Abstract
In response to stress and nutrient starvation, the Saccharomyces cerevisiae transcription factor Msn2p accumulates in the nucleus and activates expression of a broad array of genes. Here, we analyze the role of the Tor (target of rapamycin) signaling pathway in mediating these responses. Inactivation of the Tor pathway component Tap42p using tap42(Ts) alleles causes a sustained nuclear localization similar to that after the addition of the Tor kinase inhibitor rapamycin. Effects of Tap42p inactivation and rapamycin addition could be suppressed by deletion of TIP41, which encodes a Tap42p-interacting protein. These results support the notion that rapamycin affects Msn2p by inactivating Tap42p function. Tap42p interacts with the catalytic subunit of PP2A (protein phosphatase 2A) and PP2A-like phosphatases. Deletion of either the catalytic or regulatory subunit that forms the PP2A phosphatase complex prevents nuclear accumulation of Msn2p in the tap42(Ts) strain and in wild-type strains treated with rapamycin. These results suggest that Tap42p is an inhibitor of PP2A phosphatase, which in turn inhibits nuclear export of Msn2p. Interestingly, PP2A function is also required for nuclear accumulation of Msn2p in response to stresses, such as heat and osmotic shock, as well as nitrogen (but not glucose) starvation. Thus, PP2A and the Tor kinase pathway transduce stress and nitrogen starvation signals to Msn2p. Finally, Msn2p localization is unaffected by conditional loss of 14-3-3 protein function, ruling out the possibility that 14-3-3 proteins act as a scaffold to sequester Msn2p in the cytoplasm.
Eukaryotic cells, such as the yeast Saccharomyces cerevisiae, must sense and respond to a broad array of chemical and physical insults. S. cerevisiae cells have developed specialized signal transduction pathways to cope with these insults; however, they also harbor a general stress response pathway that can be stimulated by seemingly unrelated stress conditions (heat and osmotic shock, UV, oxidative stress, and nutrient depletion) and that enables them to survive aberrant protein folding, DNA damage, oxidant accumulation, and starvation. The functionally redundant transcription factors Msn2p and Msn4p are key players in the general stress response pathway. During mitotic growth, Msn2p and Msn4p reside in the cytoplasm where they are inactive. Upon exposure to stress or nutrient depletion, these transcription factors rapidly accumulate in the nucleus and stimulate expression of more than 150 genes (15, 16, 36, 39), which provide transient protection against further insult from both the same and different stresses.
At least two cis-acting domains function together to regulate Msn2p localization. A nuclear localization signal (NLS) within the carboxyl-terminal half of Msn2p responds to glucose levels but not to stress, so that glucose starvation results in rapid nuclear accumulation of a reporter protein containing the Msn2p NLS and a constitutive export signal (15, 16). In contrast, a second domain within the amino terminus of Msn2p responds to both of these stimuli. The localization function of this domain is less defined than that of the carboxyl-terminal NLS; however, it is thought to involve nuclear export. This conclusion was based on two observations. First, regulated localization of the amino-terminal domain requires an exogenous NLS, and second, the localization of this region is sensitive to mutations (msn5Δ) in the nuclear export machinery (16). The sensitivity of the amino-terminal domain to both stress and glucose depletion implies that it must contain either a single nuclear export signal (NES) that can be coregulated by glucose and stress or two separate subdomains that can be independently regulated by glucose and stress (16).
The first signaling pathway implicated in Msn2p localization was the cyclic AMP (cAMP)-dependent protein kinase A (PKA) pathway (15). PKA inactivation results in rapid nuclear accumulation of Msn2p (15); however, the pathway appears to transmit only the glucose signal, because stress does not affect the PKA-responsive carboxyl-terminal NLS. Moreover, glucose starvation, but not stress, is associated with a rapid drop in phosphorylation of several PKA-sensitive serines within the NLS (16). Thus, PKA phosphorylates and inactivates the NLS until it is unmasked by glucose depletion. Many mechanisms could explain how phosphorylation contributes to cytoplasmic retention of Msn2p; however, Beck and Hall (2) recently proposed a model in which the yeast 14-3-3 protein homologs, Bmh1p and Bmh2p, interact with and sequester phosphorylated Msn2p in the cytoplasm. Their model was based on the observation that the biochemical interaction between Bmh1p/Bmh2p and Msn2p proteins was disrupted by conditions that cause Msn2p to accumulate in the nucleus (2), as well as a generally accepted role for mammalian 14-3-3 proteins in sequestering mammalian signaling proteins. The model proposed by Beck and Hall (2) was given further support by previous suggestions that Bmh1p and Bmh2p have a functional, if poorly understood, effect on PKA-regulated processes, such as growth and pseudohyphal formation (13, 31).
Identification of the potential stress response pathway stems from the observation that inactivation of the Tor (Target of rapamycin) kinase pathway caused Msn2p to accumulate in the nucleus (2, 16, 29). Tor kinase is a phosphatidylinositol kinase-related protein kinase that was originally identified by the isolation of mutations (TOR1-1) that confer resistance to the growth inhibitory properties of the immunosuppressive drug rapamycin. Rapamycin forms a toxic complex with its intracellular receptor FKBP12, which then binds and inhibits Tor kinase (20, 27). The Tor kinase pathway plays an important, if incompletely understood, role in controlling growth in response to nutrients, because cells depleted of Tor kinase activity (by rapamycin addition or Tor inactivation) exhibit physiological characteristics of starved or stressed cells, including G1 arrest, a decrease in macromolecular synthesis, and induction of general stress response genes (1, 8). A role for the Tor kinase pathway in stress response was supported by the observation that Tor inactivation (rapamycin treatment) impinged only on the stress-sensitive amino-terminal domain of Msn2p (16).
