Abstract
Nuclear bodies contribute to nonrandom organization of the human genome and nuclear function. Using a major prototypical nuclear body, the Cajal body, as an example, we suggest that these structures assemble at specific gene loci located across the genome as a result of high transcriptional activity. Subsequently, target genes are physically clustered in close proximity in Cajal body-containing cells. However, Cajal bodies are observed in only a limited number of human cell types, including neuronal and cancer cells. Ultimately, Cajal body depletion perturbs splicing kinetics by reducing target small nuclear RNA (snRNA) transcription and limiting the levels of spliceosomal snRNPs, including their modification and turnover following each round of RNA splicing. As such, Cajal bodies are capable of shaping the chromatin interaction landscape and the transcriptome by influencing spliceosome kinetics. Future studies should concentrate on characterizing the direct influence of Cajal bodies upon small nuclear RNA gene transcriptional dynamics.
Keywords: Cajal bodies, chromatin, genome organization, nuclear bodies, RNA splicing, snRNPs, transcription
Introduction
The maintenance of spatial and temporal compartmentalization of the cell nucleus is crucial for efficient genome function (Box 1). This includes the initiation of specific metabolic and homeostatic programs at defined times and locations, and involves the formation of a number of membraneless nuclear compartments and domains [1–3]. These nuclear compartments consist of discrete regions known as chromosome territories (CTs) that demarcate interphase chromosomes [4], nuclear substructures whose structural integrity is mediated by transient protein-protein and protein-RNA interactions [5, 6], and other regions, such as the nuclear lamina. A prominent group of structurally distinguishable sub-nuclear microenvironments are nuclear bodies (NBs) [1, 3]. Examples of NBs include the largest and universally present NB, the nucleolus, as well as smaller NBs of varying number per cell such as histone locus bodies (HLBs), promyelocytic (PML) NBs and nuclear speckles. Cajal bodies (CBs) are one of the better-characterized NBs and have been studied since the early 1900’s [7]. These prominent structures, or homologous CB-like nuclear domains, are identifiable in many animals and plants. In humans, they are present in embryonic [8], fetal, neuronal [9] and, importantly, transformed cancer cells [10]. However, CBs are absent in many terminally-differentiated primary cell types. As such, CBs can be thought of as dynamic structures that only form in cells under precise physiological conditions (such as oncogenesis) or as a result of specific stimuli or genomic activity.
Box 1. Correlations between 3D genome organization and transcription dynamics.
The genome is highly spatially organized within the nucleus which lacks defining membranes. Spatial genome organization is non-random and arranged at several, hierarchical levels of chromatin fiber folding, the formation of loops and consequently to topologically associated domains culminating in their aggregation to chromosomes within defined territories (chromosome territories or CTs) (Figure 1A). In principle, each chromosome and each allele occupies certain preferred positions relative to other regions in the genome in the majority of cells within a population [11]. There are multiple layers of nuclear function that eventually may influence chromosome and gene positioning. In particular, it has been established that gene-poor chromosomes are typically located near the nuclear periphery, whilst gene-rich chromosomes tend to be located in the nuclear interior (Figure 1B). However, active genes are observed to interrupt large regions of repressed heterochromatin at the nuclear periphery. Gene-rich regions of the genome are spatially separated from gene-poor regions and repressed regions of chromosomes tend to contact other repressed regions. In contrast, active domains are preferentially located on the outside of CTs and contact other active domains on the same chromosome and other chromosomes. Thus, silent genes are preferentially located within the interior of a CT, whilst active highly expressed genes are looped out of the CT area [12]. Of note, NBs are closely related to both activation and repression of target genes through mechanisms that are not fully understood, which might contribute to the maintenance of 3D genome organization.
It is generally agreed that NBs expedite and augment the efficiency of specific molecular processes [13, 14]. To achieve this, NBs are segregated from the surrounding nucleoplasm by concentration-dependent phase-separation [13, 15]. This results in enrichment of specific macromolecules within the microenvironment where macromolecular crowding effects enhance specific biochemical and assembly reactions. Thus, as a result of enhanced RNA-protein associations, protein-protein interactions and other biophysical effects, the rate of enzymatic catalysis and substrate recycling is much higher in NBs than in the less crowded surrounding nucleoplasm. The augmentation of these biochemical processes helps to boost energetically unfavorable events and maintain optimal nuclear function. In particular, the CB is a multifunctional domain, acting as a site of transcriptional activation, RNA processing, base modification and assembly for the biogenesis of multiple classes of ribonucleoproteins (RNPs). CB function is most commonly associated with the production of the spliceosomal enzymatic backbone, known as small nuclear RNPs (snRNPs, Figure 2), which catalyze RNA splicing [16–21]. However, the CB is also believed to be a key regulatory domain in the production of telomerase RNP and several factors housed within the body are implicated in spliceosomal and telomerase RNP maturation. Both early and late-stage snRNP biogenesis components are enriched in the CB, as well as precursors extended at their 3’ ends and fully-assembled mature snRNPs [22–27]. The CB has been suggested to upregulate the expression and accelerate the 3’ end processing of small nuclear, small nucleolar and small CB-associated RNAs (snRNAs, snoRNAs and scaRNAs, respectively) through transcriptional activation and sequestration [28–30]. CBs also mediate numerous processes unrelated to transcription and are enriched with multiple RNA base modification enzymes, which regulate target sn/snoRNA dynamics by 2’-O-methylation and pseudouridinylation and influence spliceosome assembly [31, 32]. Finally, CBs have been suggested to act as a surveillance structure by helping to accelerate the step-wise assembly and recycling of spliceosomal snRNPs in times of need, such as cancer [33, 34]. However, snRNP processing and base modification also occurs in cells that lack CBs, although this is believed to be much less effective and occur at a slower rate [35]. These studies indicate that CBs influence the levels and processivity of factors crucial for efficient RNA splicing.
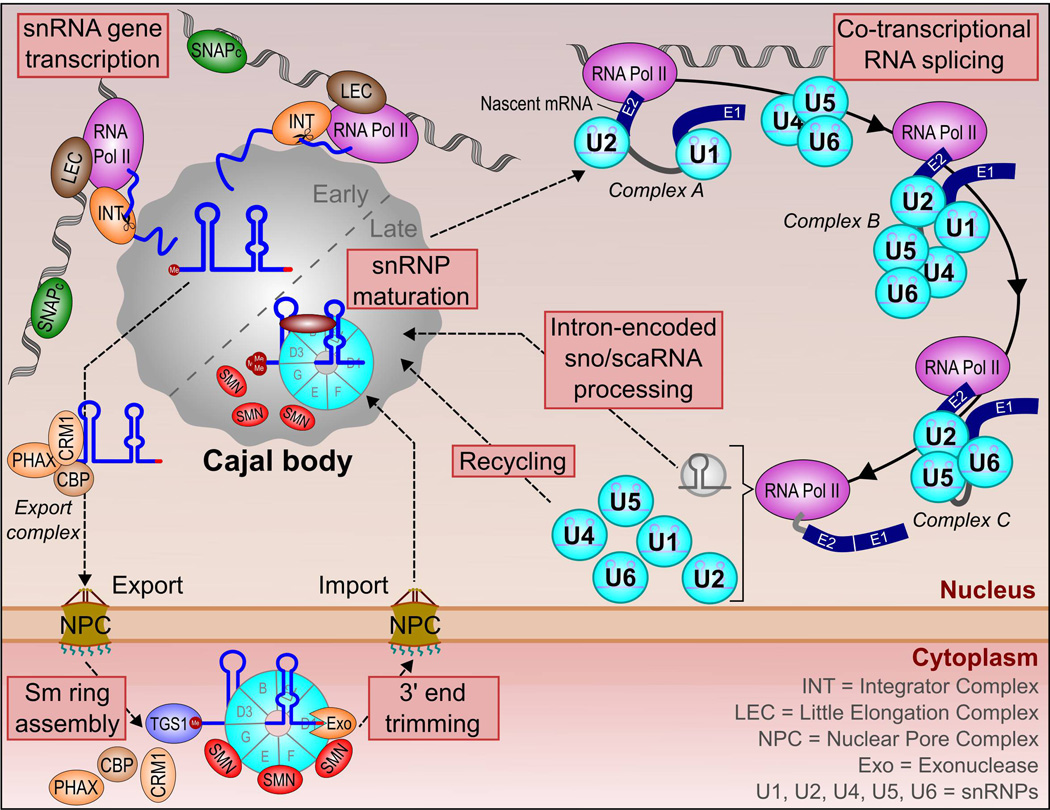
Figure 2.
