Abstract
Chlamydomonas reinhardtii controls flagellar assembly such that flagella are of an equal and predetermined length. Previous studies demonstrated that lithium, an inhibitor of glycogen synthase kinase 3 (GSK3), induced flagellar elongation, suggesting that a lithium-sensitive signal transduction pathway regulated flagellar length (S. Nakamura, H. Takino, and M. K. Kojima, Cell Struct. Funct. 12:369-374, 1987). Here, we demonstrate that lithium treatment depletes the pool of flagellar proteins from the cell body and that the heterotrimeric kinesin Fla10p accumulates in flagella. We identify GSK3 in Chlamydomonas and demonstrate that its kinase activity is inhibited by lithium in vitro. The tyrosine-phosphorylated, active form of GSK3 was enriched in flagella and GSK3 associated with the axoneme in a phosphorylation-dependent manner. The level of active GSK3 correlated with flagellar length; early during flagellar regeneration, active GSK3 increased over basal levels. This increase in active GSK3 was rapidly lost within 30 min of regeneration as the level of active GSK3 decreased relative to the predeflagellation level. Taken together, these results suggest a possible role for GSK3 in regulating the assembly and length of flagella.
How cells regulate the size of their organelles is poorly understood. Cilia and flagella are of particular interest, as the precise regulation of their length is critical to their function. The importance of the correct assembly of these organelles, including maintenance of the appropriate length, is illustrated by studies involving intraflagellar transport (IFT). IFT is required for the assembly and maintenance of cilia and flagella in organisms ranging from Chlamydomonas reinhardtii (where it was first identified) to sea urchins, Tetrahymena thermophila, Caenorhabditis elegans, and mice (10). In the absence of IFT, cilia and flagella fail to form correctly, resulting in various human diseases, including polycystic kidney disease and retinal degeneration, and anomalous determination of the left-right axis during development (51). Ultrastructural studies of children suffering from a particular form of primary ciliary dyskinesia revealed the presence of abnormally long cilia within their respiratory tracts, resulting in recurrent respiratory infections (1, 39). This observation suggests that not only do cilia and flagella need to assemble properly but that they also need to be of appropriate and defined lengths.
The green alga Chlamydomonas reinhardtii makes an ideal model organism for studies involving ciliary and flagellar assembly and length control. Chlamydomonas has two apically localized flagella that are maintained at equal lengths. Moreover, within a population of wild-type cells, flagellar lengths fall within a very narrow range. The maintenance of flagella of equal lengths as an active process was first demonstrated in the “long-zero” experiments of Rosenbaum et al. (52). In these experiments, cells were deflagellated under conditions that amputated only one flagellum. The remaining flagellum immediately began to shorten to an intermediate length, while the amputated flagellum began to regrow. After the shortening and elongating flagella reached equal lengths, they then continued to elongate at the same rate to return to their predeflagellation length. Thus, Chlamydomonas is able to detect and compare flagellar lengths and, when inequality is present, to correct discrepancies between the two flagella.
The active control of flagellar length is best demonstrated by the characterization of mutants that have lost control of their flagellar lengths. Three classes of flagellar-length mutants have been identified: short-flagellar (Shf) mutants, long-flagellar (Lf) mutants, and unequal-length-flagellar (Ulf) mutants (23, 29, 57). The functioning of an active process to control length can be seen during mating, when gametes fuse to form a temporary dikaryon. When wild-type cells are mated to Shf mutant cells, the resulting dikaryon contains two flagella of wild-type length and two half-length flagella (from the Shf cell). Shortly after cell fusion, within 10 min, the Shf flagella elongate to wild-type length (23). In dikaryons that contain two wild-type flagella and two flagella that are abnormally long (from an Lf cell), the Lf flagella shorten to wild-type length in less than 15 min (6). Interestingly, the wild-type flagella do not lengthen, even though flagellar components sufficient to assemble two or more flagella of wild-type length are resorbed into the cell body. These results demonstrate that some component of the wild-type cytoplasm can measure flagellar length and actively lengthen (in the case of Shf mutants) or shorten (in the case of Lf mutants) flagella of abnormal length. Because wild-type length flagella are not observed to undergo lengthening or shortening in dikaryons with Shf or Lf mutants, maintenance of flagella at equal lengths does not reflect simply the partitioning of flagellar components into the available flagella.
Flagellar length can also be manipulated chemically. For example, Solter and Gibor (56) demonstrated that flagellar length was inversely related to the osmolarity of the culture media. A role for calcium in the regulation of flagellar length comes from studies with fa1 mutants, which are unable to deflagellate. Treatment of fa1 cells with Ca2+ channel blockers (verapamil, brepredil, and diltiazem) or Ca2+-calmodulin antagonists (trifluoperazine or W7) induced flagella to shorten by ∼25% (60). Moreover, the inclusion of various Ca2+-chelating agents, such as EGTA, pyrophosphate, or citrate, in the culture medium of wild-type cells resulted in the disassembly and resorption of the flagella into the cell body (31). This shortening was rapidly reversed by the readdition of Ca2+ to the culture medium. Finally, the elongation of flagella was induced by the treatment of cells with lithium (38, 60). Taken together, these observations suggest that one or more signal transduction pathways control flagellar length in Chlamydomonas. Even more compelling evidence for signal transduction playing a role in the regulation of flagellar length comes from the recent identification of the LF4 gene as a microtubule-associated/mitogen-activated protein (MAP) kinase (7). Along these same lines, a MAP kinase kinase was shown to be required in the control of flagellar length for Leishmania mexicana (61).
A number of signaling molecules that are targets for regulation by lithium have been identified. Among these are inositol monophosphatases (2, 8) and glycogen synthase kinase 3β (GSK3β) (26). Lithium directly inhibits GSK3β kinase activity both in vitro and in vivo by acting as a noncompetitive inhibitor for magnesium. GSK3β, also known as tau protein kinase I, has been shown to regulate microtubule stability by phosphorylation of MAPs, such as MAP1B and tau (22, 33). Phosphorylation of MAP1B by GSK3β increases the affinity of MAP1B for binding to microtubules, resulting in an increase in microtubule stability. Conversely, the microtubule-associated protein tau loses the ability to bind microtubules when it is phosphorylated by GSK3β, decreasing the stability of microtubules. Recently, GSK3β was shown to phosphorylate the light chains of conventional kinesin, regulating both fast anterograde axonal transport and interaction with cargo (36). The inhibition of GSK3β by lithium and the ability of GSK3β to regulate microtubule stability suggest that the lithium-sensitive target involved in flagellar length regulation may be GSK3.
Here, we examine the role of lithium in flagellar length control and identify GSK3 as a target for inhibition by lithium. GSK3 is a flagellar protein whose kinase activity is inhibited by lithium. Reducing GSK3 protein levels by RNA interference (RNAi) results in aflagellate cells, phenocopying long-term lithium treatment. Taken together, these observations suggest a role for GSK3 in flagellar assembly and in the regulation of flagellar length.
MATERIALS AND METHODS
Cell culture and strains.
Chlamydomonas reinhardtii strain 21gr (mt+) (CC-1690) (Chlamydomonas Genetics Center) and E10 (arg7, mt+) (C. Silflow, University of Minnesota) were cultured with aeration at 22°C in medium I or medium II (53) on a cycle of 14 h of light and 10 h of darkness. For induction of RNAi expression, transformants were cultured with aeration at 22°C in MNO3 medium (same as medium I except that NH4NO3 is replaced with 4 mM KNO3).
Drug treatment and flagellar measurements.
Stock solutions of LiCl, NaCl, KCl, and NH4Cl at 1 M concentrations were prepared in culture medium and added to a final concentration of 25 mM. As a control for drug addition, an equal volume of culture medium was added. Cells at a concentration of 2 × 107 cells/ml were incubated on an orbital rocker under illumination. At various times following drug addition, aliquots were removed and fixed with equal volumes of 1% glutaraldehyde. Fixed cells were examined by differential interference contrast (DIC) microscopy with a Leica Diaplan microscope (100× Leitz objective; numerical aperture, 1.25) and a CCD-72 video camera (DAGE MTI, Inc.). Images of at least 100 random cells were captured with Scion Image 1.59 software (National Institutes of Health) and transferred to Adobe Photoshop, and flagellar lengths were determined by tracing flagella.
cDNA isolation and sequencing.
The Chlamydomonas reinhardtii expressed sequence tag (EST) database (http://www.kazusa.or.jp/en/plant/chlamy/EST/; Kazusa DNA Research Institute) (4, 5) was searched with the human GSK3β protein sequence. Two ESTs (AV392622 and AV388963) that represented a single cDNA sequence were identified. PCR with primers derived from the EST sequence was used to amplify a 216-bp fragment from a λScreen-1 cDNA library. The PCR fragment was cloned into the pCRII vector by using a TA cloning kit (Invitrogen) and sequenced to confirm the presence of GSK3. The insert from one clone, pGSK1, was used as a hybridization probe to screen the λScreen-1 cDNA library. A 1.6-kb partial cDNA, p4A1, was identified. The insert from p4A1 was used as a hybridization probe to screen a λZAPII cDNA library (27). Nine different cDNA clones were isolated, and the plasmid inserts were excised following the manufacturer's protocol. Both strands of the cDNA containing the longest insert, pZ1H-1BF, as well as p4A1 were sequenced (DNA Sequencing and Synthesis Facility, Iowa State University; Advanced Genetic Analysis Center, University of Minnesota).
RNA isolation and Northern analysis.
Total RNA was isolated from cells before and after deflagellation by pH shock. RNA was purified by using the method of Wilkerson et al. (62) with the modification that the lysis buffer contained 100 μg of proteinase K/ml. The 216-bp insert from pGSK1 was used to detect the GSK3 transcript by hybridization. As a loading control, the RNA blot was reprobed with a 1-kb HindIII fragment isolated from the CRY1 gene, encoding the small ribosomal protein S14. For experiments to isolate RNA from lithium-treated cells, cells were incubated with 25 mM LiCl or culture medium for 60 min prior to deflagellation. EcoRI fragments of 2.2 and 3 kb were used to detect the RSP3 and PF20 transcripts by hybridization.
