressed conditions. Water was then withheld and plant wilting and drying was followed daily.
Water loss
Rosettes of one-month old plants were cut and placed with their abaxial side on weigh boats. Samples were weighed immediately after cutting, and in ~10 min intervals. Data from each plant was normalized to its weight at time 0.
Photosynthetic efficiency
Photosynthetic efficiency of photosystem II was assayed using MINI-PAM-II fluorometer (Walz GmbH, Effeltrich, Germany). Plants were dark-adapted for 30 min. Each genotype contained 8 soil grown plants. Chlorophyl fluorescence emitted from rosette leaves of controlled and stressed plants was assayed in dark-adapted plants (Fo), and maximum fluorescence values were measured following an intensed light flash (Fm). The Fv/Fm values representing photosynthetic efficiency were calculated by (Fm-Fo)/Fm.
Transcript levels
RNA isolation, cDNA synthesis, primer design and RT-qPCR assays for determining relative steady state transcript levels were performed as previously described [36]. Primers are listed in Additional file 1: Table S1.
Recombinant protein expression
E.coli BL21 (DE3) pLYS cells were transformed with the plasmids described above. Cultures were grown at 37 °C to OD600 = 0.5. Cultures were then cooled to 16 °C, and expression of recombinant proteins was induced by adding 0.5 mM IPTG. Bacterial cells were harvested after 16 h at 16 °C and suspended in the buffer recommended by the manufacturers of the applicable affinity chromatography resins. Cells were sonicated, and the homogenates were cleared by centrifugation followed by supernatant loading onto the appropriate column. His-tagged proteins were purified on Ni-Charged resin (GenScript, New Jersey, USA), GST-tagged proteins on glutathione resin (GenScript, New Jersey, USA), and MBP-tagged proteins on amylose resin (New England BioLabs, Massachusetts, USA) according to protocols recommended by the manufacturers. Purified proteins were concentrated and chromatography elution buffers were exchanged with phosphate buffered saline (PBS) using Vivaspin 6 centrifugation ultrafilters (Sartorius, Germany). Protein aliquots were stored at −75 °C.
In vitro ubiquitylation assay
An in vitro ubiquitylation assay was performed using a modification of a previously described assay [37]. The 30 μl reaction mixtures contained 5 μg of ubiquitin (Sigma-Aldrich, USA), 100 ng of the his-tagged human E1 Ube1, 500 ng each of the indicated his-tagged Arabidopsis E2 and GST-tagged E3 in a reaction buffer containing 25 mM Tris–HCl, pH 7.5, 1 mM MgCl2, 1 mM ATP, and 0.5 mM DTT. Reactions were incubated at 30 °C for 2 h and terminated by adding SDS gel sample buffer and heating at 95 °C for 5 min. Proteins were resolved by SDS-PAGE, electroblotted onto nitrocellulose membranes, and probed by western blot analysis using an anti-ubiquitin antibody.
Statistical analyses
Each experiment was performed with at least three biological replicates with more than 50 plants in each treatment. The results are presented as mean ± SE [calculated using SPSS software version 18 (SPSS Inc, Chicago, IL). Differences between groups were analyzed by Tukey’s HSD post-hoc test (P ≥ 0.05).
Results and discussion
Gene organization of At5G18320, At5G18330 and At5G18340 and domain organization of the AtPUB46, AtPUB47 and AtPUB48 proteins they encode
We screened Arabidopsis E3 T-DNA insertion mutants for altered tolerance to water stress and found a homozygous T-DNA insertion mutant of AtPUB46 with enhanced sensitivity to water stress. This gene is a member of a cluster of 3 loci (At5G18320, At5G18330 and At5G18340) on the upper arm of chromosome 5 that encode highly homologous U-box protein ligases (AtPUB46 to AtPUB48, respectively, Fig. 1a). These three genes are in the same orientation and are encoded by the lower DNA strand. A phylogenetic tree of the 64 PUB proteins encoded by Arabidopsis indicated that AtPUB46, AtPUB47, and AtPUB48 form a small distinct cluster [13, 20]. Amino acid alignment shows that AtPUB46 shares high similarity with both AtPUB47 and AtPUB48 (63–65% identity and 73–75% similarity), whereas AtPUB47 and AtPUB48 are somewhat less similar to one another (55% identity and 65% similarity). Each protein has a U-box close to the N-terminus followed by three copies of an ARM motif (ARM1-3; Fig. 1b), which was first identified in Drosophila Armadillo protein and shown to function in protein-protein interactions [38]. The corresponding ARM motifs of the three paralogs are very similar with sequence identities of 60–71% for each of the corresponding ARM1, ARM2, and ARM3 motifs. On the other hand, there is much lower homology between the three consecutive ARM motifs (ARM1, ARM2 and ARM3) of each protein. The high degree of similarity between the corresponding ARM1-3 motifs of the three AtPUB46-48 proteins is evidence for gene duplication of a primordial PUB-ARM1-3 gene. Gene duplication is common in Arabidopsis with 15–20% of the genome comprising tandem-arrayed genes (TAG) [39, 40]. Only 17% of these duplication events have resulted in tri-genes of which a large proportion are expressed in response to abiotic stresses [41]. Furthermore, the genes encoding AtPUB46-48 have a single intron that intervenes between the codons that encode residues Q and T within the PUB motif. These correspond to residues 96 and 97 in AtPUB46, respectively (Fig. 1b, arrows). Interestingly, only five of the 64 Arabidopsis PUB genes (PUB9, 24, 46, 47, and 48) have a single intron. AtPUB9 on chromosome 3 has an intron at the same position as in AtPUB46-48 (see Fig. 1b, arrows). Interestingly, the AtPUB9 protein has the highest similarity to AtPUB46-48 on the phylogenetic tree constructed based on the U-box domain of AtPUBs [13] suggesting shared ancestry. The fifth intron-containing PUB gene, AtPUB24, has its intron at a different position and is less similar to AtPUB9 and AtPUB46-48. Thus, the first gene duplication must have given rise to AtPUB9 and the ancestor of the tri-genes. Subsequent gene multiplication leading to AtPUB46, AtPUB47 and AtPUB48 probably resulted from gene conversion or unequal crossing over. This was certainly not from retrotransposition because the three genes have retained their intron. Furthermore, we can exclude whole genome duplication since the tri-genes are located in tandem on the same chromosome. This reconstructed evolutionary history is similar to that described for the Arabidopsis MYB genes [42]. We therefore examined all three genes and the proteins they encode.