The relationship between Cajal bodies and splicing. Transcription of snRNA genes frequently occurs at the CB periphery, and is mediated by several protein-based complexes. Nascent pre-snRNA is enriched in the CB domain prior to association with an export complex and subsequent translocation to the cytoplasm. Here, the pre-snRNP undergoes 5’-cap tri-methylation by TGS1, 3’-end trimming by an exonuclease and Sm ring assembly involving the SMN protein complex. The snRNP is re-imported into the nucleus for final maturation steps, including RNA base modifications (2’-O-methylation and pseudouridinylation). Mature snRNPs are redistributed to the nuclear speckle for spliceosome formation. At the periphery of the nuclear speckle, snRNPs catalyze the excision of introns from pre-mRNAs through the formation of a number of protein complexes on the target pre-mRNA and the joining of adjacent exons (E1, E2). Most introns are degraded but a minority undergo further processing steps to produce a sno/scaRNA. These intron-encoded sno/scaRNAs are targeted to the CB, where they guide major and minor spliceosomal snRNA base-modification steps. Following splicing, snRNPs are liberated from the target pre-mRNA. The U4/U6 di-snRNP and U4/U6.U5 tri-snRNP require recycling in the CB prior to participating in another round of pre-mRNA splicing.
Intuitively, it is beneficial for the cell to limit the activity of specific and frequently limited factors to a confined area of need rather than the entire nucleoplasm [36]. This theoretically reduces off-target effects, such as incorrect target processing or spurious gene activation and repression, and reduces the energetic burden on the cell as lower quantities of NB processing factors are required. Additionally, several NBs frequently form at specific genomic loci and accelerate target molecular processes in a localized microenvironment [1]. For example, nucleoli are formed around actively transcribing, tandemly-repeated ribosomal genes concentrated on several separate chromosomes [37], HLBs form at the replication-dependent histone gene clusters [38] and PML NBs also associate with defined genomic regions (albeit in a cell type-specific manner) [39]. However, NBs are multifunctional structures that influence the DNA damage response, protein and RNA modifications, as well as gene activity [1]. Importantly, CBs are known to assemble at specific gene loci, including both spliceosomal snRNA genes and histone gene loci (through its physical association with the HLB) [22–26, 30, 40]. Thus, CBs concentrate essential but frequently limited factors for expedited snRNA transcription, extended 3’end RNA processing and snRNP maturation processes at target snRNA gene loci.
Nuclear and genomic architecture is largely non-random and is the product of many synergistic influences that act at the local (kbp) and global level (Mbp/interchromosomal), including remodeling enzymes [41], cell type-specific factors [42] and various epigenetic modifications [43]. Here, using CBs as an example prototypical NB, we propose the hypothesis that NBs may also contribute to the regulation of long-range and interchromosomal chromatin organization. Subsequently, this results in under-appreciated impacts on both coding and non-coding transcriptome diversity.
Genome organization is influenced by nuclear body function
Topological organization of the genome is structurally maintained to efficiently accommodate approximately 2 m of DNA into a cell nucleus whilst retaining functionality [44]. Firstly, 147 bp of DNA is wrapped around histone octamers in structures known as nucleosomes, which assemble into a 30 nm chromatin fiber. Higher-order DNA organization ultimately consists of highly condensed but permeable interphase chromosomes, which are partitioned into functional units, including topologically-associated domains (TADs) and chromatin loops. Furthermore, both genes and CTs occupy preferred positions within the nuclear space [4, 45–47] and this has often been linked with transcriptional activity [48]. However, the nucleus is a densely-packed space and individual CTs intermingle at their edges in highly transcriptionally active interchromosomal regions [49]. It is likely that chromosomal segregation, nonrandom CT positioning, specific transcriptional activity, gene function and stochasticity regulate physical gene pairing of most genes. However, the spatial organization of the genome within the nucleus is nonrandom [45] and frequent long-range intra- (cis) and inter-chromosomal (trans) gene pairing events are known to occur. There are multiple studies that describe nonrandom long-range gene pairing events (separated by millions of base pairs) in both cis and trans configurations between specific groups of genes that are often, but not always, coordinated by nuclear compartments [50–52]. These include associations between lamina-associated domains (LADs) [53] and immunoglobulin genes in B-cells [54, 55]. Thus, the physical association of most genes may be driven by random short-lived gene collisions. However, there is precedent for the formation of spatially-clustered gene networks that are more stable and may possess co-regulated transcriptional dynamics and RNA processing.
We envisage a simple model of genomic clustering that features a functional multi-interaction hub, which is a beneficial regulatory hotspot for target gene loci. For most genes this is potentially represented by RNA polymerase II (pol II) transcription factories, where multiple active RNA polymerases are concentrated in a nuclear substructure [56]. Indeed, it has been estimated that 10–25 distinct active gene loci may colocalize at a single transcription factory [57], which allows sharing of specific sets of transcription factors, could help co-ordinate gene expression of several distinct genes from the same pathway and potentially influences long-range chromatin looping [58]. Specific physical associations between NBs, including nucleoli [37, 59], nuclear speckles [60], PML NBs, Polycomb (PcG) bodies [61] and HLBs [62], and certain gene loci or genomic regions are also known to occur (Figure 3). Similarly, highly transcriptionally active snRNA genes have been observed to relocate and stably cluster near the CB periphery [63]. Interestingly, co-labeling of multiple CB target genes with the same fluorophore indicates that the CB simultaneously associates with several genes [22]. The frequency of these gene repositioning events and the total number of CBs per cell leads us to believe that frequent higher-order gene associations near the CB is possible.
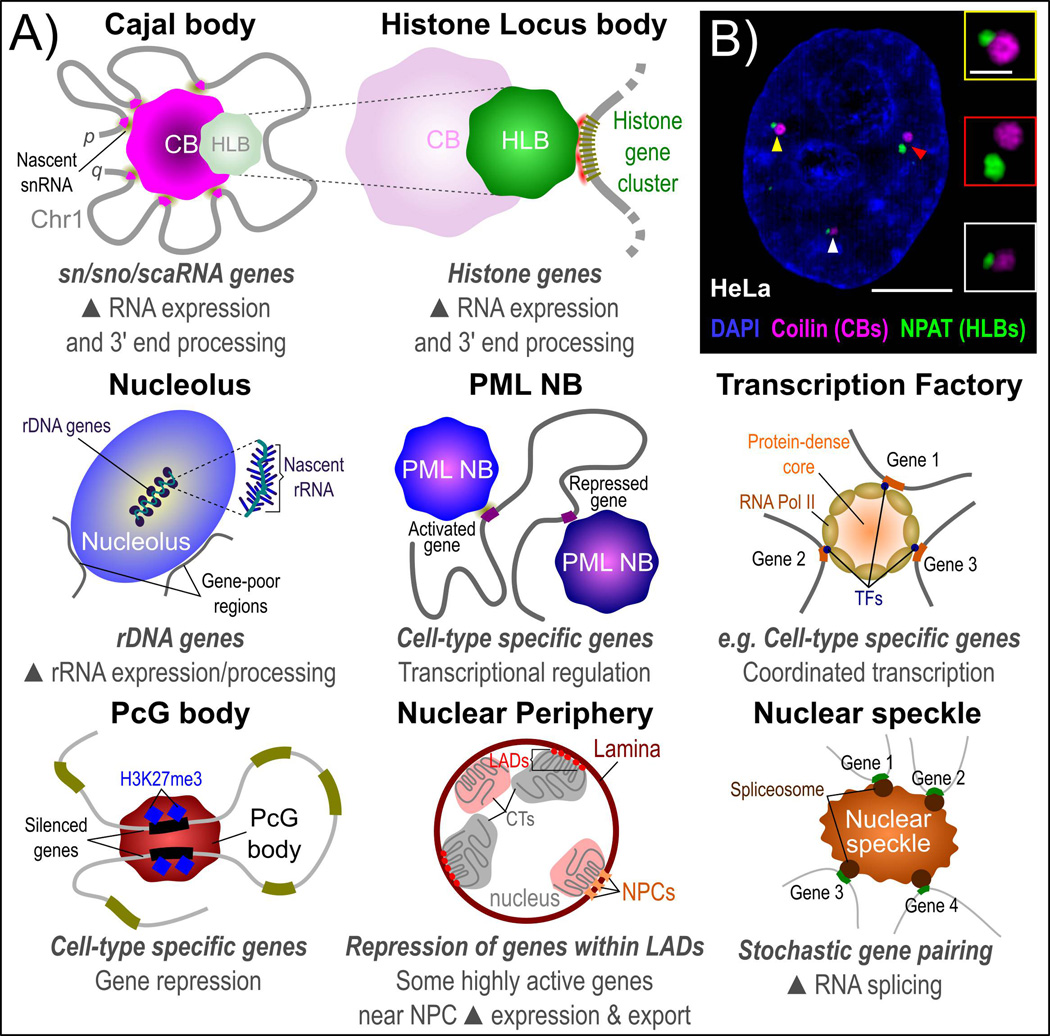
Figure 3.
