Abstract
The RABiT (Rapid Automated Biodosimetry Tool) is a dedicated Robotic platform for the automation of cytogenetics-based biodosimetry assays. The RABiT was developed to fulfill the critical requirement for triage following a mass radiological or nuclear event. Starting from well-characterized and accepted assays we developed a custom robotic platform to automate them. We present here a brief historical overview of the RABiT program at Columbia University from its inception in 2005 until the RABiT was dismantled at the end of 2015. The main focus of this paper is to demonstrate how the biological assays drove development of the custom robotic systems and in turn new advances in commercial robotic platforms inspired small modifications in the assays to allow replacing customized robotics with ‘off the shelf’ systems. Currently, a second-generation, RABiT II, system at Columbia University, consisting of a PerkinElmer cell::explorer, was programmed to perform the RABiT assays and is undergoing testing and optimization studies.
INTRODUCTION
In 2005, the US government identified the development of improved methods for radiation biodosimetry as a high priority need in an environment of heightened concern over possible nuclear or radiological terrorist attacks(1). To fulfill this need, the Columbia Center for High Throughput Minimally Invasive Radiation Biodosimetry, funded by NIAID (National Institute of Allergy and Infectious Diseases), began the development of the Rapid Automated Biodosimetry Tool (RABiT), as an end-to-end automated, ultra-high throughput robotically-based biodosimetry workstation(2–7).
Although different procedural steps involved in cytogenetic bioassays (from chromosome preparations(8), to scoring(9) and sample tracking(10)) had previously been automated, the RABiT is presumably the first end-to-end system developed thus far where the only human intervention with samples is at the sample collection step(11) and interpretation of the reported results by a qualified medical professional.
This manuscript provides a historical overview of the RABiT development and testing at Columbia University, focusing on how the assays have driven robotics development and vice versa. At the time of the RABiT program inception, the Cytokinesis Blocked Micronucleus (CBMN) assay, dicentric assay and to a lesser degree gamma-H2AX assay were already well characterized and accepted. We selected to automate the CBMN and gamma-H2AX assays as they required much of the same processing and imaging steps. The dicentric assay was considered much more complex both in terms of the sample handling (e.g. preparing the chromosome spreads) and in terms of the complex imaging required.
THE ORIGINAL ASSAYS
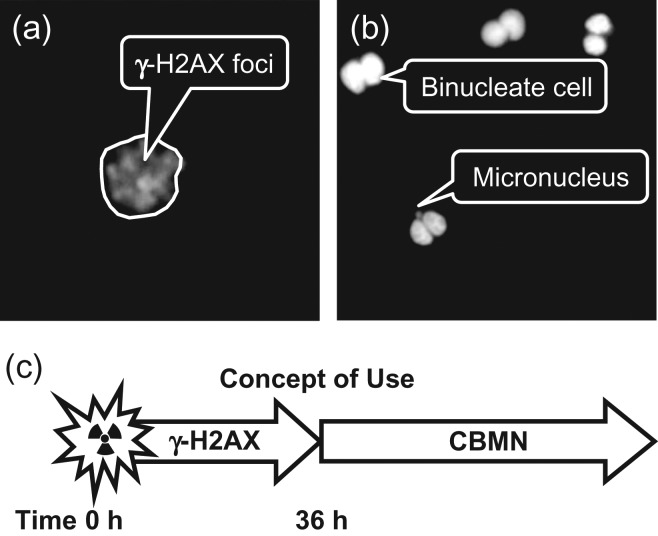
The RABiT was originally designed to automate two well-known, biodosimetry assays (Figure 1): (a) the gamma-H2AX assay(12, 13), which quantifies DNA double-strand breaks by immunolabeling of phosphorylated histone H2AX and (b) the CBMN assay(14–17), which measures persistent chromosomal aberrations as post-mitotic micronuclei. As the gamma-H2AX assay is rather rapid (requiring only a few hours to provide a dose estimate, as compared with 72 h for the CBMN assay), it is the preferred assay. Unfortunately, its time of persistence is short, with the signal fading over 24–48 h (whereas the micronucleus assay has a half-life of about a year(18)). The RABiT was therefore designed to utilize both of these bioassays depending on the time post-irradiation the samples are received: gamma-H2AX assay for samples received within 36 h of exposure and the micronucleus assay for samples received after a substantial delay (>2 days to months).
Figure 1.
The original RABiT assays. (a) Gamma-H2AX foci and DAPI counterstain in 4 Gy irradiated lymphocyte. (b) Micronucleus in binucleate cell (also in 4 Gy irradiated lymphocytes). (c) The concept of operation, gamma-H2AX analysis for all samples arriving within 36 h of irradiation, followed by a quick switchover and CBMN analysis for all subsequent samples.