Several downstream effectors of Tor kinase have been identified (7). The effector that is most relevant to the general stress response pathway is Tap42p, an essential protein that interacts with catalytic subunits of the type 2A (and 2A-like) protein phosphatases (8). Nitrogen starvation and Tor kinase inactivation result in Tap42p dephosphorylation and subsequent dissociation of Tap42-PP2A (protein phosphatase 2A) and Tap42-PP2A-like phosphatase complexes (8, 24). Yeast PP2A phosphatase is a heterotrimer that is comprised of a catalytic subunit (subunit C), regulatory subunit (subunit B), and a scaffolding subunit (subunit A). Two homologous genes, PPH21 and PPH22, redundantly encode the PP2A catalytic subunit, the loss of which results in retarded growth, defects in actin organization, and difficulty progressing through mitosis (26, 33). Regulatory subunit activity is also encoded by two genes, CDC55 and RTS1, although in this case the two proteins are thought to perform different cellular functions, targeting the phosphatase complex to distinct substrates and cellular processes (14, 37, 45). Finally, the scaffolding subunit is encoded by a single gene, TPD3 (44). S. cerevisiae also contains a PP2A-like phosphatase activity which is comprised of the catalytic subunit Sit4p and several regulatory subunits (8, 28). Although there is no general consensus as to whether Tap42p stimulates or inhibits phosphatase activity, recent results are consistent with a model in which Tap42p inhibits PP2A and PP2A-like phosphatase activity in at least some of its functions (2, 6, 35).
We recently used a tap42(Ts) mutation to show that Tap42p inactivation, like rapamycin treatment, caused Msn2p to accumulate in the nucleus (9). To examine whether the effects of rapamycin treatment are mediated by Tap42, we monitored Msn2p localization and function in strains lacking components of the Tor/Tap42/PP2A phosphatase pathway. We provide evidence to support the hypothesis that rapamycin effects on Msn2p are the result of Tap42p inactivation and show that PP2A, but not PP2A-like, catalytic activity is necessary for Msn2p nuclear accumulation and transcriptional activation in response to rapamycin treatment. Interestingly, PP2A activity is also necessary for stress-induced translocation. These results suggest that Tap42p functions in the general stress response pathway to inhibit or sequester PP2A phosphatase. In response to stress or rapamycin, Tap42 inhibition of PP2A is relieved, and dephosphorylation of Msn2p (or an Msn2p-interacting protein) stimulates Msn2p accumulation in the nucleus.
MATERIALS AND METHODS
Media and growth conditions.
Cells were grown in rich (yeast extract-peptone-dextrose [YEPD]) or synthetic complete (SC) medium lacking the appropriate amino acids (25). Temperature shifts were achieved by resuspending harvested cells into prewarmed medium. Rapamycin (Sigma) was added to a concentration of 200 ng/ml as described previously (9). Medium containing 1 M sorbitol or lacking glucose has been described elsewhere (16). Nitrogen starvation medium was made by using yeast nitrogen base lacking ammonium sulfate and amino acids.
S. cerevisiae strains.
S. cerevisiae strains used in the study are listed in Table 1.
TABLE 1.
Strains of S. cerevisiae used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| SGY446 | MATα ura3-52 his3 leu2-3,112 trp1 ade8 tpk1::ADE8 tpk2-63(Ts) tpk3::TRP1 | 39 |
| AKY54 | SGY446 (YEp352-BMH2) | This study |
| AKY53 | SGY446 (YEp352) | This study |
| AKY31 | SGY446 TPK2 (YEp352) | This study |
| AHY148 | MATaura3-52 his3 leu2-3,112 trp1 ade8 tpk1w1tpk2::HIS3 tpk3::TRP1 | 18 |
| RTF1.5-2 | MATα ura3-52 his3 leu2-3,112 trp1 ade8 tpk1w2tpk2::HIS3 tpk3::TRP1 bcy1::URA3 | 5 |
| AHY240a | MATaura3-52 his3 leu2-3,112 trp1 ade8 TPK1 tpk2::HIS3 tpk3::TRP1 bmh1::ADE8 | This study |
| AHT240b | MATaura3-52 his3 leu2-3,112 trp1 ade8 tpk1w1tpk2::HIS3 tpk3::TRP1 bmh1::ADE8 | This study |
| SL1320 | MATα ura3 leu2 his3 BMH1 bmh2::URA3 | 13 |
| SGY459 | MATα ura3 leu2 his3 pda1::TRP1 bmh1-11(Ts) bmh2::URA3 | This study |
| AKY1 | SGY459(pRS315) | This study |
| AKY2 | SGY459(pAL110) | This study |
| AKY24 | SGY459 msn2::HIS3 | This study |
| AKY25 | SGY459 msn2::HIS3 msn4::LEU2 | This study |
| AKY5 | MATα ura3 leu2 his3 bmh1-11(Ts) bmh2::KanMX | This study |
| AKY9 | AKY5(pRS315) | This study |
| AKY10 | AKY5(pAL110) | This study |
| AKY44 | AKY5 (YEp352) | This study |
| AKY45 | AKY5 (YEp352-TPK2) | This study |
| AKY46 | AKY5 (YEp352-TPK3) | This study |
| AKY32 | AKY5 MSN2::pRS306-MSN2-myc12(pAL110) | This study |
| AKY33 | AKY5 MSN2::pRS306-MSN2-myc12(pRS315) | This study |
| AKY35 | SGY559(pWB36) | This study |
| Y3033 | MATatap42::HIS3(pRS414-TAP42) | 9 |
| Y3036 | Y3033 msn2::LEU2 msn4::URA3 | This study |
| Y3034 | MATatap42::HIS3 [pRS414-tap42-106] | 9 |
| Y3037 | Y3034 msn2::LEU2 msn4::URA3 | This study |
| Y3035 | MATatap42::HIS3 [pRS414-tap42-106] | 9 |
| Y3038 | Y3035 msn2::LEU2 msn4::URA3 | This study |
| AKY38 | Y3033 MSN2::pRS306-MSN2-myc12 | 9 |
| AKY79 | Y3033 pADH-MSN2-GFP | This study |
| AKY84 | Y3034 pADH-MSN2-GFP | This study |
| AKY75 | AKY38 tip41::KanMX | This study |
| AKY142 | AKY79 tip41::KanMX | This study |
| AKY144 | AKY84 tip41::KanMX | This study |
| AKY148 | MATatap42::HIS3(pRS414-TAP42)(pAL22) | This study |
| AKY150 | MATatap42::HIS3(pRS414-tap42-106)(pAL22) | This study |
| AKY204 | AKY148 cdc55::NAT1 | This study |
| AKY205 | AKY150 cdc55::NAT1 | This study |
| Y2092 | MATα ade2 his3 leu2 trp1 ura3 GAL | This study |
| Y2762 | Y2092 pph21::LEU2 pph22::HIS3 GAL | This study |
| AKY101 | Y2092 MSN2::pRS306-MSN2-myc12 | This study |
| AKY103 | Y2762 MSN2::pRS306-MSN2-myc12 | This study |
| BY4742 | MATα his3Δ leu2Δ lys2Δ ura3Δ | Research Genetics |
| AKY198 | BY4742 pADH-MSN2-GFP | This study |
| AKY163 | Y2092 rts1::URA3 pADH-MSN2-GFP | This study |
| AKY185 | Y2092 cdc55::LEU2 MSN2::pRS306-MSN2-myc12 | This study |
| AKY191 | BY4742 tpd3::KanMX pADH-MSN2-GFP | This study |
| AKY194 | AKY79 tpd3::KanMX | This study |
| AKY195 | AKY84 tpd3::KanMX | This study |
Plasmids.