Fig. 1.
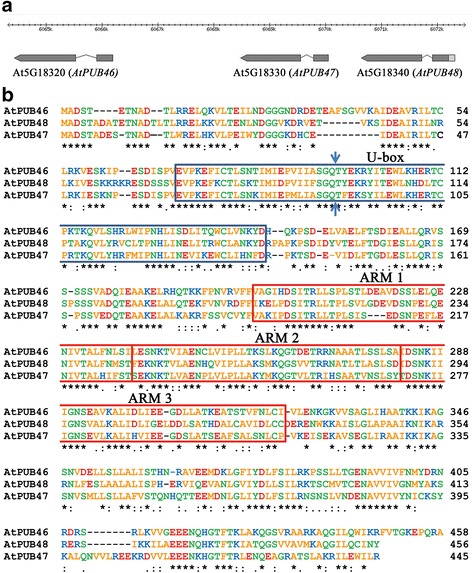
Genome organization of the AtPUB46, AtPUB47 and AtPub48 genes and the proteins they encode. a Genome organization of Arabidopsis chromosome 5 loci At5G18320, AtG18330, and At5G18340 that encode the AtPUB46, AtPUB47, and AtPUB48 genes, respectively. Exons are shown as wide and introns as narrow lines. Arrows mark gene orientation. b Amino acid sequence alignment and domain structure of AtPUB46, AtPUB47, and AtPUB48: blue (positivly charged), red (negatively charged), green (polar) and orange (non-polar) residues. U-box is shown in a blue frame, ARM motifs are marked by red frames. Arrows indicate the position corresponding to the exon-exon borders
Tissue specific expression of AtPUB46, AtPUB47 and AtPUB48
To study cell and tissue specificity of expression of the three paralogs, we transformed Arabidopsis plants with promoter::GUS constructs. Promoter sequences comprised the entire region upstream of the ATG translation start codon to the STOP codon of the adjacent upstream gene yielding 1440, 597 and 1613 bp promoter sequences for AtPUB46, AtPUB47, and AtPUB48, respectively. At least three homozygous transformants were selected for each construct, and promoter activity was determined using GUS histological staining. The expression patterns directed by each of the three promoters were very similar: they express in the vascular systems of leaves (Fig. 2a, j, s), roots (Fig. 2d-f, m-o, v-x), stem-root transition zone (Fig. 2g, p, y) and in trichomes (Fig. 2b, c, k, l, t, u). The genes also express in reproductive organs: sepals, short styles, stamen filaments (Fig. 2h, q, z), and receptacles at both the flower and fruit stages (Fig. 2h, i, q, r, z, aa). No staining was observed in root tips (Fig. 2f, o, x) or in petals and anthers (Fig. 2h, q, z). Some differences in expression patterns were observed: AtPUB46 and AtPUB48 but not AtPUB47 express in cotyledon parenchyma (Fig. 2a, j, s). The AtPUB46 is highly expressed in cotyledons and developing leaves and at lower levels in fully expanded leaves (Fig. 2a). AtPUB47 is highly expressed in petioles (Fig. 2j), and AtPUB48 is highly expressed in cotyledons and at a low level in leaves at all stages of development (Fig. 2s).
Fig. 2.
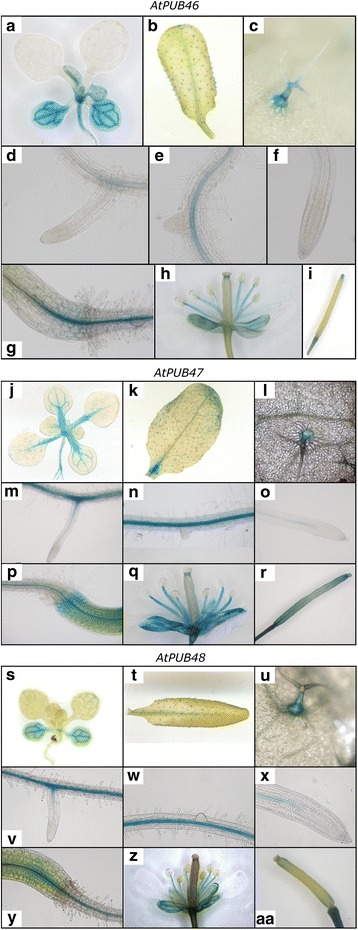
Expression pattern of the AtPUB46-48 promoters. Arabidopsis plants expressing the GUS reporter gene driven by the AtPUB46 (a-i) AtPUB47 (j-r) or AtPUB48 (s-aa) promoters were stained for GUS activity. (a, j, s), 2 week old seedlings; (b, k, t), rosette leaves of mature plants; (c, l, u), trichomes; (d, e, m, n, v, w), primary roots, root hairs and developing lateral roots; (f, o, x), root tips; (g, p, y) shoot-root transition zone; (h, q, z) flowers; (I, r, aa), siliques. At least 3 independent lines were assayed for each construct
Steady state transcript levels of AtPUB46, AtPUB47 and AtPUB48 in roots and shoots determined by RT-qPCR
The AtPUB46-48 genes are highly homologous paralogues, and thus the proteins encoded by these genes may function in a redundant fashion. Gene specific activity thus may result from differential expression patterns of each gene. Therefore, we analyzed the steady state levels of AtPUB46-48 transcripts in root and shoots under different treatments.