These two assays drove the development for an automated platform with continued interactive efforts between the biology team and the engineering team as the assays were optimized to facilitate automation in multiwell plates and the robotics to perform the assays in an optimal manner.
AUTOMATION
RABiT development was divided into two phases with a simpler phase 1 system initially developed for lower throughput (~6000 samples per day)(3) and a phase 2 system with improved performance and throughput reaching 30 000 samples per day(4).
Due to the extended nature of the assays, and the parallelized nature of sample handling in the RABiT, we define throughput as the number of samples entering/exiting the system at steady state. High throughputs are achievable, despite the long processing times because (a) 96 samples are processed per plate and (b) multiple plates passing through the system at once.
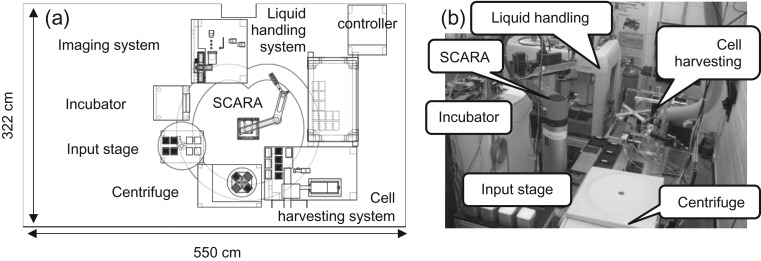
The layout of the RABiT (Figure 2) consists of several processing stations located around a central Selectively Compliant Articulated Robot Arm (SCARA).
Figure 2.
(a) Layout of the phase 1 RABiT (©2008 IEEE. Reprinted, with permission, from (2)). (b) Photo of the phase 2 RABiT.
Ideally, blood samples (30 µl) are collected in the field by finger stick into heparinized, barcoded capillary tubes and placed into the holders containing separation medium and sealing material(11). The blood-filled capillaries are shipped in these capillary holders (32 capillaries each) which, upon delivery to the RABiT, are placed onto the input stage. The SCARA robot then loads the four centrifuge buckets (containing 3 holders each, for a total of 384 capillaries) into a centrifuge, which separates the white buffy coat (lymphocyte band) from the red blood cells. The SCARA robot then transfers the buckets to the harvesting station. At this point 7 min have elapsed.
At the harvesting station, isolated lymphocytes are transferred from the capillaries to four filter-bottom 96-well plates. In the phase 1 implementation, the harvesting module used the SCARA robot to move each capillary between the different processing steps, requiring ~11 s to sequentially process each capillary (18 min per plate). This also had the effect of tying up the SCARA robot, restricting its ability to load and unload the centrifuge and to transfer multiwell plates between the harvest station, liquid handling system and incubator.
Following harvesting, each plate, containing 96 samples, is transferred to a liquid handling system, which can dispense reagents into the plate or drain reagents through the filter bottoms. This system, in conjunction with a robotic incubator, was programmed to perform the gamma-H2AX(12) and CBMN assays. It should be noted that for both the 3-h gamma-H2AX assay and the 72-h CBMN assay, the liquid handling system is idle for most of the processing time, allowing multiple plates to run in parallel.
Following the assay, the filter bottoms, containing fluorescently-labeled cells, are rapidly removed from the multiwell plates, using a custom robot(6) to facilitate imaging with high NA/short working distance lenses, in a dedicated imaging system, developed to support the RABiT. This novel automated imaging system made use of multiple simultaneous light paths, one step automated focusing and light steering(6) to accelerate grabbing multiple adjacent fields of view. Image analysis software was written to allow both on- and off-line analysis for all assays.
In 2009, the phase 1 system prototype was finalized and tested, with a throughput of up to 6000 samples per day, using the gamma-H2AX assay, which is the more complex of the two (in terms of the number of assay steps). After a successful test, development shifted gears to upgrading the system to a higher throughput of 30 000 samples per day. Following an in-depth analysis of the bottlenecks in the system, it was determined that the cell harvest station was the main limitation on throughput, since samples are handled individually rather than in parallel.
The tasks performed in this workstation are the following: (a) read the barcode on the capillary (and associate it with the barcode on a multiwell plate and the well number), (b) locate the lymphocyte band, (c) cut the capillary below the lymphocyte band (using a UV laser), (d) discard the red blood cells and (e) dispense the lymphocytes into the plate and discard the capillary.
In phase 2, the harvest station was parallelized(4) to process four capillaries at once, using dedicated robotics, with a 5-fold reduction in the processing time of 2.2 s per capillary (8.8 s for four capillaries), freeing up the SCARA robot for other tasks.
