Abstract
Mitochondrial gene expression in trypanosomes is controlled primarily at the levels of RNA processing and RNA stability. This regulation undoubtedly involves numerous ribonucleases. Here we characterize the Trypanosoma brucei homolog of the yeast DSS-1 mitochondrial exoribonuclease, which we term TbDSS-1. Biochemical fractionation indicates that TbDSS-1 is mitochondrially localized, as predicted by its N-terminal sequence. In contrast to its yeast homolog, TbDSS-1 does not appear to be associated with mitochondrial ribosomes. Targeted downregulation of TbDSS-1 by RNA interference in procyclic-form T. brucei results in a severe growth defect. In addition, TbDSS-1 depletion leads to a decrease in the levels of never edited cytochrome oxidase subunit I (COI) mRNA and both unedited and edited COIII mRNAs, indicating this enzyme functions in the control of mitochondrial RNA abundance. We also observe a considerable reduction in the level of edited apocytochrome b (CYb) mRNA and a corresponding increase in unedited CYb mRNA, suggesting that TbDSS-1 functions, either directly or indirectly, in the control of RNA editing. The abundance of both gCYb[560] and gA6[149] guide RNAs is reduced upon TbDSS-1 depletion, although the reduction in gCYb[560] is much more dramatic. The significant reduction in gCYb levels could potentially account for the observed decrease in CYb RNA editing. Western blot analyses of mitochondrial RNA editing and stability factors indicate that the perturbations of RNA levels observed in TbDSS-1 knock-downs do not result from secondary effects on other mitochondrial proteins. In all, these data demonstrate that TbDSS-1 is an essential protein that plays a role in mitochondrial RNA stability and RNA editing.
In the mitochondria of the protozoan parasite Trypanosoma brucei, posttranscriptional mechanisms of gene regulation are of major importance. Transcription of both the maxicircle (30, 48) and minicircle (22) genomes is polycistronic. This suggests that regulation of processing and stability of rRNA-, mRNA-, and guide RNA (gRNA)-containing transcripts are likely to be critical steps in gene expression. For example, both endonuclease cleavage and exonuclease trimming are presumably required for the production of mature RNAs from polycistronic precursors. Processing is a particularly critical step in the formation of those mRNAs whose genes overlap such that it is impossible to produce two mature monocistronic RNAs from the same precursor molecule (30, 48). Moreover, this overlapping gene arrangement coupled with polycistronic transcription indicates that a system must exist for the rapid degradation of nonfunctional by-products of processing reactions. The majority of mitochondrial RNAs in T. brucei also require an extensive RNA editing process involving uridine insertion and deletion to form translatable RNAs (55, 56). The steady-state RNA pool contains large amounts of improperly edited RNAs (12, 29). These RNAs may be intermediates destined to become properly edited or they may represent aberrantly processed RNAs that need to be removed. The latter scenario would require the presence of an RNA surveillance system to identify and degrade improperly edited RNAs. In addition to RNA processing, the regulation of RNA decay rates is also likely to be a major factor controlling the abundance of mature RNAs in trypanosome mitochondria (37, 38, 51). Despite the extensive requirement for ribonucleases in trypanosome mitochondrial gene expression, the nucleases that carry out the majority of these processes remain unidentified.
Several RNase activities with the potential to mediate mitochondrial RNA processing and decay in trypanosomes have been described. gRNA-directed endonuclease and U-specific exonuclease activities are associated with RNA editing complexes (42, 46, 50), and a distributive U-specific exonuclease was purified over 4,000-fold from Leishmania tarentolae mitochondria (3). In addition, purified editing complexes from both T. brucei and L. tarentolae contain proteins with exo/endo/phos and RNase III motifs that are predicted to possess RNase activity (4, 43). A T. brucei mitochondrial RNase P-like activity that presumably functions in tRNA maturation has also been reported (52). Furthermore, at least two, and possibly three, T. brucei mitochondrial endonucleases distinct from those involved in editing and tRNA processing have been partially purified and characterized (45, 53). An endoribonuclease termed MAR1, for mitochondrial associated ribonuclease, was also purified and its gene cloned from Leishmania (1). Whether there is any relationship between the MAR1 protein and the endonuclease activities described in T. brucei is unknown. Two exoribonuclease activities in addition to the U-specific nuclease were detected in the mitochondria of L. tarentolae (3). One of these is a processive hydrolytic enzyme, while the other exhibits a preference for 3′ phosphate ends. Finally, we recently described a hydrolytic exoribonuclease activity from T. brucei mitochondrial membranes that preferentially degrades polyadenylated RNAs (51).
The yeast mitochondrial degradosome (originally termed mtEXO) was first purified from Saccharomyces cerevisiae mitochondria a decade ago (39) and has been shown to play a role in multiple aspects of mitochondrial RNA turnover and processing in this organism. The degradosome comprises two proteins: the DSS-1 exoribonuclease and the SUV3 RNA helicase (17, 33). It exhibits a 3′-to-5′ exonuclease activity that is dependent on any one of the standard ribonucleosides (NTPs) or deoxyribonucleoside triphosphates (dNTPs) (39), RNA-stimulated NTPase activity (39), and RNA helicase activity (17). Isolation of the yeast degradosome by using tandem affinity purification (TAP)-tagged DSS-1 or SUV3 revealed that the complex is exclusively associated with mitochondrial ribosomes. The functions of the degradosome in mitochondrial gene expression have been explored by genetic approaches in yeast. One function appears to be related to splicing and stability of group I intron-containing transcripts. Cells that either express the SUV3-1 suppressor allele or from which the DSS-1 gene has been deleted are defective in splicing of some group I introns and undergo massive accumulation of excised group I introns (9, 16, 21). Non-splicing-related degradosome functions were specifically examined in yeast containing an intronless mitochondrial genome (16, 17). Deletion of either SUV3 or DSS-1 genes in these cells resulted in dramatic effects on the abundance of several mitochondrial RNAs. Unexpectedly, levels of mRNAs encoding both cytochrome b (COB) and 16S rRNA were dramatically decreased in both SUV3- and DSS1-null mutants. This suggests that some RNAs are either directly or indirectly stabilized by the mitochondrial degradosome. In addition, Northern blot and S1 nuclease analyses revealed a large accumulation of mitochondrial mRNA and rRNA precursors that are improperly processed at their 5′ and 3′ ends in cells with either the SUV3 or DSS-1 gene deleted (16, 17). Accumulation of precursors may be due to either a direct effect on RNA processing and/or an effect on the degradation of unprocessed transcripts. Regarding the latter possibility, it has been suggested that one function of the degradosome may be as an RNA surveillance system that targets improperly processed RNAs.
Toward our goal of functionally characterizing the RNases that mediate trypanosome mitochondrial gene expression, we searched for homologs of known exoribonucleases in the T. brucei genomic databases. We report here characterization of the T. brucei homolog of the yeast degradosome component DSS-1, which we term TbDSS-1. Western blot analysis confirms the predicted mitochondrial localization of TbDSS-1. Targeted gene depletion using RNA interference (RNAi) provides evidence that TbDSS-1 affects the stability and editing of specific mitochondrial RNAs.
MATERIALS AND METHODS
Oligonucleotides used in this study.
The oligonucleotides used in this study are listed as follows with restriction sites underlined: RXS-dT17 (5′ GAGAATTCTCGAGTCGACTTTTTTTTTTTTTTTTT 3′), ESL-22 (5′ GCGAATTCGCTATTATTAGAACAGAGTTTCTG 3′), RNase 1-1 (5′ GCGGATCCTAGTGGAGGCGGTGTGTTAC 3′), RNase 1-2 (5′ GCGGATCCACAAAGAGCTCCCCTGTG 3′), RNase 1-3 (5′ GCGGATCCGTGCCGCGAAATGGTATAG 3′), RNase 1-4 (5′ GCGGATCCTGCAACATGGTCTAAAGACAAC 3′), RNase 1-5′exp2 (5′ GCGGATCCGCTAGCATGACCCCTCGGCGCGTCGCAAA 3′), RNase 1-3′exp2 (5′ ACGCGTCGACCTACTCGAGCGAGTCCAGAGATGGAAGAAGGCACT 3′), RNase1-I-5′ (5′ CCGCTCGAGTGGAGCGCGACGGTAGCGAC 3′), RNase1-I-3′ (5′ CCCAAGCTTCCCACCTGTGCCTCCTCATC 3′), DSS1-9 (5′ GAAAATCATCTTTCTATACCATTTCGCG 3′), DSS1-10 (5′ TCGTAAGGATGTTTTGTTTAATGTGTTTC 3′), p22i-5′ (5′ CCGCTCGAGCGCAAATCCCATGGGGACGAGGA 3′), p22i-3′ (5′ CCCAAGCTTACGAAACAAATTTGTTAATGCTGCTC 3′), 12S-1 (5′ GCTTGTTAACCTGCTCGAAC 3′), 9S-1 (5′ CCGCAACGGCTGGCATCCAT 3′), Tub-RT (5′ GGGGGTCGCACTTTGTC 3′), CYb-RT (5′ CAACCTGACATTAAAAGAC 3′), COI-RT (5′ GTAATGAGTACGTTGTAAAACTG 3′), COIII-3′NE (5′ ACTTCCTACAAACTAC 3′), A6-3′NE (5′ ATTTGATCTTATTCTATAACTCC 3′), gCYb[560]A (5′ TCCCTAGAGAGTAGTTATCCTCCCCATTACTCAG 3′), and gA6[149] (5′ ATAATTTCACAGATATCTTTTC 3′).