Transcripts of AtPUB48 are the most abundant—2- and 5-fold higher than those of AtPUB46 in roots and shoots, respectively (Fig. 3a). In contrast, the levels of AtPUB47 transcripts are at least ten-fold lower than of both the other genes. The AtPUB46 and AtPUB47 mRNAs are more abundant in the roots with a shoot/root expression ratio of 0.7 and 0.3, respectively. AtPUB48 shows higher expression in shoots with a shoot/root ration of 1.8 (Fig. 3a). The low AtPUB47 expression may be due to its short promoter sequence—only 597 bp to the next upstream gene (Fig. 1a).
Fig. 3.
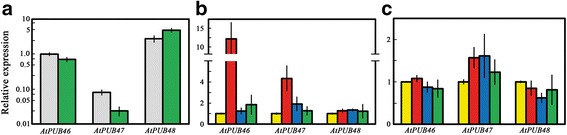
Expression levels of the AtPUB46-48 genes in roots and shoots of 10 d old seedlings untreated or treated with plant hormones. RNA was extracted from roots and shoots, cDNA was prepared and transcripts levels of the indicated genes were analyzed by RT-qPCR. a Root (gray) and shoot (green); transcript levels of all genes were normalized to that of AtPUB46 in the roots. b, c of seedlings were incubated for 6 h with 0.5 x MS without supplements (yellow), or supplemented with 10 μM each of IAA (red), zeatin (blue) or ABA (green). Transcript levels were assayed in roots (b) and shoots (c). Data shown are average ± SE. Expression levels of each gene in the respective organ of non-treated plants were defined as 1
Hormone regulation of expression of AtPUB46, AtPUB47 and AtPUB48
Plant hormones play a central role in the response to water- and salt-stress, and ABA is the major hormone involved [43]. We thus assayed the effects of exogenous ABA, auxin, and cytokinin on the expression of AtPUB46-48 in roots and shoots. The UPS system is known to be involved in hormonal signaling [8–11, 19, 20]. Thus, we assayed the steady state levels of the genes studied here in response to hormone treatment. The three genes respond differentially to application of plant hormones: expression of AtPUB46 and AtPUB47 in the roots was markedly enhanced by auxin and to a lesser extent by ABA and cytokinin (Fig. 3b). In contrast, steady state levels of AtPUB48 in root transcripts were not affected by the hormone treatments. Furthermore, the steady state levels of transcripts of these three genes in the shoots were only marginally affected by all three hormones: AtPUB47 mRNA levels were moderately induced by auxin and cytokinin, and AtPUB48 transcript levels were slightly reduced by cytokinin (Fig. 3c). Our data suggest that AtPUB46 and AtPUB47 may also be involved in modulation of target proteins whose activity/steady state levels are affected by auxin. Although auxin is mainly associated with plant growth and development, recent studies show that it also plays a role in the response to drought. For example, the rice gene TLD1/OsGH3.13 that encodes indole-3-acetic acid (IAA)-amido synthetase enhanced the expression of LEA (late embryogenesis abundant) genes, which correlated with the increased drought tolerance of rice seedlings [44]. We thus suggest that AtPUB46 and AtPUB47 may also be involved in the response to auxin.
Transcript levels of AtPUB46, AtPUB47 and AtPUB48 are differentially affected by abiotic stresses
Although salt stress generally causes osmotic-stress as well as ion toxicity, transcriptome analysis of plants exposed to salt- and osmotic-stresses revealed that most genes show a differential response to these two stresses [45]. We therefore measured steady state transcript levels of AtPUB46-48 in the roots and shoots of seedlings exposed to different abiotic stresses.
Salt stress
NaCl treatment evoked a differential response: increased transcript levels in the roots of all three genes but only elevated AtPUB46 and AtPUB47 in the shoots (Fig. 4a, b).
Fig. 4.
