Whether or not it was first said by atom-splitter Niels Bohr or splitter-ball catcher Yogi Berra, we all agree “it’s tough to make predictions- especially about the future.” In the concluding section of this monograph on the current status of predictive cancer genomics, it is appropriate to ponder the future of this translational field of medical science. As will also be addressed here, it is particularly instructive for the providers and consumers of the rapid advances in genomics and medicine to make their own predictions of the impact of “personalized genomics” on preventive oncology.
This effort to encourage introspection is meant to highlight the sea change that is shaping the way genomic predictive markers have been integrated in the practice of “precision medicine.” The elements of this sea change are multifold and have constituted a virtual “perfect storm” which is now raining down on the clinical practice of cancer genomics. As will be discussed here, these factors include: the rapid advances in genomic science and technology that allow massively parallel sequencing of both tumors and the germline1,2 a landmark shift in interpretation of statutes bearing on intellectual property of genetic discoveries3, rapid expansion of access to the internet, including mobile access to both genomic data and tools to interpret these data in a medical context, the expansion of for-profit genomic diagnostics – some masquerading as “recreational genomics,” and a worrisome view of medical professionals as barriers to rather than facilitators of understanding one’s genome. Each of these factors will impact how the discipline of predictive and preventive oncology is able to shape the translation of genomic technologies in the most responsive and responsible way. Here, we shall lay out the challenges and potential conflicts in bringing “personalized genomics” to oncology. I will use as a framework a prior essay on this topic4, updating and expanding these observations based on recent developments in the clinic, in clinical and translational laboratory research, in the courts, and in the economic and social infrastructure that impact how cancer patients and these at risk for cancer are offered genomic information.
Shifting Paradigms in Cancer Genomics: 1. Causative Events, Consequences, and Emerging Strategies
In his classic monograph The Structure of Scientific Revolutions5, the historian of science Thomas Kuhn coined the term “paradigm shift” to characterize periods of sudden departure from “normal science” when “unprecedented” discoveries shift the very practice of science in a fundamental, revolutionary way. To a real extent the rapid pace of the “genetic revolution” has impacted medicine. Perhaps in no other area has this change been more dramatically felt than clinical cancer genomics.
The preceding chapters of this monograph have updated our current knowledge of inherited mechanisms of cancer susceptibility. They have presented new information about genotype and phenotype, risk prediction, and targeted intervention. However, this monograph can only give hints at what lay ahead, since the major forces which will drive changes in clinical genomics are only now coming into maturity. Thomas Kuhn stated that to meet the bar of a paradigm shift, the new advances must be "sufficiently unprecedented to attract an enduring group of adherents away from competing modes of scientific activity." He predicted that a true paradigm shift would be "sufficiently open-ended to leave all sorts of problems for the redefined group of practitioners to resolve." Here, we will argue that the factors driving the paradigm shift in cancer genomics are not only on the verge of changing the medical model for delivering cancer genetic information but of replacing it entirely.
Consequences of Current Generation DNA Sequencing
Compared to Sanger capillary-based sequencing, massively parallel sequencing, touted as “Next Generation Sequencing” (NGS), is now part of current generation practice. NGS employs simultaneous sequencing reactions detected automatically, producing millions of sequence calls per instrument run, at a significantly lower expense. Recent advances have increased the number of nucleotides per sequence read (or read lengths) and lower cost and greater base-calling accuracy1. These technologies have been applied to sequencing of exomes, entire genomes, and exons and splice region sequences of selected genes. The research impact of NGS technologies on the pace of new syndrome identification has been remarkable. By sequencing relatively few members of families with recurrent and unexplained malignancies it has been possible over just the past few years to identify over a dozen new cancer syndromes (Table 1). Only some of these new syndromes have been included in the preceding sections of this monograph, as these discoveries are so recent that precise genotype-phenotype correlations have yet to be established. As an example of the challenges of clinical translation posed by these NGS discoveries, we described two new syndromes of predisposition to childhood acute lymphoblastic leukemia6,7, both caused by inherited mutations of transcription factors. While there was compelling functional biological evidence of “causation” behind the association of these germline mutations and the familial occurrences of leukemia, both syndromes demonstrated incomplete penetrance, and for both there was no proven preventive intervention other than pre-implantation genetics to halt transmission of the trait. Such reduced penetrance is the rule rather than exception for most if the NGS syndromic discoveries listed in Table 1.
Table 1.
