Abstract
We demonstrate live-cell super-resolution imaging using photoactivated localization microscopy (PALM). The use of photon-tolerant cell lines in combination with the high resolution and molecular sensitivity of PALM permitted us to investigate the nanoscale dynamics within individual adhesion complexes (ACs) in living cells under physiological conditions for as long as 25 min, with half of the time spent collecting the PALM images at spatial resolutions down to ~60 nm and frame rates as short as 25 s. We visualized the formation of ACs and measured the fractional gain and loss of individual paxillin molecules as each AC evolved. By allowing observation of a wide variety of nanoscale dynamics, live-cell PALM provides insights into molecular assembly during the initiation, maturation and dissolution of cellular processes.
A key advantage of using genetically expressed fluorescent proteins is that they permit the non-perturbative optical imaging of dynamic processes in living cells1. Although these markers are targeted to specific proteins with molecular precision, most of this information is lost when conventional, diffraction-limited live-cell imaging methods2 are used. In response, several super-resolution imaging modalities have been developed to image fluorescent proteins in fixed cells3–5. Extension of these methods to living cells might at last lead to nanoscale imaging under true physiological conditions. Furthermore, by capturing many super-resolution images from a single living cell at different time points, the means by which macromolecular assembly drives cellular processes might be observed directly, as opposed to the much more indirect method of inferring “the rules of the game”6 from a series of static images of different cells, each fixed at a different time.
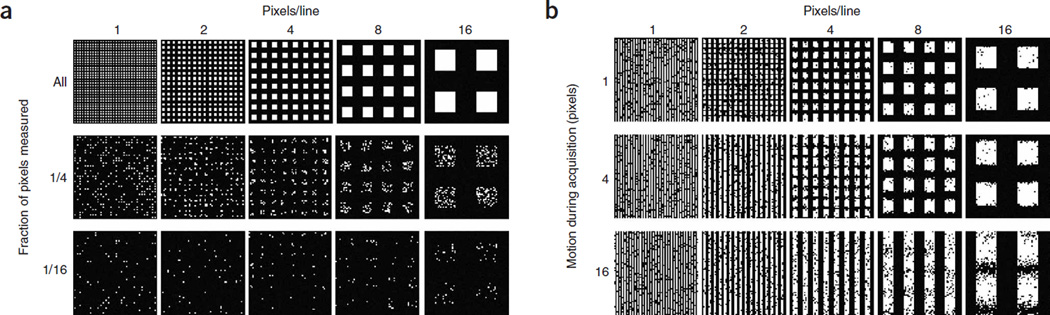
Nevertheless, reaching the longstanding goal of super-resolution live-cell imaging is far from trivial: the Nyquist criterion specifies that the image sampling interval must be smaller than half the desired resolution7, so to achieve N-fold higher resolution in D dimensions, ND-fold more pixels are required (Fig. 1a). To achieve such resolution gains without compromising either the imaging speed or signal-to-noise ratio, the signal collection rate (detected photons/s) must then increase by at least ND, requiring an ND-fold higher exposure to the excitation radiation for each image acquired. Furthermore, for point scanning (as opposed to wide-field) techniques, the intensity must increase by the even greater factor of N2D, as the pixel dwell time must additionally decrease by ND to maintain the original frame rate. Lastly, for live-cell imaging, to image moving subcellular structures without blurring, faster frame rates are required as the spatial resolution improves (Fig. 1b). Of course, whatever means is used to achieve these higher signal rates must also be demonstrably nonperturbative to the cell’s biological function, or else the goal of understanding nanoscale dynamics under physiological conditions is defeated.
Figure 1.
Factors influencing spatial resolution in live-cell PALM. (a) Resolution versus the density of localized molecules, represented here as pixels in a test pattern. Features are imaged at progressively lower signal-to-noise ratio as the molecular density decreases and become unresolvable when the mean molecular separation approaches the feature size. As more time is required to localize more molecules, this illustration implies a fundamental trade-off between spatial and temporal resolution in PALM. (b) Resolution versus feature motion. The serial, stochastic acquisition process in PALM leads to another such trade-off: the resolution limit for dynamically evolving features is comparable to the product of the feature velocity and the frame acquisition time.
Here we describe the application of PALM5 to super-resolution live-cell imaging of adhesion complexes (ACs)8—transmembrane cytoskeleton-substrate attachment points central to the process of cell migration. In PALM, numerous sparse subsets of photoactivatable fluorescent molecules are serially activated, localized and bleached. The aggregate position information from many such molecular subsets is then assembled into a super-resolution image. In spite of the challenges listed above, the application described here is made feasible by the confluence of several factors, including (i) a fast imaging speed relative to the phenomenon being imaged, (ii) efficient labeling of the target protein with a suitable photoactivatable probe and (iii) minimization of cell perturbation by use of photon-tolerant cells.
The imaging speed in PALM is aided by the fact that it is a wide-field technique that can collect information from the entire field of view at once. Furthermore EosFP9, the photoactivatable probe used here, is photostable enough that most molecules can be localized to a precision of <20 nm, yet not so stable that their persistence after activation impedes the timely acquisition of new molecular subsets. In addition, EosFP exhibits a high contrast ratio between its activated and inactive forms, which is essential for the isolation and precise localization of single molecules at the molecular densities required for such resolution (Supplementary Discussion, Supplementary Fig. 1 and Supplementary Table 1 online).