Cloning the TbDSS-1 cDNA.
A BLAST search of the T. brucei genomic database from The Institute of Genomic Research revealed a gene fragment (GATGG96TV) encoding a portion of a protein with homology to S. cerevisiae DSS-1. Based on the sequence of this genomic fragment, oligonucleotides were designed for cloning the 5′ and 3′ ends of the TbDSS-1 cDNA from RXS-dT17-primed cDNA by nested rapid amplification of cDNA ends (RACE) strategies. To clone the 5′ end, cDNA was amplified with primers ESL-22, which corresponds to the T. brucei spliced leader sequence, and RNase 1-4. Five microliters of the resulting product was then amplified with ESL-22 and RNase 1-3, and the product was cloned into the BamHI/EcoRI site of pBluescript II SK−. To obtain 3′ end sequence, cDNA was amplified with primers RXS-dT17 and RNase 1-1. Five microliters of the resulting product was then amplified with RXS-dT17 and RNase 1-2, and the product was cloned into the BamHI/EcoRI site of pBluescript II SK−. Two clones were sequenced in both directions for both 5′ and 3′ ends. Based on the 5′ and 3′ end sequences, the entire TbDSS-1 open reading frame (ORF) was amplified from cDNA with primers RNase 1-5′exp2 and RNase 1-3′exp2. The resulting product was cloned into the NheI/XhoI site of pET21a, and the sequence of the complete ORF was verified. The TbDSS-1 sequence was compared to sequences in the GenBank database, using the default BLAST algorithm. Multiple alignments were performed with CLUSTAL W.
Production of recombinant TbDSS-1 and antibody production.
TbDSS-1 was expressed as a six-His (His6) fusion protein as follows. The entire TbDSS-1 ORF was PCR amplified with oligonucleotides RNase 1-5′exp2 and RNase 1-3′exp2, and the amplified product was digested and ligated into the NheI and XhoI sites of pET21a. Escherichia coli cells carrying the pET21a-TbDSS-1 plasmid were grown in Luria-Bertani (LB) medium with 100-μg/ml ampicillin at 37°C to an A600 of 0.7. Protein production was induced with 0.3 mM isopropyl β-d-thiogalactopyranoside (IPTG) for 2 h, and cells were harvested by centrifugation at 5,000 × g for 10 min at 4°C. Cells were resuspended in lysis buffer (6 M guanidine-HCl, 20 mM Tris-Cl [pH 7.9], 500 mM NaCl, 10% glycerol) and lysed by sonication on ice (4 pulses of 30 s each). The lysed cells were rocked for 60 min at 4°C, and then centrifuged at 15,000 × g for 15 min at 4°C. The supernatant was mixed with Ni2+-nitrilotriacetic acid (NTA) resin for 3 h at 4°C, and the mixture was poured into a column. The column was washed with 10 column volumes of wash buffer A (6 M guanidine-HCl, 20 mM Tris-Cl [pH 7.9], 500 mM NaCl, 10% glycerol, 20 mM imidazole). To renature the recombinant protein, the column was then washed sequentially with 10 column volumes of 1:1, 3:1, and 7:1 (vol/vol) wash buffer B (wash buffer A lacking guanidine-HCl)-wash buffer A. The column was finally washed with 15 column volumes of wash buffer B containing 1 mM phenylmethonylsulfonyl fluoride and eluted with 10 column volumes of wash buffer B containing a gradient of 50 to 500 mM imidazole. The purified protein was dialyzed overnight at 4°C into a buffer containing 20 mM Tris [pH 8.0] and 25 mM KCl. The His6-TbDSS-1 protein yield was 6.5 mg per liter of E. coli cells. E. coli cells transformed with the empty pET21a vector were processed identically to provide a negative control for enzymatic assays. To generate anti-TBDSS-1 antibodies, His6-TbDSS1 was used as antigen for polyclonal antibody production in rabbits (Bethyl Laboratories, Inc., Mongomery, Tex.).
Trypanosome growth, mitochondrial isolation, transfection, and induction of RNAi.
The procyclic form T. brucei brucei clone IsTaR1 stock EATRO 164 was grown as described previously (7). Mitochondria were isolated by the procedure of Harris et al. (24). Procyclic T. brucei brucei strain 29-13 (kindly provided by George Cross), which contains integrated genes for T7 RNA polymerase and tetracycline repressor, were grown in SDM-79 supplemented with 15% fetal bovine serum as described previously (7, 58) in the presence of G418 (15 μg/ml) and hygromycin (50 μg/ml). To construct the pTbDSS-1i vector for RNAi, a 450-bp fragment of the TbDSS-1 gene (nucleotides [nt] 511 to 961 from the start codon) was amplified by PCR with oligonucleotides RNase1-i-5′ and RNase1-i-3′. The fragment was digested and inserted into the XhoI/HindIII sites of pZJM (kindly provided by Paul Englund) (57). For transfection, 1 × 109 cells were washed once in 100 ml of ice-cold Cytomix and resuspended in fresh Cytomix to a concentration of 2.5 × 107 cells/ml. Twenty micrograms of pTbDSS1-i linearized with NotI was then added to 0.45 ml of cells. Transfections were carried out on ice in 2-mm cuvettes using a Bio-Rad electroporator with two pulses at the following settings: 800 V, 25 μF, and 400 Ω. Following transfection, cells were transferred into 10 ml of SDM-79 supplemented with G418 and hygromycin and allowed to recuperate for 20 h. Selection was then applied by the addition of 2.5-μg/ml phleomycin, and the cells were grown for 4 weeks to obtain stable transfectants. For induction of double-stranded RNA (dsRNA), cells were cultured in the presence of 1-μg/ml tetracycline. Growth curves were obtained by plotting the total cell number (the product of the cell number and the total dilution) over a period of 11 days. Two separate inductions (including monitoring of growth and RNA isolation) were performed, and most analyses were repeated with protein and RNA from both inductions.
RNA analysis.
Total RNA was purified from 1.3 × 109 to 4.8 × 109 cells (Purescript RNA isolation kit; Gentra Systems). For PCR analysis, cDNA was synthesized from 10 μg of total RNA by using oligonucleotide RXS-dT17. Ten percent of the resulting cDNA was using as a template for amplification of the full length TbDSS-1 ORF (oligonucleotides DSS1-9 and DSS1-10) or a 500-bp fragment of the p22 cDNA (oligonucleotides p22i-5′ and p22i-3′). cDNA was titrated to ensure that PCRs were performed in the linear range (27). For Northern blot analysis, 10 μg of total RNA was electrophoresed on 1.5% formaldehyde-agarose gels and transferred to nylon membrane. Blots were probed with kinase-labeled oligonucleotide probes 12S-1 and 9S-1 for detection of 12S and 9S rRNA, respectively, as described (10). For Northern blot analysis of ND4 mRNA, an antisense riboprobe was generated by in vitro transcription with incorporation of [α-32P]UTP, and hybridization was performed as described previously (10). Primer extensions using oligonucleotides Tub-RT, CYb-RT, COI-RT, COIII-3′NE, and A6-3′NE were performed with 10 to 15 μg of total RNA as described previously (44). For primer extension of gRNAs, 25 to 35 μg of total RNA was used. Gels were analyzed either by autoradiography followed by densitometry of nonsaturated autoradiographs or by phosphoimager analysis on a Bio-Rad Personal FX Phosphoimager using Quantity One software.
Glycerol gradient fractionation.
Glycerol gradient fractionation of mitochondrial lysates was performed as previously described (11). Mitochondrial lysates from 1010 procyclic-form T. brucei cells were loaded onto each 12-ml gradient, and 500-μl fractions were collected after centrifugation. Fifteen microliters of each fraction was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (7.5% polyacrylamide) and anti-TbDSS-1 Western blot analysis. Standards (bovine serum albumin, 4S; yeast alcohol dehydrogenase, 7.4S; and thyroglobulin, 19S) were fractionated in a parallel gradient and analyzed by SDS-PAGE and Coomassie blue staining. The 40S region of the gradient was defined by the location of the REAP-1 protein (32) as determined by Western blot with antibodies generously provided by Steve Hajduk. To determine the sedimentation of mitochondrial ribosomes, RNA was isolated from 150 μl of each glycerol gradient fractionation as follows. Glycerol gradient fractions were incubated for 15 mins at 37°C with 0.5% SDS, 50-μg/ml proteinase K, and 2 U of RNaseOUT (Invitrogen). Samples were extracted twice with phenol-chloroform-isoamylalcohol (25:24:1), and RNA was precipitated with ethanol and 10 μg of glycogen as a carrier. RNAs were fractionated on a 1.5% agarose-formaldehyde gel and transferred to nylon membrane. Sedimentation of the 9S rRNA was then detected by Northern blot analysis using kinase-labeled 9S-1 oligonucleotide.