Impact of Next Generation Sequencing In Discovery Of Novel Cancer Predisposition Syndromes
| Familial Cancer Syndrome | NGS | Gene | Cases used to identify | |
|---|---|---|---|---|
| Pancreatic cancer | Jones et al., Science 200967 | Exome | PALB2 | 1 affected familial pancreatic cancer |
| Roberts et al., Can Disc 201268 | WGS | ATM | WGS/Exome:16/22 affecteds 6/10 families |
|
| Pheochromocytoma | Comino-Mendez et al., Nat Gen 201169 | Exome | MAX | 3 affecteds from 3 families |
| Wilms Tumor | Mahamdallie et al., Nat Genet 201570 | Exome | REST | 4 families |
| AML/MDS | Ostergaard et al., Nat Gen 201171 | Exome | GATA2 | 3 unrelated affecteds (2 w/ familial) |
| Familial Myeloid | Saliba et al., Nat Genet 201572 | Exome |
ATG2B GSKIP |
4 related kindreds |
| Familial Melanoma | Yokoyama et al., Nature 201173 | WGS | TIFT | 1 affected with familial melanoma |
| Horn et al., Science 201374 | Tar Seq | TERT | 4 affecteds/1 unaffected in 1 kindred | |
|
Mesothelioma/uveal melanoma/renal |
Testa et al., Nat Gen 201175 Farley et al., Mol Ca Res 201376 |
Exome | BAP1 | 2 Kindreds 1/83 kindreds |
| Colorectal adenomas/ca | Palles et al., Nat Genet 201377 Weren et al., Nat Genet 201578 |
WGS Exome |
POLE, POLD1 NTHL1 |
20 affecteds /15 families 51 affecteds/48 families |
| Non Medullary Thyroid | Gara et al., NEJM 201579 | Exome | HABP2 | 7 affected 1 kindred |
| Breast Cancer | Park et al., AJHG 201280 | Exome | XRCC2 | 5 affecteds from 2 families |
| Park et al., BCRT 201181 | Exome | FAN1 | 4 early-onset multiple-case families | |
| Ruark et al., Nature 201382 | TarSeq | PPMD1 | 1,150 with breast cancer +/− ovarian | |
| Cybulski et at., Nature Genet 201583 | Exome | RECQL | 7 cases Quebec, 30 in Poland | |
| Ovarian cancer | Rafnar et al., Nat Genet 201184 | WGS | BRIP1 | 457 Icelanders |
| ALL | Shah et al., Nat Genet 20136 | Exome | PAX5 | 2 kindreds |
| Noetzli Nat Genet85, Zhang Nat Genet86, Topka PLoS Genet 20157 | Exome | ETV6 | Multiple kindreds | |
In addition to their role powering whole genome discovery, NGS technologies have also impacted the rapid diagnosis of known syndromes by utilizing “capture” of exons and exon-intron splice regions of dozens of cancer predisposition genes, all analyzed simultaneously, as part of a new breed of multiplexed diagnostic panels8. As will be discussed, this technological innovation has stimulated the appetite of both providers as well as consumers of genetic tests, in favor of “prix fixe” menus of multiple gene tests at costs lower than that of the old “a la carte” menu of phenotype-directed genetic analysis.
Fallout of the End of Gene Patenting
Just as NGS technologies began to generate novel syndromic discoveries of potential diagnostic value, the U.S. Supreme Court ruled that isolated genomic DNA was not patent-eligible under section 101 of the Patent Act. The court, however, let stand patents for cDNA, an approach which some of us predicted before the decision, and which was to have an impact on the practice of preventive oncology3. The opinion written by Justice Thomas was unanimous and brief. The oral argument, at least to this listener, was notable for the absence of understanding both by the Justices and the U.S. Solicitor General of basic concepts of genetics (e.g. the difference between DNA and RNA), and the use of non-scientific metaphors, involving trees, baseball bats, etc. The late Justice Scalia wrote that he agreed with the majority opinion even though he admitted he did not feel educated enough on the topic to sign the recitation of “the details of molecular biology.” Within a few days of the decision, as N.Y. Times reporter Andrew Pollack sought confirmation from many of us that it would be a very short time before academic and for-profit genetic testing companies would make available NGS for panels including BRCA1/29, many also expressed concern that broad deployment of these multigene panels was premature in the absence of regulatory oversight of quality of testing, evidence of clinical utility, and strategies to interpret genetic variation8.
Awash in Variants of Familiar and Novel Genes
Despite the warnings, the rush to multigene panels left clinicians coping with interpretations of reports of variants of unknown significance (VUS), with such findings as frequent as 10–90% depending on gene and panel10. Of more concern, anecdotal experience revealed some ill-informed health practitioners recommending preventive surgeries following VUS detection. And even more challenging, the multiplex panels included genes for which mutations were only known to be associated with low to intermediate penetrance, and genes for which mutations had unclear clinical utility and were previously not recommended for clinical testing. For example CHEK2, recommended as of unclear clinical utility in the era of single gene testing,11 was now routinely included in multigene panels. Valiant efforts were made to catalogue current knowledge of disease specific gene of varying penetrance.12 As new genes came to be discovered by NGS strategies (represented in Table 1), they often were added to existing panels, even in the absence of data on associated phenotypes and penetrance.