Nuclear sub-structures and genome organization. A: Many NBs have been reported to form at specific sites of gene activity and influence gene expression. This includes transcriptional activation and repression as well as RNA processing steps. Several NBs have been associated with both gene activation and gene repression, including the role of the nucleolus in activation of ribosomal DNA (rDNA) genes and proximal positioning of gene-poor regions at the nucleolar periphery as well as gene activation and repression by the PML NB, which may be dependent on PML isoform enrichment. Nonrandom physical clustering of specific genes is also reported for non-NB-based structures, such as the nuclear lamina with lamina-associated domains (LADs) and other genes that are highly active and frequently positioned near to the nuclear pore complex (NPC) for efficient export into the cytoplasm. B: Maximally-projected super resolution structured-illumination microscopy (SIM) image of the CB structural protein coilin (far red), the histone transcription factor NPAT (HLBs, green) and DNA (DAPI, blue) in aneuploid HeLa human cervical carcinoma cells. Enlarged insets with positions marked by arrowheads demonstrate frequent physical association between CBs and HLBs. Scale bars represent 5 µm (whole cell) or 1 µm (inset). Images were acquired using a Zeiss Elyra super resolution microscope equipped with a 100× apochromatic 1.46 NA oil immersion objective. 3D-SIM images were generated using Zeiss Zen black software.
We have recently provided new evidence to support a genome-wide organizational role for the CB [30, 40]. Using a combination of advanced multicolor microscopic mapping tools and high-throughput sequencing techniques, we explored the specific clustering network, consisting of distant chromosomal targeting regions proximal to CBs [30]. Chromosomes that associate frequently with CBs display higher CB target gene density than other size-matched chromosomes (human chromosomes 1, 6 and 17 vs. 2, 7 and 18) [64]. Physical associations between specific chromosomes and CBs also correlate with major and minor spliceosomal snRNA gene clusters, such as the U1 and U2 snRNA gene arrays on chromosomes 1 and 17, respectively. These regions potentially act as primary CB nucleation sites that initiate CB assembly. In particular, the large and ubiquitously expressed U1 snRNA gene array spanning 500 kb on the p-arm of chromosome 1 is the most frequently CB-associated gene locus in HeLa cells [30]. Strikingly, using the U1 snRNA gene array as a bait region for subsequent circular chromatin conformation capture followed by deep sequencing (4C-seq) gene pair mapping experiments, we documented an overall topological rearrangement of chromosome 1 around the CB in human cancer cells. Thus, the CB forms and maintains a number of intra- and interchromosomal gene clusters in its proximity, especially between highly expressed major and minor spliceosomal snRNA genes and clustered replication-dependent histone genes. We suggest that this CB-mediated chromosome 1 rearrangement, which was not observed in CB-depleted cancer cells [30], resembles a chromosomal rosette with the CB helping to anchor the base of several large but mostly independent loops and results in a close physical association between the 5’ and 3’ ends of chromosome 1. The CB also forms a number of nonrandom interchromosomal gene clusters between major spliceosomal snRNA and intron-encoded snoRNA gene loci [30,40]. As such, the CB acts as an organizational center that targets specific genomic regions to its periphery for regulation of gene expression and expedited RNA processing.
Gene clusters near Cajal bodies are RNA-dependent
How might these higher-order gene clusters form around NBs, and the CB in particular? A clue can be found in the localization of most NBs, as they are predominantly situated in the interchromatin space at the periphery of CTs where multiple chromosomes frequently intermingle [65]. Transcriptional activation of large ubiquitously expressed gene arrays, such as the U1 and U2 snRNA arrays [64], triggers the nucleation and formation of a CB. Subsequently, other highly expressed U snRNA genes, which are positioned at the periphery of their host CT, or even looped out of the domain, associate with the growing CB. It is theorized that active snRNA genes stick to the CB periphery via nascent RNA transcripts that act as a molecular Velcro (Figures 3 and 4) [5, 22, 23, 63, 66–68]. Stabilization of stochastic gene pairing events with the CB ultimately results in the formation of long-distance intra- and interchromosomal gene clusters around the CB. Multicolor high-resolution live-cell imaging studies will be required to visualize and analyze the movement of several snRNA genes prior to and following CB association.
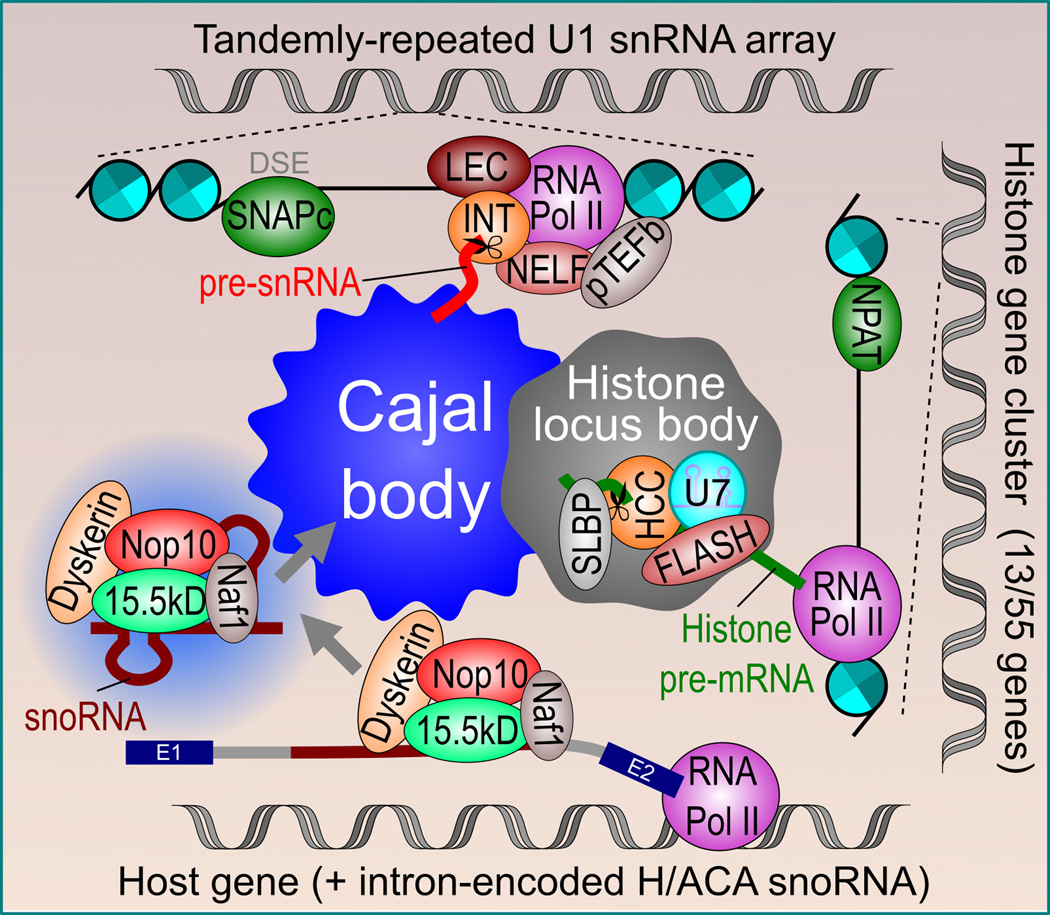
Figure 4.
Cajal body association with target sn/snoRNA and histone genes. CB nucleation and formation is most likely to occur at highly transcriptionally active snRNA gene arrays, such as the tandemly-repeated U1 snRNA gene array on chromosome 1. Transcription of snRNA genes is activated by the binding of the SNAPc transcription factor to the distal sequence element (DSE). Transcription is regulated by the Little Elongation Complex (LEC) and Integrator complex (INT), which is responsible for snRNA 3’ end trimming, as well as accessory factors such as NELF and pTEFb. Intron-encoded snoRNAs are co-transcriptionally isolated from host gene introns by the binding of CB-enriched core components of box H/ACA snoRNAs (e.g. 15.5 kD, Nop10 and dyskerin), which may physically target these regions to the CB periphery. At this point, coilin may bind to excised snoRNPs and help target these nascent snoRNPs to nearby CBs. Finally, through its physical association with the HLB, the major and minor histone gene clusters on chromosomes 6 and 1, respectively, are targeted to the CB periphery. Following transcriptional activation by NPAT, histone pre-mRNAs are processed by U7 snRNP, FLASH and the histone cleavage complex (HCC) within the HLB before binding to the stem-loop binding protein (SLBP).