ACs consist of > 90 different proteins10 in a thin plaque near the cell-substrate interface8 and are well suited for the demonstration of live-cell PALM, as they evolve at speeds (~120 nm/min in stationary fibroblasts11) slow enough to prevent blurring, even when viewed at high spatial resolution (~60 nm) yet modest frame rates (~25 s/frame). For many of these, the molecular fraction that can be labeled with EosFP by transfection is sufficient to yield PALM resolution of ~20 nm by the Nyquist criterion12.
A primary concern for dynamic super-resolution imaging is the effect of the intense illumination required. Here we identified photon-tolerant cell lines (chinese hamster ovary (CHO) and NIH 3T3 fibroblasts) that accept intensities on the order of 1 kW/cm2 for ~30 min with no apparent ill effects on the processes being observed. When these cells express moderate amounts of fluorescent proteins, such intensities are sufficient to localize EosFP molecules at rates (>100 µm−2 s−1), yielding a Nyquist-limited resolution of ~60 nm in ~25 s. Furthermore, ACs are advantageously confined near the cell-substrate interface and as such can be illuminated to a depth restricted via total internal reflection to only ~50–100 nm, vastly reducing the overall exposure of the cell to the activation and excitation light.
RESULTS
Non-perturbative live-cell PALM
To demonstrate that the conditions required for live-cell PALM are non-perturbative, an independent measure of cell viability is needed. Toward this end, we captured differential interference contrast (DIC) images every 2 s during the acquisition of the PALM image sequences herein. This permitted the motility, growth and morphology of the cells, as well as the internal transport of sub-diffractive organelles, to be monitored in real time and to be compared against the behavior of similar cells not subjected to the PALM imaging conditions.
We exposed a live NIH 3T3 cell, expressing the tandem-dimer form of EosFP fused to the AC protein paxillin (tdEosFP-paxillin) maintained at 37 °C and migrating on a fibronectin-coated glass substrate, simultaneously to 0.05 kW/cm2 of 405 nm radiation to activate single tdEosFP molecules and 1 kW/cm2 of 561 nm radiation to excite fluorescence from and eventually bleach the molecules so activated (Fig. 2). We observed no adverse effects from laser exposure over the entire course of the PALM acquisition: the cell edge continuously probed its environment (Fig. 2, Supplementary Video 1, and Supplementary Figs. 2 and 3 online), adhesions continuously formed and elongated, and the cell showed no tendency to retract from the illumination.
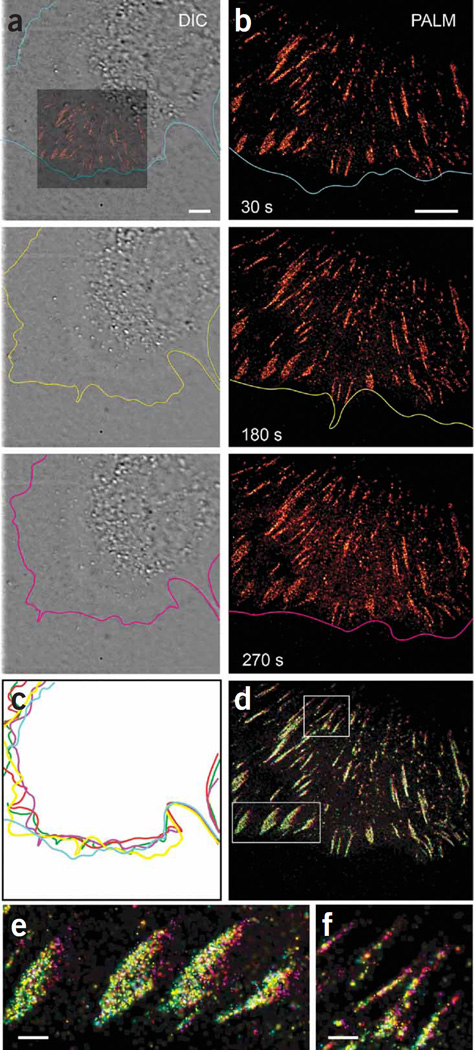
Figure 2.
Nonperturbative live-cell PALM. (a) DIC images from a time series (Supplementary Video 1) of an NIH 3T3 cell expressing tdEos-paxillin during 5 min of continuous PALM imaging. Colored inset indicates relative position of PALM image in b. Colored contours highlight the cell edge. (b) PALM images (Supplementary Video 2; middle panel) constructed from 750 single-molecule images (30 s total duration) and ending at indicated time points. (c) Overlay of cell-edge contours shows continuous movement at the leading edge during imaging. Colors correspond to labels in a and b, with red and green indicating 90 and 120 s, respectively. (d) Composite time-summed PALM image. Five PALM images corresponding to the contours in c were similarly pseudocolored and summed to provide a color map indicating the period when each tdEosFP-paxillin molecule was visualized. Molecules appeared continuously at the front of ACs (all colors), but molecules were visualized at the back of the AC only at later time points (purple). Scale bars, 5 µm. (e,f) Higher-magnification view of boxed regions in d. Scale bars, 1 µm.