The integrating Msn2-myc12 plasmid pAK3-3, full-length Msn2-green fluorescent protein (GFP) plasmid pADH-MSN2-GFP, and the put1-lacZ expression reporter plasmid pWB36 have been described previously (7, 16, 34). The BMH1 plasmid pAL110 consists of a 3-kb DNA fragment, which extends from 400 bp upstream of BMH1 to 600 bp past the downstream gene PDA1, inserted into the low-copy-number LEU2 CEN plasmid pRS315. Plasmid pAL99 was constructed by inserting a 2-kb HincII fragment containing BMH1 and PDA1 into the same site of pBSKII. Plasmid pAL100 is a pda1::TRP1 derivative of pAL99 and was constructed by inserting a Klenow fragment-treated EcoRI-BglII TRP1 fragment into the Klenow fragment-treated NcoI site within PDA1. The CTT1-lacZ fusion plasmid pAL22 was constructed by inserting a 9.2-kb HindIII fragment containing CTT1-lacZ embedded in URA3 (38) into the LEU2 CEN vector pSB32 (40).
Isolation of the bmh1-11(Ts) bmh2::LEU2 mutant.
A conditional allele of BMH1 was identified by replacing the BMH1 gene of BMH1 bmh2::URA3 strain SL1320 with hydroxylamine-mutagenized BMH1-pda1::TRP1 DNA (pAL100) and screening Trp+ colonies for growth at 23 and 36°C. Tetrad analyses showed that the temperature-sensitive growth defect of one mutant (AHY316) was tightly linked to the pda1::TRP1 marker, dependent upon the presence of the bmh2::URA3 disruption, and complemented by transformation with low-copy-number BMH1 plasmid pAL110. Moreover, sequence analysis showed that the temperature-sensitive bmh1 allele designated bmh1-11(Ts) was the result of a G-to-A substitution at codon 193 that converted an invariant Ala to Thr. This contrasts with the previously described bmh1 temperature-sensitive allele with a S189P substitution (43). Despite being implicated in several growth-related signaling pathways (12, 13, 31, 43), the only terminal phenotype associated with 14-3-3 protein dysfunction of bmh1-11(Ts) bmh2::URA3 haploid strain SGY459 was a slight (40 to 65%) increase in large budded cells. This increase was not associated with either an increase in cells with 2N DNA or discrete nuclear morphology (data not shown).
β-Galactosidase activity.
β-Galactosidase activity was measured by the permeabilized cell method (25) and are expressed as (1,000 × OD420)/(OD600 × volume [assayed in milliliters] × time [in minutes]), where OD420 and OD600 are the optical densities at 420 and 600 nm, respectively.
Fluorescence microscopy.
Cells expressing Msn2-myc12 or Msn2-GFP were grown to mid-log phase before treatment. For immunofluorescence, cells were fixed in 3.7% formaldehyde and spheroplasts were prepared (32). Msn2-myc12 was visualized using 9E10 monoclonal anti-myc antibody purchased from Covance (Richmond, Calif.). The secondary antibody was fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody from Roche Diagnostics (Indianapolis, Ind.). DNA was visualized by staining with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) from Sigma-Aldrich (St. Louis, Mo.) as described previously (9). Msn2-GFP was visualized directly after incubating cells for the final 15 min in medium containing Hoechst 33258 stain from Sigma-Aldrich.
RESULTS
Msn2p localization and 14-3-3 protein function.
A previous report postulated that yeast 14-3-3 proteins anchor Msn2p and Msn4p in the cytoplasm (2); however, the cytoplasmic localization of Msn2p was unaltered in strains (bmh1Δ bmh2Δ) lacking all 14-3-3 function (16, 29). The Σ strains used in those studies contain an uncharacterized suppressor of the bmh1Δ bmh2Δ growth defect (31). Because suppression of the bmh1Δ bmh2Δ growth defect might influence Msn2p localization, we examined Msn2p disposition after conditional loss of 14-3-3 protein function. A functional Msn2-Myc fusion protein remained in the cytoplasm of bmh1-11(Ts) bmh2 cells even after growth was completely arrested (2 h) by a shift to the nonpermissive temperature (Fig. 1A). In contrast, the Msn2-Myc fusion protein was concentrated in the nuclei of bmh1-11(Ts) bmh2 cells after 5 min of osmotic shock (Fig. 1A), indicating that the Msn2-Myc fusion was sensitive to normal physiological stimuli in that background. Thus, the rapamycin-sensitive interaction between Msn2p and Bmh2p (2) may be a simple reflection of the separation of two abundant cytoplasmic proteins by translocation of one protein to a separate compartment.
FIG. 1.