A similar series of events is likely to result in the association of CBs with intron-encoded sno/scaRNAs, which constitute 90% of all human sn/sno/scaRNAs. Here, active sno/scaRNA-containing host gene regions relocalize to the interchromosomal space upon transcriptional activation and the isolation of sno/scaRNAs from host gene introns is coupled to RNA splicing (Figure 4) [69]. The co-transcriptional assembly of sno/scaRNPs on nascent sno/scaRNA coding regions is initiated by the association of several proteins with the snoRNA that are core components of box H/ACA snoRNPs and are enriched in CBs. The snoRNA is immediately excised from the host transcript intron after splicing is completed [70]. One of these proteins, dyskerin, has previously been confirmed to be capable of nucleating a de novo CB on an engineered chromatin site [71]. It is also likely that the CB structural protein coilin binds at a relatively early stage in this process [28] and potentially helps to guide these genes towards existing CBs. However, it is unclear when 3’ end trimming of box H/ACA snoRNA precursors by poly(A)-specific ribonuclease occurs, although this protein is also enriched in CBs [72]. We believe this unique processing of intron-encoded snoRNAs and snoRNP pre-assembly may result in the co-transcriptional formation of micro- or mini-CBs [68, 73] at intron-encoded regions that ultimately fuse with larger CBs centered around major promoter-driven spliceosomal snRNA genes.
After formation at highly expressed target gene loci, CBs, and NBs in general, appear to attract other target genes that have localized to the interchromatin space and stably retain these regions in their periphery [30, 37, 39, 40, 60]. This ultimately influences global chromatin topology by forming nonrandom long-range gene pairing events. Further developments in microscopy, including more sensitive super-resolution microscopes with advanced image analyses, the synthesis of fluorescent dyes with improved spectral overlap and coverage, as well as automated high-content image acquisition techniques will improve our understanding of NB-dependent genome organization.
Nuclear bodies are beneficial for proximal target gene transcription and processing
A benefit of genomic clustering is direct spatial and temporal coordination of gene activity. NBs are thought to be optimized microenvironments that expedite and coordinate the transcription and highly efficient processing of large numbers of target RNA transcripts, including up to 106 copies of U1 and U2 snRNAs in HeLa cells [74]. Transcription at CB target genes, including the snRNA genes, is highly similar to most RNA pol II-transcribed genes. However, there are several crucial differences. Transcription at snRNA genes, which are short, relatively depleted of nucleosomes [75] and lack several canonical promoter features (e.g. TATA boxes) [76], is activated by a unique DNA binding transcription factor, known as the snRNA activating protein complex (SNAPc) [77]. RNA pol II, as well as several other protein complexes, is then recruited to perform efficient snRNA production. The first of these regulatory complexes is known as the Little Elongation Complex (LEC) [78] and the second is known as the Integrator complex [79, 80]. The LEC promotes transcription through the gene body, which continues beyond the end of the gene into the neighboring intergenic DNA. Transcription is paused by CTCF- and Integrator-dependent recruitment of negative elongation factor (NELF) and positive elongation factor b (P-TEFb) [81, 82]. This initiates phosphorylation of the C-terminal domain of RNA pol II at Serine-2 by P-TEFb and leads to the co-transcriptional recognition of the 3’ end processing element (3’ box). The Integrator complex is recruited and cleaves the nascent pre-snRNA at a site several nucleotides away from the 3’ box to generate a mature snRNA transcript, which, like histone mRNAs, is not polyadenylated [79,80]. Thus, snRNA genes display many unique characteristics and transcriptional features compared to other highly expressed genes.
CBs are highly dynamic but structurally stable structures with a continuous exchange of components into and out of the domain [83, 84]. Interestingly, components of both the SNAPc and LEC complexes have been reported to be enriched within the CB [25, 85], potentially indicating a strong link between snRNA gene transcription and CBs. By clustering of target gene loci that span large genomic distances near to CBs, this may allow a decondensed chromatin fiber harboring these genes to loop outside of the host chromosome and into an environment that is permissive to transcription, increasing their transcriptional potential. This enables quick and efficient reloading of SNAPc, LEC and Integrator components onto snRNA genes for immediate re-initiation of transcription and co-transcriptional 3’ end RNA processing. Depletion of the essential CB components TCAB1/WRAP53 or USPL1, which both lead to complete CB disassembly, decreases nascent precursor U1 and -U2 pre-snRNA levels [30], whereas coilin depletion, which does not completely abolish CBs but results in residual CB-like substructures, increases pre-U snRNA levels [86]. This provides evidence of a direct function for the CB in regulating snRNA transcription. In normal diploid cells that lack CBs, it is possible that lower snRNA levels are processed by nucleoplasmic levels of the SNAPc, LEC and Integrator complexes at slower rates without requiring the formation of a specialized microenvironment. A lack of CBs in diploid cells may also lead to random and unsynchronized production of mature snRNPs. We hypothesize that spatial clustering of snRNA genes near to the CB enables co-regulation of snRNA genes by efficient reloading of SNAPc, LEC and Integrator components. This may coordinate and enhance both snRNA transcription and 3’-end RNA processing.
Cajal bodies concentrate limited factors to clear unprocessed nascent RNA
CBs are also frequently physically associated with the HLB, a NB that is involved in histone pre-mRNA processing (Figures 2 & 3) [30, 35]. The HLB provides an optimized microenvironment for accelerated processing of the canonical 3’ region of replication-dependent histone genes [36, 87]. HLBs may benefit from a direct supply of U2 and U7 snRNPs from the CB, which are implicated in histone processing but enriched in CBs [30,88,89]. CB-dependent targeting of specific gene loci, including HLB-regulated replication-dependent histone gene clusters [62], to the highly transcriptionally active interchromatin space may also be beneficial for HLB function. However, the CB does not directly influence histone gene activity and is likely to only indirectly influence transcription at histone gene clusters [90]. An indication of the importance of NBs to gene transcription can be found in a recently described role for the HLB in histone pre-mRNA processing [87]. These data showed that HLBs not only concentrate essential limited factors for histone pre-mRNA processing but that the absence of HLBs results in specific transcriptional disruption and a small but significant level of spurious polyadenylation of unprocessed histone pre-mRNA. PML NBs and nucleoli have also been suggested to be important for transcriptional activation and repression of physically-associated gene loci, but the exact role of the PML NB in this process is still unclear [39,91]. These studies indicate that the formation of specialized nuclear microenvironments allows cells to produce large quantities of specific RNAs through enrichment of limited but essential protein factors.
The proximity of the CB to specific genomic regions and the enrichment of snRNA processing factors within the domain is a strong indicator of the role of the CB in immediate snRNA maturation. Considering the ability of specific snRNAs to induce CB assembly at an artificially engineered chromatin position, CBs may assemble in response to high local volumes of unprocessed or stalled RNA transcript [5,66,67]. This may be a common feature of several NBs and beneficial for regulating genome function [59,92,93]. Substantial advances are being made in our understanding of transcriptional dynamics [94] but it is still not fully elucidated. It will be vital to investigate the kinetics of target gene transcriptional bursting and RNA processing near to and distant from NBs and how this compares to other highly expressed but intron-containing genes. Unfortunately, sequence modifications to allow the specific binding of fluorescently-tagged proteins [95] to the distinct secondary structure of snRNAs are likely to be disruptive to their function. Therefore, it is difficult to acquire live-cell microscopic observations of sn/sno/scaRNA transcriptional events. It is still unclear what effect this may have on protein-RNA interactions that lead to CB assembly or physical gene association.
Cajal bodies help shape the transcriptome through snRNP availability
As structures that directly influence the transcription and processing of specific groups of target genes, an important aspect of NB biology is their diverse effects upon downstream molecular pathways. As an example, we will describe the effect of CB function upon RNA splicing fidelity and speculate upon the multiple layers of splicing regulation that may be influenced by CB function (Figure 5).