Western blot analysis of RNAi cells.
Aliquots (5 × 106 cells) of uninduced and induced cells were suspended in SDS-PAGE sample buffer immediately after harvesting, boiled for 10 min, and stored at −80°C until use. For Western blot analysis, 5 × 106 cells were electrophoresed by SDS-PAGE (10% polyacrylamide) (TbMP52 blot) or 1.66 × 106 cells were electrophoresed by SDS-PAGE (15% polyacrylamide) (all other blots) and transferred to nitrocellulose membrane. Blots were probed with anti-RBP16 (26), anti-p22 (27), anti-TbMP52 (54) (generously provided by Ken Stuart), or anti-gBP21 and anti-gBP25 (both generously provided by Julius Lukes). Primary antibodies were detected by incubation with horseradish peroxidase-conjugated goat anti-mouse antibodies (Pierce; 1:10,000 dilution) for TbMP52, or horseradish peroxidase-conjugated goat anti-rabbit antibodies (Pierce; 1:10,000 dilution) followed by development with the SuperSignal West Pico chemiluminescent system (Pierce).
Nucleotide sequence accession number.
The TbDSS-1 sequence has been submitted to the GenBank database under accession no. AY233297.
RESULTS
Identification of TbDSS-1.
To identify RNases that function in trypanosome mitochondrial RNA turnover and processing, we utilized the BLAST algorithm to search the available T. brucei databases for homologs to known exoribonucleases. Using this strategy, we detected a strong match between the virtual translation product of a T. brucei genomic fragment (TIGR GATGG96TV) and a segment of the S. cerevisiae DSS-1 mitochondrial exoribonuclease (14). We obtained the complete sequence of the corresponding T. brucei cDNA using nested 5′ and 3′ RACE strategies. Five prime RACE analysis using a 5′ primer corresponding to T. brucei spliced leader sequence indicated the presence of a single splice site and a 14-nt 5′-untranslated region. Two polyadenylation sites were identified by 3′ RACE, resulting in either 146 or 148 nt 3′ untranslated regions. The complete predicted ORF was subsequently amplified from procyclic-form oligo(dT)-primed cDNA, cloned into the pET21a vector, and sequenced in both directions. The cDNA sequence contained a 2,229-nt ORF that predicts a protein with a molecular mass of 83.5 kDa and a pI of 6.43. Comparison of the predicted ORF to the nonredundant databases revealed significant homology to the fungal mitochondrial exoribonucleases S. cerevisiae DSS-1 (14) and N. crassa cyt-4 (15) as well as to a wide variety of prokaryotic hydrolytic 3′-5′ exoribonucleases of the RNase II/RNase R (RNR) family (13, 60). In view of its homology to the DSS-1 protein and its mitochondrial localization (see below), we termed the T. brucei gene TbDSS-1.
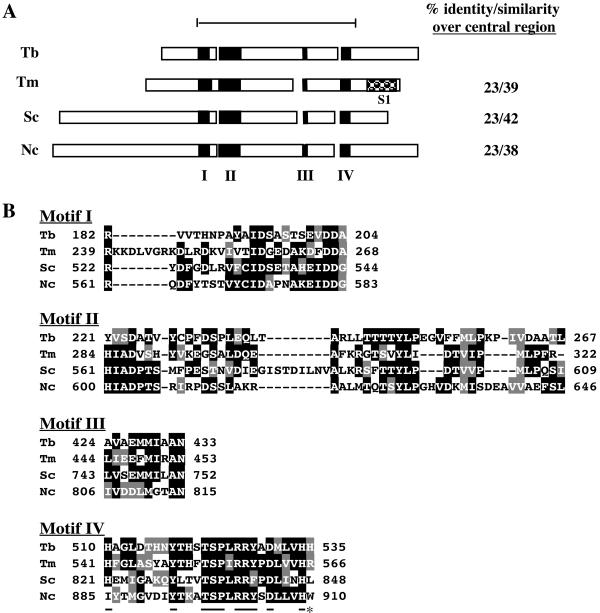
TbDSS-1 is identified as an RNR exoribonuclease family member by both the PROSITE and BLOCKS algorithms. All members of the RNR family, including prokaryotic enzymes and the mitochondrial DSS-1 and cyt-4 proteins, possess a conserved central region and a variable N-terminal extension. TbDSS-1 is 23% identical and 42% similar to S. cerevisiae DSS-1 over 430 amino acids encompassing the conserved central region (amino acids 191 to 621 of TbDSS-1) (Fig. 1A). Comparable levels of homology are observed between the TbDSS-1 amino acid sequence and those of cyt-4 and the prokaryotic Thermatoga maritima RNase R. Four conserved sequence motifs that define the RNR exoribonuclease family are contained within the central region (60). Motifs I to IV of the TbDSS-1 protein are depicted in Fig. 1A, and their sequences are aligned with the corresponding sequences of S. cerevisiae DSS-1, N. crassa cyt-4, and T. maritima RNase R proteins in Fig. 1B. Within these sequence blocks, TbDSS-1 possesses 21 of 29 amino acids that are conserved among 80% of RNR family members (60). Motif IV is the most conserved of the blocks and is often considered an RNase II signature (36, 60). TbDSS-1 contains 10 of 11 highly conserved amino acids present in motif IV (underlined in Fig. 1B). The predicted TbDSS-1 protein contains features common to both prokaryotic and mitochondrial RNR family members. Most strikingly, the majority of RNR family members, including the prokaryotic RNase R proteins, possess an S1 RNA binding motif at their C terminus (Fig. 1A) (60). However, TbDSS-1 is similar to the mitochondrial RNR family enzymes, DSS-1 and cyt-4, in lacking an S1 motif. TbDSS-1 is also more similar to mitochondrial RNR members in that the conserved arginine at the extreme C terminus of motif IV is absent (asterisk in Fig. 1B). Conversely, TbDSS-1 bears greater resemblance to prokaryotic than mitochondrial RNR family members in possessing a relatively short N-terminal variable region (Fig. 1A).
FIG. 1.
Alignment of TbDSS-1 (Tb; accession no. AY233297) with T. maritima RNase R (Tm; accession no. Q9WZI1), S. cerevisiae DSS-1 (Sc; accession no. AAC49144), and Neurospora crassa cyt-4 (Nc; accession no. P47950). (A) Schematic representation of the structure of TbDSS-1 and other RNR family members. Black boxes indicate conserved motifs I to IV. The checked box indicates an S1 RNA binding motif that is present in all RNR family exoribonucleases, with the exception of the mitochondrial members of the family. The bar at the top defines the conserved central region encompassing amino acids 191 to 621 of TbDSS-1, and amino acid conservation in this region is listed on the right. (B) Alignment of motif I to IV sequences. Amino acids that are identical in at least two of the four proteins are indicated by white letters on a black background. Conservative substitutions are indicated by white letters on a gray background. Residues that are underlined in motif IV are highly conserved RNase II signature residues. The C-terminal residue indicated by an asterisk is the highly conserved arginine that is typically absent in mitochondrial RNR family members.
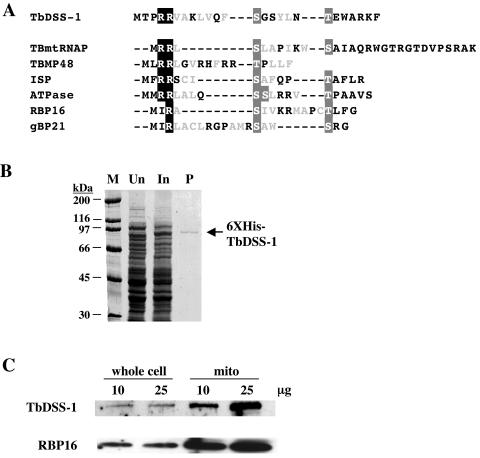
Based on its predicted function, we made several attempts to demonstrate exoribonuclease activity in bacterially expressed His6-tagged TbDSS-1 (Fig. 2B). However, we were never able to demonstrate enzymatic activity at levels above those in vector control nickel column eluates. Because solubilization of the recombinant protein required denaturation/renaturation, it is possible that only a small fraction of the protein is folded into a conformation that supports enzymatic activity. A more likely explanation is that even properly folded TbDSS-1 lacks enzymatic activity. This would be consistent with the observation that S. cerevisiae DSS-1 is completely dependent on association with the SUV3 helicase for its exoribonuclease activity (17). It is thought that the inability of DSS-1 to act as an exonuclease on its own is due to the lack of an S1 RNA binding domain and that SUV3 is required for substrate interaction. TbDSS-1 also lacks an S1 domain, and an SUV3 RNA helicase homolog is present in the T. brucei genome database.