14-3-3 protein function and the PKA pathway. A. 14-3-3 protein depletion and Msn2p localization. Wild-type BMH1 (AKY32) and conditional bmh1(Ts) (AKY33) strains containing a functional Msn2-myc12 fusion were shifted from 23 to 36°C for 30, 60, and 120 min (60 min is shown) or from 23 to 23°C plus 1 M sorbitol for 5 min, and anti-myc antibody was used to visualize Msn2p by indirect immunofluorescence. Cells were examined by epifluorescence and phase-contrast microscopy. B. 14-3-3 proteins and PKA-dependent growth. The indicated strains were incubated at 23 and 34°C for 3 days. Strains AKY54 (tpk2-63 YEp-BMH1), AKY31 (TPK2 YEp), and AKY53(tpk2-63 YEp) are shown. C. PKA pathway and bmh1-11(Ts) bmh2 growth. Isogenic derivatives of conditional bmh1-11(Ts) bmh2::URA3 strain SGY459 were incubated at 23 or 36°C for 3 days. Strains AKY1 (vector), AKY2 (BMH1), AKY24 (msn4Δ), and AKY25 (msn2Δ msn4Δ) are shown. D. PKA pathway and bmh1-11(Ts) bmh2 growth. Isogenic derivatives of conditional bmh1-11(Ts) bmh2::KanMx strain AKY5 were streaked onto agar and incubated at 23, 34, or 36°C for 3 days. Strains AKY9 (LC-BMH1), AKY10 (LC), AKY45 (HC-TPK2), AKY46 (HC-TPK3), and AKY44 (HC) are shown, where LC and HC refer to low-copy and high-copy plasmids, respectively. E. Synthetic growth defect conferred by PKA and bmh1 mutations. Strains containing the indicated genotypes were grown at 23°C and and incubated at 23 and 34°C for 3 days. Strains AHY148 (tpk1w BMH1), AHY240a (TPK1 bmh1), and AHY240b (tpk1w bmh1) are shown.
Although Bmh1p and Bmh2p did not affect Msn2p localization, both proteins have been implicated as positive activators of PKA pathway function (13, 31). However, neither BMH1 nor BMH2 suppressed the growth defect of several PKA temperature-sensitive mutants [tpk-63(Ts) and tpk2-70(Ts)] (Fig. 1B and data not shown). Moreover, the bmh1-11(Ts) bmh2 strain accumulated wild-type levels of glycogen at semipermissive temperatures (data not shown). These results suggest that 14-3-3 proteins do not act on Msn2p function or function downstream of PKA. A functional interaction between Bmh1p/Bmh2p and the PKA pathway was also suggested by the observation that elevated PKA activity could suppress the growth defect of cells depleted of 14-3-3 protein (13); however, those studies used a strain in which the only source of 14-3-3 protein was provided by BMH2 under the control of the glucose-repressed GAL1 promoter. Because PKA is known to affect carbon metabolism (4, 10), we examined whether the bmh1-11(Ts) bmh2 growth defect could be suppressed by deletion or overexpression of PKA pathway genes MSN2, MSN4, YAK1, TPK1, TPK2, and TPK3. Interestingly, none of these regimens could alleviate the bmh1-11(Ts) bmh2 growth defect (Fig. 1C and D; also data not shown). Indeed, the only genetic interaction observed was a conditional, synthetic growth defect exhibited by a strain containing a deletion of the major 14-3-3 gene, BMH1 (13), as well as a compromised (wimp) allele of the single, available PKA catalytic gene TPK1 (Fig. 1E).
Rapamycin stimulates Msn2p translocation by inhibiting Tap42p.
Several reports have indicated that rapamycin treatment, like stress, causes Msn2p to accumulate in the nucleus (2, 16); however, rapamycin treatment alone did not elicit nuclear accumulation of functional, full-length Msn2-Myc (9) or Msn2-GFP (Fig. 2A) fusions in several of our lab strains. Nevertheless, although Msn2p-GFP remained in the cytoplasm of cells that were exposed to either rapamycin or mild heat stress (36°C), it accumulated in the nuclei of cells that were transferred to 36°C in medium containing rapamycin (9; also see below). Further, Msn2-GFP rapidly accumulated in the nuclei of cells exposed to heat (38°C [Fig. 2A]) or osmotic shock (see below). This discrepancy with previous results was not due to rapamycin resistance, because cells stopped dividing (data not shown) and rapamycin treatment induced expression of a put1-lacZ gene fusion (Fig. 2B) to levels observed previously (17, 34) within 60 min of rapamycin addition.
FIG. 2.
Rapamycin and Msn2p regulation. A. Rapamycin and Msn2 localization. Wild-type strain AKY34 containing the full-length Msn2-myc12 fusion was grown at 30°C to mid-log phase and transferred to 30°C medium, 30°C medium containing 200 ng of rapamycin (rap) per ml, or 38°C medium. Cells were monitored by epifluorescence and phase-contrast light microscopy after 0, 5, 10, 15, 30, and 60 min (pictures shown are from 30 min). B. Rapamycin and PUT1 expression. The put1-lacZ fusion strain AKY35 was transferred to 30°C medium containing 200 ng of rapamycin (Rap) per ml for the indicated times, and β-galactosidase (β-Gal) activity was measured.
If rapamycin exerts its effects on Msn2p by inactivating Tap42p, Msn2p should accumulate in the nuclei of conditional tap42-106(Ts) mutants at the nonpermissive temperature. We have shown this to be true for the Msn2-Myc fusion protein (9) and confirm that observation with a full-length Msn2-GFP fusion (Fig. 3A). Yeast Tip41 protein interacts with Tap42p and has been proposed to inhibit Tap42p (23). Interestingly, all of our tap42(Ts) tip41Δ double mutants grow more slowly than the isogenic tap42(Ts) TIP41 parent at both permissive and semipermissive temperatures (Fig. 3B and data not shown). This is distinct from reported results and suggests that Tip41p function might be more complex than previously thought (23). Whatever the explanation for this discrepancy, it is not related to Msn2p function, because the tap42(Ts) growth defect was not affected by deletion of MSN2 and MSN4 (Fig. 3C). The Msn2-GFP fusion protein remains in the cytoplasm of a tap42-106(Ts) tip41Δ double mutant at 36°C (Fig. 3A). Deletion of TIP41 therefore seems to be epistatic to the tap42(Ts) mutation. Instead of being an inhibitor of Tap42p, our results suggest that Tip41p plays an active role in transducing the signal between Tap42p and the protein phosphatase complex. Consistent with this hypothesis, the tip41Δ mutation also prevented Msn2p from accumulating in the nucleus in response to rapamycin (Fig. 3D).
FIG. 3.