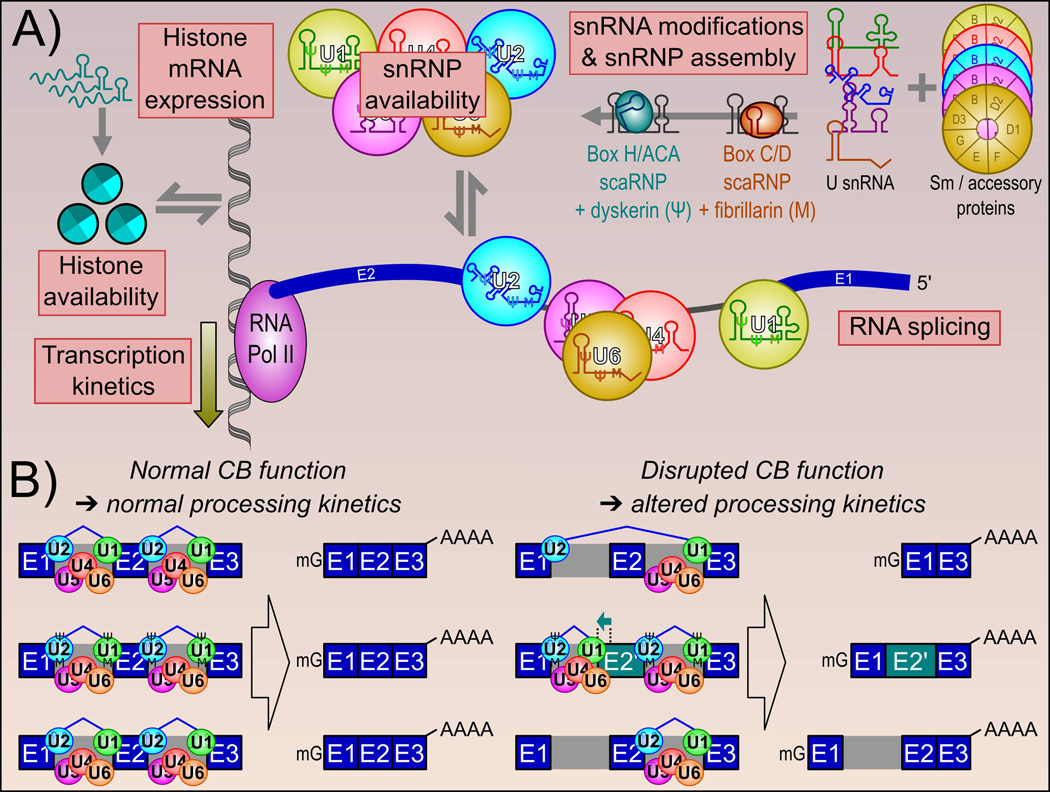
Figure 5.
Cajal bodies influence RNA splicing kinetics. A: RNA splicing by the spliceosome is dependent on many factors that are influenced by CB function. These include snRNP availability, as the CB enhances snRNA gene transcription and snRNP assembly steps, and snRNP base modifications (guided by box H/ACA and box C/D scaRNPs). Through its physical association with the HLB, the loss of CBs has been shown to reduce histone mRNA availability. We have also observed a decrease in RNA Pol II C-terminal domain inclusion following CB disassembly. Both of these changes are suggested to decrease RNA Pol II transcription rate. B: Altogether, these results show that CB-mediated functions influence RNA splicing, RNA splice site selection and splicing fidelity.
The production of mature mRNAs involves selective removal of introns from primary transcripts and splicing of adjacent exons together, with multiple possible alternative exon combinations as the outcome. Alternative splicing occurs in >90% of genes and is a major contributor to protein diversity [96], enabling the production of multiple proteins from a single gene locus. Like transcriptional regulation, splicing regulation can be mediated by cis-acting enhancers and silencers that bind to recognition motifs. This has been the basis for the search for a ‘splicing code’ to predict splicing patterns based on sequence motifs [97]. It is clear, however, that splicing is regulated by a much more diverse array of processes beyond splicing enhancers and silencers.
As mentioned above, CBs are important for the assembly of core components of the spliceosome, the macromolecular complex responsible for intron removal and splicing. Introns in the human genome are incredibly diverse, spanning five orders of magnitude in length, with degenerate recognition motifs. The spliceosome is similarly complex and flexible to accommodate this diversity, integrating multiple small RNAs and proteins into a catalytic unit. It has been established that CBs are essential players in snRNA processing and snRNP assembly to enable spliceosome function and splicing fidelity. Greater insight into splicing and the broader area of RNA processing mechanisms are coming increasingly into focus, as next-generation sequencing methods are being adapted to address them [98]. CBs, with a central role in the biogenesis of critical spliceosome components, affect splicing and other RNA processing through multiple mechanisms.
Cajal bodies may influence splicing kinetics through diverse pathways
Tissues and cell types differ in their splicing load, in terms of number of introns to remove and the complexity of alternative forms. For example, testis, neuronal and hepatic tissues have a great deal more tissue-specific splicing than does muscle tissue [99]. The complement of expressed genes can be highly variable in terms of the number of introns [100], with intron number ranging from zero to 300 [101]. To splice transcripts effectively, cells must disassemble and reassemble spliceosomes after each splicing event, and produce spliceosomal components in stoichiometrically compatible amounts [102]. Additionally, CBs perform quality control on snRNPs through the U4/U6 di-snRNP recycling factor SART3 [33]. CBs may serve as “boosters” to aid the cell in spliceosomal production to accommodate the additional load required in specific tissues.
One example is in acute forms of retinal degenerative disease, where a mutation that affects U4/U6 di-snRNP interaction in the core spliceosome machinery leads to defects not seen in other tissues [103]. The mutation leads to a defect in splicing capacity, which produces a more detrimental effect when there is a higher demand for splicing. Expression of housekeeping genes and spliceosomal snRNAs are very high in retina compared to other human tissues, with extensive transcript diversity, indicating an elevated splicing load [104,105]. This suggests that reduced splicing activity is more detrimental for regenerative tissues that require rapid cell proliferation and transcriptional diversity [102].
Several studies have observed a correlation between spliceosomal component availability and aberrant splicing. Reduction of snRNA expression (by depletion of USPL1, which influences snRNA transcription) leads to reduced snRNP availability and altered splicing patterns, while not altering general transcription [106]. Experiments in SMN-deficient mice showed that an induced decrease in snRNP production lead to widespread alternative splicing changes, particularly in genes with many introns [107]. This is in agreement with our studies in CB deficient HeLa cells, in which we observed an over-representation of aberrant splicing in genes producing many isoforms [30].
How might differences in spliceosomal components lead to different splice forms? One possible route is through alteration of co-transcriptional splicing kinetics. Transcription and splicing are tightly coupled, and the majority of splicing occurs co-transcriptionally [108, 109]. Perturbing the availability of snRNPs or the rate of snRNP assembly can alter the kinetic balance between competing splice sites, leading to the production of alternative isoforms [108, 110] (Figure 5B). A more rapid traversal of polymerase through the gene may lead to removal of introns with strong donor and acceptor sites, while a slower rate or pause may provide additional time for the spliceosome to use weaker splice sites [110] or to allow splicing factors affecting splice site choice to bind.
The CB may also influence splicing via kinetic mechanisms indirectly, for example through the expression of histones (Figures 4 and 5A). We recently demonstrated the correlation between histone gene expression and CBs in HeLa cells [30], along with aberrant splicing upon CB depletion in SNRPE, which encodes an essential component of the heptameric Sm ring. This is required for the stability of spliceosomal snRNPs as well as U7 snRNP, which specifically regulates histone 3’-end pre-mRNA processing [111]. Histone availability impacts co-transcriptional splicing through Pol II kinetics [112]. It is highly likely that, in HeLa cells at least, the HLB may be reliant on a fully functional CB for optimal productivity.
In contrast to changes in splice site choice, a defect in general intron removal could be related to spliceosomal component availability. A general intron retention (IR) increase can reduce total proteomic output via nonsense-mediated decay (NMD), or increase the diversity of intron-retained functional transcripts. Indeed, global IR increases have been seen as general phenomenon of cancers [113], and as an expression regulation strategy in differentiation [114]. An increase in general IR correlated with diminished snRNP availability in HeLa cells [115, 116]. However, a general IR effect across all introns was not observed in our CB disassembly experiments [30].
Altered RNA processing by Cajal bodies influences splicing kinetics
Within CBs, and to a lesser extent in the nucleoplasm, snRNAs undergo 2’-O-methylation and pseudouridinylation at specific nucleotides, guided by specific scaRNPs [117]. These modifications are important for RNA-RNA interactions in the spliceosome and for splicing fidelity. The extent and positions of these modifications can also be modulated in response to cellular stress [118]. This raises the possibility that spliceosomal function can be regulated by differential snRNA nucleotide modification within CBs.
One of the most widely studied RNA modifications is the methylation of adenosine (m6A), which was first identified decades ago [119]. This modification has been observed in snRNA [120, 121], and has been shown to have multiple RNA processing regulatory roles, including splicing, translational control, and modulating RNA stability [119]. A role for this modification in regulating circadian rhythm was recently discovered, where global reduction of RNA methylation lead to slower rates of RNA processing [122]. Although recent progress has been made in understanding how RNA epigenetics affects transcriptome dynamics [123], we still have a great deal to learn.
Spliceosomal snRNAs are just one class of small RNAs relevant to CBs. As a group, the importance of small RNAs to shaping the transcriptome is not completely known. Beyond the spliceosomal RNAs, additional roles for small RNAs in myriad processes from transcript stability to UTR processing have been observed [124]. Recent studies have identified hundreds of snRNAs, snoRNAs, and scaRNAs that traffic through CBs and interact with coilin [28]. Of note, we witnessed a genome-wide decrease in the expression of many intron-encoded small RNAs with no effect upon host gene expression following disassembly of CBs [30]. This may be a result of NMD-dependent decoupling of small RNA and host gene expression levels [125]. Spliceosomal sno/scaRNAs compete with other small RNAs for binding during the splicing reaction. This suggests CB regulation of snRNA production can alter competitive balances with other small RNAs to affect splicing. For example, a snoRNA was found that competes with U1 snRNA for a binding site in RNA, leading to mis-splicing for the E2F7 transcription factor [126]. As such, the CB is an important regulator of small RNA expression and function (Figures 2, 3 and 4).