In previous PALM experiments on fixed cells12, single-molecule images captured at ~10–50 ms/frame had been analyzed, and the position information obtained had been assembled into a single PALM image. Although we used similar frame-capture rates for the live-cell experiments here, we assembled the position data into several equal time interval subgroups, which then formed the individual PALM frames of a time-lapse movie. Thus, although individual single-molecule frames were captured fast enough to visualize molecular dynamics at the 20-Hz level, as in previous PALM-based diffusion studies13, here we concentrate instead on the nanoscale dynamics of the AC morphology, which occur over the time scale of minutes and require collating the molecular position information from hundreds or thousands of single-molecule frames. For example, we captured 7,500 single-molecule frames lasting 40 ms each without pause (Fig. 2) and parsed them into groups of 750 frames each, yielding the 10-frame PALM movie (Supplementary Video 2 online). During this acquisition, >25 molecules µm−2 s−1 were localized to 30 nm or better in the densest regions of the sample, a rate sufficient for a Nyquist-limited resolution of ~70 nm. The 30-s PALM frame rate obtained at this parsing was sufficiently fast compared to the evolution of most ACs in the cell that they could be tracked with minimal blurring. Binning the data into 5 frames instead of 10 yielded higher molecular densities and better Nyquist-limited resolution (~60 nm), but with a twofold reduction in temporal resolution and more blurring (Supplementary Video 2). Conversely, binning the data into 20 PALM frames doubled the temporal resolution (Supplementary Video 2) but degraded the Nyquist-limited resolution (>90 nm). Therefore, at a given rate of molecular localization, the parsing of the data across time must be carefully chosen to balance the spatial and temporal resolution in a manner best matched to the cellular processes under investigation. Furthermore, the molecular localization rate itself must be wisely selected to balance the spatiotemporal resolution against possible photodamage and the limited observation time before all available photoactivatable fluorescent proteins (PA-FPs) are bleached.
We observed the previously reported11 migration of ACs away from the cell edge and toward the cell interior (Supplementary Video 2 and Fig. 2b). Furthermore, if the frames of this video are colored to match the colors of the cell contours at the corresponding time points in Figure 2c and then combined to create the pseudocolor images in Figure 2d–f, it is evident from the broader separation of colors at the rear of each AC (closest to the cell center) that there are more paxillin molecules at this end than at the front (closest to the periphery) at later time points. As a result, most ACs lengthen as they migrate inward. We observed similar rearward-moving adhesions in 22 additional cells (data not shown).
Extended observations at high resolution
For features such as ACs that evolve on time scales slower than that necessary to construct a single PALM frame at the desired resolution, a non-imaging resting period can be included after each single-molecule collection sequence. During these periods, the cell can recover from the exposure, the ACs can evolve further, and additional labeled molecules can be recruited, extending the observation time before all the molecules photobleach. For example, we acquired single-molecule frames of a CHO cell in 11 groups of 1,000 frames, with a 60 s pause between each group (Fig. 3 and Supplementary Video 3 online). We analyzed these groups of frames to create 11 PALM and corresponding ‘summed total internal reflection fluorescence (TIRF)’ frames. The latter consist of the sum of all the single-molecule images in each group before localization and approximate the diffraction-limited images expected by TIRF microscopy. We recorded over 103 molecules/µm2 per PALM frame, corresponding to a Nyquist-limited resolution of ~60 nm. Features that appear as large homogeneous ACs in the summed TIRF microscopy frames become easily distinguished as assemblies of discrete, linear structures by PALM, and thin adhesions that appear similar as viewed by summed TIRF microscopy are revealed at their true lengths and sub-diffractive widths as well as with substantially greater internal detail (Fig. 3d).
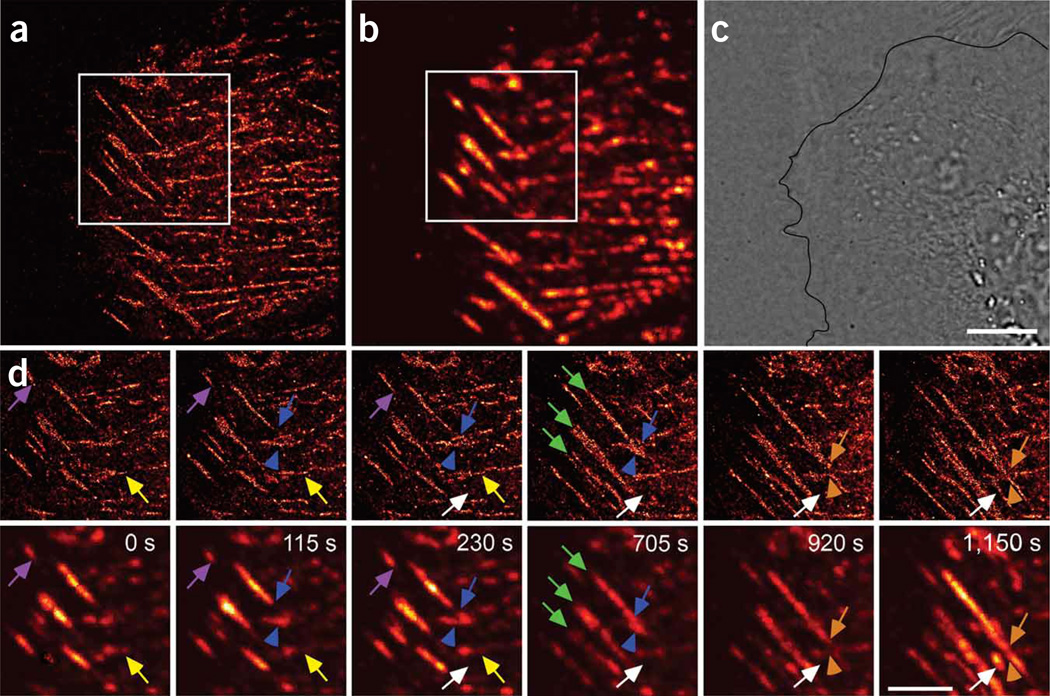
Figure 3.
