Abstract
Objective
The overall goal of this document is to demonstrate that dissemination of models and analyses for assessing the reproducibility of simulation results can be incorporated in the scientific review process in biomechanics.
Methods
As part of a special issue on model sharing and reproducibility in IEEE Transactions on Biomedical Engineering, two manuscripts on computational biomechanics were submitted: A. Rajagopal et al., IEEE Trans. Biomed. Eng., 2016 and A. Schmitz and D. Piovesan, IEEE Trans. Biomed. Eng., 2016. Models used in these studies were shared with the scientific reviewers and the public. In addition to the standard review of the manuscripts, the reviewers downloaded the models and performed simulations that reproduced results reported in the studies.
Results
There was general agreement between simulation results of the authors and those of the reviewers. Discrepancies were resolved during the necessary revisions. The manuscripts and instructions for download and simulation were updated in response to the reviewers’ feedback; changes that may otherwise have been missed if explicit model sharing and simulation reproducibility analysis were not conducted in the review process. Increased burden on the authors and the reviewers, to facilitate model sharing and to repeat simulations, were noted.
Conclusion
When the authors of computational biomechanics studies provide access to models and data, the scientific reviewers can download and thoroughly explore the model, perform simulations, and evaluate simulation reproducibility beyond the traditional manuscript-only review process.
Significance
Model sharing and reproducibility analysis in scholarly publishing will result in a more rigorous review process, which will enhance the quality of modeling and simulation studies and inform future users of computational models.
Index Terms: biomechanics, dissemination, joint, joint mechanics, mechanics, model, musculoskeletal, publishing, repeatability, reproducibility, sharing, simulation, tissue, tissue mechanics
I. Introduction
Modeling and simulation strategies have offered significant utility in biomechanics with applications ranging from rigid body dynamics based musculoskeletal modeling of locomotion, limb movement, and motor control [1]; to finite element analysis exploring deformations and stresses of joints and tissues, and medical devices [2]; to computational fluid dynamics focusing on studies in cardiovascular medicine [3]. The popularity of modeling & simulation in biomechanics is not surprising as simulation-based approaches provide for cost-effective, prompt, and systematic prediction of the mechanobiological behavior of physiological systems. Moreover, modeling & simulation permit biomechanical markers of healthy and diseased joint and organ function to be established and used for diagnosis and for a-priori assessment of performance and safety of clinical interventions [2],[3].
The increased demand to capture physiological realism for scientifically and clinically relevant simulations has resulted in highly detailed virtual representations of the human body. The modeling & simulation workflows to develop such models have become highly complicated, often integrating heterogeneous data (physiological and anatomical properties, loading and boundary conditions) while necessitating related assumptions dictated by the desired level of simulation fidelity. With increased computational capacity and through the advancement of simulation technology, coupling of different modeling modalities (musculoskeletal movements-finite element analysis [4], fluid-solid interactions in the heart valves [5]), physical domains (bioelectric-biomechanical in the heart [6]), and spatial and temporal scales (cell deformations during joint loading [7]) become possible; further complicating the appreciation and utilization of computational models.
Reproducibility of scientific studies has been challenged recently [8]. Computational approaches, and by extension modeling & simulation studies, are not exceptions [9]. The biomechanics community has been responsive in addressing the credibility problem in modeling & simulation. Strategies for verification and validation of models have been documented, e.g., for simulations of organ and tissue mechanics [10] and for musculoskeletal modeling [11]. Reporting parameters to enhance reproducibility of finite element analysis have also been provided [12]. Nonetheless, achieving the rigor to ensure a reproducible practice in computational biomechanics remains a daunting challenge.