Independent of alternative splicing, one way that transcripts from the same parental RNA and with the same splicing pattern may differ is in the location of the polyadenylation site. The length of the 3’-UTR can have significant impacts on the stability of the transcript, and hence transport and translation downstream. Widespread changes in 3’-UTR length have been observed in multiple cancer types and linked to cellular senescence [127]. The U1 snRNP is critical for alternative polyadenylation [128,129], and changes in U1 snRNA availability can lead to changes in U1 snRNP 3’-end processing [130]. This raises the possibility that snRNAs and CBs can influence polyadenylation and 3’-UTR lengths.
Conclusion
We suggest that the CB, a prototypical NB, is capable of influencing the nonrandom spatial organization of the human genome and the transcriptome. CBs are a facilitator of multiple parallel processes, serving as reaction centers and genome architecture organizers to influence transcriptional regulation and base modification of sn/snoRNAs, spliceosomal component assembly, histone gene expression, and polyadenylation. These processes likely vary significantly between cell types, aiding in the formation of transcriptome diversity. However, CBs, and NBs in general, influence many more processes than transcription and future experiments across a range of cell types with different transcriptional profiles will be critical to understand how this diversity is generated. Improvements to next-generation sequencing tools for the detection and characterization of small RNAs indicates that the future holds a great deal of promise for dissecting the multiple roles of CBs in RNA transcription, processing [131, 132], RNA base modification [133], and UTR length variation of specific genes [134].
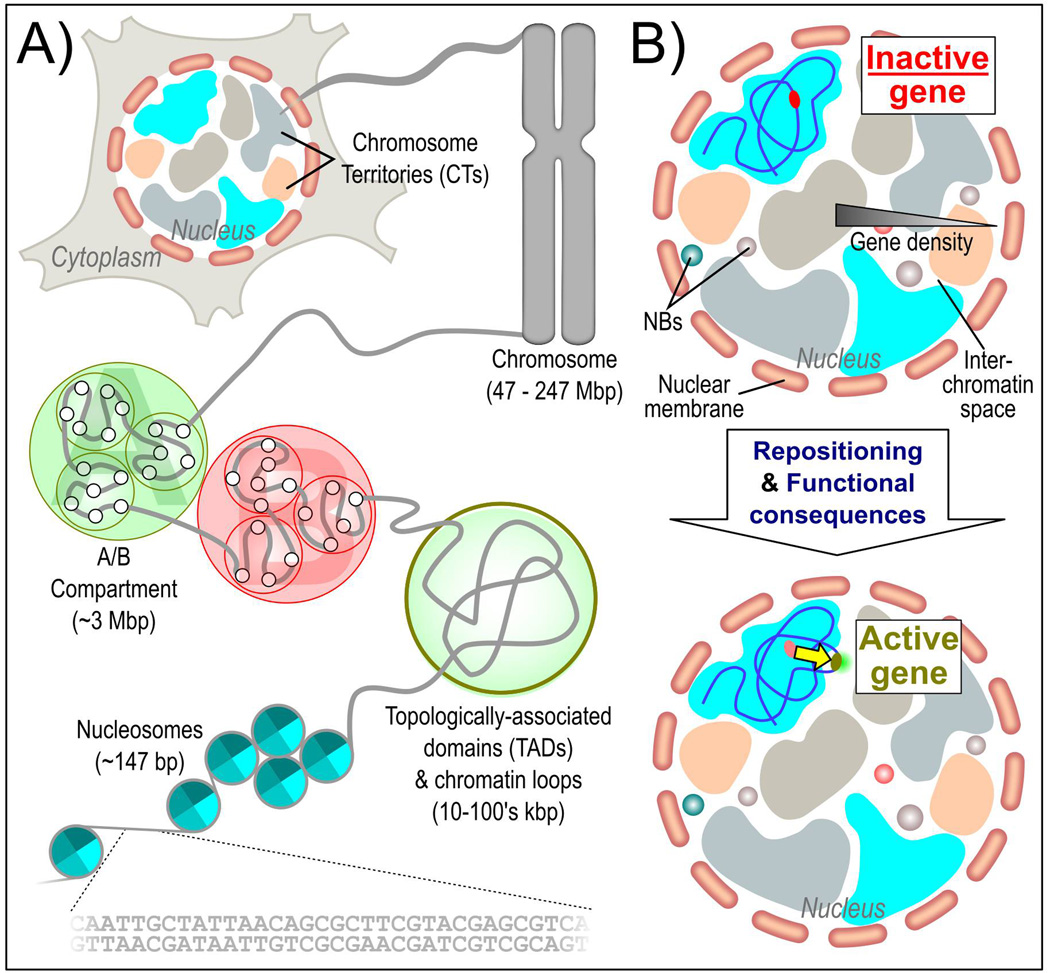
Figure 1.
Genome organization and function. A: The human genome is hierarchically organized to maintain function. Individual chromosomes, which are segregated into chromosome territories, are composed of a number of sub-compartments, loops and nucleosomes. B: Gene and chromosome position is related to activity. In general, gene dense chromosomes are more internal and gene-poor chromosomes are more peripheral within the nucleus. Also, inactive genes are frequently observed within the interior of their host chromosome. Upon activation, genes relocate to the periphery of the chromosome or looped out into the interchromatin space, where they are exposed to high local concentrations of transcriptional machinery and various nuclear bodies.
Acknowledgments
The authors are supported by a National Institutes of Health, National Institute of General Medical Sciences grant (R01GM090156, awarded to M.D.), the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (G.L.H.) and the National Institute on Aging (M-H.S.). We thank Dr. Eric Wagner and Dr. Karen Meaburn for stimulating conversations. Super-resolution structured illumination microscopy (SIM) images were acquired at the NCI Core Fluorescence Imaging Facility in Building 41, National Institutes of Health (Bethesda, MD) with the assistance of Dr. Tatiana Karpova.
Abbreviations
- CB
Cajal body
- CT
chromosome territory
- HLB
histone locus body
- LEC
little elongation complex
- (PML) NB
(promyelocytic) nuclear body
- NMD
nonsense-mediated decay
- scaRNA
small CB-associated RNA
- SNAPc
snRNA activating protein complex
- snRNA
small nuclear RNA
- snoRNA
small nucleolar RNA
- snRNP
small nuclear ribonucleoprotein
Footnotes
The authors report no conflict of interest.