Live-cell PALM allows extended observations at high resolution and over a wide range of molecular density. (a–c) PALM (a), summed TIRF (b; Supplementary Video 3) and DIC (c) images of a CHO cell expressing tdEos-paxillin, imaged for >20 min. (d) Higher-magnification views of boxed regions in a (top row) and b (bottom row) showing predominantly horizontal and stationary adhesions (yellow arrows) and diagonal inwardly moving adhesions (purple arrows). Intersecting adhesions either interacted weakly (orange arrows and arrowheads) or the moving AC bends the stationary AC (blue arrows and arrowheads). Features that appeared homogeneous in summed TIRF microscopy were revealed at their true size with substantially greater internal detail by PALM (green arrows). PALM also revealed nascent adhesions initially not seen by summed TIRF microscopy that arose in the cell interior (white arrows). Indicated times are final time points of each 60-s acquisition, with the first time in the image series set to 0 s. Scale bars, 5 µm.
The PALM frames in Figure 3 also exhibit many sub-diffractive ACs and nano-clusters of paxillin that are not readily apparent when viewed by summed TIRF microscopy: their density is relatively constant over diffraction-limited distances, and thus they form a general, indistinct background when viewed by diffraction-limited means. Likewise, the PALM image of a live CHO cell in Supplementary Figure 4 online reveals a broad pool of paxillin near the cell edge that is not apparent by summed TIRF microscopy images, as the individual paxillin molecules obvious in the former are averaged in the latter to yield a dim background easily overlooked compared to the much brighter, molecularly dense ACs.
Applied to living cells, this combination of high-resolution and single-molecule sensitivity permitted us to visualize intracellular movements by PALM that would be obscured when viewed by conventional means. For example, most ACs in the image in Figure 3 and in Supplementary Video 3 are either horizontally oriented and predominantly stationary, or diagonally oriented and moving inward from the cell edge. In the latter case, the rear of each AC moved faster than the front (~140 nm/min versus ~90 nm/min for the AC marked in purple in Fig. 3d), causing them to elongate as they translocated. As the two types of ACs intersect, they either interacted weakly, or the moving AC bent the stationary one, suggesting that the traction forces exerted by the moving adhesion against the fibronectin matrix are much greater than the traction forces exerted by the stationary AC.
The interpretation that the leading ends of the diagonal adhesions shown in Figure 3 are retracting needs to be carefully examined. An alternative explanation is that, as they have existed longer than the trailing ends (which are growing), they may be disappearing rather than retracting, owing to exhaustion of the local supply of PA-FPs. However, this retraction is likely real, as the static horizontal ACs have similar molecular densities yet persist through all frames.
Finally, analysis of the images in Figure 3 reveals that, unlike in the image in Figure 2, not all adhesions form at the cell edge. Instead, nascent adhesions, initially too small to be detected by summed TIRF microscopy, arise within the cell interior and thereafter grow in the same direction as the pre-existing mobile adhesions. PALM allowed us to observe higher-resolution features not detectable by summed TIRF microscopy in 27 additional cells (data not shown).
Paxillin flow to and from adhesion complexes
Using live-cell PALM, we also observed recruitment of paxillin at established ACs interior to an NIH 3T3 cell (Fig. 4a,b and Supplementary Video 4 online): the number of molecules detected in each of the three ACs (Fig. 4b) increased 1.3–2.9-fold as the adhesions matured, lengthened and migrated before subsiding again, even though the overall number of activated molecules per single-molecule frame was held constant. This 3T3 cell and most other 3T3 cells we studied did not exhibit new ACs forming at the cell edge when observed with our imaging parameters. In contrast, in the CHO cell (Fig. 4c,d and Supplementary Video 5 online), we observed the formation of such peripheral ACs. The number of unbleached molecules in these ACs was substantially depleted by the end of each PALM frame, but then recovered during the 60-s rest period between frames, suggesting that the turnover of paxillin within such ACs occurs on this time scale. These differences between ACs in the two cell types may reflect differences in the rate of recruitment or turnover of paxillin because of the greater protrusive motion of 3T3 cells versus CHO cells (Supplementary Fig. 2).