In the general community of biomedical disciplines, independent review of models and their dissemination have been recognized as important steps for establishing the quality of a modeling & simulation study, i.e., by the committee on Credible Practice of Modeling & Simulation in Healthcare [13]. Funding programs have also started to request model sharing as a requirement and third-party review as part of a model credibility plan [14]. The ultimate goal of these initiatives has been to promote quality assurance in computational modeling and subsequently, to permit re-use or re-purposing of these models by the community, therefore enabling advancement of biomedical sciences and healthcare delivery. It would be reasonable to assume that model sharing will facilitate any analysis that is aimed to understand the reproducibility of a modeling & simulation study and that the documented quality of a computational model, obtained from a reproducibility analysis, will promote its appropriate re-use in prospective scientific and clinical studies. However, implementing model sharing and reproducibility analysis introduces scientific, technological, and cultural challenges to the academic enterprise, which has traditionally relied on exchange of knowledge through scholarly publishing. Strategies need to be developed and tested in order to integrate systematic sharing and evaluation of computational models in the academic publication workflow.
The primary goal of this article was to document our experiences to integrate simulation reproducibility analyses, supported by model sharing, in the publication review workflow. An added benefit was the demonstration of the reproducibility potential of two musculoskeletal models, which were disseminated and published as part of this activity: one on movement simulations of the full body [15], another on a computational representation of the knee joint [16]. While this report focused on musculoskeletal modeling, it can be utilized as an example in any area of computational biomechanics. Similarly, although the document provides a biomechanics perspective, it will likely inform other scientific disciplines tackling the challenging problems of model sharing and simulation reproducibility.
II. Methods
For a special issue on model sharing and reproducibility, to be published in IEEE Transactions on Biomedical Engineering, manuscripts were invited for submissions. Unsolicited submissions were accepted along with those acquired through targeted invitations. Some of these manuscripts were preferred to be modeling & simulation studies in the area of biomechanics, which would describe relevant computational models and their utilization. The special issue aimed to promote dissemination of the computational models as part of the scholarly publication process. An additional goal was to leverage model sharing in the review process, during which the reviewers can download and use the models to assess and comment on the reproducibility of simulation results presented in the manuscripts. As a by-product, dissemination was also anticipated to facilitate evaluation of model robustness and the potential impact of model sharing on the discipline.
The authors of the modeling & simulation studies were expected to give public access to the computational models, at least after the confirmation of acceptance of their manuscripts for publication. A staged dissemination was anticipated, i.e., only the associate editor and the reviewers would be given access during the review process of the manuscript. The authors were requested to document in the manuscript the location of the model on the web, ideally in an online repository. Supplementary information included download instructions and guidance on how to reproduce simulations described in the manuscript, either provided as part of the submission or at the dissemination site. The authors were informed that the dissemination and reproducibility of the models would be evaluated in addition to the scientific review.
The reviewers of the modeling & simulation studies (co-authors of this document) were instructed to download the models and conduct simulations to reproduce results reported in the manuscripts. They were also asked to provide feedback on the adequacy of download and simulation instructions, and on the likely reproducibility of the whole modeling & simulation study. The reviewers were informed that the review process extended beyond the routine scientific review in biomechanics. As such, they were asked to comment on the incorporation of model sharing and reproducibility analysis to the review workflow; including its value and its challenges.
The associate editor of the special issue (Ahmet Erdemir) was responsible for the oversight of the review process. He ensured that the initial manuscript submissions included the necessary information to access the models and provided instructions to the authors and reviewers for single-sided blind reviews of the manuscripts, i.e., the names of the reviewers were hidden from the authors (until publication of this document).
III. Results
Two manuscripts were submitted to the special issue on model sharing and reproducibility. Both studies dealt with modeling & simulation of the musculoskeletal system; one specifically on a full body model for simulating human gait [15]; the other on a knee model created for incorporation in musculoskeletal models [16]. Both models relied on OpenSim (available at http://opensim.stanford.edu/), a freely available, open source simulation software for musculoskeletal modeling [17]. The studies utilized SimTK (available at https://simtk.org/) as the dissemination platform. For each manuscript, the review process started with an initial submission, which was followed by two cycle of revisions to address reviewers’ comments and editorial issues. Both submissions included detailed supplementary material on model parameters, on a variety of analyses indicating the quality of model predictions, and on sensitivity to various model parameters.