References
- 1.Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol. 2012;24:415–422. doi: 10.1016/j.ceb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dundr M. Seed and grow: a two-step model for nuclear body biogenesis. J Cell Biol. 2011;193:605–606. doi: 10.1083/jcb.201104087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chujo T, Yamazaki T, Hirose T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Biophys Acta. 2016;1859:139–146. doi: 10.1016/j.bbagrm.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- 8.Young PJ, Le TT, Dunckley M, Nguyen TM, et al. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res. 2001;265:252–261. doi: 10.1006/excr.2001.5186. [DOI] [PubMed] [Google Scholar]
- 9.Pena E, Berciano MT, Fernandez R, Ojeda JL, et al. Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J Comp Neurol. 2001;430:250–263. doi: 10.1002/1096-9861(20010205)430:2<250::aid-cne1029>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 10.Hearst SM, Gilder AS, Negi SS, Davis MD, et al. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci. 2009;122:1872–1881. doi: 10.1242/jcs.044040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams RRE. Transcription and the territory: the ins and outs of gene positioning. Trends Genet. 2013;19:298–302. doi: 10.1016/S0168-9525(03)00109-4. [DOI] [PubMed] [Google Scholar]
- 13.Cho EJ, Kim JS. Crowding effects on the formation and maintenance of nuclear bodies: insights from molecular-dynamics simulations of simple spherical model particles. Biophys J. 2012;103:424–433. doi: 10.1016/j.bpj.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan C, Saurabh S, Bruchez MP, Schwartz R, et al. Molecular crowding shapes gene expression in synthetic cellular nanosystems. Nat Nanotechnol. 2013;8:602–608. doi: 10.1038/nnano.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu L, Brangwynne CP. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol. 2015;34:23–30. doi: 10.1016/j.ceb.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis--evidence that the coiled body is a kinetic nuclear structure. J Cell Biol. 1993;120:841–852. doi: 10.1083/jcb.120.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol. 1992;117:1–14. doi: 10.1083/jcb.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira JA, Carmo-Fonseca M, Lamond AI. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol. 1994;126:11–23. doi: 10.1083/jcb.126.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemm I, Girard C, Kuhn AN, Watkins NJ, et al. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novotny I, Blazikova M, Stanek D, Herman P, et al. In vivo kinetics of U4/U6.U5 tri-snRNP formation in Cajal bodies. Mol Biol Cell. 2011;22:513–523. doi: 10.1091/mbc.E10-07-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanek D, Neugebauer KM. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol. 2004;166:1015–1025. doi: 10.1083/jcb.200405160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol. 1999;9:126–135. doi: 10.1016/s0960-9822(99)80066-9. [DOI] [PubMed] [Google Scholar]
- 23.Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol. 2001;154:499–509. doi: 10.1083/jcb.200105084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schul W, van Driel R, de Jong L. Coiled bodies and U2 snRNA genes adjacent to coiled bodies are enriched in factors required for snRNA transcription. Mol Biol Cell. 1998;9:1025–1036. doi: 10.1091/mbc.9.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem. 1995;59:473–485. doi: 10.1002/jcb.240590408. [DOI] [PubMed] [Google Scholar]
- 27.Smith KP, Lawrence JB. Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol Biol Cell. 2000;11:2987–2998. doi: 10.1091/mbc.11.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machyna M, Kehr S, Straube K, Kappei D, et al. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell. 2014;56:389–399. doi: 10.1016/j.molcel.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Kiss T, Fayet E, Jady BE, Richard P, et al. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Sawyer IA, Sung MH, Sturgill D, et al. Cajal bodies are linked to genome conformation. Nat Commun. 2016;7:10966. doi: 10.1038/ncomms10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raska I, Andrade LE, Ochs RL, Chan EK, et al. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res. 1991;195:27–37. doi: 10.1016/0014-4827(91)90496-h. [DOI] [PubMed] [Google Scholar]
- 32.Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol. 1994;127:1505–1514. doi: 10.1083/jcb.127.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novotný I, Malinová A, Stejskalová E, Matějů D, et al. SART3-Dependent Accumulation of Incomplete Spliceosomal snRNPs in Cajal Bodies. Cell Rep. 2015;10:429–440. doi: 10.1016/j.celrep.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 34.Stanek D, Rader SD, Klingauf M, Neugebauer KM. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol. 2003;160:505–516. doi: 10.1083/jcb.200210087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawyer IA, Dundr M. Nuclear bodies: Built to boost. J Cell Biol. 2016;213:509–511. doi: 10.1083/jcb.201605049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kalmarova M, Smirnov E, Masata M, Koberna K, et al. Positioning of NORs and NOR-bearing chromosomes in relation to nucleoli. J Struct Biol. 2007;160:49–56. doi: 10.1016/j.jsb.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salzler HR, Tatomer DC, Malek PY, McDaniel SL, et al. A sequence in the Drosophila H3-H4 Promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Dev Cell. 2013;24:623–634. doi: 10.1016/j.devcel.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ching RW, Ahmed K, Boutros PC, Penn LZ, et al. Identifying gene locus associations with promyelocytic leukemia nuclear bodies using immuno-TRAP. J Cell Biol. 2013;201:325–335. doi: 10.1083/jcb.201211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawyer IA, Shevtsov SP, Dundr M. Spectral imaging to visualize higher-order genomic organization. Nucleus (Austin, Tex) 2016;7 doi: 10.1080/19491034.2016.1187344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagano T, Lubling Y, Yaffe E, Wingett SW, et al. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protocols. 2015;10:1986–2003. doi: 10.1038/nprot.2015.127. [DOI] [PubMed] [Google Scholar]
- 43.Mourad R, Cuvier O. Predicting the spatial organization of chromosomes using epigenetic data. Genome Biol. 2015;16:1–3. doi: 10.1186/s13059-015-0752-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramani V, Shendure J, Duan Z. Understanding Spatial Genome Organization: Methods and Insights. Genomics Proteomics Bioinformatics. 2016;14:7–20. doi: 10.1016/j.gpb.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leshner M, Devine M, Roloff GW, True LD, et al. Locus-specific gene repositioning in prostate cancer. Mol Biol Cell. 2016;27:236–246. doi: 10.1091/mbc.E15-05-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marella NV, Bhattacharya S, Mukherjee L, Xu J, et al. Cell type specific chromosome territory organization in the interphase nucleus of normal and cancer cells. J Cell Physiol. 2009;221:130–138. doi: 10.1002/jcp.21836. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen HQ, Bosco G. Gene positioning effects on expression in eukaryotes. Annu Rev Genet. 2015;49:627–646. doi: 10.1146/annurev-genet-112414-055008. [DOI] [PubMed] [Google Scholar]
- 49.Aten JA, Kanaar R. Chromosomal organization: mingling with the neighbors. PLoS Biol. 2006;4:e155. doi: 10.1371/journal.pbio.0040155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hakim O, Sung M-H, Hager GL. 3D shortcuts to gene regulation. Curr Opin Cell Biol. 2010;22:305–313. doi: 10.1016/j.ceb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams A, Spilianakis CG, Flavell RA. Interchromosomal association and gene regulation in trans. Trends Genet. 2010;26:188–197. doi: 10.1016/j.tig.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tjong H, Li W, Kalhor R, Dai C, et al. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proc Natl Acad Sci USA. 2016;113:E1663–E1672. doi: 10.1073/pnas.1512577113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 54.Park SK, Xiang Y, Feng X, Garrard WT. Pronounced cohabitation of active immunoglobulin genes from three different chromosomes in transcription factories during maximal antibody synthesis. Genes Dev. 2014;28:1159–1164. doi: 10.1101/gad.237479.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hewitt SL, Farmer D, Marszalek K, Cadera E, et al. Association between the Igk and Igh immunoglobulin loci mediated by the 3' Igk enhancer induces 'decontraction' of the Igh locus in pre-B cells. Nat Immunol. 2008;9:396–404. doi: 10.1038/ni1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edelman LB, Fraser P. Transcription factories: genetic programming in three dimensions. Curr Opin Genet Dev. 2012;22:110–114. doi: 10.1016/j.gde.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 57.Eskiw CH, Fraser P. Ultrastructural study of transcription factories in mouse erythroblasts. J Cell Sci. 2011;124:3676–3683. doi: 10.1242/jcs.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rieder D, Trajanoski Z, McNally JG. Transcription factories. Front Genet. 2012;3:221. doi: 10.3389/fgene.2012.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr Biol. 2016;26:277–285. doi: 10.1016/j.cub.2015.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rieder D, Ploner C, Krogsdam AM, Stocker G, et al. Co-expressed genes prepositioned in spatial neighborhoods stochastically associate with SC35 speckles and RNA polymerase II factories. Cell Mol Life Sci. 2014;71:1741–1759. doi: 10.1007/s00018-013-1465-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 62.Romeo V, Schumperli D. Cycling in the nucleus: regulation of RNA 3' processing and nuclear organization of replication-dependent histone genes. Curr Opin Cell Biol. 2016;40:23–31. doi: 10.1016/j.ceb.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 63.Dundr M, Ospina JK, Sung MH, John S, et al. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sawyer IA, Hager GL, Dundr M. Specific genomic cues regulate Cajal body assembly. RNA Biol. 2016 doi: 10.1080/15476286.2016.1243648. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- 66.Dundr M. Nucleation of nuclear bodies. Methods Mol Biol (Clifton, NJ) 2013;1042:351–364. doi: 10.1007/978-1-62703-526-2_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 68.Neugebauer KM. RNA: master or servant? RNA (New York, NY) 2015;21:701–702. doi: 10.1261/rna.051250.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richard P, Kiss T. Integrating snoRNP assembly with mRNA biogenesis. EMBO Rep. 2006;7:590–592. doi: 10.1038/sj.embor.7400715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darzacq X, Kittur N, Roy S, Shav-Tal Y, et al. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 72.Berndt H, Harnisch C, Rammelt C, Stohr N, et al. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA (New York, NY) 2012;18:958–972. doi: 10.1261/rna.032292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marz M, Mosig A, Stadler BM, Stadler PF. U7 snRNAs: a computational survey. Genomics Proteomics Bioinformatics. 2007;5:187–195. doi: 10.1016/S1672-0229(08)60006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O'Reilly D, Kuznetsova OV, Laitem C, Zaborowska J, et al. Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Res. 2014;42:264–275. doi: 10.1093/nar/gkt892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Egloff S, O'Reilly D, Murphy S. Expression of human snRNA genes from beginning to end. Biochem Soc Trans. 2008;36:590–594. doi: 10.1042/BST0360590. [DOI] [PubMed] [Google Scholar]