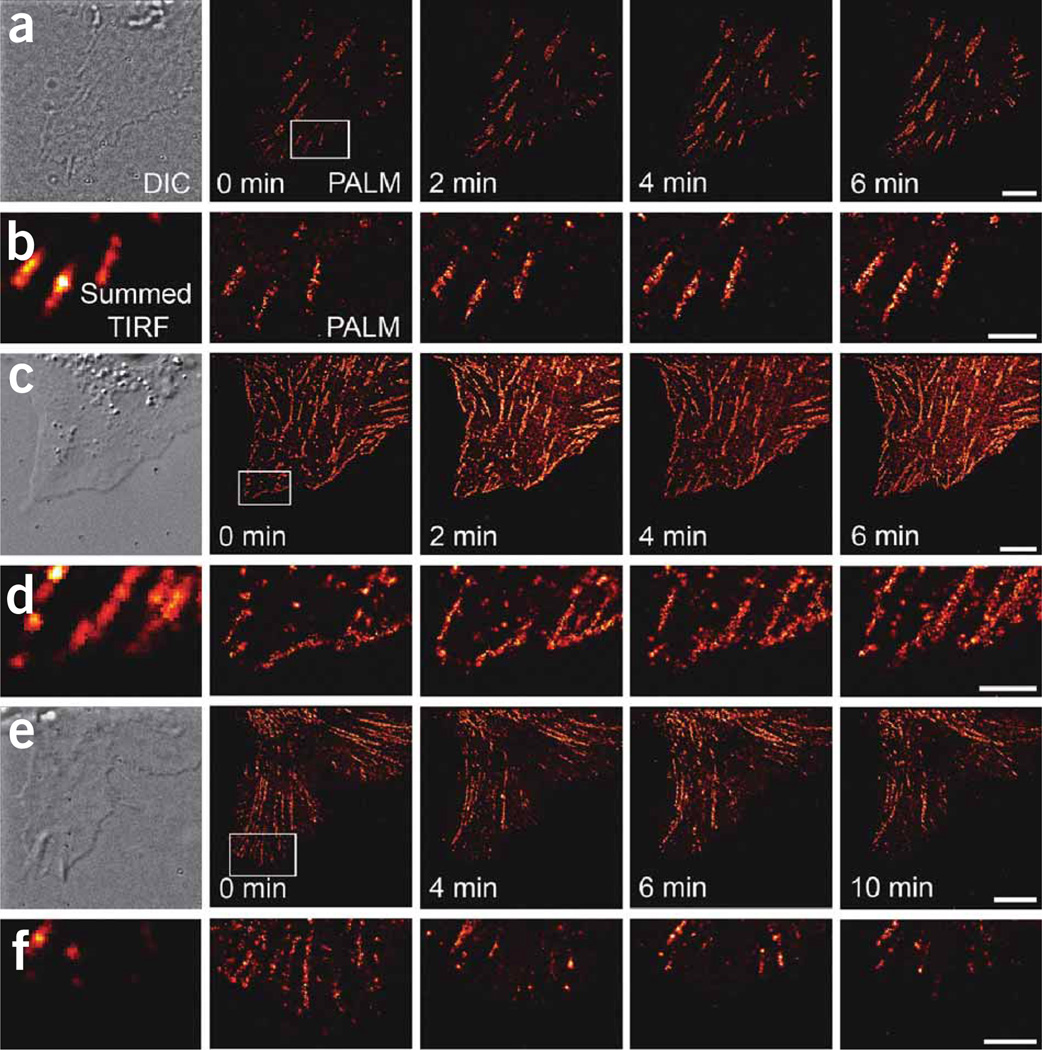
Figure 4.
Paxillin flow to and from ACs. (a–f) Selected live-cell PALM images (a,c,e; scale bars, 5 µm) and higher-magnification views (b,d,f; scale bars, 2 µm) of boxed regions of tdEos-paxillin distributions in several cells. DIC and summed TIRF microscopy images are shown at left. The NIH 3T3 cell in a,b and Supplementary Video 4 showed inward AC motion (upward, toward the cell center) but no new AC formation at the cell periphery, similar to the adhesions we observed in 12 additional cells (data not shown). In contrast, the CHO cell in c,d and Supplementary Video 5 displayed inward motion and a constant replenishment of paxillin molecules at the cell edge, typical of the adhesions in 8 additional cells we observed (data not shown). The NIH 3T3 cell in e,f retracted with a resulting dissolution of ACs (Supplementary Video 6), similar to the dissolving ACs we observed in 8 additional cells (data not shown). Indicated times are final time points of each 30 s acquisition (a,b) or 60 s acquisition (c–f), with the first time in the image series set to 0 s.