Fractionation of cells.
Flagella were removed from cell bodies by either pH shock or the addition of 5 mM dibucaine (19, 50). Briefly, cells were resuspended in 10 mM HEPES (pH 7.5) containing 7% sucrose. For dibucaine treatment, dibucaine was added to a final concentration of 5 mM and incubated on ice for 1 to 2 min. For pH shock, the pH was decreased to 4.4 to 4.5 by the dropwise addition of 5 M HAc. Cells were examined by phase contrast microscopy to ensure that deflagellation had occurred, and the pH was returned to 7 by the dropwise addition of 5 M KOH. To separate cell bodies from flagella, the pH-shocked or dibucaine-treated cells were transferred to graduated 50-ml centrifuge tubes, underlaid twice with 25% sucrose, 10 mM HEPES (pH 7.5) and centrifuged at 2,200 × g for 15 min. Flagella, which were present in the interphase between sucrose layers, were harvested by centrifugation at 32,000 × g for 20 min in a Sorvall RC-5B centrifuge. Cell bodies, which pellet through the 25% sucrose cushion, were washed twice in 10 mM HEPES (pH 7.5). Cell bodies and flagella were resuspended in either a small volume of HMDEK buffer (5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, 25 mM KCl, and 10 mM HEPES [pH 7.5]) or 10 mM HEPES (pH 7.5) containing a phosphatase inhibitor cocktail (50 mM NaF, 25 mM β-glycerolphosphate, and 1 mM Na orthovanadate) and 1× protease inhibitor cocktail (P-8340; Sigma Aldrich), flash frozen in liquid N2, and stored at −80°C until used.
Fractionation of flagella.
Flagella were resuspended at a concentration of 2 mg/ml in HMDEK buffer or 10 mM HEPES (pH 7.5) containing the 1× protease inhibitor cocktail with and without the phosphatase inhibitor cocktail, and NP-40 was added to a final concentration of 1%. After 20 min on ice, the samples were centrifuged at 16,000 × g for 20 min at 4°C. The supernatant fraction, which contains the membrane+matrix fraction of flagella, was transferred to a microfuge tube and the axonemal pellet was solubilized with 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer.
Immunoblot analysis.
Samples for immunoblot analysis were solubilized with SDS-PAGE sample buffer. After electrophoresis, proteins were transferred to membranes and probed with antibodies against Xenopus GSK-3β (Santa Cruz Biotechnology), phosphotyrosine GSK-3β (Clone 5G-2F; Upstate Biotechnology), Fla10p and IFT172 (11), IC140 (44), and MBO2p (58). Membranes were developed for enhanced chemiluminescence as described by the manufacturer (Amersham Pharmacia Biotech).
Kinase assay.
To immunoprecipitate GSK3 for kinase assays, flagella were incubated with 0.5% NP-40 for 20 min on ice followed by centrifugation at 16,000 × g for 20 min. The supernatant containing GSK3 was incubated with anti-Xenopus GSK-3β antibody. After 1 h at 4°C, the supernatant was divided into equal aliquots, protein G-Sepharose (Santa Cruz Biotechnology) was added to each sample, and the incubation was continued for an additional 1 h. The beads containing GSK3 were collected and washed four times in GSK3 wash buffer (0.1 mM EDTA, 20 mM MgCl2, and 4 mM MOPS [pH 7.4]), with and without 25 mM LiCl, by centrifugation. The final bead pellet was resuspended in 12.5 μl of GSK3 wash buffer with and without 25 mM LiCl.
For the kinase assay, either 12.5 μl of crude NP-40 supernatant or the immunoprecipitated GSK3 was incubated with 250 μM phospho-GS peptide (Upstate Biotechnology). The kinase assay was initiated by the addition of 28 μl of kinase reaction mix (0.1 mM EDTA, 20 mM MgCl2, 100 μM unlabeled ATP, 250 μM [γ-32P]ATP, and 4 mM MOPS [morpholinepropanesulfonic acid; pH 7.4]) with and without 25 mM LiCl. The reaction was stopped after 15 min at 30°C by the addition of 10 μl of 25% trichloroacetic acid, and 10 μl of the reaction mix was spotted onto a P81 phosphocellulose filter. Filters were washed on ice twice for 5 min with 0.75% phosphoric acid, washed once for 5 min with acetone, and air dried.
RNAi and transformation of Chlamydomonas.
To knock down the expression of GSK3, a plasmid was constructed to produce double-stranded RNA by using the inducible nitrate reductase (NIT1) promoter. The promoter of NIT1 is negatively regulated by ammonium; in the presence of ammonium, NIT1 transcripts do not accumulate (16). NIT1-regulated transcripts are induced in ammonium-free medium and enhanced by nitrate (40, 49). The NIT1 promoter and genomic and cDNA fragments of GSK3 were obtained by using PCR. The NIT1 promoter was amplified with primers that added KpnI and HindIII to the 5′ and 3′ ends, respectively (forward, 5′-ATCGGGTACCTAGATATGCACGCC; reverse, 5′-AAGTGAAGCTTCGTGTATGGCTTTGG). The amplified promoter was TA cloned, sequenced, and subsequently cloned into pBS (Stratagene) by using the added restriction sites to create the plasmid pNIT1prom. A 1.0-kb genomic fragment of GSK3 was amplified from 21gr DNA with primers containing BamHI and SfiI sites at the 5′ and 3′ ends, respectively. This genomic fragment contained two introns to enhance transcript stability. A 1.2-kb cDNA fragment containing EcoRI and SfiI sites at the 5′ and 3′ ends, respectively, was amplified from the GSK3 cDNA. All PCRs for GSK3 were TA cloned, sequenced, and subsequently ligated to the pNIT1prom plasmid to generate the inverted repeat. The plasmid, pGSK3IR, was sequenced to confirm the presence of the GSK3 inverted repeat. The final construct contained 800 bp of inverted repeat separated by a loop of 400 bp, which was required for the stability of the plasmid in Escherichia coli. The inverted repeat was not maintained in E. coli until the spacer sequence between repeats was increased to 400 bp (55). Prior to use in transformation, the pGSK3IR plasmid was linearized with AflIII.
For transformation into Chlamydomonas, an arg7 strain, E10, was used. Following the removal of cell walls, 2 μg (each) of linearized plasmid pGSK3IR and the plasmid pARG7.8 was transformed into Chlamydomonas by using the glass bead method (57). Arg+ transformants, identified by the ability to grow in the absence of exogenous arginine, were screened for the presence of pGSK3IR by PCR. RNAi-positive transformants were screened for a flagellar defect that was dependent on induction with nitrate.
RESULTS
Lithium induces changes in flagellar length.
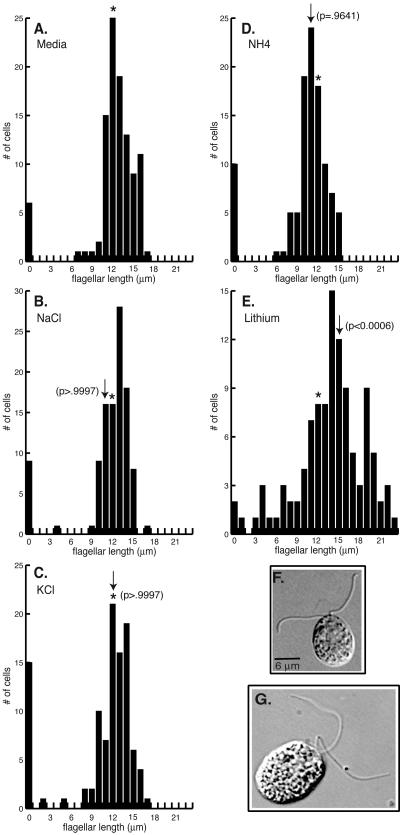
Earlier reports demonstrating a lithium-sensitive component involved in length control (38, 60) suggested the possibility that GSK3 signaling plays a role in regulating flagellar length. Treatment with 25 mM LiCl for 60 min resulted in a statistically significant (P < 0.0006) elongation of flagella (Fig. 1E and G) (38, 60). Lithium-treated cells also developed small bulb-like structures at the flagellar tips. Following lithium treatment for 60 min, 19% of the population had flagella with bulbs, while bulbs were present on only 1% of control cells (Table 1). To determine if the flagellar bulbs were caused by the rapid growth of flagella, cells undergoing flagellar regeneration were examined. Fewer than 5% of cells regenerating flagella had bulbs at the flagellar tips. This result suggests that flagellar bulbs do not simply reflect growth of the flagellum. Interestingly, flagellar bulbs have been observed on mutants defective in length control (Ulf, Lf) and flagellar assembly (42, 43, 46, 59).
FIG. 1.
Lithium chloride induces elongation of flagella. (A-E) Histograms (n = 104 to 107 cells for each measurement) show distributions of flagellar lengths following drug treatment. Wild-type cells were incubated for 60 min with M media (A), 25 mM NaCl (B), 25 mM KCl (C), 25 mM NH4Cl (D), or 25 mM LiCl (E). Asterisks in this and subsequent histograms indicate the average lengths of flagella on control cells. Arrows in each histogram indicate the average lengths of flagella for each cell population. (F-G) DIC images of wild-type cells incubated for 60 min with M media (F) or 25 mM LiCl (G). Bars: 6 μm.
TABLE 1.