- 77.Hung KH, Stumph WE. Regulation of snRNA gene expression by the Drosophila melanogaster small nuclear RNA activating protein complex (DmSNAPc) Crit Rev Biochem Mol Biol. 2011;46:11–26. doi: 10.3109/10409238.2010.518136. [DOI] [PubMed] [Google Scholar]
- 78.Takahashi H, Takigawa I, Watanabe M, Anwar D, et al. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat Commun. 2015;6:5941. doi: 10.1038/ncomms6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen J, Wagner EJ. snRNA 3' end formation: the dawn of the Integrator complex. Biochem Soc Trans. 2010;38:1082–1087. doi: 10.1042/BST0381082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takata H, Nishijima H, Maeshima K, Shibahara K. The integrator complex is required for integrity of Cajal bodies. J Cell Sci. 2012;125:166–175. doi: 10.1242/jcs.090837. [DOI] [PubMed] [Google Scholar]
- 81.Stadelmayer B, Micas G, Gamot A, Martin P, et al. Integrator complex regulates NELF-mediated RNA polymerase II pause/release and processivity at coding genes. Nat Commun. 2014;5:5531. doi: 10.1038/ncomms6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laitem C, Zaborowska J, Tellier M, Yamaguchi Y, et al. CTCF regulates NELF, DSIF and P-TEFb recruitment during transcription. Transcription. 2015;6:79–90. doi: 10.1080/21541264.2015.1095269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dundr M, Hebert MD, Karpova TS, Stanek D, et al. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Misteli T. Physiological importance of RNA and protein mobility in the cell nucleus. Histochemistry and cell biology. 2008;129:5–11. doi: 10.1007/s00418-007-0355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu D, Smith Edwin R, Garruss Alexander S, Mohaghegh N, et al. The little elongation complex functions at initiation and elongation phases of snRNA gene transcription. Mol Cell. 2013;51:493–505. doi: 10.1016/j.molcel.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Broome HJ, Hebert MD. In vitro RNase and nucleic acid binding activities implicate Coilin in U snRNA processing. PLoS One. 2012;7:e36300. doi: 10.1371/journal.pone.0036300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tatomer DC, Terzo E, Curry KP, Salzler H, et al. Concentrating pre-mRNA processing factors in the histone locus body facilitates efficient histone mRNA biogenesis. J Cell Biol. 2016;213:557–570. doi: 10.1083/jcb.201504043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Friend K, Lovejoy AF, Steitz JA. U2 snRNP binds intronless histone pre-mRNAs to facilitate U7-snRNP-dependent 3' end formation. Mol Cell. 2007;28:240–252. doi: 10.1016/j.molcel.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci USA. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shopland LS, Byron M, Stein JL, Lian JB, et al. Replication-dependent histone gene expression is related to Cajal body (CB) association but does not require sustained CB contact. Mol Biol Cell. 2001;12:565–576. doi: 10.1091/mbc.12.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Olson MOJ, Dundr M. eLS. John Wiley & Sons, Ltd; 2001. Nucleolus: Structure and Function. [Google Scholar]
- 92.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lenstra TL, Rodriguez J, Chen H, Larson DR. Transcription dynamics in living cells. Annu Rev Biophys. 2016;45:25–47. doi: 10.1146/annurev-biophys-062215-010838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weil TT, Parton RM, Davis I. Making the message clear: visualizing mRNA localization. Trends Cell Biol. 2010;20:380–390. doi: 10.1016/j.tcb.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang ET, Sandberg R, Luo S, Khrebtukova I, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barash Y, Calarco JA, Gao W, Pan Q, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 98.de Klerk E, t Hoen PA. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 2015;31:128–139. doi: 10.1016/j.tig.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 99.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heyn P, Kalinka AT, Tomancak P, Neugebauer KM. Introns and gene expression: cellular constraints, transcriptional regulation, and evolutionary consequences. BioEssays. 2015;37:148–154. doi: 10.1002/bies.201400138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gorlova O, Fedorov A, Logothetis C, Amos C, et al. Genes with a large intronic burden show greater evolutionary conservation on the protein level. BMC Evol Biol. 2014;14:50. doi: 10.1186/1471-2148-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Papasaikas P, Valcarcel J. The spliceosome: The ultimate RNA chaperone and sculptor. Trends Biochem Sci. 2016;41:33–45. doi: 10.1016/j.tibs.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 103.Mozaffari-Jovin S, Wandersleben T, Santos KF, Will CL, et al. Novel regulatory principles of the spliceosomal Brr2 RNA helicase and links to retinal disease in humans. RNA Biol. 2014;11:298–312. doi: 10.4161/rna.28353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Farkas MH, Grant GR, White JA, Sousa ME, et al. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hutten S, Chachami G, Winter U, Melchior F, et al. A role for the Cajal-body-associated SUMO isopeptidase USPL1 in snRNA transcription mediated by RNA polymerase II. J Cell Sci. 2014;127:1065–1078. doi: 10.1242/jcs.141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang Z, Lotti F, Dittmar K, Younis I, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Carrillo Oesterreich F, Herzel L, Straube K, Hujer K, et al. Splicing of Nascent RNA Coincides with Intron Exit from RNA Polymerase II. Cell. 2016;165:372–381. doi: 10.1016/j.cell.2016.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 2014;15:163–175. doi: 10.1038/nrg3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shukla S, Kavak E, Gregory M, Imashimizu M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pillai RS, Will CL, Luhrmann R, Schumperli D, et al. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jimeno-Gonzalez S, Payan-Bravo L, Munoz-Cabello AM, Guijo M, et al. Defective histone supply causes changes in RNA polymerase II elongation rate and cotranscriptional pre-mRNA splicing. Proc Natl Acad Sci USA. 2015;112:14840–14845. doi: 10.1073/pnas.1506760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med. 2015;7:45. doi: 10.1186/s13073-015-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wong JJ, Ritchie W, Ebner OA, Selbach M, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 115.Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, et al. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res. 2014;24:1774–1786. doi: 10.1101/gr.177790.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saltzman AL, Kim YK, Pan Q, Fagnani MM, et al. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jady BE, Kiss T. A small nucleolar guide RNA functions both in 2'-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 2001;20:541–551. doi: 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Adachi H, Yu YT. Insight into the mechanisms and functions of spliceosomal snRNA pseudouridylation. World J Biol Chem. 2014;5:398–408. doi: 10.4331/wjbc.v5.i4.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Niu Y, Zhao X, Wu YS, Li MM, et al. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11:8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bringmann P, Luhrmann R. Antibodies specific for N6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett. 1987;213:309–315. doi: 10.1016/0014-5793(87)81512-0. [DOI] [PubMed] [Google Scholar]
- 121.Patel SB, Bellini M. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res. 2008;36:6482–6493. doi: 10.1093/nar/gkn658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fustin JM, Doi M, Yamaguchi Y, Hida H, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 123.Cao G, Li HB, Yin Z, Flavell RA. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6:160003. doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Valadkhan S, Gunawardane LS. Role of small nuclear RNAs in eukaryotic gene expression. Essays Biochem. 2013;54:79–90. doi: 10.1042/bse0540079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lykke-Andersen S, Chen Y, Ardal BR, Lilje B, et al. Human nonsense-mediated RNA decay initiates widely by endonucleolysis and targets snoRNA host genes. Genes Dev. 2014;28:2498–2517. doi: 10.1101/gad.246538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Falaleeva M, Pages A, Matuszek Z, Hidmi S, et al. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci USA. 2016;113:E1625–E1634. doi: 10.1073/pnas.1519292113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gruber AR, Martin G, Keller W, Zavolan M. Means to an end: mechanisms of alternative polyadenylation of messenger RNA precursors. Wiley Interdiscip Reviews RNA. 2014;5:183–196. doi: 10.1002/wrna.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kaida D. The reciprocal regulation between splicing and 3'-end processing. Wiley Interdiscip Rev RNA. 2016;7:499–511. doi: 10.1002/wrna.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Workman E, Veith A, Battle DJ. U1A regulates 3' processing of the survival motor neuron mRNA. Journal Biol Chem. 2014;289:3703–3712. doi: 10.1074/jbc.M113.538264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Locati MD, Terpstra I, de Leeuw WC, Kuzak M, et al. Improving small RNA-seq by using a synthetic spike-in set for size-range quality control together with a set for data normalization. Nucleic Acids Res. 2015;43:e89. doi: 10.1093/nar/gkv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baran-Gale J, Kurtz CL, Erdos MR, Sison C, et al. Addressing Bias in Small RNA Library Preparation for Sequencing: A New Protocol Recovers MicroRNAs that Evade Capture by Current Methods. Frontiers Genet. 2015;6:352. doi: 10.3389/fgene.2015.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 134.Xia Z, Donehower LA, Cooper TA, Neilson JR, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3'-UTR landscape across seven tumour types. Nat Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]