An additional change required for the RABiT to achieve high throughput was to increase the incubator capacity, allowing storage of more samples during the culturing step of the micronucleus assay and this could not be accomplished due to financial constraints.
THE SECOND FUNDING CYCLE (2010–2015)
In 2010, the Center was funded for an additional 5 years. At this point developments transitioned to optimization of the existing system(7, 19), developing novel bioassays could be performed on the RABiT and testing the existing assays under complex radiation exposure scenarios(20–22). The gamma-H2AX and CBMN RABiT assays were transitioned to BARDA funding for further development and rigorous validation in preparation for licensing by the Food and Drug Administration.
New cytogenetic assays
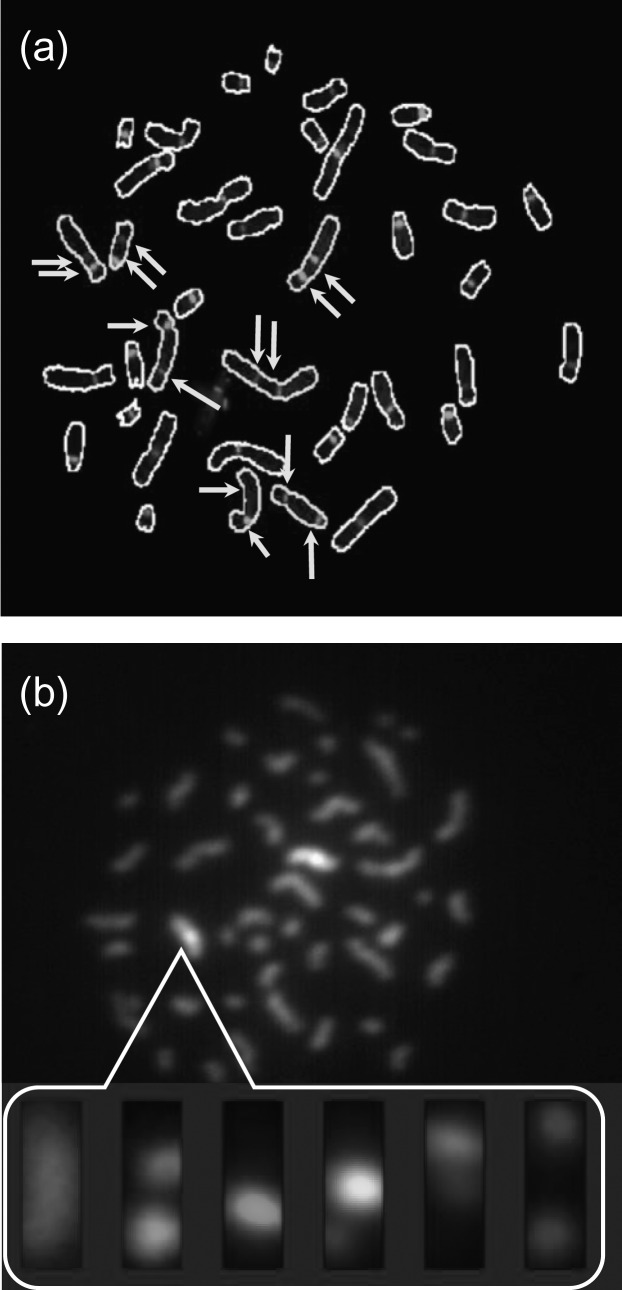
In this cycle, two additional assays, based on partial chromosome painting, were developed for incorporation into the RABiT: the Dicentric assay(23) (Figure 3a), which is the gold standard for radiation biodosimetry. Here, dicentric chromosomes are rapidly identified and scored by hybridizing to a centromeric PNA probe and identifying chromosomes with two (dicentric) vs one centromeric (monocentric) spot. This greatly facilitates the scoring, replacing complex morphometric analysis with spot counting.
Figure 3.
Chromosome-based assays developed for the RABiT: (a) Dicentrics and (b) mBAND. Detected chromosomes in (a) are circled. Arrows denote the centromeres in dicentric chromosomes. The inset in (b) shows the individual images for the six dyes used to determine the band structure in the indicated chromosome.
The mBAND (multicolor band) assay(24, 25) (Figure 3b). Here intra-arm and inter-arm chromosomal rearrangements, specific to densely ionizing radiations (e.g. neutrons), are scored by ‘barcoding’ the entire chromosome length using partial paints, scoring deviations from the normal pattern of bands.