The model and a sample simulation package relevant to the study on full body musculoskeletal modeling [15] were provided at the website https://simtk.org/home/full_body. During the initial submission, the dissemination was private, i.e., access was provided only to project members. To permit reviewers to download the materials, an account was created and login information was provided to the reviewers as part of the submission. Upon acceptance of the article for publication, the dissemination site was made public for anyone to download and access the model. Relevant information on model sharing, e.g., location, was provided in the abstract and the body of the manuscript. Supplementary material included detailed download instructions and guidance to re-generate sample simulation results. All the reviewers (a total of four), were able to download the model and re-run the simulations. Many reviewers were already experienced in OpenSim. Nevertheless, some challenges were noted. While the instructions were adequate, a need to provide a more streamlined and documented process to re-run simulations was apparent. In addition, some of the reviewers had to utilize different operating platforms, different versions of OpenSim, and most importantly many did not have access to a certain optimization algorithm used by the authors of the model and used alternative algorithms available in OpenSim.
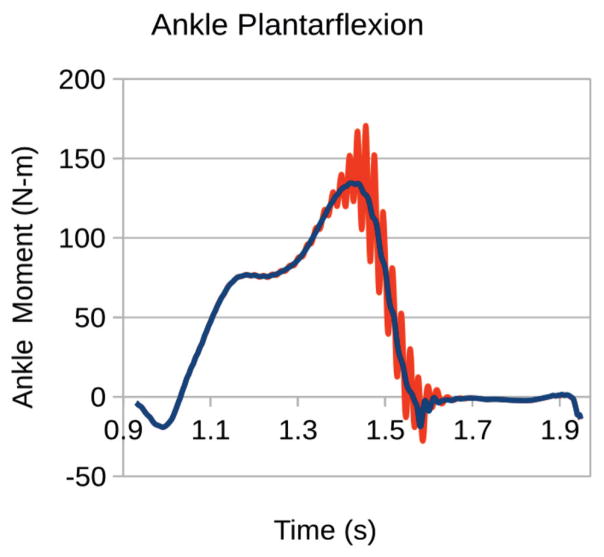
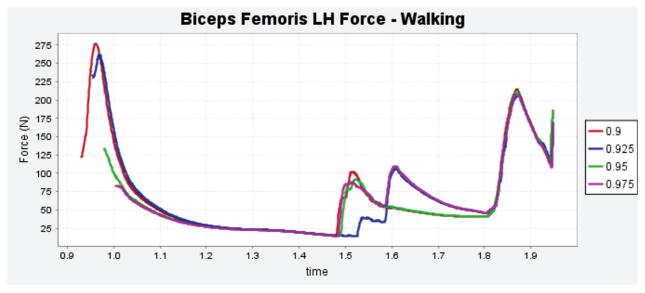
From the perspective of reproducibility of simulation results, all reviewers indicated a general agreement between their results and those reported in the manuscript. Most of these simulations explored the use of the full body model to simulate walking and running and report muscle activations, joint kinematics and kinetics, etc. While the reviewers were generally able to reproduce the reported simulation results, some discrepancies were observed with the materials provided on the initial submission. This was true especially for the simulations of walking, e.g., a phase lag in time histories of muscle forces and joint kinematics, oscillations in joint kinetics that were not reported in the manuscript (Fig. 1), and differences in computed muscle activations (Fig. 2). Leveraging the availability of the model and relevant data for sample simulations, one reviewer expanded the study to conduct a simple sensitivity analysis on the selection of control algorithm parameters to calculate muscle force trajectories (Fig. 3). In subsequent revisions, the authors of the full body modeling study [15], created a MATLAB (The Mathworks, Natick, MA, USA) script to auto-generate simulation results, which was confirmed by the reviewers as a useful addition to dissemination. Upon resubmission, the authors also switched to an optimization algorithm that is more generally available to the users of OpenSim. In response to simulation result discrepancies, the authors attempted to reproduce the noted oscillation (Fig. 1 and Fig. 2), this time with different versions of OpenSim, on different operating systems, and by changing some of the control algorithm parameters. They concluded that the oscillatory behavior was a result of the experimental data used for generating the walking simulation. Subsequently, a more recently collected and higher quality walking data set was used for simulations in the revisions. In follow-up reviews, the reviewers were able to obtain visibly similar results as the authors of the model.