The Tap42p-interacting protein Tip41p influences Msn2p localization. A. tip41Δ prevents Msn2p nuclear accumulation in a tap42(Ts) mutant. Strains containing full-length Msn2-GFP fusion were grown at 23°C to mid-log phase and transferred to 36°C medium. After 30 min, cells were viewed by epifluorescence and phase-contrast light microscopy. In panels A and B, strains AKY79 (TAP42 TIP41), AKY84 [tap42-106(Ts) TIP41], AKY142 (TAP42 tip41Δ), and AKY144 [tap42-106(Ts) tip41Δ] are shown. B. tip41Δ and tap42(Ts) growth. Strains from panel A were incubated for 3 days at 23 and 30°C. C. Msn2 and tap42(Ts) growth. Strains were incubated for 3 days at 23, 30, and 36°C. Strains Y3033 (TAP42 MSN2,4), Y3036 (TAP42 msn2Δ,4Δ), Y3034 (tap42-106 MSN2,4), Y3037 (tap42-106 msn2Δ,4Δ), Y3034 (tap42-109 MSN2,4), Y3038 (tap42-109 msn2Δ,4Δ) are shown. D. tip41Δ prevents Msn2p nuclear accumulation in response to rapamycin. Strains were grown at to mid-log phase at 23°C and transferred to 36°C medium either lacking or containing rapamycin (200 ng/ml). Strains AKY36 (TAP42 TIP41) and AKY75 (TAP42 tip41Δ) are shown.
Tap42p regulation of Msn2p is mediated by PP2A.
In contrast to its effects on Msn2p translocation in response to rapamycin and Tap42p depletion, the tip41Δ mutation did not affect either conditional growth or Msn2p translocation of the conditional PKA tpk2-63(Ts) strain (data not shown). This result suggests that Tap42p affects Msn2p by a PKA-independent mechanism. This conclusion was supported by the observation that a C-terminal Msn2-GFP fusion, which is regulated by PKA but not rapamycin or stress (16), remained in the cytoplasm of tap42(Ts) cells even after 60 min at the nonpermissive temperature (data not shown).
If Tap42p inhibits PP2A phosphatase catalytic activity, inactivation of the PP2A catalytic subunits, encoded by redundant genes PPH21 and PPH22, should block Msn2p translocation in rapamycin-treated cells. Interestingly, full-length Msn2-GFP remained in the cytoplasm of the pph21Δ pph22Δ double mutant when cells were transferred to 36°C in the presence of rapamycin, whereas the same treatment caused Msn2-GFP to accumulate in the nuclei of isogenic single mutants, as well as the wild-type parent (Fig. 4A). The pph21 pph22Δ mutant also failed to concentrate Msn2-GFP in the nucleus in response to 1 M sorbitol (Fig. 4B) and heat (38°C [data not shown]), suggesting that Pph21/Pph22 protein phosphatase activity is required for both rapamycin- and stress-stimulated Msn2p translocation. This translocation defect was not due to a general perturbation of Msn2p localization, because Msn2-GFP accumulated in the nucleus of the pph21Δ pph22Δ strain upon glucose depletion (Fig. 4B). Finally, although Tap42p also regulates the PP2A-like phosphatase catalytic subunit Sit4p (8), the Msn2-GFP fusion protein accumulated in the nuclei of rapamycin-treated sit4(Ts) and sit4Δ ssd1v strains that were shifted to 36°C (data not shown), in agreement with a previous claim that Sit4p does not play a role in Msn2p localization (2).
FIG. 4.
PP2A activity and Msn2p translocation in response to rapamycin and stress. A. PP2A catalytic activity is required for Msn2p translocation in response to rapamycin. Strains containing the Msn2-myc12 fusion were grown to mid-log phase at 23°C and transferred to 36°C medium lacking or containing 200 ng of rapamycin per ml. After 30 min, cells were viewed by epifluorescence and phase-contrast light microscopy. Strains AKY101 (wild type [WT]) and AKY103 (pph21Δ pph22Δ) are shown. B. PP2A catalytic activity is required for Msn2p translocation in response to stress. Strains shown in panel A were grown to mid-log phase in medium containing 2% glucose and then transferred to the same medium, the same medium containing 1 M sorbitol, or the same medium lacking glucose. After 30 min (10 min for glucose depletion), cells were visualized by epifluorescence, phase-contrast light, and differential-contrast (DIC) microscopy.
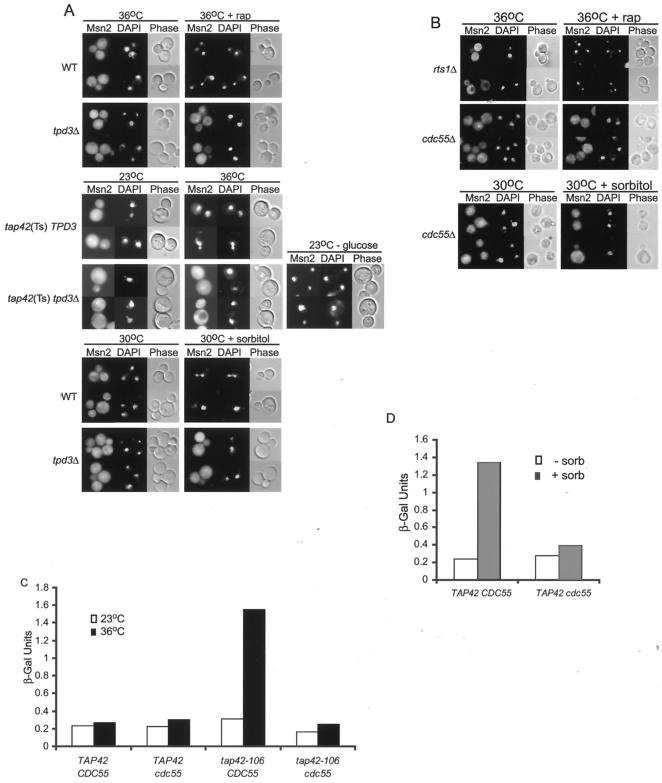
Msn2p accumulation requires the PP2A regulatory subunit Cdc55p.
In addition to the catalytic subunits, yeast PP2A phosphatase is comprised of a regulatory subunit, which can be either Rts1p or Cdc55p, and a scaffolding protein, Tpd3p. Strains lacking either of the regulatory subunits exhibit distinct phenotypes, whereas a tpd3Δ mutant displays phenotypes that appear to be a composite of the two regulatory mutants (44). Mutants lacking individual subunits were treated with rapamycin or stress to determine their contribution to Msn2p localization. Both tpd3Δ (Fig. 5A) and cdc55Δ (Fig. 5B) cells retained Msn2p in the cytoplasm upon treatment with rapamycin (rapamycin and 36°C) or 1 M sorbitol, whereas Msn2p quickly translocated to the nuclei of rts1Δ and wild-type cells under the same regimens (Fig. 5A and B). Thus, scaffolding protein Tpd3p and regulatory protein Cdc55p are required for rapamycin- and stress-induced Msn2p translocation.
