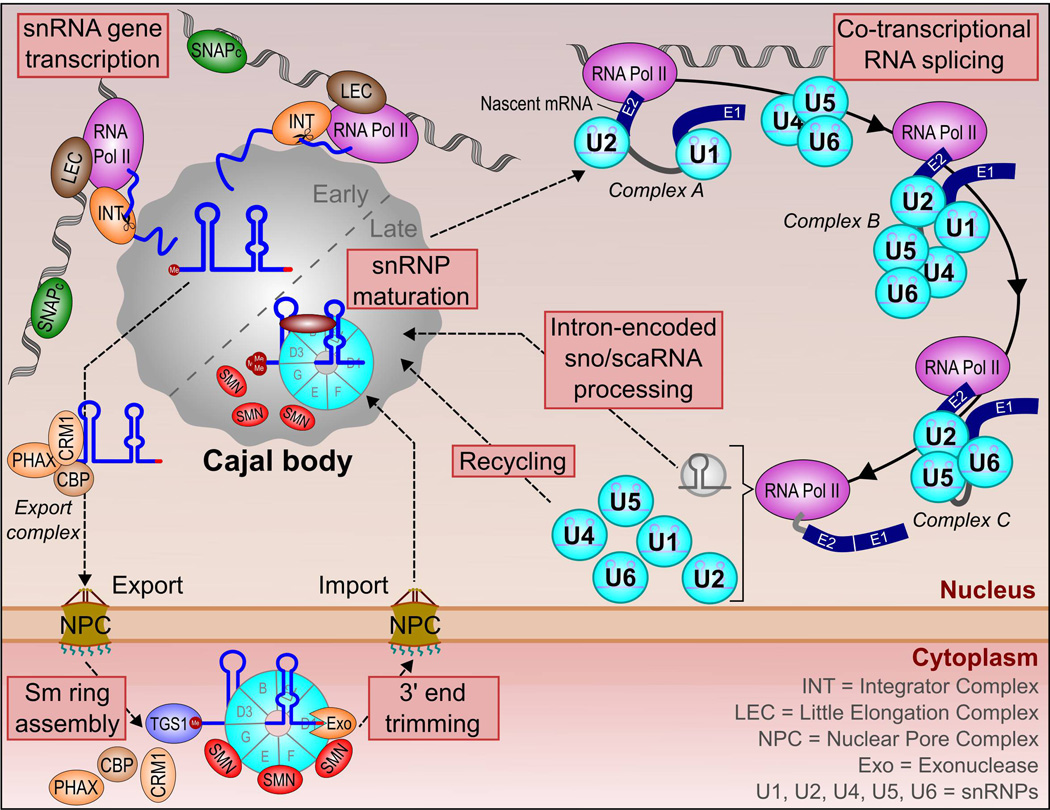
Figure 2.
The relationship between Cajal bodies and splicing. Transcription of snRNA genes frequently occurs at the CB periphery, and is mediated by several protein-based complexes. Nascent pre-snRNA is enriched in the CB domain prior to association with an export complex and subsequent translocation to the cytoplasm. Here, the pre-snRNP undergoes 5’-cap tri-methylation by TGS1, 3’-end trimming by an exonuclease and Sm ring assembly involving the SMN protein complex. The snRNP is re-imported into the nucleus for final maturation steps, including RNA base modifications (2’-O-methylation and pseudouridinylation). Mature snRNPs are redistributed to the nuclear speckle for spliceosome formation. At the periphery of the nuclear speckle, snRNPs catalyze the excision of introns from pre-mRNAs through the formation of a number of protein complexes on the target pre-mRNA and the joining of adjacent exons (E1, E2). Most introns are degraded but a minority undergo further processing steps to produce a sno/scaRNA. These intron-encoded sno/scaRNAs are targeted to the CB, where they guide major and minor spliceosomal snRNA base-modification steps. Following splicing, snRNPs are liberated from the target pre-mRNA. The U4/U6 di-snRNP and U4/U6.U5 tri-snRNP require recycling in the CB prior to participating in another round of pre-mRNA splicing.