Lithium induces bulbs at flagellar tipsa
| Time point | % Cells with flagellar bulbs after treatment indicated
|
|||
|---|---|---|---|---|
| None | Cycloheximide only | Lithium only | Cycloheximide and lithium | |
| Predeflagellation | 1 | 5 | 19 | 29 |
| 5 min | 1 | 0 | 0 | NDb |
| 15 min | 0 | 0 | 0 | ND |
| 30 min | 0 | 0 | 0 | ND |
| 60 min | 0 | 0 | 1 | ND |
| 120 min | 0 | 3 | 2 | ND |
| 18 h | 2 | 3 | 5 | ND |
The numbers of cells examined for each condition were 105 to 115. Data are percentages of cells with bulbs at flagellar tips.
ND, the percentage of cells with flagellar bulbs was not determined, as cells did not have flagella.
In addition to the lengthening of the flagella, lithium treatment also resulted in a rapid loss of motility. Both flagellar elongation and paralysis of flagellar motility were dose dependent (38 and data not shown). In contrast to the elongation of flagella seen with lithium treatment, no significant effect on either flagellar length (P > 0.9997) or motility was observed after treatment with comparable concentrations of NaCl, KCl, or NH4Cl (Fig. 1A to D and F). These results demonstrate that the LiCl effect was specific and not caused by other monovalent cations.
Incubation of cells with 25 mM LiCl for longer time periods resulted in a shift in phenotype from long flagella to a short-flagellar or aflagellate phenotype. More than 60% of cells cultured with LiCl for 48 h had no flagella. Among those cells that retained flagella, a broad distribution of lengths was seen (Fig. 2B). Many cells had flagella of unequal lengths and/or flagella with small bulbs at their tips (Fig. 2C). These results suggest that a lithium-sensitive component is required for the assembly and maintenance of flagella.
FIG. 2.
Prolonged incubation with lithium induces an aflagellate phenotype. (A, B) Histograms show the distribution of flagellar lengths following incubation with lithium. Cells were incubated without (A) (n = 109 cells) or with (B) 25 mM LiCl (n = 108 cells) for 48 h. Arrows indicate the average lengths of flagella for each cell population. (C) DIC images of control or lithium-treated cells illustrate effects on flagellar length. Arrows identify small bulbs at flagellar tips. Bars: 6 μm.
Lithium reduces the cell body pool of flagellar precursors.
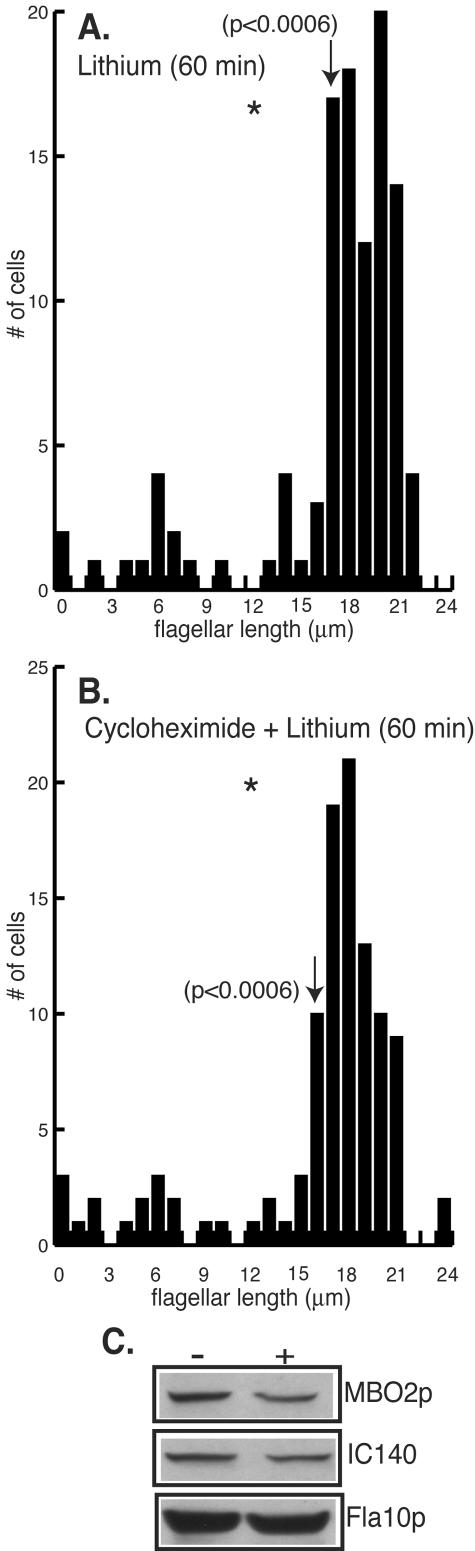
In Chlamydomonas, the cell body contains a pool of flagellar precursor proteins sufficient for regeneration of half-length flagella in the absence of new protein synthesis (52). Nakamura et al. (38) proposed that the growth of flagella induced by lithium treatment could reflect the recruitment of flagellar proteins from the cell body. To examine this model, we determined whether new protein synthesis was required for flagellar elongation in the presence of lithium. Cells were incubated with and without the protein synthesis inhibitor cycloheximide (10 μg/ml) for 10 min prior to treatment with lithium. The lithium-treated cells underwent flagellar elongation to the same extent in the absence and presence of cycloheximide (Fig. 3A and B). While lithium treatment induced a statistically significant elongation of flagella (P < 0.0006) (Fig. 3A and B), statistical analysis of the effect of cycloheximide on the lithium-induced elongation of flagella revealed no significant difference in flagellar lengths (P = 0.9821). Cells incubated with cycloheximide alone did not change flagellar length (31 and data not shown). This result suggests that new protein synthesis is not required for the elongation of flagella in response to lithium.
FIG. 3.
Protein synthesis is not required for lithium-induced elongation of flagella. (A-B) Histograms (n = 105 to 111 cells per measurement) show the distribution of flagellar lengths following treatment with 25 mM LiCl for 60 min. Wild-type cells were pretreated without (A) or with (B) 10 μg of cycloheximide/ml. After 10 min, LiCl was added to a final concentration of 25 mM. Arrows indicate the average lengths of flagella for each cell population. (C) Immunoblots reveal the lithium-induced depletion of flagellar precursors from cell bodies. Equal numbers of cell bodies from lithium (+) and control treated (−) cells were probed with antibodies against axonemal structural proteins (MBO2p, IC140), an axonemally associated protein (Fla10p), and the chloroplast protein OEE1.
We also examined the effect of lithium on the size of the precursor pool of flagellar proteins. Treatment of cells with lithium decreased the amounts of several axonemal proteins in cell bodies. In multiple experiments, the axonemal structural proteins MBO2p and IC140 were reduced to 61 and 65%, respectively, of control levels (Fig. 3C). In contrast, the anterograde IFT motor protein Fla10p did not undergo significant change in the presence of lithium (97% of that of the control) (Fig. 3C). To demonstrate that the same amount of protein was present in each lane, the blot was reprobed with an antibody to the chloroplast protein OEE1 (Fig. 3C) (35). In addition, multiple exposures of the immunoblots were performed to ensure that the signal for each band was in the linear range. Although the pool of flagellar precursors in the cell body decreased, it was not totally depleted under the conditions examined in this experiment (Fig. 3C). To ensure that the cell body pool was not replenished through new protein synthesis during the course of the experiment, cells were pretreated with cycloheximide. Under these conditions, lithium again partially decreased the cell body pool (data not shown). Taken together, these results demonstrate that the elongation of flagella induced by lithium does not require new protein synthesis. Flagellar elongation occurs, at least in part, through the recruitment of flagellar proteins from the cell body pool.
Lithium inhibits flagellar regeneration.
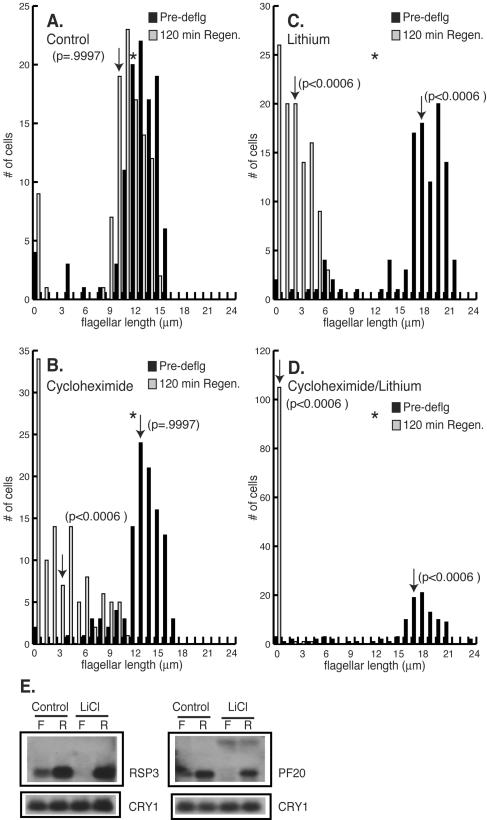
For Chlamydomonas, deflagellation results in the rapid regrowth of flagella. Within 120 min of flagellar amputation, control cells regrew flagella to their predeflagellation length (P = 0.9997) (Fig. 4A). Cycloheximide pretreatment had no effect on flagellar length (P = 0.9997). Following deflagellation in cycloheximide, however, cells regenerated flagella that were one-half of their predeflagellation length (P < 0.0006) (Fig. 4B and C) (38, 52). The incubation of cells with lithium induced flagellar elongation as previously described (P < 0.0006) (Fig. 1E and 2) (38). Lithium treatment has been reported to inhibit the regeneration of flagella in a manner similar to that of cells treated with cycloheximide. As seen with the regeneration of cycloheximide-treated cells, cells incubated with lithium regrew flagella that were one-half of their predeflagellation length (P < 0.0006) (Fig. 4B and C) (38). The fact that cells regenerated similar flagellar lengths when treated with lithium or cycloheximide (P > 0.9997) (Fig. 4C and D) led to the proposal that lithium affected protein synthesis (38). We examined this possibility by pretreating cells with cycloheximide followed by a 60-min incubation with lithium. Under these conditions, cells were unable to regenerate flagella at all (P < 0.0006) (Fig. 4D). These results suggest that new protein synthesis is required for flagellar regeneration in the presence of lithium. Moreover, the process of flagellar elongation induced by lithium, which does not require new protein synthesis (Fig. 3B), is fundamentally different from the initiation of flagellar growth following deflagellation in lithium, which does require new protein synthesis (Fig. 4D).