Initial Response of Federal Agencies and the Academy to the Genomic Tsunami
Just as the “tsunami” from the perfect storm of NGS breakthroughs, internet marketing, and the lifting of IP restrictions hit clinical oncology, one federally supported body charged with interpreting the evidence basis for genomic diagnostics, including those for cancer susceptibility, experienced a 95% budget reduction. This group, called The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative, funded largely by the CDC, had produced a number of evidence reviews bearing on cancer13–16. But EGAPP was not to be fully available for the sudden commercial proliferation of multigene panels in cancer risk testing. To address the most pressing need for cross-sectional databases, to document genetic variation and curation, and in the absence of a unified strategy from the for-profit laboratories to address the consequences of premature deployment of multigene panels, spontaneous initiatives were launched by other stakeholders. The BRCA Global Challenge was organized by a combination of governmental, commercial, and academic groups to seek to establish a universal database of BRCA variants17. The NHGRI organized investigators through ClinGen18 to form a Cancer Working Group to establish databases and strategies to curate key cancer susceptibility genes such as PTEN, and deposit these data into ClinVar19. At the same time, we at Memorial Sloan Kettering Cancer Center, and colleagues at the University of Pennsylvania, the Mayo Clinic and the Dana Farber Cancer Institute, built an on-line portal open to all individuals who had multigene panel testing. This initiative, called the Prospective Registry for Multiplex Testing (PROMPT) aimed to create a cohort for study of penetrance and outcome, and has over two thousand participants and is growing. The effort was joined by the seven largest commercial laboratories, who added onto their reports the link for patients with VUS and/or mutations in intermediate penetrance genes to consider joining this registry20. Indeed, all oncologists, genetic counselors, and others ordering multigene panel tests are encouraged here to provide their patients with links to join the PROMPT registry (www.promptstudy.org) An immediate observation of the PROMPT registry, presented as a 2016 ASCO abstract, was a substantial rate of divergent reports among the commercial laboratories. Such a finding is consistent with reports at recent meetings of the Clinical Sequencing Exploratory Research groups of the NHGRI documenting divergent results of “bake off” exercises of carefully blinded variant curation comparisons among experts21. These findings underscore the risks of the premature deployment of these technologies.
Response from Payers
Some third party payers had already recognized that the increasing cost of cancer diagnostic testing could be manipulated by decreasing access. Based on perceptions of the need for continued physician education in the realm of cancer genetic testing and the effectiveness of genetic counseling, one insurance carrier put in place policies to deter licensed oncologists from ordering BRCA tests. In the name of “quality improvement,” such tests were approved only if patients were first screened by genetic counselors who were either funded by the insurer, or accessed via directed consultation to determine if a test was indicated22,23. This strategy established a de facto filter to access to cancer genetic testing24 and also raised a potential challenge of restraint of the practice of cancer medicine by oncologists seeking to order genetic tests to guide therapy (e.g. PARP inhibitors) as well as prevention.23 With the planned expansion of these policies, a prediction for the future is a confrontation between practitioners and at least one large payer over the issue of scope of practice.
Less May Be More In Germline Genomic Scans
The advent of multigene testing served to galvanize some payers to seek to limit their use, on the basis of the unproven clinical utility of all gene tests included on the panels25. Within the “expert committees” such as the National Comprehensive Cancer Network, NCCN, there has been healthy and ongoing debate and efforts to ensure that guidelines, monitored closely by insurers, reflect the rapidly changing evidence base. At present and going forward, there will be a trend to reimburse only those tests for which there is proven clinical utility, with clinicians and patients facing a web of different thresholds for testing varying by laboratory or type of third party provider.
There is also an emerging “push back” among both patients and providers against the obligate “prix fixe” model of multiplex testing. It was a hallmark observation of clinical genetics that from a third to a half of patients counseled for suspected Li Fraumeni syndrome would defer p53 testing. TP53 testing, if offered as an option, was not desired for inclusion on the test panel by a proportion of patients offered multi-gene testing (Robson M, personal communication). Many clinicians would also like to be able to exclude or include specific genes depending on phenotype, and lack of clinical utility for some genes on “panels.” (e.g. CDH1 in non-lobular breast cancer). Thus, another prediction for the future is the movement toward physician and provider selected panel compositions.
Going forward, increasing numbers of labs are offering “custom” gene panels, in most cases running the larger panels internally, but “filtering” reported results only to what is requested. This strategy allows both consumers and genetics professionals to request only those tests which have evidence of clinical utility, or which are based on phenotype. Clinicians (and their patients) can then defer other results until data on clinical utility emerges, thus reserving for the laboratories the future option to report additional results - and perhaps recover costs. Such a strategy of maintaining identified potentially actionable genomic data will likely require documented prospective consent. Unlike the “diagnostic odyssey” which often justifies whole exome testing in the evaluation of some dysmorphic children, and will be documented by NHGRI funded studies in progress, the burden of “duty to warn” of non-cancer predispositions raises significant challenges for adults subjected to whole exome germline tumor-normal screens26,27
Shifting Paradigms In Cancer Genomics: 2. The Tale of Two Genomes, Screening, and Pharmcogenomics
Just as NGS technology allowed multiplex gene-panel testing, the second wave of the NGS tsunami impacted the clinical application of genome-wide re-sequencing of tumors to guide targeted therapies. In the process, the patients’ “normal” or inherited DNA is typically also scanned, raising immediate medical as well as ethical challenges.26,27 While one commercial laboratory and many academic laboratories purposely avoid sequencing normal DNA as a comparator for the tumor DNA, it is now clear that inclusion of such reference normal sequence adds to the sensitivity of the assay28. Initial tumor sequencing strategies have simply “subtracted” inherited variation from the tumor genomic reports, resolving some of the ethical and medical complexity surrounding consent for familial cancer risk testing at time of diagnosis of malignancy.26 Anonymized retrospective analyses of tumor-normal genomic data from several large centers28–32, published at the outset of 2016 (Table 2) demonstrated that within the germline compartment of tumor-normal sequence data, is a trove of clinically relevant information. While many of these early reports haves focused on common adult and pediatric cancers, a number of other studies will appear in 2016 and 2017. In our early series of adult cancer cases, inherited mutations of cancer susceptibility genes which were clinically “actionable” were noted in about 10 % of cases. Importantly, these studies and others to be published later in 2016, will identify significant proportions of cases of breast, ovarian, prostate, and other cancers with inherited mutations of DNA homologous repair genes, potentially amenable to therapy with PARP inhibitors, as well as subsets with Lynch associated mutations, potentially amenable to immunotherapy. In the MSK series, and others, there were also a fascinating set of cases where germline mutations in known genes (e.g. BRCA1/2, RET) occurred in tumors not part of known syndromes.