Fig. 1.
Predictions of ankle plantar flexion moment during walking, as reported in the initial submission of the full body musculoskeletal model [15] (blue solid line) and as reported by a reviewer repeating simulations (red solid line). The discrepancies in simulation results, which utilized the same model, were resolved in revisions of the manuscript by relying on higher quality experimental walking data. The plot is a direct copy from the reviewer’s response.
Fig. 2.
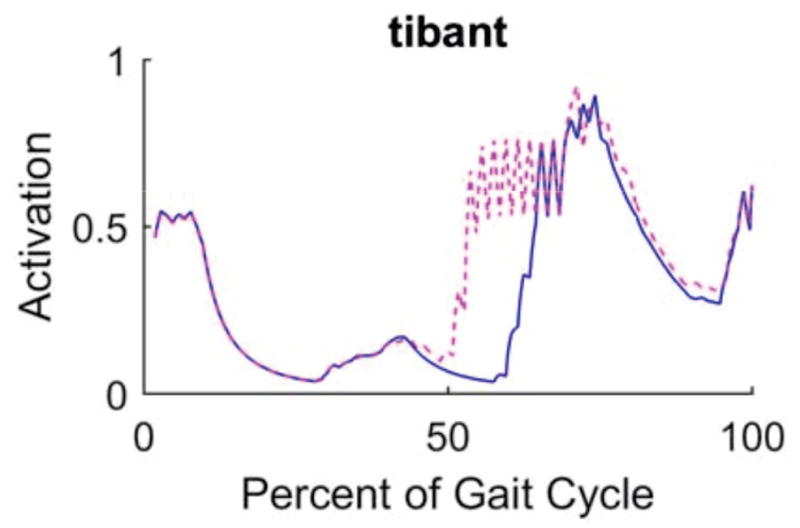
Predictions of tibialis anterior activation during walking, as reported in the initial submission of the full body musculoskeletal model [15] (blue solid line) and as reported by a reviewer repeating simulations (red dashed line). The discrepancies in simulation results, which utilized the same model, were resolved in revisions of the manuscript by relying on higher quality experimental walking data (please, also see Fig. 1). The plot is a direct copy from the reviewer’s response. It should be noted that the reviewer who provided this plot was different than the one who provided the simulation results for Fig. 1.
Fig. 3.
A reviewer of the manuscript on the full body musculoskeletal model [15], extended the analysis to understand the sensitivity of muscle force trajectory prediction on the selection of a control algorithm parameter (specifically the Computed Muscle Control start time). Biceps femoris forces during walking are shown. This analysis was facilitated by the model sharing approach adapted during scholarly publication. The plot is a direct copy from the reviewer’s response. The reviewer who provided this analysis was different than those who provided plots for Fig. 1 and Fig. 2.
The model relevant to the study on knee joint representation for musculoskeletal modeling [16] was provided at the website https://simtk.org/home/kneemodel. Relevant information on the online location of the model was provided in the body of the manuscript. Dissemination was public, including the initial submission of the manuscript. The dissemination site included a presentation on instructions about model use. The manuscript dealt with the specific problem of including a higher fidelity representation of the knee in musculoskeletal models. The article included comparisons of predicted knee joint response to literature data available for passive flexion and for laxity. Passive flexion simulations were aimed to demonstrate coupling between knee joint degrees of freedom, whereas laxity simulations illustrated the overall mobility of the joint under prescribed loads. Detailed supplementary information was provided to describe ligament properties and relevant sources and to quantify sensitivity of knee joint response to ligament properties. A total of two reviewers assessed the model. In the review of the initial submission, downloading the model and running an OpenSim simulation was found to be straightforward. Nonetheless, the reviewers requested additional instructions on using the model and on evaluating simulation results. In particular, one reviewer, who did not have extensive experience in OpenSim, noted some difficulties to manipulate the model and reproduce the results. In subsequent revisions, the authors of the manuscript provided additional files (for input, setup, and batch processing) and expanded upon instructions provided in a presentation at the dissemination site to reflect the workflow of reproducing simulation results.