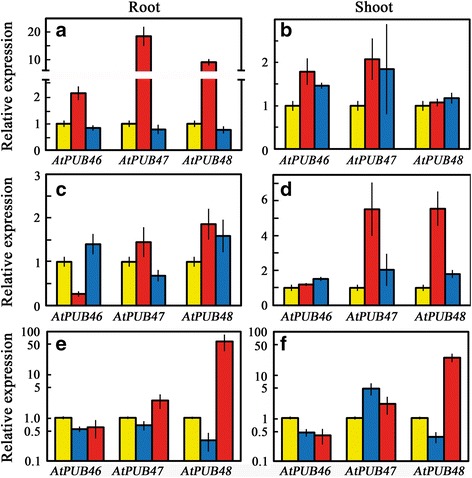
Expression levels of AtPUB46-AtPUB48 genes in response to abiotic stress analyzed by RT-qPCR. Ten day old seedlings were exposed to the following treatments: (a, b) salt- and osmotic stress: control (yellow); 0.2 M NaCl (red); 0.4 M mannitol (blue) for 6 h. c & d, oxidative stress: control (yellow); 100 mM H2O2 (red); 1 μM methyl viologen (blue) for 3 h in the light. e, f temperature stress: 25 °C (yellow); 3 h at 4 °C (blue); 15 min at 45 °C (red). Data shown are average ± SE
Osmotic stress
Mannitol did not affect transcript levels of the three studied AtPUB genes in the roots, but did enhance the levels of AtPUB46 and AtPUB47 in the shoots (Fig. 4a, b).
Oxidative stress
The H2O2-treated seedlings displayed elevated mRNA levels of AtPUB47 and AtPUB48 in both roots and shoots; AtPUB46 showed reduced transcript levels of AtPUB46 in the roots and unchanged levels in the shoots (Fig. 4c, d). Similarly, methyl viologen (MV) enhanced the expression of AtPUB47 and AtPUB48 in the shoots (Fig. 4d) and to a lesser extent of AtPUB46 in roots and shoots and of AtPUB48 in the roots. Arabidopsis plants exposed to H2O2 or MV show very different gene profiles for each treatment [46, 47]. In agreement, the steady-state levels of AtPUB46-48 transcripts were induced more by NaCl than by a similar osmotic stress administrated by mannitol (Fig. 4a, b).
Heat stress
The AtPUB48 was markedly induced in roots and shoots following heat exposure (Fig. 4e, f); AtPUB47 was also induced but to a lesser extent. On the other hand, the expression of AtPUB46 was reduced in all vegetative parts following heat treatment (Fig. 4e, f). AtPUB48 and AtPUB46 transcript levels were reduced by low temperatures in both roots and shoots, whereas cold treatment increased the levels of AtPUB47 transcripts in the shoots (Fig. 4e, f).
Heat shock transcription factors (HSFs) are major players in the induction of heat-responsive genes [48]. Analysis of the putative promoter sequence of the AtPUB48 gene for HSF binding sites (HSE) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) revealed a putative HSE element: CTCGAAGTTTCTAG in the 5' UTR, which is −65 to −53 bases upstream of the translation ATG codon. This matches the HSE consensus sequence CTNGAANNTTCNAG first identified in Drosophila and shown to function in plants [49].
Thus, although the three genes are expressed for the most part in the same cell types (Fig. 2), their differential response to plant hormones and abiotic stresses (Figs. 3 and 4) and the identification of a HSE uniquely in the promoter of AtPUB48 indicates that their activity does not entirely overlap.
AtPUB46, AtPUB47 and AtPUB48 encode catalytically active E3s
Bioinformatics analysis indicated that AtPUB46, AtPUB47, and AtPUB48 are PUB-ARM E3s ([10, 12–14, 16] and www.Arabidopsis.org). To test this directly we produced recombinant GST-tagged AtPUBs, His-tagged human E1, and His-tagged Arabidopsis E2 in E. coli. Recombinant proteins were purified by affinity chromatography, and E3 activity was assayed by auto-ubiquitylation of the E3. High MW ubiquitylated proteins were observed in reaction mixtures that contained E1, E2, and E3 (Fig. 5), indicating that all three recombinant AtPUB proteins possess E3 activity. Lower levels of protein polyubiquitylation could also be detected in reaction mixes containing two of the three enzymes in this short metabolic pathway. Similar partial ubiquitylation activities were reported over 30 years ago where mixes containing 2 of the E1, E2 and E3 enzymes yielded 20–44% of the activity obtained in a full reaction mix [50]. Polyubiquitylation by E1 + E3 without E2 or by E1 + E2 without E3 was also recently reported [51–53].
Fig. 5.
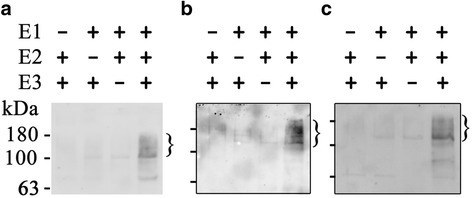
AtPUB46-48 possesses E3 activity. Self-ubiquitylation of each E3 was assayed in vitro using purified recombinant proteins. Uniquitylated protein (marked by } sign) were detected by western blot using anti-ubiquitin antiserum. a AtPUB46; b AtPUB47; c AtPUB48. a & b had the E2, AtPUBC10 (At5G53300); c had AtUBC8 (At5G41700)
Homozygous Atpub46 and Atpub48 T-DNA insertion mutants are hypersensitive to water stress
Our original screen for Arabidopsis homozygous T-DNA insertion mutant plants for altered water stress sensitivity identified the T-DNA insertion mutant Atpub46-plants (SALK_096071) as hypersensitive to water stress. To extend this observation to the adjacent E3s, we used six T-DNA insertion lines—two for each of the AtPUB46-48 genes: Atpub46-1 (SALK_096071), Atpub46-2 (SALK 109233), Atpub47-1 (SALK_018208), Atpub47-2 (SALK_056774), Atpub48-1 (SALK_086659) and Atpub48-2 (SALK_057909) (Fig. 6a). The T-DNA insertion sites in the Atpub46-1, Atpub47-2 and Atpub48-2 mutants disrupt the sequences that encode the U-box suggesting that these are loss-of-function mutants. The Atpub47-1 mutant has a T-DNA insert in exon2 and probably encodes the U-box domain. It may act as a dominant positive mutant. In the Atpub46-2 and Atpub48-1 mutants, the T-DNA is inserted in the 5' UTR. The T-DNA insertions in the 5’ UTRs have been proposed to result from reduced gene expression [54] and also significantly affect protein translation [55]. Thus, Atpub46-2 and Atpub48-1 can be regarded as knockdown mutants [56]. The RT-PCR analyses of these mutants confirmed that their transcripts are affected by the T-DNA insertions at the respective sites (Fig. 6b).