Table 2.
| Institutional Series (references 28–32) |
Johns Hopkins University |
University of Michigan |
Memorial Sloan Kettering |
St. Jude Children's Research Hospital |
Baylor College of Medicine |
|---|---|---|---|---|---|
| Number Sequenced | 815 | 91 | 1566 | 1120 | 150 |
| Number of Germline Actionable Findings (N) |
27 | 9 | 198 | 95 | 13 |
| Germline Actionable Findings (%) | 3% | 10% | 12.6% | 8.5% | 8.6% |
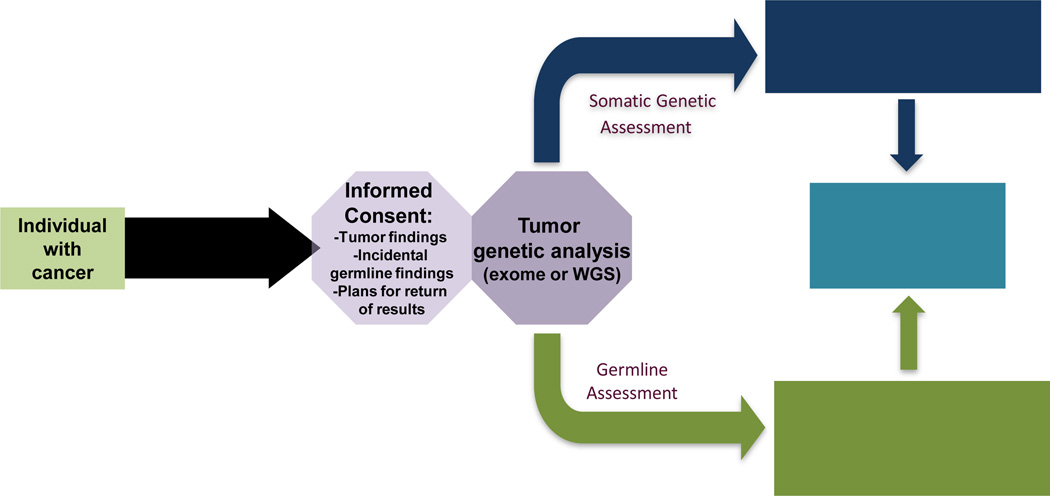
These findings have led our institution to collect tumor-normal DNA sequence in the setting of a consent process which explains that if inherited markers of cancer susceptibility are found, and if the patient desires, these results will be communicated in the context of genetic counseling. This so called “opt in” approach may soon be replaced by an “opt out” approach, where communication of germline findings is the “default” setting, unless the patient does not wish to know this information. Pending these refinements, a tiered approach to informed consent for NGS studies (Figure 1) will provide germline cancer risk assessment at the same time as tumor mutations are assessed as therapeutic targets. Thus, one of the evident future scenarios for clinical cancer genomics is a “tale of two genomes” where both tumor and inherited information is made available to all cancer patients at the time of diagnosis, with the “cascade” of genomic information to unaffected relatives for use in targeted prevention and even reproductive planning.
Figure 1.
Next-Generation Sequencing of Tumors with Incorporation of Incidental Germline Findings (adapted from Stadler, 2014)
Population Screening
It was evident even at close of the first wave of cancer predisposition gene discovery in the 1990’s that genetic testing could lead to early diagnosis and prevention of many breast, ovarian, colon, thyroid, stomach, and pediatric cancers33. For breast and ovarian cancer evidence supported decreased mortality due to breast and ovarian cancer.34 However, current guidelines limit BRCA testing to those with strong family histories of breast or ovarian cancer and/or early age of onset of disease, “triple negative” breast cancer affected before age 60, those with invasive ovarian cancer, and individuals of Ashkenazi origin with breast cancer.35 The U.S. Preventive Services Task Force has not endorsed population based BRCA screening.36,37 Nonetheless, with the advent of NGS technologies, during the past year some have come to call for population based BRCA testing.38,39 A thoughtful discussion concluded that population-based BRCA screening would likely accentuate health access and resource limitations, particularly for minority women, and could result in false-negative results in the absence of professional genetic counseling, false positives due to incorrect interpretation of variants of uncertain significance, as well as other potential harms due to psychosocial factors40. However, a different argument can be made for genetic screening in “founder” populations such as is the Ashkenazi Jews, where we described a single BRCA2 mutation present in over 1%, and 1 in 40 individuals carrying one of three BRCA1/2 mutations.41–44 Strikingly, 26–55% of individuals with BRCA mutations will be missed if testing is limited to criteria based on family history,45–51 and some of these cases invariably will represent potential lives lost if BRCA-based surgical or medical interventions are not initiated.52 Indeed BRCA population based screening in Ashkenazi Jews (AJ) has been performed in pilot studies53,54 and is cost effective; the cost per cancer detected was nearly 40 fold less expensive in Ashkenazi Jews compared to non Ashkenazim.55 A group of us are committed to initiating a population based study to offer BRCA testing to Ashkenazi Jews in the context of a “medical model” that provides appropriate counseling.56 The results of this trial may offer important guidance into the integration of genomics into mainstream medical practice. Other large scale studies are underway to provide genomic sequencing in 100,000 individuals57 as well as other studies as part of federal as well as academic “precision medicine” initiatives.