Opportunities and challenges of model sharing and reproducibility analysis as part of scholarly publication and scientific review can be best attested by the comments and the sentiment of the reviewers. A sampling of such commentary are provided in the following sections.
A Reviewer Comments on Model Download
“I was able to download the model and other associated files from SimTK without any issues. It is hard to see how this aspect could be improved upon, as long as the location of the files remains consistent with the URL provided in the published paper.”
“In my opinion, in the context of an anonymized reviewing process, models and simulation data should be provided together with the manuscript through the journal editorial system directly.”
B. Reviewer Comments on Simulation Instructions
“For someone who is not that proficient in OpenSim, this reviewer could not figure out what to look at or how to manipulate the model to reproduce results. There were a few details missing on the specifics of how the boundary conditions were applied. While some of these issues may be inherently obvious for someone more proficient in OpenSim, a simple set of instructions on reproducing simulations would have really been helpful.”
“Instruction accompanying the simulations could mention possible sources of variation from the ‘original’ simulation results.”
“The Matlab script that runs all of the simulations and generates the results figures is a nice addition. That will allow users to either simply generate the final results, or it will assist them with drilling down in detail on a specific aspect of interest.”
C. Reviewer Comments on Discrepancies in Simulation Results
“Given the focus on reproducibility, I should note that I was again able to reproduce the authors’ results in general, though still with some minor discrepancies (different than before) in the predicted muscle activations. I am willing to believe that this does not represent any shortcoming in the work the authors have done, but rather could reflect the complexities of using sophisticated models and control algorithms across different operating systems and computer architectures. This special issue is a perfect venue in which to bring these issues to light.”
D. Reviewer Comments on Reporting
“I was able to thoroughly explore the model in the OpenSim environment. This allowed me to easily find some cases where there were inconsistencies or omissions between the descriptions in the manuscript and the actual model. This allowed me to provide feedback that the authors can respond to when they revise their manuscript. Without access to the model, I would have been guessing at some of these issues and completely unaware of others.”
“A good journal policy would ensure that, if a computational tool or simulations are made available, enough material is included to allow the reader to reproduce at least the manuscript figures from the input data.”
“In my opinion the journals should decide some minimum requirements or relative strict guidelines regarding the organization of the material provided for reproducibility purposes.”
E. Reviewer Comments on Burden of Model Sharing & Reproducibility Analysis
“It is not easy task to replicate a simulation, even when the original data and setups files are available.”
“If the model had not been shared, assessment of reproducibility would have been considerably more difficult and possibly time prohibitive.”
“CMC simulations were relatively long (around 15 minutes). It is easy to realize that repeatability assessment within the reviewing process will not be feasible for simulations requiring longer times.”
“The reviewing process was significantly longer than it would have been otherwise and it took a reasonable time only because I was already familiar with the software used for the simulations.”
“It is not difficult to imagine less skilled authors giving the reviewers a hard time submitting badly organized material.”
“Will submitting a journal manuscript also require writing an extensive set of documentation? Will the only possible reviewers be people who have the necessary expertise with the modeling environment or programming language used for the simulations? This could create an additional burden on writing and reviewing simulation papers that does not exist for experimental studies.”
F. Reviewer Comments on Premise of Model Sharing & Reproducibility Analysis
“A critical consideration when doing a repeatability analysis of this kind is distinguishing between pure repeatability (the reviewer runs the simulations, and if they match the results presented in the paper that’s the end of the task), or using the reproduced simulations to judge the correctness of the work under revision.”