FIG. 2.
Mitochondrial localization of TbDSS-1. (A) A putative mitochondrial targeting sequence is present in TbDSS-1. The putative N-terminal mitochondrial import sequence of TbDSS-1 was predicted by Target P v1.0. This sequence is aligned with the known and predicted targeting sequences from the T. brucei mitochondrial proteins TBmtRNAP, TBMP48, ISP, ATPase, RBP16, and gBP21 (8, 23, 26, 28, 35, 47). The characteristic one or two N-terminal arginine residues are shown as white letters on a black background. Serine and threonine residues (white letters on a gray background) flank hydrophobic residues that are shown in light gray. (B) Production of bacterially expressed His6-TbDSS-1 (6XHis-TbDSS-1). Proteins were electrophoresed on SDS-PAGE and stained with Coomassie blue. M, molecular mass markers; Un, extracts from uninduced cells; In, extracts from induced cells; P, nickel column-purified His6-TbDSS-1. (C) TbDSS-1 is enriched in mitochondria. Anti-TbDSS-1 antibodies were used to probe blots containing 10 or 25 μg of procyclic-form T. brucei whole-cell or mitochondrial extracts. Anti-RBP16 antibodies (26) were used as a positive control for a known mitochondrial protein.
Southern blot analysis indicates that the TbDSS-1 gene is present as a single copy in the T. brucei genome (data not shown). The genome of the related trypanosomatid, Leishmania major, encodes a predicted protein with 50% amino acid identity to TbDSS-1 (GenBank accession no. AL389894.4), indicating that this mitochondrial exoribonuclease homolog is highly conserved among kinetoplastid species.
Mitochondrial localization of TbDSS-1.
The high degree of homology between TbDSS-1 and known mitochondrial exoribonucleases suggested that TbDSS-1 might be localized to the mitochondria of T. brucei. To further address this issue, we examined the TbDSS-1 amino acid sequence for regions of homology to known trypanosome mitochondrial import sequences (25, 35). The amino terminus of the TbDSS-1 ORF exhibits several characteristics of such sequences, including tandem arginine residues followed by multiple hydrophobic amino acids with interspersed and flanking serine and threonine residues (Fig. 2A). Furthermore, both the TargetP and PSORTII programs predict a mitochondrial localization for TbDSS-1. To biochemically define the subcellular localization of TbDSS-1, we expressed His6-tagged TbDSS-1 in E. coli and purified the protein by denaturing nickel chelate chromatography (Fig. 2B). Antibodies generated against the renatured protein were then used to analyze the TbDSS-1 protein in T. brucei whole-cell and mitochondrial extracts by Western blot (Fig. 2C). Anti-TbDSS-1 antibodies recognized a protein with an apparent molecular mass of approximately 90 kDa in whole-cell and mitochondrial extracts, in agreement with the predicted mass of 83.5 kDa. Comparison of TbDSS-1 levels in whole-cell and mitochondrial extracts reveals that TbDSS-1 is enriched approximately 10-fold in mitochondrial lysates. This is similar to the degree of enrichment observed for the mitochondrial RNA binding protein, RBP16, which was probed on the same blot. Based on the sequence and biochemical evidence, we conclude that the TbDSS-1 protein is a nuclearly encoded, mitochondrially localized protein.
TbDSS-1 is does not appear to be associated with mitochondrial ribosomes.
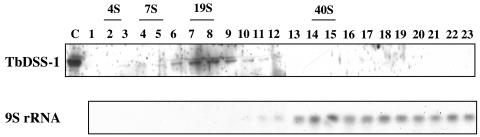
Sucrose gradient analysis of yeast mitochondria indicated that degradosomes, including their DSS-1 component, are entirely associated with mitochondrial ribosomes (17). To determine if TbDSS-1 is similarly ribosome associated in trypanosomes and to assess its potential association with other mitochondrial components, we fractionated T. brucei mitochondrial extracts on 10 to 40% glycerol gradients (11) (Fig. 3). Gradient fractions were analyzed by Western blot to determine the sedimentation of TbDSS-1. RNA was isolated from gradient fractions, and 9S rRNA was analyzed by Northern blot to determine the sedimentation of mitochondrial ribosomes. TbDSS-1 protein exhibited a broad distribution in fractions 3 to 12, which correspond to S values of approximately 5 to 30S. These data suggest that at least some fraction of TbDSS-1 is present in multicomponent complexes, as expected if it is a constituent of the mitochondrial degradosome. In contrast to the sedimentation of TbDSS-1, 9S rRNA was detected in the bottom half of the gradient, primarily in fractions corresponding to S values of 35S or greater. We observed almost no overlap in the distribution of TbDSS-1 and mitochondrial ribosomes. Thus, our initial experiments suggest that TbDSS-1 is complexed with other mitochondrial proteins, but is not stably associated with ribosomes.
FIG. 3.
Glycerol gradient analysis of TbDSS-1. Mitochondrial extract from procyclic-form T. brucei was fractionated on 10 to 40% glycerol gradients. The sedimentation of TbDSS-1 protein was determined by Western blot. The position of mitochondrial ribosomes was identified by Northern blot analysis of 9S rRNA. Fraction numbers and the positions of size standards are indicated above the figure. C, His6-TbDSS-1.
TbDSS-1 is essential in procyclic T. brucei.
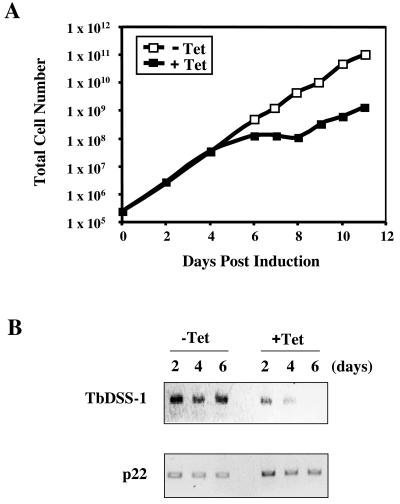
To begin to assess the function of TbDSS-1 in vivo, we used RNAi to selectively down-regulate TbDSS-1 mRNA levels. A 450-bp fragment of the TbDSS-1 coding region corresponding to nt 511 to 961 from the start codon was cloned into the pZJM vector between opposing T7 promoters (57). The resulting vector was transformed into T. brucei strain 29-13, which expresses both T7 polymerase and the tetracycline repressor protein (58). Two cultures that had integrated the TbDSS-1 fragment into the rDNA spacer were selected by phleomycin treatment, and one of these cultures was used for subsequent detailed analyses. RNAi was induced by addition of 1-μg/ml tetracyline, and cell growth was monitored in induced versus uninduced cells (Fig. 4A). Cells grew normally for the first 4 days of induction. However, cell growth ceased by day 6 after tetracycline addition, indicating that TbDSS-1 is essential for growth of procyclic-form T. brucei (Fig. 4A). Beyond day 9, cells escaped RNAi and growth resumed, as is commonly observed in T. brucei. To confirm the downregulation of TbDSS-1 mRNA, steady-state TbDSS-1 mRNA levels were analyzed by reverse transcription (RT)-PCR on days 2, 4, and 6 following induction of RNAi and compared to those in uninduced cells. TbDSS-1 mRNA levels were reduced to 43 and 23% of uninduced levels by days 2 and 4, respectively. By 6 days after RNAi induction, TbDSS-1 mRNA was undetectable. mRNA encoding the p22 protein (27) was analyzed as a control for RNA recovery and integrity. Together, these results indicate that TbDSS-1 is an essential gene in procyclic-form T. brucei.
FIG. 4.
Effect of TbDSS-1 RNAi on cell growth. (A) Growth of procyclic-form T. brucei TbDSS-1 RNAi cells either uninduced (open squares) or induced with 1 μg of tetracycline (closed squares). Growth curves were obtained by plotting the total cell number as the product of the cell density and the total dilution. (B) TbDSS-1 mRNA levels were monitored by PCR amplification of full-length TbDSS-1 RNA in induced and uninduced cells on days 2, 4, and 6 after tetracycline addition. p22 mRNA levels were monitored as a control.
Effect of TbDSS-1 downregulation on rRNAs and never edited mRNAs.