Fig. 6.
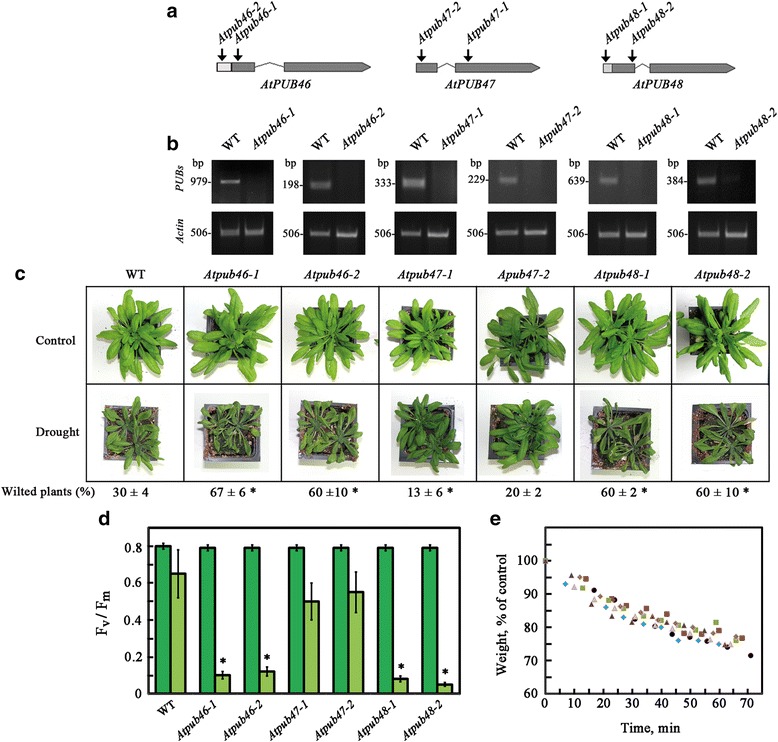
Water stress performance of pot-grown Atpub46-48 mutant plants. a Location of the T-DNA insertion in the studied mutants. Exons and introns are shown as wide and narrow lines, respectively. Arrows mark gene orientation. The location of the T-DNA insertions in the mutant lines used in this study are marked by arrows. b Upper panels: analysis of the indicated AtPUB46-48 genes in 1-week old wild type (WT) and the indicated mutants using gene-specific primers that anneal to sequences on both sides of the T-DNA insertions. Lower panels: the expression of ACTIN2 was used as an internal control. c, d Water stress performance of Atpub46-48 mutant plants. Plants were grown in pots for 3 (d) or 4 (c) weeks and then water was withheld from drought treated plants. c, Plants were photographed after 10 days. d Photosynthetic efficiency was assayed 20 days after water withheld. e Detached rosettes of 1 month old pot grown plants were assayed for water loss. Data shown are average ± SE. Statistically significant changes from WT plants (P < 0.05) are marked with asterisks
Pot-grown plants of these T-DNA insertion mutants were assayed for water stress sensitivity (Fig. 6c). All four Atpub46 and Atpub48 mutants were hypersensitive to a lack of water (Fig. 6c). The Atpub47-2 mutant did not significantly alter plant survival under water deficit, whereas the Atpub47-1 mutant displayed slightly increased tolerance (Fig. 6c). These differences may be attributed to the location of the T-DNA insertion (above), which may allow expression of the U-box domain in the Atpub47-1 mutant but not in the Atpub47-2 mutant.
We have measured chlorophyll fluorescence during the process of water deprivation. A decrease in chlorophyll fluorescence was used for quantitative assessment of drought survival in agreement with previous reports showing a sharp decrease in Fv/Fm values only when Arabidopsis plants were irreversibly affected by drought [57]. Figure 6d shows that Atpub48 and Atpub46 mutants completely lost their photosynthetic potential at the same time where type plants and Atpub47 mutants were only slightly affected. This confirms the wilting experiments shown in Fig. 6c. The reduction in chlorophyll fluorescence were seen when leaves of the mutants became necrotic.
Water loss experiments resulted in detached rosettes and showed that the water-loss rates in the wild type and the mutant plants were similar (Fig. 6e). This suggests that that the hyper-drought sensitivity of Atpub46 and Atpub48 mutants do not result from impaired stomata function.
The above mutants were assayed for germination and seedling establishment under control conditions and abiotic stresses (Fig. 7). When germinated on standard medium, seeds of all tested lines (WT and mutants) were fully germinated suggesting that the mutations do not affect seed viability or germination. The Atpub46-1 and Atpub46-2 mutants were hypersensitive to MV-promoted oxidative stress, whereas the extent of inhibition of seedling greening of the Atpub47 and Atpub48 mutants did not differ or was only marginally different from that of WT seedlings, respectively (Fig. 7a). Germination in the presence of NaCl or mannitol was not affected in any of the tested mutants (Fig. 7b, c). Moreover, the inhibition of seed germination of the mutants by ABA was not significantly different than that of WT (Fig. 7d). Although mannitol treatment is often used as an osmotic stressor, exposing plants to osmotic shock via high mannitol concentrations may differ from gradually increased water stress induced by withholding water from plants growing in soil [58, 59].