“Without providing the model and results files, troubleshooting my results would have been very challenging. I believe this supports the benefit of dissemination and I appreciate that the authors are willing to provide this level of information. While dissemination of model and results is not common practice during publication, supporting this culture shift should contribute to both the credibility and validity of future simulation studies.”
“As a final remark, I think that reproducibility as part of the publication process would be highly beneficial to the field of computational biomechanics, acting as a natural filter towards publication of dubious results, reducing the amount of low quality submissions (because of the amount of work necessary to prepare a submission) and enhancing the quality and quantity of publicly available computational tools.”
IV. Discussion
This document provided a summary of our experience incorporating strategies for sharing and evaluating models in computational biomechanics. The experience presented here illustrate that dissemination and reproducibility analysis of computational models can be part of the scientific review and publication process, albeit at an increased workload indicated by the reviewers. In their communications with the associate editor and the reviewers, the authors of the modeling & simulation studies, e.g., [15], also recognized the value of dissemination and reproducibility analysis, and the efforts associated to accomplish these.
The modeling & simulation studies, which went through the more rigorous review process described herein, benefited from the assessments of model sharing and simulation reproducibility. The science of the studies improved, i.e., by utilization of higher quality data sets in revisions and by additional sensitivity analysis conducted by the authors and by the reviewers. Reporting of the studies were enhanced as well; additional details on the models and simulation cases were provided, both in the manuscript and in the material for dissemination. Adequacy of the dissemination approach was tested. This resulted in adapting generally available numerical algorithms, evaluation of simulation performance in different operating systems with different simulation software versions, and accommodating the anonymity of the reviewers when accessing the models. More importantly, additional scripts and instructional materials to facilitate reproduction of simulation results were provided. All these improvements will likely benefit future users of these specific models by facilitating their re-use and re-purposing.
Our experimentation with model sharing and simulation reproducibility also identified areas of improvement to incorporate such analysis in the scientific review and publication workflow. Utilization of OpenSim [17], a freely available and open source software, permitted access to simulation software. A software with limited availability to the reviewers may have prevented them to execute simulations for the reproducibility analysis. The reviewers were experienced in musculoskeletal modeling & simulation and the majority have had exposure to OpenSim. Identification of reviewers with matching expertise, not only possessing the scientific insight but also the technical capability, was necessary. Otherwise, the demanding tasks to navigate and review the models and to conduct simulations may have not been completed. Standards, when and if available to describe models and simulation workflows, will likely be helpful. Similarly, automation of some tedious tasks of modeling & simulation workflow, e.g., model preparation, simulation, post-processing of simulation results, etc., may facilitate model evaluation process. In some cases, recognizing computational cost may be necessary as this constraint may prevent others to re-run simulations. Under such circumstances, delivery of simplified yet representative simulation test cases can be necessary and sufficient in order to evaluate the assumptions of the modeling & simulation. To mitigate all these issues, clear guidelines should be provided and pragmatic strategies will need to be implemented. While these are beyond the scope of this document, potential mechanisms outlined in literature, e.g., [18], and work conducted by interdisciplinary committees, e.g., [13], will likely be instrumental.
Overall, the documented activities will provide a template for the biomechanics community to appropriately leverage modeling & simulation for scientific discovery. They will also inform journals, communities on modeling & simulation practices, and funding agencies to accommodate independent reviews of models and simulation results.