FIG. 5.
PP2A regulatory subunits and Msn2p translocation. A. PP2A scaffolding subunit Tpd3p is required for Msn2p translocation in response to rapamycin (rap), Tap42p depletion, and stress. Strains containing the full-length Msn2-GFP fusion protein were treated as described in the legend to Fig. 4 (23°C - glucose, 23°C medium lacking glucose) and visualized by epifluorescence and phase-contrast light microscopy. Strains AKY198 (wild type [WT]), AKY191 (tpd3Δ), AKY194 [tap42(Ts) TPD3], and AKY195 [tap42(Ts) tpd3Δ] are shown. B. PP2A regulatory subunit Cdc55p is required for Msn2p translocation in response to rapamycin and stress. Strains AKY163 (rts1Δ) and AKY185 (cdc55Δ) containing full-length Msn2-GFP and Msn2-myc12 fusion proteins, respectively, were treated as described in the legend to Fig. 4 and visualized by epifluorescence and phase-contrast light microscopy. C. PP2A function and Msn2-dependent gene expression in response to Tap42p inactivation.β-Galactosidase (β-Gal) activities of the indicated CTT1-lacZ strains were measured during exponential growth at 23°C and 30 and 45 min (shown) after the shift to 36°C. Values shown are the mean of four measurements from two separate experiments. All standard deviations were less than 15%, with the exception of the 23°C measurement of the tap42-106 cdc55 strain, which was less than 35%. Strains AKY148 (TAP42 CDC55), AKY204 (TAP42 cdc55), AKY150 (tap42-106 CDC55), and AKY205 (tap42-106 cdc55) are shown. D. PP2A function and Msn2-dependent gene expression in response to osmotic shock. β-Galactosidase activities of the indicated CTT1-lacZ strains were measured during exponential growth in minimal medium and 30 min after the shift to minimal medium containing 1 M sorbitol (sorb). Values shown are the mean of four measurements from two separate experiments. All standard deviations were less than 15%. Strains AKY148 (TAP42 CDC55) and AKY205 (TAP42 cdc55) are shown.
Expression of Msn2-dependent genes is elevated upon stress or nutrient starvation as a result of the accumulation of Msn2p in the nucleus. To determine whether PP2A-deficient cells are refractory to stress-induced stimulation of Msn2-dependent genes, we monitored expression levels of a CTT1-lacZ reporter fusion. Whereas Tap42p inactivation normally resulted in fivefold induction of CTT1-lacZ expression, this increase was totally dependent upon Cdc55p function (Fig. 5C). Similar results were observed when wild-type and cdc55Δ cells were treated with osmotic shock (Fig. 5D), implying that PP2A activity is required for stress-activated localization and function of Msn2p.
Nitrogen starvation regulates Msn2p localization by a PP2A-dependent mechanism.
In contrast to glucose availability, which is generally thought to modulate Msn2p localization through its influence on the cAMP-dependent protein kinase signaling pathway (4, 16, 29), there is no generally accepted model that accounts for the effects of nitrogen starvation on Msn2p localization. One model posits that nitrogen levels are sensed and transmitted to the PKA catalytic subunit by a poorly defined, cAMP-independent mechanism referred to as the FGM (fermentable-growth-medium-induced) pathway. The FGM model was proposed to explain how nitrogen starvation could affect Msn2-regulated processes in the absence of an effect on cAMP levels; however, there is no direct evidence to suggest that either PKA activity or Msn2p phosphorylation corresponds to nitrogen availability.
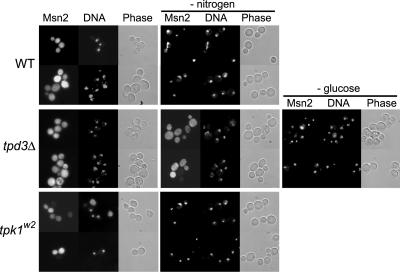
An alternative model is that the TOR kinase pathway, which is known to respond to nitrogen levels, transmits the nitrogen starvation signal to Msn2p (2, 16). Despite the attraction of the model, Msn2p localization has not been examined in either wild-type or mutant cells starved of nitrogen. Interestingly, full-length Msn2-GFP accumulated in the nuclei of wild-type cells within 30 min of nitrogen starvation (Fig. 6), whereas it took at least 2 h to exhibit nuclear accumulation of the carboxyl-terminal Msn2-GFP fusion in a minor fraction of the cells (data not shown). The full-length Msn2-GFP fusion also accumulated in the nuclei of tpk1w2 bcy1 cells after 30 min of nitrogen starvation (Fig. 6), whereas tpd3Δ cells displayed cytoplasmic accumulation at least 2 h after nitrogen starvation (Fig. 6). In contrast, the full-length Msn2-GFP fusion quickly accumulated in the nuclei of glucose-depleted tpd3Δ cells (Fig. 6), indicating that the strain's lack of response to nitrogen was not a general defect in nutrient sensing or response. Thus, nitrogen limitation affects the amino-terminal, stress-sensitive domain of Msn2p by a mechanism that requires PP2A phosphatase activity and is independent of PKA regulatory protein Bcy1p.
FIG. 6.
PP2A activity is essential for Msn2p nuclear accumulation in response to nitrogen starvation. Strains containing the full-length Msn2-GFP fusion protein were grown in selective minimal medium at 30°C before they were transferred to the same medium or the same medium lacking (−) nitrogen or glucose for 30 min. Strains AKY198 (wild type [WT]), AKY191 (tpd3Δ), and RTF1.5-2 (tpk1w2) are shown. Cells were examined by epifluorescence or phase-contrast microscopy.
DISCUSSION
Accumulation of Msn2p in the nucleus and subsequent activation of stress response genes results from a balance between nuclear import and nuclear export. Görner and colleagues recently identified a carboxyl-terminal domain that is responsible for glucose-responsive nuclear import, as well as an amino-terminal region that is thought to promote nuclear export in response to glucose and stress (16). Our results help clarify the signaling pathway that impinges on the export process.