Fig. 7.
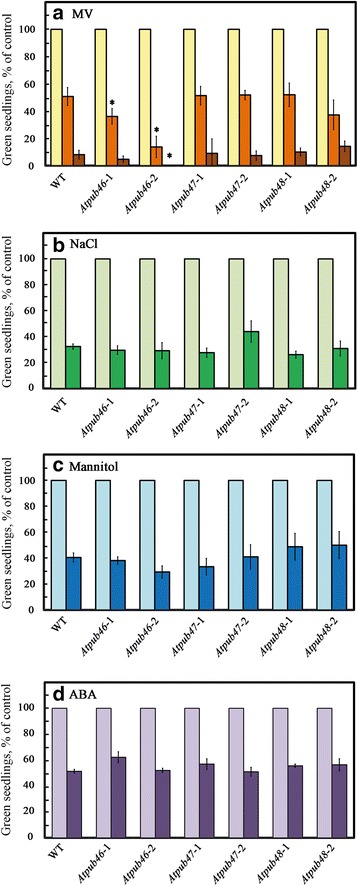
Effects of oxidative, salt and osmotic stresses on seedling germination. Surface sterilized cold treated seeds of the indicated plant lines were plated on agar media containing 0.5 x MS, 0.5% sucrose (control) supplemented with: a methyl viologen (MV) at 0 (yellow bars), 0.5 μM (orange bars) or 1 μM (brown bars); b NaCl at 0 (light green) or 150 mM (green) NaCl; c mannitol at 0 (light blue) or 300 mM (blue); d ABA at 0 (light purple) or 1 μM (purple). Green seedlings were scored 5 (a-c) or 6 (d) days later. Data shown are average ± SE. Statistically significant changes from WT plants (P < 0.05) are marked with asterisks
The water stress hypersensitivity observed for T-DNA insertion mutants of the AtPUB46 and AtPUB48 genes suggests that the biological activities of these genes do not fully overlap. On the other hand, the sensitivity of the Atpub47 mutants to water stress was probably not altered because their expression levels are negligible compared with those of AtPUB46 and AtPUB48 (Fig. 3a) or because of functional redundancy with other E3(s). Gene redundancy is observed when the respective gene products share activity as well as temporal and cell-type expression. Thus, expression in different cell types at different developmental stages or in response to different cues is expected to appear as non-redundant even if the protein activity is identical. Gene families are very common in plants, and the resulting functional redundancy means that most single loss-of-function mutants do not have a phenotype [60].
AtPUB46 and AtPUB48 have a distinct response to water stress compared with other PUB genes involved in the response to drought
A number of PUB genes are involved in the response to drought: CaPUB1 from the hot pepper Capsicum annuum as well as AtPUB18, AtPU19, AtPUB22 and AtPUB23 [21–24]. However, the role of these genes in the response to water stress is opposite that of AtPUB46 and AtPUB48. The Atpub19, Atpub22 and Atpub23 mutants showed enhanced tolerance to drought; in contrast, Atpub46 and Atpub48 mutant plants were hypersensitive to water stress (Fig. 6). These results suggest that at least some of the protein targets of AtPUB46 and AtPUB48 E3 activity are degraded in water stress conditions. Our data suggests that protein targets of AtPUB46 and AtPUB48 are likely to negatively regulate the water stress response because their expected accumulation in Atpub46 and Atpub48 mutants decreases plants tolerance to water stress.
Conclusions
The paralogous AtPUB46-48 genes located in tandem on Arabidopsis chromosome 5 resulted from gene duplication. We showed that these genes have a unique function in response to water stress because single homozygous mutants of AtPUB46 and AtPUB48 are hypersensitive to water stress. Our results suggest that the biological activities of AtPUB46-48 genes are at least partially gene specific. This specificity may result from differential spatial and/or temporal expression and from possible differences in their substrate specificity. Protein targets of AtPUB46 and AtPUB48 are likely to negatively regulate the water stress response. The Identification of these target proteins will enhance our understanding of their role in the control of specific protein levels under non-stress and stress conditions.
Acknowledgements
We thank Gali Prag for fruitful discussions. DBZ is the incumbent of The Israel and Bernard Nichunsky Chair in Desert Agriculture, Ben-Gurion University.
Funding
This work is supported by a grant 692/10 from the Israel Science Foundation (to DR and DBZ) and by the I-CORE Program of the Planning and Budgeting Committee and the Israel Science Foundation (Center No. 757, to DBZ). The funding agencies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All supportive data are presented as supplement.
Authors’ contributions
GA, DR and DBZ designed the experiments, generated data, analyzed the data and drafted the manuscript. GA carried out most experiments. ZK did some constructs and protein expression. LZ screened mutants for drought tolerance. AKM participated in drought tolerance experiments. GA, DR and DBZ wrote the paper. All authors read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
Not applicable.
Abbreviations
- ABA
abscisic acid
- ARM
Armadillo domain
- E1
ubiquitin activating enzyme
- E2
ubiquitin conjugating enzyme
- E3
ubiquitin ligase
- IAA
Indole-3-acetic acid
- PUB
Plant U-box
- UPS
ubiquitin proteasome system
Additional file
List of primers used in this study. (DOCX 18 kb)
Contributor Information
Guy Adler, Email: adlergu@gmail.com.