The Belated Arrival of Pharmacogenomics
Pharmacogenomics assesses inherited (or acquired) genetic abnormalities to predict treatment response or outcome. Despite anticipation a decade ago of its explosive impact on the field of oncology, the clinical utility established by pharmacogenetic studies in cancer have been limited to a handful of variants linked to treatment response (e.g. UGT1A1, CYP2D6) and a plethora of genome-wide studies of response and toxicity58, including some with a high level of interest in clinical application59. Recently rare variants have been associated with cardiotoxicity following anthracycline based chemotherapy, an issue of pressing clinical relevance for those planning adjuvant and/or curative treatments of a number of hematopoietic and solid tumors. One such finding was that a coding variant in RARG appeared to be associated with cardiotoxicity following childhood cancer.60 A major challenge of pharmacogenomic studies remains the need for large numbers of well-phenotyped patients treated with the same dosage and type of chemotherapy. Successful pharmacogenomic studies conducted in in vitro cell-based models, with confirmation of findings in vivo, now provide an important approach to move this field forward, with initial GWAS data providing identification of SNPs predicting, for example, response to platinum in patients with urothelial (or other) carcinoma, and in colorectal and prostate cancer.58 One would clearly anticipate that an inevitable result of the era of expanded tumor-normal sequencing, will be the identification of variants associated with treatment out come and toxicity.
Shifting Paradigms in Cancer Genomics: 3. Concluding Comments on the “Demedicalizing” of Cancer Genomic Testing
As mentioned at the outset, a “perfect storm” of factors, including scientific discovery of new cancer susceptibility genes, the availability of large scale genomic sequence data unfettered by intellectual property limitations, mobile access to the internet, entrepreneurial investment in for-profit genomics, and exhortations to end “genetic exceptionalism” by non-clinician enthusiasts of direct to consumer genetic testing, have led to a view of medical professionals as barriers to rather than facilitators of understanding one’s genome. Each of these factors will impact predictive and preventive oncology.
To illustrate the scope of these challenges to predictive oncology, Table 3 lists potential future scenarios for clinical cancer genomics. Underlying the future path chosen will be a need to understand the conflation of terminology which seeks to cast health professionals as barriers to rather than trusted guides to accessing the personal genome. On one hand, there is clear trend in biomedical disciplines for greater empowerment and participation of the patient in all aspects of research and care.61 Medical records will increasingly reflect genomic data.62 At the same time, while some have cast doubt on the speed of the impact of “precision oncology”63, and characterized this set of changes as part of the continuum of positive but “disruptive” technologic innovation64, most would predict that in person, phenotype-driven genetic testing will soon be replaced. Instead of extended genetic counseling sessions, there will be ”automated” pretest introduction to panels of genes, with results provided by “alternative” strategies to decrease reliance on in-person communication. However, it is unclear as to the tempo of this transition. Will this paradigm shift be complete by 2020? Or 2040? Why does this matter?
Table 3.
2020 Foresight: Future Paradigms for Clinical Cancer Genomics
| If you would be willing to anonymously reply to this opinion survey online, please go to https://www.surveymonkey.com/r/6XHXS5J. Results may be posted in the future. |
In your opinion, which response best characterizes the future state of predictive cancer genomics:
|
Certainly the tempo of this shift to “high throughput genetic counseling and testing” matters economically for those for-profit entrepreneurs who have invested. Already some genetic testing companies have failed, while other large corporations, particularly search engines and information technology firms, have committed substantial sums toward on-line delivery of “personalized genomics.” Other than its economic fallout, the tempo of this transition to more direct, unfiltered access to individual genomic sequence matters for society. Indeed, the public health may be as much at risk from the premature deployment of de-medicalized, commercialized testing for genetic predisposition, as it is from the health threats of the syndromes of cancer predisposition themselves. According to this view, while claiming to “empower” individuals to seize rightful control of their personal genomes, for-profit companies, abetted by some fervent but clinically naïve basic scientists, are “commoditizing” the genome. By implying that healthcare professionals are now coming between the individual and the right to “know” their personalized genome, commercial companies and their distinguished (and sometime co-invested) consultants, are de facto seeking to exclude the one group with an explicit fiduciary responsibility to the patient, family, and individual. When independent health care providers- physicians, genetic counselors, and other health care providers- are removed from the individual’s quest for self genetic knowledge, there may be no one else to turn to except an employee of the testing organization itself, incentivized to profit from increased utilization of its services.
In de-medicalizing genomic direct to consumer testing, there was an initial appeal to the broader concept of “recreational genomics.65” However recreational cancer genetic testing may be more similar to recreational drug use than commercial purveyors would advertise. The important distinctions lay in the medical implications of the test, and not simply the access to the test; TP53 germline testing for the risk of lethal- and mostly unpreventable- malignancies is quite different from testing for a predisposition to ear wax formation.