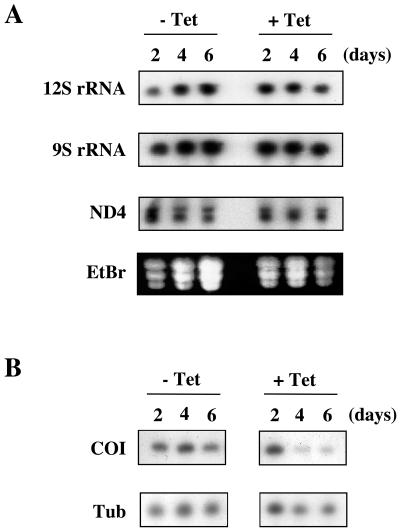
To assess the role of TbDSS-1 in mitochondrial RNA metabolism, we asked whether steady-state levels of specific mitochondrial RNAs were perturbed in TbDSS-1 RNAi cells. We began by looking at RNAs that do not undergo RNA editing, including both rRNAs and never edited mRNAs. In this way, we could determine the role of TbDSS-1 in RNA stability uncomplicated by any potential effects on the editing process. When the abundance of mitochondrial rRNAs was examined by Northern blot analysis, no significant changes in the levels of either mature 9S or 12S rRNA were observed (Fig. 5A). In DSS-1-null mutants in S. cerevisiae, mature mitochondrial 15S rRNAs were significantly decreased, and accumulation of unprocessed precursor RNAs was evident by Northern blot (17). However, even upon long exposures of 9S and 12S Northern blots from TbDSS-1 RNAi cells, we did not observe any larger transcripts indicative of precursor RNAs. Thus, TbDSS-1 does not appear to be involved in rRNA processing in T. brucei mitochondria.
FIG. 5.
Analysis of the effect of TbDSS-1 RNAi on rRNA and never edited mRNA levels. (A) Ten micrograms of RNA isolated from uninduced (−Tet) and induced (+Tet) cells on days 2, 4, and 6 after tetracycline addition was subjected to Northern blot analysis. rRNAs and ND4 mRNA were detected by radiolabeled oligonucleotide probes or a riboprobe, respectively. Ethidium bromide staining is shown to indicate loading. (B) COI mRNA levels were detected by poisoned primer extension. Tubulin (Tub) levels were similarly analyzed as a control.
We also examined the effect of TbDSS-1 downregulation on two never edited mRNAs, NADH dehydrogenase subunit 4 (ND4), and cytochrome oxidase subunit I (COI). As with the rRNAs, ND4 levels were essentially unaffected and no precursor transcripts were detected by Northern blot in TbDSS-1 RNAi cells (Fig. 5A). In contrast, COI mRNA levels, as detected by poisoned primer extension, were dramatically decreased by TbDSS-1 downregulation (Fig. 5B). Taking into account the levels of the tubulin control RNA, COI mRNA in induced cells was reduced to 26% of the levels in uninduced cells by day 4. These results are similar to what was observed in DSS-1-null mutants in yeast, where the COB mRNA was found to be unstable (17). The significantly reduced COI mRNA levels observed upon TbDSS-1 downregulation indicate that this protein plays a role in regulating mitochondrial RNA abundance in T. brucei.
Effect of TbDSS-1 downregulation on edited RNAs.
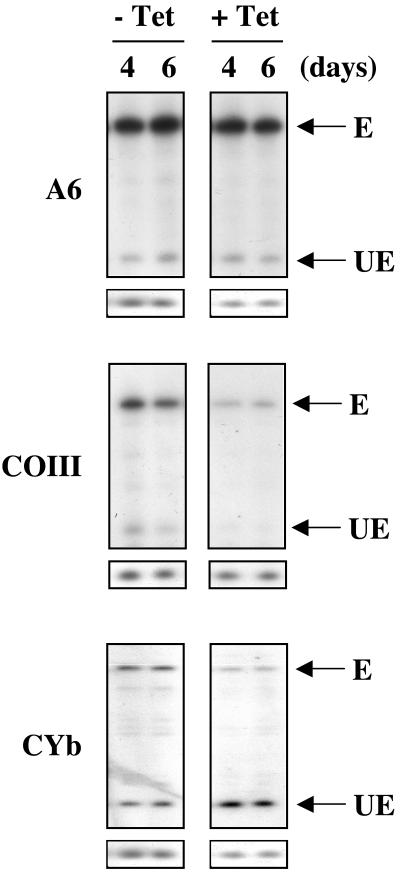
We next wanted to determine whether TbDSS-1 RNAi affected mRNAs that require editing for their maturation. We used a poisoned primer extension assay that allows visualization of both edited and unedited versions of a given RNA in the same assay (41). Interestingly, we observed three different phenotypes regarding edited RNAs in TbDSS-1 RNAi cells as shown in Fig. 6. ATPase subunit 6 (A6) and COIII mRNAs are extensively edited in both the insect (procyclic) and mammalian life cycle stages of T. brucei (5, 19). Neither edited nor unedited A6 mRNA abundance was significantly affected by downregulation of TbDSS-1. On the other hand, we observed that both the unedited and edited versions of COIII mRNAs were dramatically reduced on days 4 and 6 after TbDSS-1 RNAi induction. Accounting for RNA recovery as determined by the tubulin primer extension standard, both edited and unedited COIII mRNA levels were reduced to 20% of those in uninduced cells by day 4. Similar results were observed in RNA preparations from two separate TbDSS-1 RNAi inductions. The corresponding decrease in both edited and unedited mRNA may indicate that TbDSS-1 downregulation directly affects the abundance of unedited COIII mRNA. The decrease in edited RNA levels may then be an indirect effect of decreased editing substrate. It is also possible that both edited and unedited COIII mRNAs are directly destabilized to a similar degree by TbDSS-1 depletion. We cannot currently distinguish between these two possibilities.
FIG. 6.
Analysis of the effect of TbDSS-1 RNAi on edited RNAs. Ten to 15 μg of RNA isolated from uninduced (−Tet) and induced (+Tet) cells on days 4 and 6 after tetracycline addition was subjected to poisoned primer extension analysis. Unedited (UE) and edited (E) A6, COIII, and CYb RNAs are indicated by arrows. The small boxes below each primer extension are the corresponding tubulin control poisoned primer extensions.
Accumulation of edited apocytochrome b (CYb) mRNA is strictly developmentally regulated, with the edited mRNA being present only in procyclic-form cells (18). When the levels of CYb mRNA were examined in TbDSS-1 RNAi cells, we observed both a decrease in edited RNA levels and an increase in unedited CYb mRNA (Fig. 6). Taking into account RNA recovery, edited CYb mRNA levels were reduced to 59 and 47% of uninduced levels on days 4 and 6 after induction, respectively. At the same time, unedited CYb mRNA levels increased by 2.3- and 1.8-fold, on days 4 and 6. We observed similar results in four separate primer extensions: two from each of two individual RNAi inductions. Two different scenarios could account for the reduction in edited CYb mRNA and increase in unedited CYb mRNA upon TbDSS-1 downregulation. First, TbDSS-1 may have opposite effects on the stabilization of edited and unedited CYb mRNAs. Edited CYb mRNA may be stabilized by TbDSS-1 in a manner similar to that observed for COI and COIII mRNAs. On the other hand, TbDSS-1 may normally degrade some percentage of unedited CYb mRNA, leading to an accumulation of this RNA when TbDSS-1 levels are reduced. A second possibility that would simultaneously explain both the decrease in edited RNA and increase in unedited RNA is that TbDSS-1 plays a role in facilitating CYb mRNA editing.
Effect of TbDSS-1 downregulation on gRNA levels.
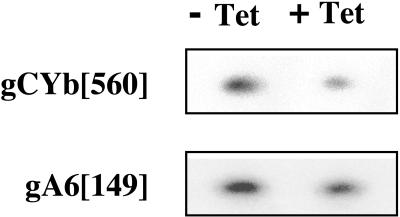
One potential mechanism by which TbDSS-1 could affect CYb mRNA editing is through regulation of gRNA abundance. That is, the decrease in CYb mRNA editing in TbDSS-1 knock-down cells (Fig. 6) may be a direct result of gCYb depletion. To determine whether gRNA levels are affected in TbDSS-1 RNAi cells, we used poisoned primer extension to quantify the levels gRNAs that specify editing of CYb and A6 mRNA in cells that were either uninduced or induced with tetracycline for 4 days (Fig. 7). gCYb[560] directs editing of the 3′ end of the CYb editing domain, which is the same region whose editing was monitored in the experiment presented in Fig. 6. The region of editing specified by gA6[149] is 5′ of the edited region monitored by poisoned primer extension in Fig. 6. We found that both gCYb[560] and gA6[149] levels were decreased in tetracycline-induced cells, but to different extents. In duplicate experiments, we found that gCYb[560] levels in induced cells were 48.9% ± 3.1% of those in uninduced cells. A second gRNA, gCYb[558], can also specify editing of the 3′ region of the CYb edited domain (49). However, we were unable to detect gCYb[558] by poisoned primer extension, Northern blot, or PCR and so could not determine whether this gRNA is affected by TbDSS-1 depletion. The decrease in gCYb[560] is similar in extent to the decrease we observed in CYb mRNA editing on the same day after tetracycline induction (compare Fig. 6 and 7). These results suggest that CYb gRNAs may be limiting and that their depletion leads to a downregulation of CYb RNA editing. gA6[149] levels in induced cells were decreased to 82.0% ± 2.8% of those in uninduced cells. The modest reduction that we observe in gA6[149] compared to gCYb[560] is consistent with the absence of any A6 editing defect upon TbDSS-1 depletion. However, our poisoned primer extension analysis of A6 mRNA does not specifically monitor the region whose editing is directed by gA6[149], so we cannot rule out that a modest reduction in editing is observed in this region of the mRNA in TbDSS-1 knock-down cells. In all, our results indicate that TbDSS-1 downregulation leads to a decrease in the abundance of at least some gRNAs, and the magnitude of the decrease is gRNA specific.