Zvia Konrad, Email: conrad@bgu.ac.il.
Lyad Zamir, Email: lyadza@gmail.com.
Amit Kumar Mishra, Email: amit.bhu.bot@gmail.com.
Dina Raveh, Email: dina.raveh@gmail.com.
Dudy Bar-Zvi, Phone: +972-8-6461365, Email: barzvi@bgu.ac.il.
References
- 1.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Ann Rev Plant Biol. 2004;55:373–99. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 2.Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143:291–9. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Breusegem F, Dat JF. Reactive oxygen species in plant cell death. Plant Physiol. 2006;141:384–90. doi: 10.1104/pp.106.078295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacques S, Ghesquière B, Van Breusegem F, Gevaert K. Plant proteins under oxidative attack. Proteomics. 2013;13:932–40. doi: 10.1002/pmic.201200237. [DOI] [PubMed] [Google Scholar]
- 5.Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–97. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- 6.Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S. The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol. 2012;196:13–28. doi: 10.1111/j.1469-8137.2012.04266.x. [DOI] [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Stone SL. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front Plant Sci. 2014;5:135. doi: 10.3389/fpls.2014.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyzenga WJ, Stone SL. Abiotic stress tolerance mediated by protein ubiquitination. J Exper Bot. 2012;63:599–616. doi: 10.1093/jxb/err310. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Hellmann H. Plant E3 ligases: flexible enzymes in a sessile world. Mol Plant. 2013;6:1388–404. doi: 10.1093/mp/sst005. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Kim WT. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol Cells. 2011;31:201–8. doi: 10.1007/s10059-011-0031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azevedo C, Santos-Rosa MJ, Shirasu K. The U-box protein family in plants. Trend Plant Sci. 2001;6:354–8. doi: 10.1016/S1360-1385(01)01960-4. [DOI] [PubMed] [Google Scholar]
- 13.Wiborg J, O'Shea C, Skriver K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem J. 2008;413:447–57. [DOI] [PubMed]
- 14.Aravind L, Koonin EV. The U box is a modified RING finger — a common domain in ubiquitination. Curr Biol. 2000;10:R132–R134. doi: 10.1016/S0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- 15.Andersen P, Kragelund BM, Olsen AN, Larsen FH, Chua NH, Poulsen FM, Skriver K. Structure and biochemical function of a prototypical Arabidopsis U-box domain. J Biol Chem. 2004;279:40053–61. doi: 10.1074/jbc.M405057200. [DOI] [PubMed] [Google Scholar]
- 16.Zeng LR, Park CH, Venu RC, Gough J, Wang GL. Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol Plant. 2008;1:800–15. doi: 10.1093/mp/ssn044. [DOI] [PubMed] [Google Scholar]
- 17.Peifer M, Berg S, Reynolds AB. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–91. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 18.Mudgil Y, Shiu SH, Stone SL, Salt JN, Goring DR. A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 2004;134:59–66. doi: 10.1104/pp.103.029553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duplan V, Rivas S. E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front Plant Sci. 2014;5:42. doi: 10.3389/fpls.2014.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yee D, Goring DR. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot. 2009;60:1109–21. doi: 10.1093/jxb/ern369. [DOI] [PubMed] [Google Scholar]
- 21.Cho SK, Chung HS, Ryu MY, Park MJ, Lee MM, Bahk YY, Kim J, Pai HS, Kim WT. Heterologous expression and molecular and cellular characterization of CaPUB1 encoding a hot pepper U-box E3 ubiquitin ligase homolog. Plant Physiol. 2006;142:1664–82. doi: 10.1104/pp.106.087965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho SK, Ryu MY, Song C, Kwak JM, Kim WT. Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell. 2008;20:1899–914. doi: 10.1105/tpc.108.060699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu YC, Wu YR, Huang XH, Sun J, Xie Q. AtPUB19, a U-Box E3 ubiquitin ligase, negatively regulates abscisic acid and drought responses in Arabidopsis thaliana. Mol Plant. 2011;4:938–46. [DOI] [PMC free article] [PubMed]
- 24.Seo DH, Ryu MY, Jammes F, Hwang JH, Turek M, Kang BG, Kwak JM, Kim WT. Roles of four Arabidopsis U-Box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 2012;160:556–68. [DOI] [PMC free article] [PubMed]
- 25.Ni XM, Tian ZD, Liu J, Song BT, Li JC, Shi XL, Xie CH. StPUB17, a novel potato UND/PUB/ARM repeat type gene, is associated with late blight resistance and NaCl stress. Plant Sci. 2010;178:158–69. doi: 10.1016/j.plantsci.2009.12.002. [DOI] [Google Scholar]
- 26.Bergler J, Hoth S. Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol. 2011;13:725–30. doi: 10.1111/j.1438-8677.2010.00431.x. [DOI] [PubMed] [Google Scholar]
- 27.Stone SL, Anderson EM, Mullen RT, Goring DR. ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell. 2003;15:885–98. [DOI] [PMC free article] [PubMed]
- 28.Park JJ, Yi J, Yoon J, Cho LH, Ping J, Jeong HJ, Cho SK, Kim WT, An G. OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant J. 2011;65:194–205. doi: 10.1111/j.1365-313X.2010.04416.x. [DOI] [PubMed] [Google Scholar]
- 29.Hur YJ, Yi YB, Lee JH, Chung YS, Jung HW, Yun DJ, Kim KM, Park DS, Kim DH. Molecular cloning and characterization of OsUPS, a U-box containing E3 ligase gene that respond to phosphate starvation in rice (Oryza sativa). Mol Biol Rep. 2012;39:5883–8. [DOI] [PubMed]
- 30.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–7. [DOI] [PubMed]