Our results indicate that the Tor kinase pathway regulates Msn2p localization through its effects on PP2A protein phosphatase. Because Tor kinase function does not affect the carboxyl-terminal NLS (16; data not shown), these results are most consistent with a model in which Tor kinase and PP2A regulate nuclear export of Msn2p. That is, the PP2A catalytic subunit encoded by PPH21/PPH22 in a complex with the regulatory subunit Cdc55p and the scaffold protein Tpd3p inhibits export of Msn2p from the nucleus. During mitotic growth, Tor-activated Tap42p suppresses the activity of this phosphatase complex and stimulates Msn2p export from the nucleus. When Tor kinase is inactivated by the addition of rapamycin, PP2A activity is derepressed, and Msn2p export is inhibited. Thus, Tor inactivation causes Msn2p to accumulate in the nucleus through the trapping of a normal low-level flux of Msn2p into the nucleus. There is at least one other example of a phosphatase modulating the localization of a yeast transcription factor. In response to mating pheromone, cytosolic calcium levels increase and activate the calcium-calmodulin-dependent phosphatase calcineurin. Activated calcineurin then dephosphorylates the transcription factor Crz1p, which accumulates in the nucleus and stimulates expression of genes needed for cell wall remodeling and survival (3).
Tor inactivation, stress, and nitrogen starvation result in accumulation of Msn2p in the nucleus and prolonged activation of stress response genes (2, 9, 16, 29) (Fig. 6). The parsimonious conclusion is that all of these conditions act on the same mechanism of nuclear export. In contrast to glucose depletion, which regulates both the amino- and carboxyl-terminal domains of Msn2p, Tor inactivation, stress, and nitrogen depletion affect only the amino-terminal domain (9, 16). This correlation is further strengthened by our observation that PP2A activity is essential to Msn2p regulation by the latter stimuli, whereas Msn2p responds to glucose depletion independent of PP2A function (Fig. 4 and 5). This specificity also argues against the possibility that PP2A phosphatase activity plays an indirect role in Msn2p localization. Thus, we propose that cells sense and respond to stress (and Tor inactivation) by stimulating PP2A-dependent dephosphorylation and inhibiting export of Msn2p. Finally, although glucose depletion could also affect nuclear export of Msn2p by a PP2A-independent mechanism, a simpler model would be one in which glucose depletion regulates nuclear import of the amino-terminal region in much the same way that it regulates the carboxyl-terminal NLS (16). In this latter scenario, the amino-terminal domain would contain a glucose-sensitive NLS in addition to the stress-sensitive NES.
Even if Tor kinase does not transmit the stress signal, stress conditions could still be transduced to PP2A phosphatase by a separate signaling pathway. Little is known about the regulation or function of the individual PP2A subunits; however, it is interesting that Cdc55p, but not Rts1p, is required for Msn2p translocation (Fig. 5). These results imply that Cdc55p performs a PP2A targeting function that is separate from that of Rts1p, a conclusion that is consistent with the notion that these two proteins influence independent cellular processes (14, 19, 37). The independence of these regulatory subunits underscores the possibility that they are regulated by distinct physiological stimuli and regulatory mechanisms. Thus, stress and rapamycin (Tor kinase) could impinge upon PP2A phosphatase activity by convergent mechanisms, rather than by a linear pathway in which stress inhibits Tor kinase activity.
Bmh1p/Bmh2p and the PKA pathway.
Beck and Hall (2) presented a model in which yeast 14-3-3 protein (Bmh1p/Bmh2p) acted as a cytoplasmic anchor for the transcription factor Msn2p. Evidence supporting this model included genetic interactions between 14-3-3 genes BMH1/BMH2 and the PKA pathway (13, 31), a rapamycin-sensitive interaction between Bmh2p and Msn2p (2), and a previously documented role for mammalian 14-3-3 proteins in cytoplasmic sequestration of signaling proteins (30). However, this model was inconsistent with several recent observations that Msn2p remained in the cytoplasm of a strain (Σ bmh1Δ bmh2Δ) lacking all 14-3-3 proteins (16, 29). Yeast 14-3-3 function is normally essential for viability (13, 31, 43), implying that the Σ bmh1Δ bmh2Δ strain used in the Msn2p localization studies (16, 29) contained a suppressor of the growth defect. Although the bmh1Δ bmh2Δ growth suppressor might have influenced Msn2p localization, our results (Fig. 1A) are consistent with the conclusion that 14-3-3 protein does not act as a cytoplasmic anchor of Msn2p. Yeast 14-3-3 proteins are relatively abundant and interact with a large number of proteins by both biochemical and two-hybrid techniques (11, 21, 22, 42). Thus, their association with Msn2p might simply reflect a nonspecific interaction, and the rapamycin sensitivity of this interaction could, in turn, be rationalized by a change in cellular distribution of one (Msn2p) of these factors.
If 14-3-3 proteins do not anchor Msn2p in the cytoplasm, what is their role in PKA-dependent processes? Genetic studies showed that 14-3-3 proteins stimulated PKA-dependent growth and pseudohyphal formation (13, 31), and several observations were consistent with the notion that they impinged upon the pathway between the upstream components Cdc25p and Ras2p. For example, while Bmh1/Bmh2 overexpression suppressed the growth defect of a cdc25(Ts) mutant, lesions in the downstream Ras2p protein were unaffected by overproduction of either 14-3-3 protein (13). Moreover, PKA-regulated processes remained responsive to the activated Ras2G2V allele in a bmh1Δ bmh2Δ strain, implying that 14-3-3 proteins do not act downstream of Ras2p protein function (31). Although our observation that BMH1/BMH2 overexpression cannot suppress the tpk2(Ts) growth defect is consistent with this interpretation, it is hard to reconcile such a model with our observation that Bmh1/Bmh2 inactivation did not affect Msn2p localization (Fig. 1). Because Ras and PKA function are required to maintain Msn2p in the cytoplasm (15), this result would seem to rule out a role for yeast 14-3-3 proteins in regulating Ras/PKA function. Taken together, these results are most consistent with a model in which 14-3-3 proteins impinge upon several PKA-regulated processes by an unknown, or at least PKA-independent, pathway. Support for this conclusion also comes from our observation that PKA activation (by overexpression of each of the three TPK genes or inactivation of MSN2/MSN4 or YAK1) failed to alleviate the bmh1(Ts) bmh2 growth defect (Fig. 1).
Acknowledgments
We thank Gustav Ammerer, Deepti Saxena, and Marjorie Brandriss for providing strains and plasmids and Lorena Equez and Eric White for help with fluorescence microscopy.