The “de-medicalizing” of cancer genetic testing is not a requirement for its increased uptake66. There is no question that cancer predisposition testing will be more accessible in the future; it remains to be determined how fast and to what extent it should be de-medicalized. As has been shown, interpretation of variants, indications for preventive surgeries, discussions of reproductive options, to name a few, are aspects of this discipline not casually considered. The issue is whether the “inevitable” future of cancer genomics will be thrust upon us by commercial interests, or whether that future course can be modulated in a responsible way that protects the public health while implementing powerful new medical tools for cancer prevention and early detection.
Acknowledgments
Supported by the Robert and Kate Niehaus Center for Inherited Cancer Genomics, the Andrew Sabin Family Foundation, and the Sharon Corzine Research Foundation. The views expressed are those of the author and do not represent those of federal or professional advisory groups on which he serves (National Cancer Institute, Centers for Disease Control, American Society of Clinical Oncology) or of Memorial Sloan Kettering Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The author discloses no conflicts of interest.
References
- 1.Stadler ZK, Schrader KA, Vijai J, Robson ME, Offit K. Cancer genomics and inherited risk. J Clin Oncol. 2014;32:687–698. doi: 10.1200/JCO.2013.49.7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weitzel JN, Blazer KR, MacDonald DJ, Culver JO, Offit K. Genetics, genomics, and cancer risk assessment: State of the Art and Future Directions in the Era of Personalized Medicine. CA Cancer J Clin. 2011;61:327–359. doi: 10.3322/caac.20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Offit K, et al. Gene patents and personalized cancer care: impact of the Myriad case on clinical oncology. J Clin Oncol. 2013;31:2743–2748. doi: 10.1200/JCO.2013.49.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Offit K. Personalized medicine: new genomics, old lessons. Hum Genet. 2011;130:3–14. doi: 10.1007/s00439-011-1028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn TS. The Structure of Scientific Revolutions. 1996 [Google Scholar]
- 6.Shah S, et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet. 2013;45:1226–1231. doi: 10.1038/ng.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topka S, et al. Germline ETV6 Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia. PLoS Genet. 2015;11:e1005262. doi: 10.1371/journal.pgen.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domchek SM, Bradbury A, Garber JE, Offit K, Robson ME. Multiplex genetic testing for cancer susceptibility: out on the high wire without a net? J Clin Oncol. 2013;31:1267–1270. doi: 10.1200/JCO.2012.46.9403. [DOI] [PubMed] [Google Scholar]
- 9.Pollack A. The New York Times. U.S.A: The New York Times; 2013. After Patent Ruling, Availability of Gene Tests Could Broaden; p. A16. [Google Scholar]
- 10.Kurian AW, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32:2001–2009. doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Offit K, Garber JE. Time to check CHEK2 in families with breast cancer? J Clin Oncol. 2008;26:519–520. doi: 10.1200/JCO.2007.13.8503. [DOI] [PubMed] [Google Scholar]
- 12.Easton DF, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evaluation of Genomic Applications in, P. & Prevention Working, G. The EGAPP initiative: lessons learned. Genet Med. 2014;16:217–224. doi: 10.1038/gim.2013.110. [DOI] [PubMed] [Google Scholar]
- 14.Evaluation of Genomic Applications in, P. & Prevention Working, G. Recommendations from the EGAPP Working Group: does the use of Oncotype DX tumor gene expression profiling to guide treatment decisions improve outcomes in patients with breast cancer? Genet Med. 2015 doi: 10.1038/gim.2015.173. [DOI] [PubMed] [Google Scholar]
- 15.Evaluation of Genomic Applications in, P. & Prevention Working, G. Recommendations from the EGAPP Working Group: can UGT1A1 genotyping reduce morbidity and mortality in patients with metastatic colorectal cancer treated with irinotecan? Genet Med. 2009;11:15–20. doi: 10.1097/GIM.0b013e31818efd9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evaluation of Genomic Applications in, P. & Prevention Working, G. Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11:35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Global Alliance for Genomics and Health. Website; 2016. [Google Scholar]
- 18.Rehm HL, et al. ClinGen--the Clinical Genome Resource. N Engl J Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ClinGen - Clinical Genome Resource. 2016 [Google Scholar]
- 20.PROMPT. Vol. 2016. PatientCrossroads; [Google Scholar]
- 21.Gail Jarvik HR, Dan Roden . Panel 2: Consistency of Interpretation of Variants Across Expert Labs / Groups, ClinVar Submissions? Genomic Medicine VIII; 2015. [Google Scholar]
- 22.Genetic Testing and Couseling Program. Vol. 2016. Cigna; [Google Scholar]
- 23.Lee J. Cigna to require counseling prior to some genetic tests. Modern Healthcare; 2013. [Google Scholar]
- 24.New Cigna Policy on Cancer Genetic Testing Poses Risks to High Quality Cancer Care. ASCO; 2013. [Google Scholar]
- 25.Zweig D. Aetna, Anthem and Cigna don't cover genetic tests made popular by 'Angelina effect'. FierceHealthPayer; 2015. [Google Scholar]