FIG. 4.
Lithium inhibits regeneration of flagella. (A-D) Histograms (n = 105 to 111 cells per measurement) show distribution of flagellar lengths before and after regeneration. Wild-type cells were preincubated without (A, C) or with (B, D) 10 μg of cycloheximide/ml. After 10 min, 25 mM LiCl was added (C, D) and and the incubation was continued for an additional 60 min. Arrows indicate the average lengths of flagella for each cell population. (E) RNA blot analysis of flagellar transcripts during regeneration. Twenty micrograms of total RNA from either flagellated cells (lanes F) incubated without (Control) or with LiCl and cells regenerating flagella (lanes R) in the absence (Control) or presence of LiCl was loaded in each lane. The transcripts for RSP3 and PF20 were detected by a hybridization probe from the corresponding gene. Blots were reprobed with CRY1 (the gene encoding ribosomal protein S14) to demonstrate equal loading. A faint second message that is unaffected by LiCl was identified with the PF20 probe.
Deflagellation has been shown to increase rapidly the accumulation of RNA transcripts of genes encoding flagellar proteins (28, 54). One possible reason that lithium blocked much of the flagellar regeneration may be that lithium prevented the stimulation of transcript accumulation of genes encoding flagellar proteins. To test this possibility, we examined transcript levels in control and lithium-treated cells. Cells treated with lithium upregulated flagellar messages in a manner similar to control cells in response to flagellar amputation (Fig. 4E). This result suggests that the signal transduction pathway(s) between the detection of flagellar loss and the upregulation of flagellar messages is intact in lithium-treated cells. Interestingly, treatment with lithium caused a striking reduction in the basal levels of transcripts for flagellar genes (Fig. 4E and F). Thus, even though lithium did not prevent the increase in flagellar gene transcripts seen after flagellar amputation, it clearly reduced the level of these transcripts in flagellated cells.
Cloning and sequencing of GSK3.
We examined Chlamydomonas GSK3 as a possible target for lithium in affecting the length and maintenance of flagella. The kinase activity of GSK3 has been shown both in vivo and in vitro to be directly and indirectly inhibited by lithium (26, 45). An examination of the Chlamydomonas EST database revealed the presence of a GSK3 transcript. Primers were designed from EST sequences for use in generating a hybridization probe, which was used to obtain a full-length cDNA from a λZAPII library (27). This 2.1-kb cDNA has a 318-bp 5′ untranslated region containing several in-frame stop codons upstream of the putative initiating AUG and 658 bp of the 3′ untranslated region containing the Chlamydomonas polyadenylation signal TGTAA 12 bases upstream of the polyadenine tract.
The cDNA contains a single open reading frame of 1,044 nucleotides encoding a 347-amino-acid protein with a predicted pI of 8.65 and a predicted molecular mass of 43.5 kDa. GSK3 is encoded by a single gene, based on a Southern analysis of Chlamydomonas genomic DNA (data not shown) as well as an examination of the complete sequence of the nuclear genome (http://genome.jgi-psf.org/chlre2/chlre2.home.html). The gene contains six exons separated by five introns. Hybridization of the cDNA to a Chlamydomonas BAC library (24) identified nine BAC clones representing a single contig mapping near the molecular marker CNC53 on linkage group XII/XIII between LF2 and ODA6. Given GSK3's essential housekeeping function in other systems and the fact that GSK3 is a single-copy gene in Chlamydomonas, mutations abolishing its kinase activity would most likely be lethal. It is not surprising, therefore, that we were unable to identify a GSK3 knockout from several insertional mutant collections (data not shown).
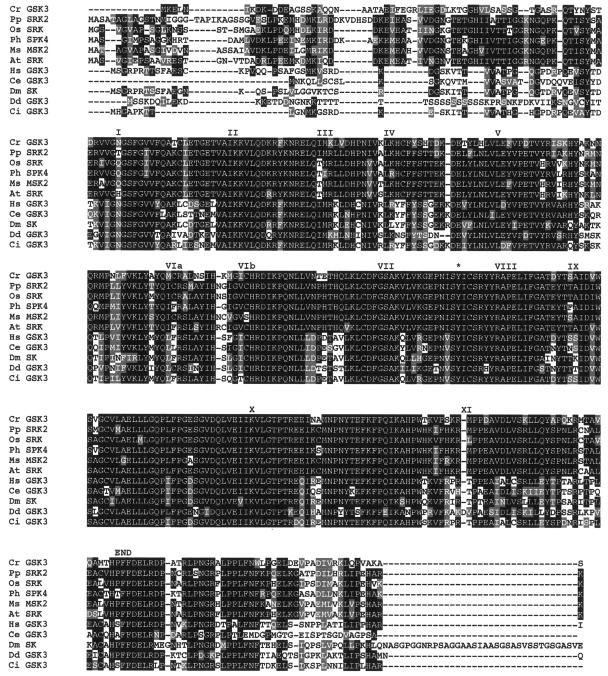
Searches of public databases revealed that the GSK3 genes from plants are the closest homologues for Chlamydomonas GSK3. Chlamydomonas GSK3 is 71 and 74% identical (262 of 370 and 262 of 356 amino acids) and 83 and 86% conserved (306 of 370 and 306 of 356 amino acids), respectively, to GSK3 from Physcomitrella patens and Petunia hybrida. GSK3 is one of the most highly conserved of protein kinases, as evidenced by the alignment of GSK3 from Chlamydomonas and a variety of other organisms (Fig. 5). Like all other kinases, GSK3 contains the 11 conserved subdomains that are essential for kinase activity (18). GSK3 from Chlamydomonas also has an invariant lysine residue at amino acid 88 within subdomain II that is required for ATP binding. These observations suggest that GSK3 from Chlamydomonas is a functioning kinase.
FIG. 5.
Alignment of GSK3 from Chlamydomonas with various GSK3 amino acid sequences. Cr, C. reinhardtii GSK3 (accession no. AY621077); Pp, Physcomitrella patens shaggy-related protein kinase 2 (accession no. AAQ23107.1); Os, Oryza sativa, shaggy-related protein kinase γ (accession no. BAB40983.1); Ph, petunia hybrid shaggy kinase 4 (accession no. S5115); Ms, Medicago sativa MSK2 (accession no. P51138); At, Arabidopsis thaliana shaggy-related protein kinase γ (accession no. NP_187235.1); Hs, Homo sapiens GSK3β (accession no. P49841); Ce, C. elegans (accession no. AD45354); Dm, Drosophila melanogaster sgg protein kinase (accession no. CAA37419.1); Dd, Dictyostelium discoideum (accession no. P51136); and Ci, Ciona intestinalis (accession no. BAA92186). Roman numerals indicate the 11 kinase subdomains. The regulatory tyrosine that must be phosphorylated for GSK3 activity is identified with an asterisk.
In higher eukaryotes, the kinase activity of GSK3 is regulated by two different phosphorylation events. The phosphorylation of a tyrosine residue located in the activation loop is required for kinase activity (9, 20). This tyrosine residue is present in GSK3 from Chlamydomonas (Fig. 5). The kinase activity of tyrosine-phosphorylated GSK3 can be inhibited by the phosphorylation of an N-terminal serine residue, which then acts as a pseudosubstrate (13, 15). Unlike GSK3 from mammals, this serine residue is not conserved in Chlamydomonas GSK3 or in higher plants. These observations suggest that the regulation of the kinase activity of GSK3 from plants and Chlamydomonas is fundamentally different from that described for mammalian GSK3.
GSK3 is a flagellar protein.
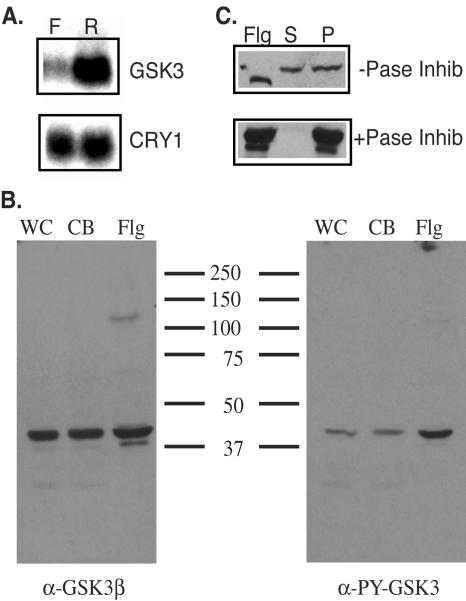
Analysis of the RNA transcript for GSK3 and localization of the protein suggest that GSK3 is a flagellar protein. The cDNA hybridized to a 2.1-kb transcript (Fig. 6A), consistent with the size of the full-length cDNA. This transcript was upregulated in cells regenerating flagella, as has been seen for many flagellar proteins (28), suggesting that GSK3 is a flagellar protein. The suggestion that GSK3 localizes to the flagella was confirmed by immunoblot analysis. The highly conserved nature of the GSK3 amino acid sequence (Fig. 5) allowed us to use commercially available antibodies generated against full-length recombinant GSK3β from Xenopus. To identify active GSK3, we employed an antibody generated against a tyrosine-phosphorylated peptide derived from the activation lip and therefore providing a marker for active GSK3 (17). The anti-Xenopus GSK3 antibody identified a 48-kDa protein in wild-type cells (Fig. 6B). A protein of the same size was identified with the phosphorylation-specific anti-GSK3 antibody (Fig. 6B). Analysis of equal amounts of protein from whole cells, cell bodies, and isolated flagella revealed that the majority of GSK3, both total and tyrosine phosphorylated, is present in the cell body. The active form of GSK3, however, was highly enriched in the flagella.