- 31.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–43. [DOI] [PubMed]
- 32.Adler G, Blumwald E, Bar-Zvi D. The sugar beet gene encoding the sodium/proton exchanger 1 (BvNHX1) is regulated by a MYB transcription factor. Planta. 2010;232:187–95. doi: 10.1007/s00425-010-1160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzfira T, Tian G-W, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V. pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol. 2005;57:503–16. doi: 10.1007/s11103-005-0340-5. [DOI] [PubMed] [Google Scholar]
- 34.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr Purif. 1999;15:34–9. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 35.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plantar. 1962;15:473–97. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 36.Shkolnik-Inbar D, Adler G, Bar-Zvi D. ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J. 2013;73:993–1005. doi: 10.1111/tpj.12091. [DOI] [PubMed] [Google Scholar]
- 37.Mbengue M, Camut S, de Carvalho-Niebel F, Deslandes L, Froidure S, Klaus-Heisen D, Moreau S, Rivas S, Timmers T, Hervé C, et al. The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell. 2010;22:3474–88. [DOI] [PMC free article] [PubMed]
- 38.Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–82. doi: 10.1016/S0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 39.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–64. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 40.Cannon S, Mitra A, Baumgarten A, Young N, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. [DOI] [PMC free article] [PubMed]
- 41.Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comp Biol. 2006;2 doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang C, Gu X, Peterson T. Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol. 2004;5:R46. [DOI] [PMC free article] [PubMed]
- 43.Peleg Z, Blumwald E. Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol. 2011;14:290–5. doi: 10.1016/j.pbi.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 44.Zhang S-W, Li C-H, Cao J, Zhang Y-C, Zhang S-Q, Xia Y-F, Sun D-Y, Sun Y. Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol. 2009;151:1889–901. doi: 10.1104/pp.109.146803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kreps JA, Wu Y, Chang H-S, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–41. [DOI] [PMC free article] [PubMed]
- 46.Scarpeci TE, Zanor MI, Carrillo N, Mueller-Roeber B, Valle EM. Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol Biol. 2008;66:361–78. [DOI] [PMC free article] [PubMed]
- 47.Davletova S, Schlauch K, Coutu J, Mittler R. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 2005;139:847–56. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schöffl F, Prändl R, Reindl A. Regulation of the heat-shock response. Plant Physiol. 1998;117:1135–41. doi: 10.1104/pp.117.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wing D, Koncz C, Schell J. Conserved function in Nicotiana tabacum of a single Drosophila hsp70 promoter heat shock element when fused to a minimal T-DNA promoter. Molec Gen Genet. 1989;219:9–16. [DOI] [PubMed]
- 50.Hershko A, Heller H, Elias S, Ciechanover A, Components of ubiquitin-protein ligase system Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–14. [PubMed] [Google Scholar]
- 51.David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285:8595–604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banka PA, Behera AP, Sarkar S, Datta AB. RING E3-catalyzed E2 self-ubiquitination attenuates the activity of Ube2E ubiquitin-conjugating enzymes. J Mol Biol. 2015;427:2290–304. doi: 10.1016/j.jmb.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 53.Park S, Krist DT, Statsyuk AV. Protein ubiquitination and formation of polyubiquitin chains without ATP, E1 and E2 enzymes. Chem Sci. 2015;6:1770–9. doi: 10.1039/C4SC02340D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krysan PJ, Young JC, Sussman MR. T-DNA as an insertional mutagen in Arabidopsis. Plant Cell. 1999;11:2283–90. doi: 10.1105/tpc.11.12.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Busi MV, Maliandi MV, Valdez H, Clemente M, Zabaleta EJ, Araya A, Gomez-Casati DF. Deficiency of Arabidopsis thaliana frataxin alters activity of mitochondrial Fe–S proteins and induces oxidative stress. Plant J. 2006;48:873–82. [DOI] [PubMed]
- 56.Bolle C, Schneider A, Leister D. Perspectives on systematic analyses of gene function in Arabidopsis thaliana: new tools, topics and trends. Curr Genom. 2011;12:1–14. [DOI] [PMC free article] [PubMed]
- 57.Woo NS, Badger MR, Pogson BJ. A rapid, non-invasive procedure for quantitative assessment of drought survival using chlorophyll fluorescence. Plant Methods. 2008;4:1–14. doi: 10.1186/1746-4811-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J Exp Bot. 2004;55:2331–2341. doi: 10.1093/jxb/erh270. [DOI] [PubMed] [Google Scholar]
- 59.Alcázar R, Planas J, Saxena T, Zarza X, Bortolotti C, Cuevas J, Bitrián M, Tiburcio AF, Altabella T. Putrescine accumulation confers drought tolerance in transgenic Arabidopsis plants over-expressing the homologous Arginine decarboxylase 2 gene. Plant Physiol Biochem. 2010;48:547–552. doi: 10.1016/j.plaphy.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 60.Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS. Unequal genetic redundancies in Arabidopsis – a neglected phenomenon? Trend Plant Sci. 2006;11:492–8. doi: 10.1016/j.tplants.2006.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All supportive data are presented as supplement.