- 26.Bombard Y, Robson M, Offit K. Revealing the incidentalome when targeting the tumor genome. JAMA. 2013;310:795–796. doi: 10.1001/jama.2013.276573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bombard Y, Bach PB, Offit K. Translating genomics in cancer care. J Natl Compr Canc Netw. 2013;11:1343–1353. doi: 10.6004/jnccn.2013.0158. [DOI] [PubMed] [Google Scholar]
- 28.Jones S, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53. doi: 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med. 2015;373:2336–2346. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrader KA, et al. Germline Variants in Targeted Tumor Sequencing Using Matched Normal DNA. JAMA Oncol. 2016;2:104–111. doi: 10.1001/jamaoncol.2015.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons DW, et al. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2015.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mody RJ, et al. Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. Jama. 2015;314:913–925. doi: 10.1001/jama.2015.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Offit K. Clinical Cancer Genetics: Risk Counseling and Management. Wiley; 1998. [Google Scholar]
- 34.Couch FJ, Nathanson KL, Offit K. Two decades after BRCA: setting paradigms in personalized cancer care and prevention. Science. 2014;343:1466–1470. doi: 10.1126/science.1251827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daly MB, et al. Genetic/familial high-risk assessment: breast and ovarian, version 1.2014. J Natl Compr Canc Netw. 2014;12:1326–1338. doi: 10.6004/jnccn.2014.0127. [DOI] [PubMed] [Google Scholar]
- 36.Nelson HD, et al. Risk Assessment, Genetic Counseling, and Genetic Testing for BRCA-Related Cancer: Systematic Review to Update the US. Preventive Services Task Force Recommendation. Rockville (MD): 2013. [PubMed] [Google Scholar]
- 37.Force, U.S.P.S.T. Genetic risk assessment and BRCA mutation testing for breast and ovarian cancer susceptibility: recommendation statement. Ann Intern Med. 2005;143:355–361. doi: 10.7326/0003-4819-143-5-200509060-00011. [DOI] [PubMed] [Google Scholar]
- 38.King MC, Levy-Lahad E, Lahad A. Population-based screening for BRCA1 and BRCA2: 2014 Lasker Award. JAMA. 2014;312:1091–1092. doi: 10.1001/jama.2014.12483. [DOI] [PubMed] [Google Scholar]
- 39.Levy-Lahad E, Lahad A, King MC. Precision medicine meets public health: population screening for BRCA1 and BRCA2. J Natl Cancer Inst. 2015;107:420. doi: 10.1093/jnci/dju420. [DOI] [PubMed] [Google Scholar]
- 40.Yurgelun MB, Hiller E, Garber JE. Population-Wide Screening for Germline BRCA1 and BRCA2 Mutations: Too Much of a Good Thing? J Clin Oncol. 2015;33:3092–3095. doi: 10.1200/JCO.2015.60.8596. [DOI] [PubMed] [Google Scholar]
- 41.Neuhausen S, et al. Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat Genet. 1996;13:126–128. doi: 10.1038/ng0596-126. [DOI] [PubMed] [Google Scholar]
- 42.Roa BB, Boyd AA, Volcik K, Richards CS. Ashkenazi Jewish population frequencies for common mutations in BRCA1 and BRCA2. Nat Genet. 1996;14:185–187. doi: 10.1038/ng1096-185. [DOI] [PubMed] [Google Scholar]
- 43.Struewing JP, et al. The carrier frequency of the BRCA1 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals. Nat Genet. 1995;11:198–200. doi: 10.1038/ng1095-198. [DOI] [PubMed] [Google Scholar]
- 44.Oddoux C, et al. The carrier frequency of the BRCA2 6174delT mutation among Ashkenazi Jewish individuals is approximately 1% Nat Genet. 1996;14:188–190. doi: 10.1038/ng1096-188. [DOI] [PubMed] [Google Scholar]
- 45.Struewing JP, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336:1401–1408. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 46.Moslehi R, et al. BRCA1 and BRCA2 mutation analysis of 208 Ashkenazi Jewish women with ovarian cancer. Am J Hum Genet. 2000;66:1259–1272. doi: 10.1086/302853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King MC, Marks JH, Mandell JB New York Breast Cancer Study, G. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 48.Risch HA, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet. 2001;68:700–710. doi: 10.1086/318787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartman AR, et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer. 2012;118:2787–2795. doi: 10.1002/cncr.26576. [DOI] [PubMed] [Google Scholar]
- 50.Metcalfe KA, et al. A comparison of the detection of BRCA mutation carriers through the provision of Jewish population-based genetic testing compared with clinic-based genetic testing. Br J Cancer. 2013;109:777–779. doi: 10.1038/bjc.2013.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manchanda R, et al. Population testing for cancer predisposing BRCA1/BRCA2 mutations in the Ashkenazi-Jewish community: a randomized controlled trial. J Natl Cancer Inst. 2015;107:379. doi: 10.1093/jnci/dju379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finch AP, et al. Impact of oophorectomy on cancer incidence and mortality in women with a BRCA1 or BRCA2 mutation. J Clin Oncol. 2014;32:1547–1553. doi: 10.1200/JCO.2013.53.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gabai-Kapara E, et al. Population-based screening for breast and ovarian cancer risk due to BRCA1 and BRCA2. Proc Natl Acad Sci U S A. 2014;111:14205–14210. doi: 10.1073/pnas.1415979111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warner E, et al. Prevalence and penetrance of BRCA1 and BRCA2 gene mutations in unselected Ashkenazi Jewish women with breast cancer. J Natl Cancer Inst. 1999;91:1241–1247. doi: 10.1093/jnci/91.14.1241. [DOI] [PubMed] [Google Scholar]