FIG. 6.
GSK3 is a flagellar protein. (A) RNA blot analysis of the GSK3 transcript. Twenty micrograms of RNA from flagellated cells (F) or cells regenerating flagella (R) was loaded on each lane, and the GSK3 transcript was identified with a probe corresponding to the 3′ region of the gene. (B) Immunoblot analysis of the GSK3 protein. The same amount of protein from whole cells (WC), cell bodies (CB), or flagella (Flg) was loaded in each lane, and the protein was identified with either an antibody generated against recombinant Xenopus GSK3β (α-GSK3β) or an antibody generated from a tyrosine-phosphorylated peptide from the activation loop (α-PY-GSK3). (C) Identification of axonemal location of GSK3. Flagella (Flg) were fractionated into the membrane+matrix fraction (S) or the axonemes (P) in the absence (−Pase Inhib) or presence (+Pase Inhib) of phosphatase inhibitors. Equal amounts of proteins were loaded in each lane. Proteins were visualized after immunoblots were probed with the anti-Xenopus GSK3β antibody.
Treatment with 1% NP-40 fractionates flagella into a soluble membrane+matrix fraction and insoluble axonemes. An immunoblot analysis of NP-40 supernatants and pellets revealed that 50% of GSK3 was present in the membrane-matrix fraction of flagella (Fig. 6C). An identical pattern of solubilization was seen for the tyrosine-phosphorylated form of GSK3 (data not shown). Upon addition of NP-40, 50% of tyrosine-phosphorylated GSK3 was present in the membrane-matrix fraction and 50% remained associated with the axoneme. This result suggests that the tyrosine phosphorylation of GSK3 is not responsible for its association with the axoneme. Interestingly, the addition of 1% NP-40 to the flagella resulted in a reproducible shift in mobility upon SDS-PAGE analysis, suggesting a detergent-induced modification of GSK3 (Fig. 6C). The inclusion of phosphatase inhibitors during the NP-40 extraction resulted in 100% of GSK3 remaining associated with the axoneme (Fig. 6C). Under these conditions, GSK3 frequently migrated as a doublet on SDS-PAGE. This doublet comigrated with the two forms of GSK3 seen before and after NP-40 extraction in the absence of phosphatase inhibitors. Taken together, these observations suggest that the flagellar form of GSK3 is active and that GSK3 associates with the axoneme in a phosphorylation-dependent manner that is independent of tyrosine phosphorylation.
GSK3 kinase activity is inhibited by lithium.
To confirm that Chlamydomonas GSK3 activity is sensitive to inhibition with lithium, we used an in vitro kinase assay (41). Although there are many kinases present in the membrane+matrix fraction of flagella in Chlamydomonas, lithium had no effect on their ability to phosphorylate a phosphopeptide substrate (Fig. 7A). We looked at the lithium sensitivity of flagellar GSK3 directly by purifying the enzyme by immunoprecipitation from the membrane+matrix fraction of flagella that contained approximately 50% of the total GSK3 in the flagella. The kinase activity of flagellar GSK3 was significantly inhibited by the inclusion of lithium in the kinase reaction (Fig. 7B). Lithium routinely caused a fourfold reduction in the phosphorylation of the phosphopeptide substrate by GSK3. These results demonstrate that lithium does not broadly inhibit the activity of kinases present in the membrane-matrix fraction of flagella. In fact, lithium had no detectable effect on kinase activity until GSK3 was purified from this fraction. The kinase activity of GSK3, however, was specifically inhibited by lithium.
FIG. 7.
Lithium inhibits GSK3 kinase activity in vitro. The membrane+matrix fraction or GSK3 immunoprecipitated from the membrane+matrix fraction of flagella was assayed in the absence (Media) or presence (Lithium) of lithium (25 mM) in an in vitro kinase assay using a phosphopeptide from glycogen synthase as a substrate. Results shown are representative of at least three independent experiments.
Fla10p and tyrosine-phosphorylated GSK3 accumulate in the flagella of lithium-treated cells.
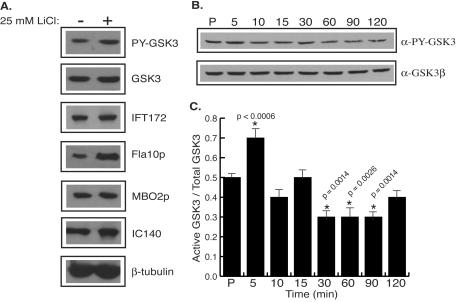
Marshall and Rosenbaum (34) proposed that flagellar length is determined by regulating the ratio of assembly and disassembly of axonemal microtubules by the action of a discrete amount of IFT particles. Tam et al. (59) and Perrone et al. (43) showed an accumulation of IFT components in the flagella of mutants that have abnormally long flagella. We examined the flagella from lithium-treated cells to determine if flagellar growth was associated with an increased recruitment of IFT particles into the flagellum. Flagella from lithium-treated cells had a threefold increase in the amount of Fla10p (the 90-kDa motor subunit of the kinesin II responsible for anterograde IFT motility) relative to control cells (Fig. 8A). Although lithium treatment increased the amount of Fla10p in the flagella, another IFT particle component, IFT172, accumulated to a lesser degree (Fig. 8A). Structural proteins of the axoneme did not accumulate in the flagella of lithium-treated cells (Fig. 8A, lanes MBO2p, IC140, and β-tubulin). The total amount of GSK3 in flagella did not change appreciably following lithium treatment (Fig. 8A). Interestingly, lithium treatment increased the amount of tyrosine-phosphorylated GSK3 in flagella twofold compared to control levels (Fig. 8A). Lithium inhibits GSK3 kinase activity in vivo by acting as a noncompetitive inhibitor for magnesium. Thus, lithium should not necessarily decrease the level of tyrosine phosphorylation of GSK3. Lithium-treated cells may recognize the reduced level of GSK3 activity in flagella and respond either by activating a tyrosine protein kinase or by inactivating a tyrosine protein phosphatase.
FIG. 8.
Levels of active GSK3 change with lithium treatment or flagellar regeneration. (A) Immunoblots reveal accumulation of Fla10p and tyrosine-phosphorylated GSK3 in flagella of lithium-treated cells. The same amount of flagellar protein from control-treated cells (−) or cells treated with 25 mM LiCl for 60 min (+) was loaded into each well. Blots were probed with antibodies against GSK3 (GSK3 and PY-GSK3), IFT components (IFT172 and Fla10p), and structural axonemal components (MBO2p, IC140, and β-tubulin). (B, C) The active form of GSK3 decreases during flagellar regeneration. (B) Cells were incubated with 25 mM LiCl for 60 min and deflagellated by pH shock, and the flagella were removed by centrifugation. Cells were allowed to regenerate flagella for 5 min (lane 5), 10 min (lane 10), 15 min (lane 15), 30 min (lane 30), 60 min (lane 60), 90 min (lane 90), and 120 min (lane 120). Protein from equal numbers of cells from each time point were loaded into wells, and the levels of active GSK3 were determined by immunoblotting with the anti-phosphotyrosine GSK3 antibody. The blot was reprobed with the anti-Xenopus GSK3β antibody to show total GSK3 levels. (C) The histogram shows the ratio of active GSK3/total GSK3 from the blot shown in panel B. Statistical analysis was preformed to determine the significance in the change of active GSK3/total GSK3 compared to control length during flagellar regeneration. Asterisks indicate P values of 0.0014 to <0.0006.
The ability of lithium to inhibit the kinase activity of GSK3 and induce flagellar elongation suggests that perhaps GSK3 activity correlates with flagellar length. To test this possibility, we examined levels of active GSK3 in cells with flagella of different lengths isolated at different times during the regrowth of flagella after amputation. Cells were deflagellated by pH shock and allowed to regenerate flagella for 120 min. During this time, the total amount of GSK3 did not change (Fig. 8B). During flagellar regeneration, tyrosine-phosphorylated GSK3 levels initially increased. This increase in tyrosine-phosphorylated GSK3 was statistically significant (P < 0.0006) and reproducibly peaked at 5 min of regeneration (Fig. 8B and C). The level of tyrosine-phosphorylated GSK3 then began to decrease within the next 5 min. The decrease in tyrosine-phosphorylated GSK3 peaked within 30 min of regeneration, when flagella had regrown to half of their predeflagellation length (Fig. 8B). The ratio of tyrosine-phosphorylated GSK3 to total GSK3 during flagellar regeneration confirms that tyrosine-phosphorylated GSK3 increases early during regeneration (Fig. 8C). As flagella reached their full lengths, the levels of tyrosine-phosphorylated GSK3 decreased, suggesting that GSK3 kinase activity correlates with flagellar length. The change in levels of tyrosine-phosphorylated GSK3 suggests that regulation of GSK3 kinase activity is required for flagellar assembly.
GSK3 regulates flagellar assembly and maintenance.