FIG. 7.
Analysis of the effect of TbDSS-1 RNAi on gRNA levels. RNA was isolated from uninduced (−Tet) and induced (+Tet) cells on day 4 after tetracycline addition. Twenty-five or 35 μg of RNA was subjected to poisoned primer extension analysis of gA6[149] and gCYb[560] levels, respectively.
TbDSS-1 does not act indirectly through modulation of known mitochondrial editing and stability factors.
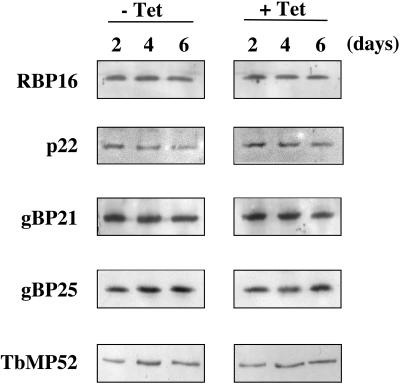
The phenotype of TbDSS-1 RNAi cells is reminiscent of, although not identical to, the phenotype reported for RNAi of several mitochondrial RNA binding proteins. One such protein is RBP16, whose downregulation in procyclic-form T. brucei leads to a significant reduction in the levels of COI and ND4 RNAs, as well as a decrease in edited CYb and a corresponding increase in unedited CYb mRNA (44). Based on these observations, we wanted to determine if TbDSS-1 was acting, at least in part, indirectly through a decrease in RBP16. We performed Western blot analysis of TbDSS-1 RNAi cells both uninduced and induced with tetracycline for 2, 4, or 6 days (Fig. 8). We observed no significant perturbation of RBP16 levels upon TbDSS-1 downregulation. p22 is a T. brucei mitochondrial protein that interacts with RBP16 and can dramatically increase its RNA binding capacity in vitro (27). Thus, it is also possible that RBP16 function could be compromised in vivo by decreased p22 levels. However, Western blot analysis indicated that p22 levels were also unchanged in TbDSS-1 RNAi cells. gBP21 and gBP25 are two RNA binding proteins from T. brucei that bind gRNA with high affinity, and gBP21 may interact transiently with the editosome (2, 6, 28). Bloodstream-form gBP21 knock-outs display greatly decreased levels of COI mRNA as well as several unedited RNAs (31). In addition, simultaneous elimination of gBP21 and gBP25 in procyclic forms results in decreased COI mRNA and edited CYb mRNA levels, among other alterations in mitochondrial RNA abundance (J. Lukes and K. Stuart, personal communication). However, Western blot analysis of gBP21 and gBP25 indicated that the levels of these proteins were also unaffected by TbDSS-1 downregulation. Finally, because the alterations of CYb mRNA levels in TbDSS-1 RNAi cells suggested a possible editing defect, we determined whether levels of an editosome component were altered. Analysis of TbMP52 RNA ligase abundance by Western blot in TbDSS-1 RNAi cells revealed no changes in the levels of this core editosome component. Taken together, the Western blot data indicate that the perturbations of mitochondrial RNA metabolism observed upon downregulation of TbDSS-1 are not secondary effects attributable to known mitochondrial RNA processing factors. Thus, TbDSS-1 appears to directly affect the abundance of several classes of mitochondrial RNAs in procyclic-form T. brucei.
FIG. 8.
Western blot analysis of known mitochondrial RNA stability and editing factors in TbDSS-1 RNAi cells. Extracts from TbDSS-1 RNAi cells either uninduced or induced with tetracycline for 2, 4, or 6 days were electrophoresed on an SDS-PAGE (10% polyacrylamide) gel (TbMP52 blot) or an SDS-PAGE (15% polyacrylamide) gel (all other blots) and transferred to nitrocellulose membrane. Blots were probed with antibodies against RBP16, p22, gBP21, gBP25, and TbMP52.
DISCUSSION
In this study, we present the characterization of a putative exoribonuclease from T. brucei mitochondria, termed TbDSS-1. TbDSS-1 was identified through a BLAST search of the T. brucei genome databases as an RNR exoribonuclease family member and a homolog of the S. cerevisiae DSS-1 and N. crassa cyt-4 mitochondrial 3′-to-5′ exoribonucleases. TbDSS-1 possesses a predicted N-terminal mitochondrial import sequence, and Western blot analysis of whole-cell and mitochondrial extracts indicates the protein is highly enriched in mitochondria. In yeast, the DSS-1 exoribonuclease associates with the SUV3 RNA helicase to form the mitochondrial degradosome (17). Although SUV3 homologs are present in human (40) and Arabidopsis (20) mitochondria, DSS-1 homologs have not been identified in these organisms. Thus, this study marks the first report of a DSS-1 exoribonuclease homolog outside of the fungal kingdom. A protein with high homology to the S. cerevisiae SUV3 RNA helicase is also present in the T. brucei database. However, we do not yet know if this protein is associated with TbDSS-1 in a degradosome complex. It is of interest in this regard that we were unable to demonstrate exoribonuclease activity in recombinant TbDSS-1. Yeast DSS-1 is strictly dependent on association with SUV3 for its exoribonuclease activity, probably because it is unable to bind RNA with high affinity. Mitochondrial RNR exoribonuclease family members, including TbDSS-1, differ from their prokaryotic counterparts in that they lack a C-terminal S1 RNA binding domain. It is thought that the SUV3 helicase is required for both substrate recognition and unwinding in the yeast degradosome complex. Studies are under way to determine the protein composition and enzymatic activities of the T. brucei mitochondrial degradosome.
Targeted disruption of TbDSS-1 using RNAi demonstrates that this protein is essential for growth in procyclic-form T. brucei. Western blot analyses of several known mitochondrial RNA stability and editing factors indicate that this is not an indirect effect due to alterations in the abundance of these proteins. The essential nature of TbDSS-1 is in contrast to the phenotype of S. cerevisiae DSS-1-null mutants which are viable, albeit respiratory incompetent (14). It will be of interest to determine the phenotype of TbDSS-1 disruption in bloodstream-form trypanosomes, which rely primarily on glycolysis for energy generation. Because bloodstream forms do not depend on cytochrome-mediated respiration, TbDSS-1 may not be as critical in this life cycle stage.
In procyclic trypanosomes, TbDSS-1 depletion results in aberrant levels of several mitochondrial RNA species, including never edited, unedited, and edited mRNAs as well as gRNAs. The diversity of phenotypes associated with TbDSS-1 downregulation suggests participation of this protein in multiple aspects of mitochondrial RNA metabolism. We showed that never edited COI and both unedited and edited COIII mRNAs are significantly reduced in abundance after TbDSS-1 RNAi. It may seem paradoxical that depletion of a putative exoribonuclease would lead to decreased RNA levels. However, this is similar to what was observed in both DSS-1- and SUV3-null mutants in yeast, where mature 15S rRNA and COB mRNAs were significantly reduced (17). There are several potential explanations for the observation of decreased RNA levels in TbDSS-1 RNAi cells. First, since trypanosome mitochondrial RNAs are polycistronically transcribed and require 5′ and 3′ processing for maturation, it is possible that TbDSS-1 is involved in these processing events. If precursor transcripts are improperly processed, this could result in lower levels of mature mRNA species. Future experiments will be aimed at characterization of precursor RNAs in TbDSS-1-depleted cells. A second possibility is that TbDSS-1 and/or a degradosome complex of which it is a component acts to stabilize mRNAs, either directly or indirectly. In E. coli, depletion of RNase II leads to destabilization of mRNAs ending in hairpins, whereas RNase II overproduction stabilizes some RNA species (34). RNase II preferentially degrades poly(A) tails, and its depletion leads to an increase in polyadenylated mRNAs. Because poly(A) tails are destabilizing elements in prokaryotes, the expanded population of polyadenylated RNAs is then subject to increased rates of degradation by a second exonuclease (34). We recently reported that polyadenylation destabilizes unedited RNAs in partially purified T. brucei mitochondrial extracts (51). If TbDSS-1 normally degrades the poly(A) tails of some mitochondrial RNA species, its downregulation could result in increased polyadenylation and subsequent RNA destabilization, analogous to E. coli RNase II. A third potential scenario is that when TbDSS-1 levels are depleted by RNAi, the levels of other mitochondrial ribonucleases are increased in an effort to compensate. This would be reminiscent of the reported situation in E. coli, where polynucleotide phosphorylase (PNPase) levels are increased about twofold in RNase II-deficient strains, and RNase II activity is increased about twofold when the PNPase gene is deleted (59). Testing of this hypothesis awaits identification of additional mitochondrial exoribonuclease genes and production of antibodies against the encoded proteins. Finally, the mitochondrial degradosome may be comprised of multiple exoribonucleases, similar to the cytoplasmic and nuclear exosome, some of which become deregulated upon depletion of TbDSS-1. Identification of mitochondrial degradosome components will begin to address this possibility.