This work was supported in part by NIH grants GM44666 to S.G. and CA41086 to J.R.B.
REFERENCES
- 1.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 3.Boustany, L., and M. S. Cyert. 2002. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site. Genes Dev. 16:608-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broach, J. R., and R. J. Deschenes. 1990. The function of ras genes in Saccharomyces cerevisiae. Adv. Cancer Res. 54:79-139. [DOI] [PubMed] [Google Scholar]
- 5.Cameron, S., L. Levin, M. Zoller, and M. Wigler. 1988. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell 53:555-566. [DOI] [PubMed] [Google Scholar]
- 6.Cherkasova, V. A., and A. G. Hinnebusch. 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 17:859-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crespo, J. L., and M. N. Hall. 2002. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 66:579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10:1904-1916. [DOI] [PubMed] [Google Scholar]
- 9.Düvel, K., A. Santhanam, S. Garrett, L. Schneper, and J. R. Broach. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11:1467-1478. [DOI] [PubMed] [Google Scholar]
- 10.Fraenkel, D. G. 1985. On ras gene function in yeast. Proc. Natl. Acad. Sci. USA 82:4740-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 12.Gelperin, D., L. Horton, A. DeChant, J. Hensold, and S. K. Lemmon. 2002. Loss of ypk1 function causes rapamycin sensitivity, inhibition of translation initiation and synthetic lethality in 14-3-3-deficient yeast. Genetics 161:1453-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelperin, D., J. Weigle, K. Nelson, P. Roseboom, K. Irie, K. Matsumoto, and S. Lemmon. 1995. 14-3-3 proteins: potential roles in vesicular transport and Ras signaling in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 92:11539-11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentry, M. S., and R. L. Hallberg. 2002. Localization of Saccharomyces cerevisiae protein phosphatase 2A subunits throughout mitotic cell cycle. Mol. Biol. Cell 13:3477-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Görner, W., E. Durchschlag, M. T. Martinez-Pastor, F. Estruch, G. Ammerer, B. Hamilton, H. Ruis, and C. Schüller. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Görner, W., E. Durchschlag, J. Wolf, E. L. Brown, G. Ammerer, H. Ruis, and C. Schüller. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21:135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartley, A. D., S. Bogaerts, and S. Garrett. 1996. cAMP inhibits bud growth in a yeast strain compromised for Ca2+ influx into the Golgi. Mol. Gen. Genet. 251:556-564. [DOI] [PubMed] [Google Scholar]
- 19.Healy, A. M., S. Zolnierowicz, A. E. Stapleton, M. Goebl, A. A. DePaoli-Roach, and J. R. Pringle. 1991. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol. Cell. Biol. 11:5767-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 21.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 22.Ito, T., T. Chiba, R. Ozawa, M. Yoshida, M. Hattori, and Y. Sakaki. 2001. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98:4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacinto, E., B. Guo, K. T. Arndt, T. Schmelzle, and M. N. Hall. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8:1017-1026. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling growth in yeast. EMBO J. 18:2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Lin, F. C., and K. T. Arndt. 1995. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 14:2745-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorenz, M. C., and J. Heitman. 1995. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 270:27531-27537. [DOI] [PubMed] [Google Scholar]
- 28.Luke, M. M., F. Della Seta, C. J. Di Como, H. Sugimoto, R. Kobayashi, and K. T. Arndt. 1996. The SAPs, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16:2744-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayordomo, I., F. Estruch, and P. Sanz. 2002. Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (stress response element)-regulated genes. J. Biol. Chem. 277:35650-35656. [DOI] [PubMed] [Google Scholar]
- 30.Muslin, A. J., and H. Xing. 2000. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 12:703-709. [DOI] [PubMed] [Google Scholar]
- 31.Roberts, R. L., H. U. Mosch, and G. R. Fink. 1997. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell 89:1055-1065. [DOI] [PubMed] [Google Scholar]
- 32.Robinson, M. K., W. H. van Zyl, E. M. Phizicky, and J. R. Broach. 1994. TPD1 of Saccharomyces cerevisiae encodes a protein phosphatase 2C-like activity implicated in tRNA splicing and cell separation. Mol. Cell. Biol. 14:3634-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ronne, H., M. Carlberg, G. Z. Hu, and J. O. Nehlin. 1991. Protein phosphatase 2A in Saccharomyces cerevisiae: effects on cell growth and bud morphogenesis. Mol. Cell. Biol. 11:4876-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxena, D., K. B. Kannan, and M. C. Brandriss. 2003. Rapamycin treatment results in GATA factor-independent hyperphosphorylation of the proline utilization pathway activator in Saccharomyces cerevisiae. Eukaryot. Cell 2:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmelzle, T., T. Beck, D. E. Martin, and M. N. Hall. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24:338-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shu, Y., H. Yang, E. Hallberg, and R. Hallberg. 1997. Molecular genetic analysis of Rts1p, a B′ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol. Cell. Biol. 17:3242-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon, M., G. Adam, W. Rapatz, W. Spevak, and H. Ruis. 1991. The Saccharomyces cerevisiae ADR1 gene is a positive regulator of transcription of genes encoding peroxisomal proteins. Mol. Cell. Biol. 11:699-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, A., M. P. Ward, and S. Garrett. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spencer, F., C. Connelly, S. Lee, and P. Hieter. 1989. Isolation and cloning of conditionally lethal chromosome transmission fidelity genes in Saccharomyces cerevisiae. Cancer Cells (Eukaryotic DNA Replication) 6:441-452. [Google Scholar]
- 41.Thevelein, J. M., and J. H. de Winde. 1999. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33:904-918. [DOI] [PubMed] [Google Scholar]
- 42.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 43.van Heusden, G. P., and H. Y. Steensma. 2001. 14-3-3 proteins are essential for regulation of RTG3-dependent transcription in Saccharomyces cerevisiae. Yeast 18:1479-1491. [DOI] [PubMed] [Google Scholar]
- 44.van Zyl, W., W. Huang, A. A. Sneddon, M. Stark, S. Camier, M. Werner, C. Marck, A. Sentenac, and J. R. Broach. 1992. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:4946-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Y., and D. J. Burke. 1997. Cdc55p, the B-type regulatory subunit of protein phosphatase 2A, has multiple functions in mitosis and is required for the kinetochore/spindle checkpoint in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]