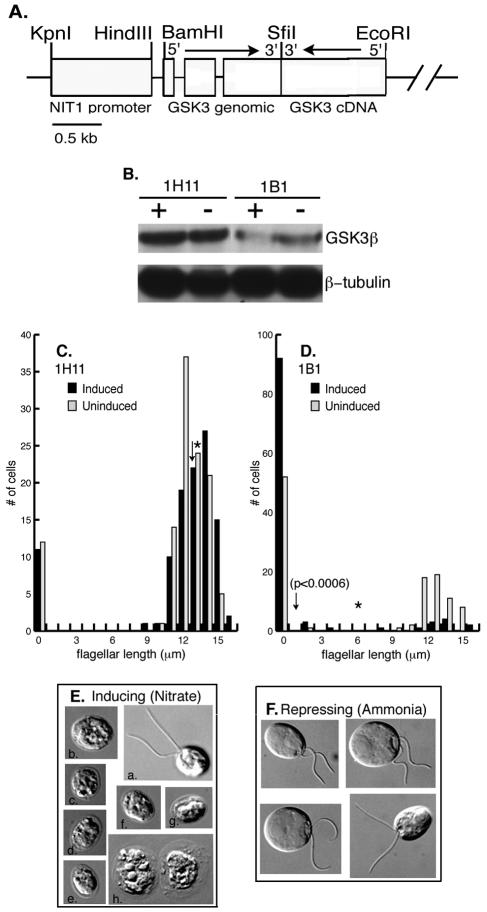
RNAi knockdown experiments were used to confirm that the lithium-induced changes in flagellar phenotypes were due to the inhibition of GSK3. A plasmid containing the inducible NIT1 promoter, a fragment of the GSK3 gene, and a GSK3 cDNA fragment in the 3′ to 5′ orientation relative to the genomic fragment was transformed into wild-type cells (Fig. 9A). Transformants containing the RNAi plasmid were identified by PCR. The protein levels of GSK3 were reduced to various amounts under inducing conditions (the presence of nitrate). In a few transformants, such as 1H11, there was no appreciable change in GSK3 protein levels and no effect on flagellar phenotypes in the presence of nitrate (Fig. 9B, inducing conditions). Twelve percent of 1H11 cells have no flagella under inducing conditions, compared to 10% of aflagellate cells without induction (Fig. 9C). In addition, the average length of flagella upon induction is 13 μm, compared to 14 μm under noninducing conditions (Fig. 9C). The majority of transformants, however, had clear decreases in GSK3 protein levels upon induction of expression of the RNAi plasmid. In some transformants, such as 1B1, the NIT1 promoter was leaky, as evidenced by the threefold decrease in GSK3 protein levels under noninducing conditions (the presence of ammonia) compared to control levels (Fig. 9B, compare lanes “−” for 1B1 and 1H11). Under inducing conditions (the presence of nitrate), the RNAi plasmid reduced expression of GSK3 by 20 to 35% in the majority of transformants (data not shown). In the transformant 1B1, the GSK3 protein level was reduced to 40% of the noninduced levels (Fig. 9B). This relatively small decrease in GSK3, however, was sufficient to induce a change in flagellar phenotype. Although 1B1 transformants had an increase in the numbers of aflagellate cells under noninducing conditions (the presence of ammonia), flagellated cells had wild-type length flagella and swam normally (Fig. 9D and F). Under inducing conditions (nitrate), a few cells (13%) within a population of 1B1 transformants had flagella of wild-type lengths (Fig. 9E, panel a). The majority of cells, however, were aflagellate (Fig. 9E, panels b to g), reminiscent of long-term incubations with lithium (Fig. 2). The aflagellate phenotype was only observed under inducing conditions, indicating that a secondary mutation caused by the transformation with the transgene was not responsible for the RNAi phenotype. With increasing lengths of time of RNAi induction, increasing numbers of dead cells were apparent in the cultures (Fig. 9E, panel h). In addition, prolonged induction (i.e., longer than 4 days) also increased the numbers of flagellated cells present in the culture. The most likely explanation for these results is that even small decreases in GSK3 levels (60 to 80% of wild-type levels) was lethal, resulting either in cell death or cell survival by silencing of the GSK3 RNAi transgene.
FIG. 9.
GSK3 RNAi phenocopies the lithium treatment of cells. (A) A schematic representation of the plasmid pGSK3IR. Open boxes indicate the NIT1 promoter and genomic and cDNA fragments of GSK3, respectively. The arrows indicate the 5′ to 3′ direction of genomic and cDNA fragments of GSK3. (B) Immunoblot analysis of GSK3 RNAi transformants. Cells were cultured for 4 days in media containing ammonia (lane −, RNAi repressing) or nitrate (lane +, RNAi inducing). Equal numbers of cells were loaded in each lane, and the proteins were identified with an antibody against GSK3β. The blot was reprobed with an antibody against β-tubulin to demonstrate equal loading of protein. (C, D) Histograms show the distribution of flagellar lengths from RNAi transformants 1H11 and 1B1. Cells were incubated under inducing (NO3, black bars) or repressing (NH4, gray bars) conditions. Asterisks and arrows indicate the average lengths of flagella in each cell population under noninducing and inducing conditions, respectively. (E) DIC images of RNAi transformant 1B1 under RNAi inducing conditions (nitrate). (F) DIC images of RNAi transformant 1B1 under RNAi repressing conditions (ammonia). Bars: 6 μm.
DISCUSSION
Lithium effects on flagellar length and regeneration.
Wild-type cells maintain flagella at a median length of 10 to 13 μm. Cells incubated with lithium elongated their flagella to a median length of 15 to 18 μm. Individual cells with flagellar lengths up to 22 μm, almost twice the normal length, were detected after lithium treatment. The increase in flagellar length was maintained for up to 12 h in lithium. Lithium is the only drug identified to date that induces substantial elongation of flagella. Prolonged treatment of cells with lithium resulted in a change in phenotype from long flagella to cells with no flagella. This aflagellate phenotype was maintained for up to 72 h in lithium. In addition to effects on flagellar length, lithium also affected the regeneration of flagella. Cells treated with lithium prior to deflagellation regenerated flagella of only one-half of their predeflagellation length. The similarity of flagellar regeneration following lithium treatment to regeneration of flagella in cycloheximide led to the proposal that lithium inhibited protein synthesis (38). The pretreatment of cells with cycloheximide prior to lithium treatment, however, resulted in a complete inhibition of flagellar regeneration. This observation suggests that lithium does not prevent regeneration by inhibiting protein synthesis.
Lithium treatment for 60 min had no effect on the ability of cells to upregulate flagellar gene transcript levels following deflagellation. An increase in the rate of transcription as well as an increase in message stability has been shown to be responsible for the accumulation of mRNAs for flagellar genes following deflagellation (reviewed in reference 32). Surprisingly, the incubation of cells with lithium led to the almost complete loss of basal mRNA levels for flagellar genes. A similar loss of mRNAs for flagellar genes was observed following treatment of cells with amiprophosmethyl, 3-isobutyl-1-methylxanthine, or pyrophosphate (32). Treatment of cells with these chemicals, however, results in flagellar resorption, effectively increasing the cytoplasmic pool of flagellar proteins. This increase in the cytoplasmic pool is thought to destabilize flagellar transcripts, thus decreasing flagellar mRNA levels (32). While a similar loss of basal mRNAs for flagellar genes was seen following lithium treatment, during the time that flagellar transcripts are lost there was no resorption of flagella. Instead, the flagella elongated. It is possible that lithium exerts its effects on basal mRNAs for flagellar genes by decreasing transcription or message stability. In either case, in the absence of mRNAs for flagellar genes, it is unlikely that new protein synthesis is contributing to the growth of flagella induced by lithium. Consistent with this model, flagellar elongation did not require new protein synthesis; flagella elongated to comparable lengths in the presence or absence of cycloheximide.
Interestingly, the lithium-induced increase in flagellar length is equivalent to 50% of the original length, which is the size of the pool of flagellar precursors in the cell body (52). Nakamura et al. (38) proposed that flagellar elongation induced by lithium occurred by recruiting flagellar proteins from the cell body pool out into the flagella, resulting in flagellar growth. An immunoblot analysis of cell bodies isolated from lithium-treated cells revealed a significant, but not complete, depletion of flagellar proteins from the cell body pool. These results suggest that a lithium-sensitive component regulates the partitioning of flagellar proteins between the flagella and cell body, thereby regulating flagellar length.
Lithium inhibits GSK3 kinase activity.
Lithium inhibits many signaling molecules, including GSK3 (26). GSK3 is a serine/threonine protein kinase that has been evolutionarily conserved. In mammals, the kinase activity of GSK3 is both stimulated and inhibited by the phosphorylation of different amino acid residues. GSK3 requires phosphorylation of a tyrosine residue within the activation loop between subdomains VII and VIII for kinase activity. This tyrosine residue is analogous to the tyrosine of the TEY sequence in the activation loop of MAP kinase (20, 21). Numerous studies have shown that phosphorylation and dephosphorylation of tyrosine occur concomitant with increases and decreases in GSK3 kinase activity, respectively (25, 37).
GSK3 has an unique substrate specificity in that it requires some substrates, such as glycogen synthase, to have a priming phosphorylation of a serine or threonine four residues C terminal to the site of phosphorylation by GSK3 (14). The substrates axin and β-catenin, on the other hand, do not require this priming phosphorylation (21, 47). Lithium inhibits the kinase activity of GSK3 directly by acting as a noncompetitive inhibitor of magnesium binding. Consistent with this mechanism of inhibition by lithium, the kinase activity of GSK3 immunoprecipitated from flagella was significantly inhibited by lithium in an in vitro kinase assay.
Lithium also inhibits GSK3 indirectly by activating kinases, such as protein kinase B (12), that phosphorylate a serine residue at position 9 of GSK3, inhibiting the kinase activity of tyrosine-phosphorylated GSK3. Once phosphorylated, the serine residue acts as a pseudosubstrate for primed substrates, occluding their binding site (13, 15). Phosphorylation of S9 therefore inhibits only the activity toward primed substrates. S9 phosphorylation, however, probably does not represent a universal mode of regulation of GSK3. In fact, mammals and Drosophila appear to be the only species that utilize this form of regulation for GSK3. An examination of the GSK3 sequence suggests that the inhibitory phosphorylation site at S9 is not conserved. This regulatory serine residue is missing in GSK3 from a wide variety of organisms, including Toxoplasma gondii, Saccharomyces cerevisiae, Chlamydomonas reinhardtii, Ciona intestinalis, Dictyostelium, Caenorhabditis elegans, and plants. In these organisms, it seems likely that regulation of GSK3 will occur through the phosphorylation and dephosphorylation of the tyrosine residue. Studies of Chlamydomonas GSK3 should allow dissection of the tyrosine kinase signaling pathways that regulate GSK3.