- 55.Manchanda R, et al. Cost-effectiveness of population screening for BRCA mutations in Ashkenazi jewish women compared with family history-based testing. J Natl Cancer Inst. 2015;107:380. doi: 10.1093/jnci/dju380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Offit K. The BRCA Gene and Breast Cancer. In: Garber J, Karlan B, Tung N, Domchek S, Nathanson K, Robson M, editors. The New York Times. 2015. The Opinion Pages, Letters. [Google Scholar]
- 57.Regeneron Launches 100K-Patient Genomics Study with Geisinger, Forms New Genetics Center. GenomeWeb; 2014. [Google Scholar]
- 58.Wheeler HE, Maitland ML, Dolan ME, Cox NJ, Ratain MJ. Cancer pharmacogenomics: strategies and challenges. Nat Rev Genet. 2013;14:23–34. doi: 10.1038/nrg3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Offit K, Robson ME. New pharmacogenomic paradigm in breast cancer treatment. J Clin Oncol. 2010;28:4665–4666. doi: 10.1200/JCO.2010.31.2926. [DOI] [PubMed] [Google Scholar]
- 60.Aminkeng F, Bhavsar AP. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. 2015;47:1079–1084. doi: 10.1038/ng.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richards T, Montori VM, Godlee F, Lapsley P, Paul D. Let the patient revolution begin. Bmj. 2013;346:f2614. doi: 10.1136/bmj.f2614. [DOI] [PubMed] [Google Scholar]
- 62.Shirts BH, et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc. 2015;22:1231–1242. doi: 10.1093/jamia/ocv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prasad V, Fojo T, Brada M. Precision oncology: origins, optimism, and potential. Lancet Oncol. 2016;17:e81–e86. doi: 10.1016/S1470-2045(15)00620-8. [DOI] [PubMed] [Google Scholar]
- 64.Williams MS. Is the genomic translational pipeline being disrupted? Hum Genomics. 2015;9:9. doi: 10.1186/s40246-015-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Offit K. Genomic profiles for disease risk: predictive or premature? JAMA. 2008;299:1353–1355. doi: 10.1001/jama.299.11.1353. [DOI] [PubMed] [Google Scholar]
- 66.Offit K. Decade in review--genomics: a decade of discovery in cancer genomics. Nat Rev Clin Oncol. 2014;11:632–634. doi: 10.1038/nrclinonc.2014.170. [DOI] [PubMed] [Google Scholar]
- 67.Jones S, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roberts NJ, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2:41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Comino-Mendez I, et al. Exome sequencing identifies MAX mutations as a cause of hereditary pheochromocytoma. Nat Genet. 2011;43:663–667. doi: 10.1038/ng.861. [DOI] [PubMed] [Google Scholar]
- 70.Mahamdallie SS, et al. Mutations in the transcriptional repressor REST predispose to Wilms tumor. Nat Genet. 2015;47:1471–1474. doi: 10.1038/ng.3440. [DOI] [PubMed] [Google Scholar]
- 71.Ostergaard P, et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome) Nat Genet. 2011;43:929–931. doi: 10.1038/ng.923. [DOI] [PubMed] [Google Scholar]
- 72.Saliba J, et al. Germline duplication of ATG2B and GSKIP predisposes to familial myeloid malignancies. Nat Genet. 2015;47:1131–1140. doi: 10.1038/ng.3380. [DOI] [PubMed] [Google Scholar]
- 73.Yokoyama S, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 75.Testa JR, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farley MN, et al. A novel germline mutation in BAP1 predisposes to familial clear-cell renal cell carcinoma. Mol Cancer Res. 2013;11:1061–1071. doi: 10.1158/1541-7786.MCR-13-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weren RD, et al. A germline homozygous mutation in the base-excision repair gene NTHL1 causes adenomatous polyposis and colorectal cancer. Nat Genet. 2015;47:668–671. doi: 10.1038/ng.3287. [DOI] [PubMed] [Google Scholar]
- 79.Gara SK, et al. Germline HABP2 Mutation Causing Familial Nonmedullary Thyroid Cancer. N Engl J Med. 2015;373:448–455. doi: 10.1056/NEJMoa1502449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park DJ, et al. Rare mutations in XRCC2 increase the risk of breast cancer. Am J Hum Genet. 2012;90:734–739. doi: 10.1016/j.ajhg.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park DJ, et al. FAN1 variants identified in multiple-case early-onset breast cancer families via exome sequencing: no evidence for association with risk for breast cancer. Breast Cancer Res Treat. 2011;130:1043–1049. doi: 10.1007/s10549-011-1704-y. [DOI] [PubMed] [Google Scholar]
- 82.Ruark E, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493:406–410. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cybulski C, et al. Germline RECQL mutations are associated with breast cancer susceptibility. Nat Genet. 2015;47:643–646. doi: 10.1038/ng.3284. [DOI] [PubMed] [Google Scholar]
- 84.Rafnar T, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011;43:1104–1107. doi: 10.1038/ng.955. [DOI] [PubMed] [Google Scholar]
- 85.Noetzli L, et al. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nat Genet. 2015;47:535–538. doi: 10.1038/ng.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang MY, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet. 2015;47:180–185. doi: 10.1038/ng.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]