We also used live-cell PALM to monitor the disassembly of adhesions during cell retraction. We observed an NIH 3T3 cell with a retracting lamellum (Fig. 4e,f and Supplementary Video 6 online). Several ACs near the periphery of the lamellum initially displayed motion toward the interior but then dissolved as the cell retracted further. These adhesions lost ~50% of their tdEos-paxillin molecules after the first PALM frame (2 min), and contained progressively fewer molecules as the acquisition proceeded until, by the last frame (10 min), their molecular densities had dropped to ~10% of their initial values. However, the relatively constant molecular densities observed in other adhesions outside the retracting lamellum suggest that the loss of molecules in the ACs (Fig. 4f) arises from the dissolution of the adhesions rather than photobleaching-induced depletion of the tdEos marker molecules therein.
Finally, we used live-cell PALM to study the relative mobility of AC-associated and non-AC-associated molecules in both cell types. The molecular density in ACs decreased over the acquisition period, as molecules in ACs were progressively activated and bleached (Fig. 4c and Supplementary Video 5, and to a lesser extent in Supplementary Videos 2 and 4). This suggests that the addition of new tdEos-paxillin molecules into the ACs was slower than the rate of molecular bleaching, and although the pool of PA-FPs in the ACs diminished, the non-AC-associated tdEos-paxillin molecules visible in TIRF microscopy images appeared to be readily replenished from the large soluble pool. Thus, as we increased the activation intensity in an attempt to keep the number of activated AC-associated molecules constant, the number of activated molecules in the unbound pool further increased, contributing to the observed increase in non-AC-associated molecules.
Diversity of adhesion complex dynamics within a single cell
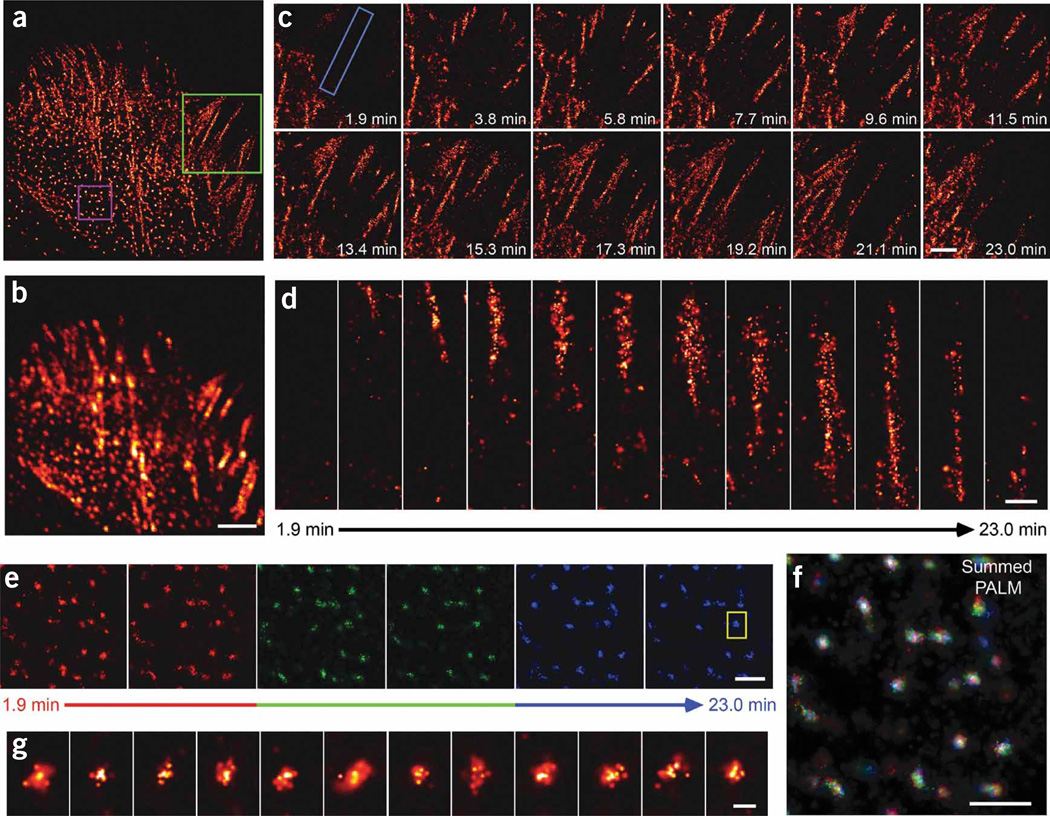
Even within a single cell, ACs can exhibit a wide range of morphologies and dynamic behaviors. For example, the live CHO cell expressing tdEos-paxillin imaged by PALM (Fig. 5a and Supplementary Video 7 online) and by summed TIRF microscopy (Fig. 5b) illustrated three different behaviors: initiation of ACs (Fig. 5c); elongation of ACs (Fig. 5c,d); and comparatively stable ACs (Fig. 5e,f) that appear as diffraction limited points by conventional fluorescence microscopy.
Figure 5.