The effect of TbDSS-1 downregulation on edited mRNAs differs, depending on the RNA analyzed. Whereas both edited and unedited COIII mRNAs are decreased, in the case of CYb only the edited mRNA is decreased. Coincident with the decrease in edited CYb mRNA, unedited CYb mRNA levels are increased in TbDSS-1-depleted cells. The CYb phenotype could result from opposite effects on edited and unedited mRNA stabilization. That is, the abundance of edited CYb mRNA may be decreased through one or more of the mechanisms described above. Conversely, TbDSS-1 may normally regulate unedited CYb mRNA levels by degrading transcript in excess of what is required, leading to accumulation of the unedited transcript when the enzyme is depleted. Alternatively, TbDSS-1 may act at the level of CYb RNA editing. We favor the latter model, since our analysis of gRNA levels in TbDSS-1 knock-down cells reveals a corresponding decrease in gCYb[560] and edited CYb mRNA. Interestingly, the regulation of gRNA levels by TbDSS-1 appears to be somewhat gRNA specific. While gCYb[560] levels in induced TbDSS-1 knock-down cells were less than 50% of those in uninduced cells, gA6[149] was still present at greater than 80% of uninduced levels. The absence of a dramatic decrease in gA4[149] levels is consistent with unaltered A6 editing in TbDSS-1 knock-down cells.
How could TbDSS-1 specificity for regulating certain gRNAs, in this case gCYb[560], be achieved? A potential point of regulation is gRNA processing. gRNAs are polycistronically transcribed from their minicircle-encoded genes (22). Results based on an in vitro processing system suggest that mature gRNAs are produced by accurate transcription initiation at the 5′ end and ribonucleolytic processing at the 3′ end (22). If TbDSS-1 is involved in gRNA 3′ end processing, this could result in a decrease in mature gRNAs and a subsequent decrease in RNA editing. Intriguingly, the 3′-most CYb gRNAs, gCYb[558] and gCYb[560], are present in an unusual minicircle location (49). While the vast majority of gRNAs are encoded in cassettes defined by conserved inverted 18-bp repeats, the known CYb gRNA genes are present not within a cassette, but between cassettes encoding other gRNAs. This suggests that their processing may differ somewhat from that of most gRNAs. In support of this hypothesis, gCYb[560]-containing chimeras with 19- to 20-nt 3′ extensions beyond that predicted to guide accurate editing have been reported (49). Thus, gCYbs may be inherently more subject to inaccurate 3′ processing than other gRNAs. Future experiments will be aimed at determining if TbDSS-1 plays a role in gRNA maturation.
In summary, the results presented here indicate that TbDSS-1, the trypanosome homolog of the yeast mitochondrial DSS-1 exoribonuclease, is mitochondrially localized and has effects on the steady-state levels of mature RNAs, potentially at the levels of RNA stability and RNA editing. The presence of both DSS-1 exoribonuclease and SUV3 helicase homologs in T. brucei suggests that trypanosomes may possess a mitochondrial degradosome similar in structure to that described in S. cerevisiae (17). This is in contrast to plants and mammals, which apparently lack DSS-1 homologs. Nevertheless, our results suggest that structural and functional differences exist between yeast and T. brucei DSS-1 homologs. TbDSS-1 does not appear to be stably associated with ribosomes and does not participate in rRNA maturation like its yeast counterpart. Moreover, the role of TbDSS-1 in modulating levels of specific edited RNAs, whether direct or indirect, constitutes a novel function for this family of enzymes.
Acknowledgments
This work was supported by National Institutes of Health grant AI47329 to L.K.R. C.M.R. was supported in part by National Institutes of Health training grant AI07614.
REFERENCES
- 1.Alfonzo, J. D., O. H. Thiemann, and L. Simpson. 1998. Purification and characterization of MAR1, a mitochondrial associated ribonuclease from Leishmania tarentolae. J. Biol. Chem. 273:30003-30011. [DOI] [PubMed] [Google Scholar]
- 2.Allen, T. E., S. Heidmann, R. Reed, P. J. Myler, H. U. Göringer, and K. D. Stuart. 1998. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol. Cell. Biol. 18:6014-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aphasizhev, R., and L. Simpson. 2001. Isolation and characterization of a U-specific 3′-5′-exonuclease from mitochondria of Leishmania tarentolae. J. Biol. Chem. 276:21280-21284. [DOI] [PubMed] [Google Scholar]
- 4.Aphasizhev, R., I. Aphasizheva, R. E. Nelson, G. Gao, A. M. Simpson, X. Kang, A. M. Falick, S. Sbicego, and L. Simpson. 2003. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 22:913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhat, G. J., D. J. Koslowsky, J. E. Feagin, B. L. Smiley, and K. Stuart. 1988. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell 61:885-894. [DOI] [PubMed] [Google Scholar]
- 6.Blom, D., J. V. Burg, C. K. Breek, D. Speijer, A. O. Muijsers, and R. Benne. 2001. Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21. Nucleic Acids Res. 29:2950-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brun, R., and M. Schonenberg. 1979. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 8.Chi, T. B., S. V. Brown, and N. Williams. 1998. Subunit 9 of the mitochondrial ATP synthase of Trypanosoma brucei is nuclearly encoded and developmentally regulated. Mol. Biochem. Parasitol. 92:29-38. [DOI] [PubMed] [Google Scholar]
- 9.Conrad-Webb, H., P. S. Perlman, H. Zhu, and R. A. Butow. 1990. The nuclear SUV3-1 mutation affects a variety of post-transcriptional processes in yeast mitochondria. Nucleic Acids Res. 18:1369-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corell, R. A., P. Myler, and K. Stuart. 1994. Trypanosoma brucei mitochondrial CR4 gene encodes an extensively edited mRNA with completely edited sequence only in bloodstream forms. Mol. Biochem. Parasitol. 64:65-74. [DOI] [PubMed] [Google Scholar]
- 11.Corell, R. A., L. K. Read, G. R. Riley, J. K. Nellissery, T. E. Allen, M. L. Kable, M. D. Wachal, S. D. Seiwert, P. J. Myler, and K. Stuart. 1996. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol. Cell. Biol. 16:1410-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decker, C. J., and B. Sollner-Webb. 1990. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell 61:1001-1011. [DOI] [PubMed] [Google Scholar]
- 13.Deutscher, M. P., and Z. Li. 2001. Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 66:67-105. [DOI] [PubMed] [Google Scholar]
- 14.Dmochowska, A., P. Golik, and P. P. Stepien. 1995. A novel nuclear gene DSS-1 of Saccharomyces cerevisiae is necessary for mitochondrial biogenesis. Curr. Genet. 28:108-112. [DOI] [PubMed] [Google Scholar]
- 15.Dobinson, K. F., M. Henderson, R. L. Kelley, R. A. Collins, and A. M. Lambowitz. 1989. Mutations in nuclear gene cyt-4 of Neurospora crassa result in pleiotropic defects in processing and splicing of mitochondrial RNAs. Genetics 123:97-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dziembowski, A., M. Malewicz, M. Minczuk, P. Golik, A. Dmochowska, and P. P. Stepien. 1998. The yeast nuclear gene DSS1, which codes for a putative RNase II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol. Gen. Genet. 260:108-114. [DOI] [PubMed] [Google Scholar]
- 17.Dziembowski, A., J. Piwowarski, R. Hoser, M. Minczuk, A. Dmoshowska, M. Siep, H. van der Spek, L. Grivell, and P. P. Stepien. 2003. The yeast mitochondrial degradosome, its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 278:1603-1611. [DOI] [PubMed] [Google Scholar]
- 18.Feagin, J. E., D. P. Jasmer, and K. Stuart. 1987. Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell 49:337-345. [DOI] [PubMed] [Google Scholar]
- 19.Feagin, J. E., J. M. Abraham, and K. Stuart. 1988. Extensive editing of the cytochrome c oxidase III in Trypanosoma brucei. Cell 53:413-422. [DOI] [PubMed] [Google Scholar]
- 20.Gagliardi, D., J. Kuhn, U. Spadinger, A. Brennicke, C. J. Leaver, and S. Binder. 1999. An RNA helicase (AtSUV3) is present in Arabidopsis thaliana mitochondria. FEBS Lett. 458:337-342. [DOI] [PubMed] [Google Scholar]