V. Conclusion
In biomechanics, public sharing of models and the analysis of simulation reproducibility can be incorporated in the scholarly publication workflow at manuscript submission and during the scientific review processes. Consequently, an increase in the quality of computational models can be expected, as well as the modeling & simulation studies that rely on them. Demonstration of the utility of a computational model and its documentation, to recreate published simulations, can also identify potential hurdles and opportunities that may be experienced by future users. Nonetheless, certain assumptions should be met to carry-out such endeavors. Model sharing may clash with cultural perceptions within the academic enterprise and constraints of intellectual property rights. Computational biomechanics heavily rely on stand-alone simulation software, which may not be available for reviewers and future users of the models, particularly when products with restrictive licensing are utilized. Even at times when the model and the simulation software are accessible, the burden on the authors and the reviewers of modeling & simulation studies should not be underestimated. Preparing a model for distribution and providing necessary information to enhance its reproducibility potential are activities that require significant effort and time beyond what is expected from authors in traditional publishing. Similarly, attempting to reproduce simulations by downloading models and simulation software, trying to evaluate the sources of discrepancies, and documenting results of simulation reproducibility add to the tasks of reviewers who are already burdened by scientific review requests. As the discipline of biomechanics has evolved, the need for more complicated models depending on heterogeneous data and utilizing a variety of sophisticated simulation strategies emerged. Balancing the prompt delivery of biomechanical discoveries with the burden associated with dissemination and third-party quality assurance in computational biomechanics will likely be a challenging task. On the other hand, the rewards will be substantial, i.e., in the form of increased credibility of modeling & simulation studies and by the general availability of high quality, robust, and reusable computational models. The experience documented in this article demonstrated that a rigorous dissemination and scientific review can be conducted successfully. In doing so, this experience provides a foundation to focus on model sharing and simulation reproducibility in biomechanics.
Acknowledgments
Contributions by Ahmet Erdemir was supported in part by the National Institutes of Health under Grant R01GM104139; Trent M. Guess in part by the National Institutes of Health under Grant RAR061698; Luca Modenese in part by the EPSRC under Grant EP/K03877X/1 and Grant FP7-ICT-600932; Jeffrey A. Reinbolt in part by the National Science Foundation under Grant CAREER 1253317; Darryl G. Thelen in part by the National Institutes of Health under Grant EB015410; and Brian R. Umberger in part by the National Science Foundation under Grant IIS-1526986.
The authors would like to thank Apoorva Rajagopal, Christopher L. Dembia, Matthew S. DeMers, Denny D. Delp, Jennifer L. Hicks, Scott L. Delp, authors of the study on full body musculoskeletal modeling [15] and Anne Schmitz, Davide Piovesan, authors of the study on knee joint modeling [16]. Without their willingness to disseminate their computational models and to go through the rigorous review process, the presented work would not be possible. A. Erdemir served as the associate editor during the review of the aforementioned modeling studies and the co-authors of this document served as the reviewers. A. Erdemir would like to recognize the diligent work of the co-authors, who took responsibilities beyond those expected in a traditional scientific review process. Assistance of Michael Vignos in the review process is also appreciated. The insights from the co-organizers of the special issue on model sharing and reproducibility, Herbert Sauro and Grace Peng, and from the leadership at the IEEE Transactions on Biomedical Engineering, Bin He, are also appreciated. A. Erdemir would like to acknowledge the IEEE-EMBS Technical Committee on Computational Biology and the Physiome and the Interagency Modeling and Analysis Group (IMAG) and Multiscale Modeling (MSM) Consortium, where many ideas on model sharing and reproducibility were seeded and have been discussed continuously.
Contributor Information
Ahmet Erdemir, Department of Biomedical Engineering and the Computational Biomodeling (CoBi) Core, Cleveland Clinic, Cleveland, OH, USA.
Trent M. Guess, Departments of Physical Therapy, Orthopaedic Surgery, and Bioengineering, University of Missouri, Columbia, MO, USA
Jason P. Halloran, Department of Mechanical Engineering, Cleveland State University, Cleveland, OH, USA
Luca Modenese, Department of Mechanical Engineering and INSIGNEO Institute for in silico Medicine, University of Sheffield, Sheffield, UK.