GSK3 regulates the assembly and maintenance of flagella.
Using the NIT1 inducible promoter, we expressed an RNAi construct for GSK3 in wild-type cells. The expression of NIT1-regulated transcripts is repressed by ammonium and enhanced by nitrate. Consistent with the aflagellate phenotype of lithium-treated cells, those cells transformed with an inducible RNAi construct for GSK3 also had no flagella. When the expression of GSK3 RNAi was repressed (by the inclusion of ammonium in the culture medium), cells swam normally and had flagella of wild-type lengths. Our results demonstrate the usefulness of the NIT1 promoter for the controlled expression of RNAi constructs to downregulate the expression of essential genes.
The aflagellate phenotype of cells following prolonged treatment with lithium or by induction of GSK3 RNAi may suggest a cell cycle-dependent effect of GSK3 on flagellar assembly. During the cell cycle, cells must resorb their flagella, remodel existing cellular microtubules to undergo mitosis and cytokinesis, and, finally, reassemble their flagella. mRNAs encoding tubulin were shown to accumulate prior to or during cell division (3). It is possible that the inhibition of GSK3 activity, either by treatment with lithium or by knockdown of protein levels by RNAi, affects the upregulation of tubulin expression during cell division. The signal transduction pathway responsible for the upregulation of tubulin mRNAs during the cell cycle is unknown; however, it is likely different from the signaling pathway that regulates the upregulation of flagellar genes during the flagellar regeneration following amputation. Support for this possibility comes from the isolation of mutants that are defective in the regeneration of flagella following amputation yet assemble flagella during the cell cycle (30). Whether the cell cycle-specific upregulation of tubulin is required for assembly of the mitotic spindle or the assembly of flagella following completion of cell division is unknown. A defect in either the completion of mitosis or the flagellar assembly following cell division, however, could account for the aflagellate phenotype seen with extended treatment with lithium as well as the RNAi-induced knockdown of GSK3.
The observation that lithium induces flagellar elongation suggests that GSK3 functions to limit flagellar growth. If so, then the kinase activity of GSK3 might be expected to change during flagellar regeneration. Our regeneration data partially fits this model. In whole cell extracts, the levels of tyrosine-phosphorylated, active GSK3 decreased below basal levels within 30 min of flagellar regeneration. At 120 min, when flagella were full length, the levels of active GSK3 began to increase although they were not yet back to the basal level. Surprisingly, the levels of active GSK3 increased early during flagellar regeneration (5 min). This increase in active GSK3 occurred during the lag period between flagellar amputation and regrowth of flagella (52), perhaps reflecting a second, as-yet-unknown function of GSK3 during flagellar regeneration.
Interestingly, the level of GSK3 in RNAi transformants was decreased to only 20 to 40% of that of the uninduced controls, suggesting that flagellar assembly and maintenance require a very narrow window of GSK3 activity levels. Consistent with this possibility, flagella from lithium-treated cells had significant increases in the levels of tyrosine-phosphorylated GSK3. One model for the role of GSK3 in regulating flagellar assembly involves the tight regulation of GSK3 activity. First, flagellar growth, either during regeneration or lithium-induced elongation, is accompanied by decreased levels of active GSK3. Second, when flagella are full length, a narrow window of active GSK3 may be required to maintain a static flagellar length. It is likely that the cell monitors the level of active GSK3, maintaining it within a narrow range of activity. If active GSK3 levels fall below the threshold, the cell activates a tyrosine kinase that phosphorylates GSK3, increasing the levels of active GSK3. These trends are consistent with the observation that lithium-treated cells overcompensate for the decrease in active GSK3 by stimulating the tyrosine phosphorylation of GSK3.
The enrichment of the tyrosine-phosphorylated, active form of GSK3 in the flagella suggests that a major site of action for GSK3 is in the flagella (Fig. 7). GSK3 associates with the axoneme in a phosphorylation-dependent manner. In addition, the rigid paralysis of flagella seen with lithium treatment suggests that GSK3 could function in flagella to regulate flagellar motility. Consistent with this model, a preliminary analysis of the phosphorylation pattern of flagellar proteins revealed a limited number of proteins whose phosphorylation state changed following lithium treatment (data not shown).
It is likely that GSK3 also functions in the cell body to regulate flagellar assembly and length. GSK3 in the cell body could regulate signaling pathways involved in the regulation of the levels of tubulin and/or other flagellar gene products during cell division or the cell cycle. In addition, GSK3 could regulate flagellar assembly and length through the regulation of IFT. IFT has been shown to be required for the assembly and maintenance of cilia and flagella (10, 51). In addition, flagellar proteins have been shown to be transported in and out of the flagella by IFT particles (48). IFT particles accumulate in flagellar swellings (i.e., bulges along the length or bulbs at the tips of the flagella) in mutants defective in length control (Ulf and Lf mutants) or flagellar assembly (42, 43, 46, 59). A microscopic examination of lithium-treated cells revealed small bulbs at the flagellar tips. An immunoblot analysis of flagella from lithium-treated cells revealed an increase in Fla10p, the anterograde motor for IFT particles. Interestingly, IFT172, a member of complex B, did not increase to the same extent as Fla10p in lithium-treated flagella. This discrepancy between Fla10p and IFT172 levels in lithium-treated flagella could reflect an increased recruitment of Fla10p into the flagella to restore IFT172 levels. This observation suggests a model where GSK3 could directly or indirectly regulate the interaction of Fla10p with its cargo, including IFT172. Precedent for a role for GSK3 in regulating cargo-motor interactions comes from neurons in which GSK3 phosphorylation of kinesin light chains results in the release of kinesin from its cargo (36).
Acknowledgments
This work was supported by National Institutes of General Medical Sciences grant GM34437 to P.A.L., National Institutes of Health postdoctoral fellowship 5F32-GM20149 to N.F.W., and the Plant Molecular Genetics Institute of the University of Minnesota.
We thank Nichja Heimbuch for her technical assistance and Nancy Hass, Steve Berman, Rachel Nguyen, Lai-Wa Tam, and Carolyn Silflow for helpful discussions.
REFERENCES
- 1.Afzelius, B. A., G. Gargani, and C. Romano. 1985. Abnormal length of cilia as a possible cause of defective mucociliary clearance. Eur. J. Respir. Dis. 66:173-180. [PubMed] [Google Scholar]
- 2.Allison, J. H., and M. A. Stewart. 1971. Reduced brain inositol in lithium-treated rats. Nat. New Biol. 233:267-268. [DOI] [PubMed] [Google Scholar]
- 3.Ares, M., Jr., and S. H. Howell. 1982. Cell cycle stage-specific accumulation of mRNAs encoding tubulin and other polypeptides in Chlamydomonas. Proc. Natl. Acad. Sci. USA 79:5577-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asamizu, E., K. Miura, K. Kucho, Y. Inoue, H. Fukuzawa, K. Ohyama, Y. Nakamura, and S. Tabata. 2000. Generation of expressed sequence tags from low-CO2 and high-CO2 adapted cells of Chlamydomonas reinhardtii. DNA Res. 7:305-307. [DOI] [PubMed] [Google Scholar]
- 5.Asamizu, E., Y. Nakamura, S. Sato, H. Fukuzawa, and S. Tabata. 1999. A large scale structural analysis of cDNAs in a unicellular alga, Chlamydomonas reinhardtii. I. Generation of 3,433 non-redundant expressed sequence tags. DNA Res. 6:369-373. [DOI] [PubMed] [Google Scholar]
- 6.Barsel, S.-E, D. E. Wexler, and P. A. Lefebvre. 1988. Genetic analysis of long-flagella mutants of Chlamydomonas reinhardtii. Genetics 118:637-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman, S. A., N. F. Wilson, N. A. Haas, and P. A. Lefebvre. 2003. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr. Biol. 13:1145-1149. [DOI] [PubMed] [Google Scholar]
- 8.Berridge, M. J., C. P. Downes, and M. R. Hanley. 1989. Neural and developmental actions of lithium: a unifying hypothesis. Cell 59:411-419. [DOI] [PubMed] [Google Scholar]
- 9.Bhat, R. V., J. Shanley, M. P. Correll, W. E. Fieles, R. A. Keith, C. W. Scott, and C.-M. Lee. 2000. Regulation and localization of tyrosine216 phosphorylation of glycogen synthase kinase 3β in cellular and animal models of neuronal degeneration. Proc. Natl. Acad. Sci. USA 97:11074-11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, D. G. 2003. The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic 4:435-442. [DOI] [PubMed] [Google Scholar]
- 11.Cole, D. G., D. R. Diener, A. L. Himelblau, P. L. Beech, J. C. Fuster, and J. L. Rosenbaum. 1998. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 141:993-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross, D. A., D. R. Alessi, P. Cohen, M. Andjelkovich, and B. A. Hemmings. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378:785-789. [DOI] [PubMed] [Google Scholar]
- 13.Dajani, R., E. Fraser, S. M. Roe, N. Young, V. Good, T. C. Dale, and L. H. Pearl. 2001. Crystal structure of glycogen synthase kinase 3β: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 105:721-732. [DOI] [PubMed] [Google Scholar]
- 14.Fiol, C. J., A. M. Mahrenholz, Y. Wang, R. W. Roeske, and P. J. Roach. 1987. Formation of protein kinase recognition sites by covalent modification of the substrate. Molecular mechanism for the synergistic action of casein kinase II and glycogen synthase kinase 3. J. Biol. Chem. 262:14042-14048. [PubMed] [Google Scholar]
- 15.Frame, S., P. Cohen, and R. M. Biondi. 2001. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell 7:1321-1327. [DOI] [PubMed] [Google Scholar]
- 16.Franco, A. R., J. Cárdenas, and E. Fernández. 1988. Regulation by ammonium of nitrate and nitrite assimilation in Chlamydomonas reinhardtii. Biochim. Biophys. Acta 951:98-103. [DOI] [PubMed] [Google Scholar]
- 17.Goold, R. G., and P. R. Gordon-Weeks. 2001. Microtubule-associated protein 1B phosphorylation by glycogen synthase kinase 3β is induced during PC12 cell differentiation. J. Cell Sci. 114:4273-4284. [DOI] [PubMed] [Google Scholar]
- 18.Hanks, S. K., and T. Hunter. 1995. Protein kinases. 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9:576-596. [PubMed] [Google Scholar]
- 19.Harris, E. H. 1989. The Chlamydomonas sourcebook. Academic Press, Inc., San Diego, Calif.