A diversity of AC dynamics in a single cell. (a,b) PALM (a) and summed TIRF microscopy (b) images of a CHO cell expressing tdEos-paxillin from a 23-min time series (Supplementary Video 7). Scale bar, 5 µm. (c) Higher magnification of the green-boxed region in a to illustrate AC initiation and elongation. Note the established ACs at lower left in addition to paxillin molecules that are less well organized in regions at upper right. Scale bar, 3 µm. (d) Higher magnification of a single AC (boxed region in c) to visualize molecular organization during the elongation process. Scale bar, 0.5 µm. (e–g) The motion of subdiffractive ACs (magenta box in a) is highlighted in the time series. The initial, middle and late frames in e were colored red, green and blue, and then added to create the time-summed pseudocolor image in f. Scale bars, 0.5 µm. The mostly white central cores in f, the higher magnification view of the single AC (g; close-up of boxed region in e) and Supplementary Video 8 suggest the central portions of ACs are constrained to regions that are small compared to AC diameters. Indicated times are final time points of each 55 s acquisition, with the first time in the image series set to 0 s. Scale bar, 0.05 µm.
To visualize adhesion initiation and elongation, we used PALM to image a region near the cell edge of a CHO cell expressing tdEos-paxillin (Fig. 5a,c). We observed several established ACs as well as regions in which paxillin molecules were less well organized. These molecules may represent the initial response of a probing cellular extension that is just beginning to make contact with the substrate14. In the second frame of Figure 5c (3.8 min), these molecules have organized into easily discernable nascent ACs. More ACs then developed and matured, as illustrated in subsequent frames. For example, the three small ACs at upper center (3.8 min) elongate (5.8 min), merge (7.7–13.4 min) and begin to pull back into the cell interior (19.2–23.0 min).
We observed the evolution of another AC (Fig. 5d). Again, the formation of the nascent adhesion was evident only by 3.8 min. The AC initially widened as it elongated inward (5.8–13.4 min), while its outward end appeared stationary throughout this period. Thereafter, the outward end also appeared to retract, while the AC narrowed and continued to lengthen until it merged with the established ACs in the interior. However, even later (17.3–23.0 min) there remained a stochastically varying sparse subset of molecules extending out to the initial position, suggesting that the AC was not retracting, but only appeared to do so under the influence of photobleaching at the outward end and the recruitment of paxillin at the lengthening, inward end.
We also observed many point-like adhesions in the interior of the cell (Fig. 5e,f). Although most of these were individually resolvable by summed TIRF microscopy (Fig. 5b), only PALM revealed their true sub-diffractive widths of ~120–300 nm. To readily visualize the dynamics of these ACs we colored the initial, middle and later frames red, green and blue, respectively (Fig. 5e), to obtain a time-integrated pseudocolor image (Fig. 5f). The point-like ACs are mostly white at their centers in this image, whereas elongated adhesions have color variation. Thus, unlike the elongating adhesions (Fig. 5c,d), the cores of point-like ACs were largely stationary during the entire 23 min acquisition period (Supplementary Video 8 online). Furthermore, the mean number of molecules observed in these ACs (~40) was remarkably uniform, both from frame to frame (Fig. 5g) and adhesion to adhesion, suggesting an underlying morphological similarity between ACs of this type. We observed initiating adhesions and constrained sub-diffractive ACs in 13 and 8 cells, respectively (data not shown).
DISCUSSION
Although the PALM frame rate of 25–60 s at ~60 nm resolution demonstrated here is sufficient for our study of adhesion dynamics, it is clearly inadequate to study many dynamic nanoscale cellular processes. Faster imaging will depend on increasing the number of molecules localized per unit area per unit time. This can occur either by increasing the bleaching rate of activated molecules (so that new subsets of molecules can be activated and localized in turn), or by increasing the density of activated molecules in each single molecule frame. Bleaching can be expedited by increasing the excitation intensity or using photoactivatable molecules of higher excitation cross-section and/or lesser photostability. However, intensities ~2–5× beyond the ~1 kW/cm2 applied to the CHO and NIH 3T3 cells here led to non-physiological morphological changes (for example, cellular retraction or lamella collapse); EosFP is already one of the brighter PA-FPs currently available; and the median photostabilty can be reduced only ~3× from that of EosFP (750 photons/molecule) before the localization precision limits resolution to a greater degree than the molecular density. Similarly, the ability to increase the activation density well beyond the ~2–4 molecules µm−2 frame−1 used in the densest regions here is limited: the localization precision quickly declines as the images of activated molecules begin to overlap (Supplementary Fig. 5 online). Thus, although a combination of factors may eventually yield PALM frame rates of 2–10 s at a Nyquist-limited resolution of 50 nm, gains beyond this would likely require major advances in molecular brightness and/or reduced cellular phototoxicity.
Another limitation of live-cell PALM is that as the pool of unactivated, unbleached PA-FP molecules is depleted, the molecular density per PALM frame will eventually decline despite efforts to maintain a constant density by increasing the activation intensity. As both the Nyquist-limited resolution and the molecular signal-to-noise ratio at all longer length scales depend upon this density (Supplementary Discussion and Supplementary Fig. 1), the resolution will inevitably degrade and the stochastic noise will increase the longer the cell is under observation, provided that new PA-FP tagged molecules are not recruited to the features of interest. For example, in the fixed CHO cell shown in Supplementary Figure 6 and Supplementary Video 9 online, low-density ACs initially exhibit poor molecular signal-to-noise ratios and become undetectable after ~20 min as a result of bleaching of the available EosFP. In contrast, high-density ACs display higher signal-to-noise ratios initially, so although they too degrade in later frames, they consist of enough molecules to persist throughout the entire acquisition. The signal-to-noise ratios can be improved by increasing the number of molecules binned into each PALM frame but at the cost of the number of such frames and hence, in a live-cell experiment, at the cost of the temporal resolution (Supplementary Fig. 6).