Jeffrey A. Reinbolt, Department of Mechanical, Aerospace and Biomedical Engineering, University of Tennessee, Knoxville, TN, USA
Darryl G. Thelen, Departments of Mechanical Engineering, Biomedical Engineering, and Orthopaedics and Rehabilitation, University of Wisconsin-Madison, Madison, WI, USA
Brian R. Umberger, Department of Kinesiology, University of Massachusetts, Amherst, MA, USA
References
- 1.Erdemir A, et al. Model-based estimation of muscle forces exerted during movements. Clin Biomech. 2007 Feb;22(2):131–154. doi: 10.1016/j.clinbiomech.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Taylor M, Prendergast PJ. Four decades of finite element analysis of orthopaedic devices: where are we now and what are the opportunities? J Biomech. 2015 Mar;48(5):767–778. doi: 10.1016/j.jbiomech.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 3.Morris PD, et al. Computational fluid dynamics modelling in cardiovascular medicine. Heart. 2016 Jan;102(1):18–28. doi: 10.1136/heartjnl-2015-308044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halloran JP, et al. Concurrent musculoskeletal dynamics and finite element analysis predicts altered gait patterns to reduce foot tissue loading. J Biomech. 2010 Oct;43(14):2810–2815. doi: 10.1016/j.jbiomech.2010.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toma M, et al. Fluid-structure interaction analysis of papillary muscle forces using a comprehensive mitral valve model with 3D chordal structure. Ann Biomed Eng. 2016 Apr;44(4):942–953. doi: 10.1007/s10439-015-1385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baillargeon B, et al. The Living Heart Project: A robust and integrative simulator for human heart function. Eur J Mech A Solids. 2014 Nov;48:38–47. doi: 10.1016/j.euromechsol.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sibole SC, Erdemir A. Chondrocyte deformations as a function of tibiofemoral joint loading predicted by a generalized high-throughput pipeline of multi-scale simulations. PLoS One. 2012;7(5):e37538. doi: 10.1371/journal.pone.0037538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker M. 1,500 scientists lift the lid on reproducibility. Nature. 2016 May;533(7604):452–454. doi: 10.1038/533452a. [DOI] [PubMed] [Google Scholar]
- 9.Ince DC, et al. The case for open computer programs. Nature. 2012 Feb;482(7386):485–488. doi: 10.1038/nature10836. [DOI] [PubMed] [Google Scholar]
- 10.Anderson AE, et al. Verification, validation and sensitivity studies in computational biomechanics. Comput Methods Biomech Biomed Engin. 2007 Jun;10(3):171–184. doi: 10.1080/10255840601160484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hicks JL, et al. Is my model good enough? Best practices for verification and validation of musculoskeletal models and simulations of movement. J Biomech Eng. 2015 Feb;137(2):020905. doi: 10.1115/1.4029304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erdemir A, et al. Considerations for reporting finite element analysis studies in biomechanics. J Biomech. 2012 Feb;45(4):625–633. doi: 10.1016/j.jbiomech.2011.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erdemir A, et al. Ten “not so” simple rules for credible practice of modeling and simulation in healthcare: a multidisciplinary committee perspective. presented at the 2015 Biomedical Engineering Society/Food and Drug Administration Frontiers in Medical Devices Conference: Innovations in Modeling and Simulation; Washington, DC, USA. May 18–20, 2015. [Google Scholar]
- 14.Department of Health and Human Services, USA. Predictive Multiscale Models for Biomedical, Biological, Behavioral, Environmental and Clinical Research (U01) [Online]. Available: http://grants.nih.gov/grants/guide/pa-files/PAR-15-085.html.
- 15.Rajagopal A, et al. Full body musculoskeletal model for muscle-driven simulation of human gait. IEEE Trans Biomed Eng. 2016 doi: 10.1109/TBME.2016.2586891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitz A, Piovesan D. Development of an open-source, discrete element knee model. IEEE Trans Biomed Eng. 2016 doi: 10.1109/TBME.2016.2585926. [DOI] [PubMed] [Google Scholar]
- 17.Delp SL, et al. OpenSim: open-source software to create and analyze dynamic simulations of movement. IEEE Trans Biomed Eng. 2007 Nov;54(11):1940–1950. doi: 10.1109/TBME.2007.901024. [DOI] [PubMed] [Google Scholar]
- 18.Yilmaz L. Reproducibility in M&S research: issues, strategies, and implications for model development environments. J Exp Theor Artif Intell. 2012 Sep;24(4):457–474. [Google Scholar]