- 20.Hughes, K., E. Nikolakaki, S. E. Plyte, N. F. Totty, and J. R. Woodgett. 1993. Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J. 12:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda, S., S. Kishida, H. Yamamoto, H. Murai, S. Koyama, and A. Kikuchi. 1998. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and promotes GSK-3β-dependent phosphorylation of β-catenin. EMBO J. 17:1371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imahori, K., and T. Uchida. 1997. Physiology and pathology of tau protein kinases in relation to Alzheimer's disease. J. Biochem. 121:179-188. [PubMed] [Google Scholar]
- 23.Jarvik, J. W., F. D. Reinhart, M. R. Kuchka, and S. A. Adler. 1984. Altered flagellar size-control in Shf-1 short flagella mutants of Chlamydomonas reinhardtii. J. Protozool. 31:119-204. [Google Scholar]
- 24.Kathir, P., M. Lavoie, W. J. Brazelton, N. A. Haas, P. A. Lefebvre, and C. D. Silflow. 2003. Molecular map of the Chlamydomonas reinhardtii nuclear genome. Eukaryot. Cell 2:362-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, L., J. Liu, and A. R. Kimmel. 1999. The novel tyrosine kinase ZAK1 activates GSK3 to direct cell fate specification. Cell 99:399-408. [DOI] [PubMed] [Google Scholar]
- 26.Klein, P. S., and D. A. Melton. 1996. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. USA 93:8455-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurvari, V., F. Qian, and W. J. Snell. 1995. Increased transcript levels of a methionine synthase during adhesion-induced activation of Chlamydomonas reinhardtii gametes. Plant Mol. Biol. 29:1235-1252. [DOI] [PubMed] [Google Scholar]
- 28.Lefebvre. P. A., C. D. Silflow, E. D. Wieben, and J. L. Rosenbaum. 1980. Increased levels of mRNA for tubulin and other flagellar proteins after amputation or shortening of Chlamydomonas flagella. Cell 20:469-477. [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre, P. A., C. M. Asleson, and L.-W. Tam. 1995. Control of flagellar length in Chlamydomonas. Semin. Dev. Biol. 6:317-323. [Google Scholar]
- 30.Lefebvre, P. A., S.-E. Barsel, and D. E. Wexler. 1988. Isolation and characterization of Chlamydomonas reinhardtii mutants with defects in the induction of flagellar protein synthesis after deflagellation. J. Protozool. 35:559-564. [DOI] [PubMed] [Google Scholar]
- 31.Lefebvre, P. A., S. A. Nordstrom, J. E. Moulder, and J. L. Rosenbaum. 1978. Flagellar elongation and shortening in Chlamydomonas. IV. Effects of flagellar detachment, regeneration, and resorption on the induction of flagellar protein synthesis. J. Cell Biol. 78:3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lefebvre, P. A., and J. L. Rosenbaum. 1986. Regulation of the synthesis and assembly of ciliary and flagellar proteins during regeneration. Annu. Rev. Cell Biol. 2:517-546. [DOI] [PubMed] [Google Scholar]
- 33.Lucas, F. R., R. G. Goold, P. R. Gordon-Weeks, and P. C. Salinas. 1998. Inhibition of GSK-3β leading to the loss of phosphorylated MAP-1B is an early event in axonal remodeling induced by WNT-71 or lithium. J. Cell Sci. 111:1351-1361. [DOI] [PubMed] [Google Scholar]
- 34.Marshall, W. F., and J. L. Rosenbaum. 2001. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol. 155:405-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayfield, S. P., P. Bennoun, and J. D. Rochaix. 1987. Expression of the nuclear encoded OEE1 protein is required for oxygen evolution and stability of photosystem II particles in Chlamydomonas reinhardtii. EMBO J. 6:313-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morfini, G., G. Szebenyi, R. Elluru, N. Ratner, and S. T. Brady. 2002. Glycogen synthase kinase 3 phosphorylates kinesin light chains and negatively regulates kinesin-based motility. EMBO J. 21:281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murai, H., M. Okazaki, and A. Kikuchi. 1996. Tyrosine dephosphorylation of glycogen synthase kinase-3 is involved in its extracellular signal-dependent inactivation. FEBS Lett. 392:153-160. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura, S., H. Takino, and M. K. Kojima. 1987. Effect of lithium on flagellar length in Chlamydomonas reinhardtii. Cell Struct. Funct. 12:369-374. [Google Scholar]
- 39.Niggemann, B., A. Muller, A. Nolte, N. Schnoy, and U. Wahn. 1992. Abnormal length of cilia-a cause of primary ciliary dyskinesia-a case report. Eur. J. Pediatr. 151:73-75. [DOI] [PubMed] [Google Scholar]
- 40.Ohresser, M., R. F. Matagne, and R. Loppes. 1997. Expression of the arylsulphatase reporter gene under the control of the nit1 promoter in Chlamydomonas reinhardtii. Curr. Genet. 31:264-271. [DOI] [PubMed] [Google Scholar]
- 41.Pap, M., and G. M. Cooper. 1998. Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-kinase/Akt cell survival pathway. J. Biol. Chem. 273:19929-19932. [DOI] [PubMed] [Google Scholar]
- 42.Pazour, G. J., C. G. Wilkerson, and G. B. Witman. 1998. A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J. Cell Biol. 141:979-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perrone, C. A., D. Tritschler, P. Taulman, R. Bower, B. K. Yoder, and M. E. Porter. 2003. A novel dynein light intermediate chain colocalizes with the retrograde motor for intraflagellar transport at sites of axoneme assembly in Chlamydomonas and mammalian cells. Mol. Biol. Cell 14:2041-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perrone, C. A., P. Yang, E. O'Toole, W. S. Sale, and M. E. Porter. 1998. The Chlamydomonas IDA7 locus encodes a 140 kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol. Biol. Cell 9:3351-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phiel, C. J., and P. S. Klein. 2001. Molecular targets of lithium action. Annu. Rev. Pharmacol. Toxicol. 41:789-813. [DOI] [PubMed] [Google Scholar]
- 46.Piperno, G., E. Siuda, S. Henderson, M. Segil, H. Vaananen, and M. Sassaroli. 1998. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J. Cell Biol. 143:1591-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polakis, P. 1999. The oncogenic activation of β-catenin. Curr. Opin. Genet. Dev. 9:15-21. [DOI] [PubMed] [Google Scholar]
- 48.Qin, H., D. R. Diener, S. Geimer, D. G. Cole, and J. L. Rosenbaum. 2004. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J. Cell Biol. 164:255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quesada, A., and E. Fernandez. 1994. Expression of nitrate assimilation related genes in Chlamydomonas reinhardtii. Plant Mol. Biol. 24:185-194. [DOI] [PubMed] [Google Scholar]
- 50.Rosenbaum, J. L., and F. M. Child. 1967. Flagellar regeneration in protozoan flagellates. J. Cell Biol. 34:345-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenbaum, J. L., and G. B. Witman. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813-825. [DOI] [PubMed] [Google Scholar]
- 52.Rosenbaum, J. L., J. E. Moulder, and D. L. Ringo. 1969. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J. Cell Biol. 41:600-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sager, R., and S. Granick. 1954. Nutritional control of sexuality in Chlamydomonas reinhardi. J. Gen. Physiol. 37:729-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schloss. J. A., C. D. Silflow, and J. L. Rosenbaum. 1984. mRNA abundance changes during flagellar regeneration in Chlamydomonas reinhardtii. Mol. Cell. Biol. 4:424-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinden, R. R., G. X. Zheng, R. G. Brankamp, and K. N. Allen. 1991. Deletion of inverted repeated DNA in Escherichia coli: effects of length, thermal stability, and cruciform formation in vivo. Genetics 129:991-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solter, K. M., and A. Gibor. 1978. The relationship between tonicity and flagellar length. Nature 275:651-652. [DOI] [PubMed] [Google Scholar]
- 57.Tam, L.-W., and P. A. Lefebvre. 1993. Cloning of flagellar genes in Chlamydomonas reinhardtii by insertional mutagenesis. Genetics 135:375-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tam, L.-W., and P. A. Lefebvre. 2002. The Chlamydomonas MBO2 locus encodes a conserved coiled-coil protein important for flagellar waveform conversion. Cell Motil. Cytoskelet. 51:197-212. [DOI] [PubMed] [Google Scholar]
- 59.Tam, L.-W., W. L. Dentler, and P. A. Lefebvre. 2003. Defective flagellar assembly and length regulation in LF3 null mutants in Chlamydomonas. J. Cell Biol. 163:597-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tuxhorn, J., T. Daise, and W. L. Dentler. 1998. Regulation of flagellar length in Chlamydomonas. Cell Motil. Cytoskelet. 40:133-146. [DOI] [PubMed] [Google Scholar]
- 61.Wiese, M., D. Kuhn, and C. G. Grunfelder. 2003. Protein kinase involved in flagellar-length control. Eukaryot. Cell 2:769-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilkerson, C. G., S. M. King, and G. B. Witman. 1994. Molecular analysis of the gamma heavy chain of Chlamydomonas flagellar outer arm dynein. J. Cell Sci. 107:497-506. [DOI] [PubMed] [Google Scholar]