- 21.Golik, P., T. Szczcepancek, E. Bartnik, P. P. Stepien, and J. Lasowska. 1995. The S. cerevisiae nuclear gene SUV3 encoding a putative RNA helicase is necessary for the stability of mitochondrial transcripts containing multiple introns. Curr. Genet. 28:217-224. [DOI] [PubMed] [Google Scholar]
- 22.Grams, J., M. T. McManus, and S. L. Hajduk. 2000. Processing of polycistronic guide RNAs is associated with RNA editing complexes in Trypanosoma brucei. EMBO J. 19:5525-5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grams, J., J. C. Morris, M. E. Drew, Z. Wang, P. T. Englund, and S. L. Hajduk. 2002. A trypanosome mitochondrial RNA polymerase is required for transcription and replication. J. Biol. Chem. 277:16952-16959. [DOI] [PubMed] [Google Scholar]
- 24.Harris, M. E., D. R. Moore, and S. L. Hajduk. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 267:23963-23971. [PubMed] [Google Scholar]
- 25.Hausler, T., Y. D. Stierhof, J. Blattner, and C. Clayton. 1997. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur. J. Cell Biol. 73:240-251. [PubMed] [Google Scholar]
- 26.Hayman, M. L., and L. K. Read. 1999. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem. 274:12067-12074. [DOI] [PubMed] [Google Scholar]
- 27.Hayman, M. L., M. M. Miller, D. M. Chandler, C. G. Goulah, and L. K. Read. 2001. The trypanosome homolog of human p32 interacts with RBP16 and stimulates its gRNA binding activity. Nucleic Acids Res. 29:5216-5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Köller, J., U. F. Müller, B. Schmid, A. Missel, V. Kruft, K. Stuart, and H. U. Göringer. 1997. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 272:3749-3757. [DOI] [PubMed] [Google Scholar]
- 29.Koslowsky, D. J., G. J. Bhat, L. K. Read, and K. Stuart. 1991. Cycles of progressive realignment of gRNA with mRNA in RNA editing. Cell 67:537-546. [DOI] [PubMed] [Google Scholar]
- 30.Koslowsky, D. J., and G. Yahampath. 1997. Mitochondrial mRNA 3′ cleavage/polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol. Biochem. Parasitol. 90:81-94. [DOI] [PubMed] [Google Scholar]
- 31.Lambert, L., U. F. Müller, A. E. Souza, and H. U. Göringer. 1999. The involvement of gRNA-binding protein gBP21 in RNA editing—an in vitro and in vivo analysis. Nucleic Acids Res. 27:1429-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madison-Antenucci, S., R. S. Sabatini, V. W. Pollard, and S. L. Hajduk. 1998. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J. 17:6368-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margossian, S. P., H. Li, H. P. Zassenhous, and R. A. Butow. 1996. The DexH box protein SUV3p is a component of a yeast mitochondrial 3′-to-5′ exoribonuclease that suppresses group I intron toxicity. Cell 84:199-209. [DOI] [PubMed] [Google Scholar]
- 34.Marujo, P. E., E. Hajnsdorf, J. Le Derout, R. Andrade, C. M. Arraiano, and P. Regnier. 2000. RNase II removes oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA 6:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McManus, M. T., M. Shimamura, J. Grams, and S. L. Hajduk. 2001. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA 7:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mian, I. S. 1997. Comparative sequence analysis of ribonucleases HII, III, II, PH, and D. Nucleic Acids Res. 25:3187-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michelotti, E. F., M. E. Harris, B. Adler, A. F. Torri, S. L. Hajduk. 1992. Trypanosoma brucei mitochondrial ribosomal RNA synthesis, processsing and developmentally regulated expression. Mol. Biochem. Parasitol. 54:31-42. [DOI] [PubMed] [Google Scholar]
- 38.Militello, K. T., and L. K. Read. 2000. UTP-dependent and -independent pathways of mRNA turnover in Trypanosoma brucei mitochondria. Mol. Cell. Biol. 20:2308-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min, J., R. M. Heuertz, and H. P. Zassenhaus. 1993. Isolation and characterization of an NTP-dependent 3′-exoribonuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 268:7350-7357. [PubMed] [Google Scholar]
- 40.Minczuk, M., J. Piwowarski, M. A. Papworth, K. Awiszus, S. Schalinski, A. Dziembowski, A. Dmochowska, E. Bartnik, K. Tokatlidis, P. P. Stepien, and P. Borowski. 2002. Localisation of the human hSuv3p helicase in the mitochondrial matrix and its preferential unwinding of dsDNA. Nucleic Acids Res. 30:5074-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Missel, A., A. E. Souza, G. Nörskau, and H. U. Göringer. 1997. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 17:4895-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panigrahi, A. K., S. P. Gygi, N. L. Ernst, R. P. Igo, Jr., S. S. Palazzo, A. Schnaufer, D. S. Weston, N. Carmean, R. Salavati, R. Aebersold, and K. D. Stuart. 2001. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:380-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panigrahi, A. K., A. Schnaufer, N. L. Ernst, B. Wang, N. Carmean, R. Salavati, and K. Stuart. 2003. Identification of novel components of Trypanosoma brucei editosomes. RNA 9:484-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelletier, M., and L. K. Read. 2003. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial RNAs in Trypanosoma brucei. RNA 9:457-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piller, K. J., L. N. Rusché, J. Cruz-Reyes, and B. Sollner-Webb. 1997. Resolution of the RNA editing gRNA-directed endonuclease from two other endonucleases of Trypanosoma brucei mitochondria. RNA 3:279-290. [PMC free article] [PubMed] [Google Scholar]
- 46.Pollard, V. W., M. E. Harris, and S. L. Hajduk. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11:4429-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Priest, J. W., and S. L. Hajduk. 1995. The trypanosomatid Rieske iron-sulfur proteins have a cleaved presequence that may direct mitochondrial import. Biochim. Biophys. Acta 1269:201-204. [DOI] [PubMed] [Google Scholar]
- 48.Read, L. K., P. J. Myler, and K. Stuart. 1992. Extensive editing of both processed and preprocessed maxicircle CR6 transcripts in Trypanosoma brucei. J. Biol. Chem. 267:1123-1128. [PubMed] [Google Scholar]
- 49.Riley, G. R., R. A. Corell, and K. Stuart. 1994. Multiple guide RNAs for identical editing of Trypanosoma brucei apocytochrome b mRNA have an unusual minicircle location and are developmentally regulated. J. Biol. Chem. 269:6101-6108. [PubMed] [Google Scholar]
- 50.Rusché, L. N., J. Cruz-Reyes, K. J. Piller, and B. Sollner-Webb. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16:4069-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryan, C. M., K. T. Militello, and L. K. Read. 2003. Polyadenylation regulates the stability of Trypanosoma brucei mitochondrial RNAs. J. Biol. Chem. 278:32753-32762. [DOI] [PubMed] [Google Scholar]
- 52.Salavati, R., A. Panigrahi, and K. D. Stuart. 2001. Mitochondrial ribonuclease P activity of Trypanosoma brucei. Mol. Biochem. Parasitol. 115:109-117. [DOI] [PubMed] [Google Scholar]
- 53.Salavati, R., A. K. Panigrahi, B. A. Morach, S. S. Palazzo, R. P. Igo, and K. Stuart. 2002. Endoribonuclease activities of Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol. 120:23-31. [DOI] [PubMed] [Google Scholar]
- 54.Schnaufer, A., A. K. Panigrahi, B. Panicucci, R. P. Igo, R. Salavati, and K. Stuart. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291:2159-2162. [DOI] [PubMed] [Google Scholar]
- 55.Simpson, L., R. Aphasizhev, G. Gao, and X. Kang. 2004. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA 10:159-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart, K., and A. K. Panigrahi. 2002. RNA editing: complexity and complications. Mol. Microbiol. 45:591-596. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:174-179. [DOI] [PubMed] [Google Scholar]
- 58.Wirtz, E., S. Leal, C. Ochatt, and G. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 59.Zilhão, R., F. Cairrão, P. Régnier, and C. M. Arraiano. 1996. PNPase modulates RNase II expression in Escherichia coli: implications for mRNA decay and cell metabolism. Mol. Microbiol. 20:1033-1042. [DOI] [PubMed] [Google Scholar]
- 60.Zuo, Y., and M. P. Deutscher. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]