Thus, live-cell PALM is used most advantageously when the total budget of PA-FP–labeled molecules is parsed over time in a manner well matched to the dynamics under investigation, that is, when just enough molecules are localized per frame to achieve the desired resolution, when the acquisition time for each frame is short enough to mitigate motion-induced blurring at this resolution, and when the interval between PALM frames is short enough to capture the dynamics but long enough to study the dynamics for an extended period of time.
Despite these concerns, time-resolved PALM can be applied in several ways. Already, serial photoactivated localization of single PA-FP tagged molecules has been used to track the diffusion of up to ~100× more protein molecules in the membranes of living cells than is possible with conventional fluorescent probes13,15. In vitro experiments can also be contemplated at much higher temporal resolution than demonstrated here, as many such experiments involve much lower molecular densities. Even when higher densities are required to meet the Nyquist criterion for true live-cell imaging beyond the diffraction limit, the spatial and temporal resolution can be balanced against one another to match the requirements of different cellular processes of interest.
METHODS
Instrumentation
We performed PALM imaging on an Olympus IX81 inverted microscope as previously described12. Several modifications to the system enabled live-cell PALM. First, we kept the cells at 37 °C by addition of a heater unit (Air-Therm ATX; World Precision Instruments) that supplied temperature-controlled air to an enclosed volume surrounding the microscope stage and objective. Second, we used a 60×, 1.49 numerical aperture (NA) objective (Olympus) and 2× magnification optic instead of the earlier 100×, 1.65 NA objective, in part to add DIC capability and in part because the immersion oil of the 1.49 NA objective is more suitable for extended observations at physiological temperatures. Third, we used a commercial beamsplitting device (Dual-Cam; Photometrics) to separate simultaneously acquired DIC and EosFP fluorescence signals before their measurement at independent electron-multiplying charge-coupled device (CCD) cameras (DV887ECS-BV; Andor Technology). A 532-nm shortpass filter (SP01-532RU-25; Semrock) placed in the transmitted light pathway provided DIC illumination that was spectrally distinct from EosFP fluorescence, while other filters (Supplementary Table 2 online) isolated the DIC and EosFP fluorescence signals from one another as well as from the activation (405 nm) and excitation (561 nm) light. Finally, we used a shutter (IQ35-SA; Sutter Instruments) to ensure that the DIC illumination was present only during the DIC frame acquisition. Details of the sample preparation are available in Supplementary Methods online.
Supplementary Material
Acknowledgments
We thank H. White for assistance with cell culture, M. Davidson (Florida State University) for the tdEos-paxillin plasmid and H. Peng (Janelia Farm Research Campus) for creating the simulation leading to Supplementary Fig. 5. This research was supported in part by the Intramural Research Programs of the US National Institutes of Health, the National Institute of Dental and Craniofacial Research, and the National Institute of Neurological Disorders and Stroke.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
References
- 1.Lippincott-Schwartz J, Patterson GH. Development and use of fluorescent protein markers in living cells. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 2.Stephens DJ, Allan VJ. Light microscopy techniques for live cell imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- 3.de Lange F, et al. Cell biology beyond the diffraction limit: near-field scanning optical microscopy. J. Cell Sci. 2001;114:4153–4160. doi: 10.1242/jcs.114.23.4153. [DOI] [PubMed] [Google Scholar]
- 4.Willig KI, et al. Nanoscale resolution in GFP-based microscopy. Nat. Methods. 2006;3:721–723. doi: 10.1038/nmeth922. [DOI] [PubMed] [Google Scholar]
- 5.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 6.Lichtman JW, Fraser SE. The neuronal naturalist: watching neurons in their native habitat. Nat. Neurosci. 2001;4:1215–1220. doi: 10.1038/nn754. [DOI] [PubMed] [Google Scholar]
- 7.Shannon CE. Communication in the presence of noise. Proc. Inst. Radio Eng. 1949;37:10–21. [Google Scholar]
- 8.Zamir E, Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 9.Wiedenmann J, et al. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci USA. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Functional atlas of the integrin adhesome. Nat. Cell Biol. 2007;9:858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smilenov LB, Mikhailov A, Pelham RJ, Jr, Marcantonio EE, Gundersen GG. Focal adhesion motility revealed in stationary fibroblasts. Science. 1999;286:1172–1174. doi: 10.1126/science.286.5442.1172. [DOI] [PubMed] [Google Scholar]
- 12.Shroff H, et al. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc. Natl. Acad. Sci. USA. 2007;104:20308–20313. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 14.Galbraith CG, Yamada KM, Galbraith JA. Polymerizing actin fibers position integrins primed to probe for adhesion sites. Science. 2007;315:992–995. doi: 10.1126/science.1137904. [DOI] [PubMed] [Google Scholar]
- 15.Hess ST, et al. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. USA. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.