Both of these assays require much higher resolution imaging than the gamma-H2AX and CBMN assays. The imaging system was therefore equipped with a high NA 60× oil immersion lens and a high sensitivity, high-resolution scientific CMOS camera(26). The multiple optics paths were eliminated as it was deemed unfeasible to expand this feature to support six fluorochromes(27). The filter-bottom plates were also eliminated at this stage as they are incompatible with chromosome processing and imaging due to their 1 µm pore size and surface roughness. We therefore developed these assays entirely in glass-bottom plates. Rather than filtering liquid through the plate to wash the cells, we now sediment the cells and aspirate the supernatant.
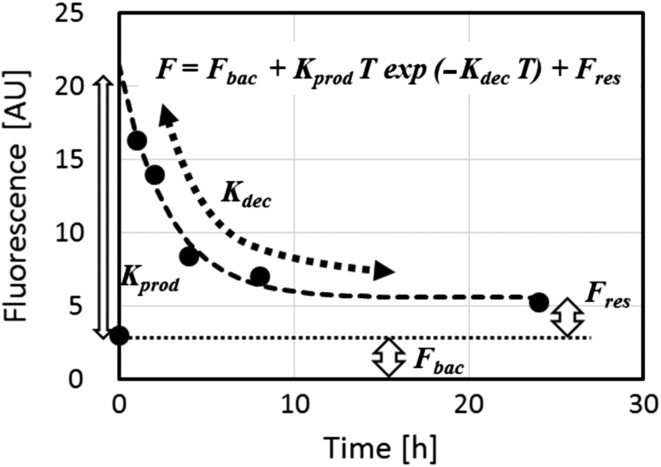
An assay for quantifying DNA repair kinetics(28, 29) was also developed (under NIEHS funding) by modifying the gamma-H2AX RABiT assay. In the modified assay, an irradiated sample is split and analyzed at multiple time points post irradiation, generating a curve as seen in Figure 4. A model fit provides parameters that can potentially be used to predict an individual's susceptibility to late sequelae.
Figure 4.
Repair kinetics assay. Gamma-H2AX yields are measured at various times following a 4-Gy irradiation.
RABIT II – LOOKING FORWARD BEYOND 2015
When this program started in 2005, commercial robotically-based platforms for high content and cellular screening were rare, which motivated us to build the RABiT prototype, specifically tailored for cell-based bioassays. In the subsequent years, high throughput/high content screening (HTS/HCS) has become a widely-used technique in both academia and industry(30) and has evolved as sets of standardized tools and techniques, covering the palette of tools in the RABiT. HTS systems generally include(31): robotic plate-handling systems, liquid handling systems, an automated microscope and control software. Similar to the RABiT, plates are moved through the system by robotic handling. They are filled via the liquid handling systems, and evaluated, often after a period of incubation. Control software choreographs the entire process, ensuring accuracy within the process and repeatability between processes. Assay automation has become modular, creating systems that can be easily expanded and modified, typically costing under $1 M. The greatest advances have not been the introduction of more complex robotics but rather simplification and increased reliability(32).
In the pharmaceutical industry, HTS systems are widely used to rapidly screen large libraries of drug candidates for biochemical activity, on a specific protein or cellular target, whereas in academia, the same systems are used to investigate more fundamental biological targets(30). In the past, such screening was a slow, manual process that relied on human labor to perform the screening and human observation to measure results. Automation has allowed biomedical researchers to greatly increase the number of evaluations that they can perform in parallel while maintaining high levels of quality. The quality of HTS data is very high and often better controlled than data generated by lower-throughput biological tests(33).
There are many advantages in transitioning the RABiT platform from a custom designed and homebuilt prototype to a commercial ‘off the shelf’ system(34). The major advantage is that of deployment. An estimate of the number of systems in university, industry and clinical testing laboratories in the New York City metropolitan area was 20, with 400 systems nationwide.
A second major advantage is that of reliability. A commercial system with such wide deployment capabilities undergoes a more rigorous quality control during development, manufacture and most importantly maintenance, than a university-built system. Due to the fact that these systems are in continuous operation, they also have a broad base of trained users and maintenance personnel ensuring operation in time of crisis.
Our future goals are to transition the RABiT assay protocols for operation on commercial systems, eliminating the ‘customized’ robotics in favor of more standard systems, eliminating, for example, the lymphocyte harvest, in favor of whole blood processing(14). The gamma-H2AX, Dicentric and CBMN assays have been implemented on the PerkinElmer cell::explorer HTS platform, and are undergoing optimization and validation studies.
In December 2015, the Columbia University RABiT system was decommissioned.
FUNDING
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) [U19-AI067773 to the Center for High-Throughput Minimally Invasive Radiation Biodosimetry], and the National Institute of Environmental Health Sciences (NIEHS) [R21-ES019494]. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIAID, NIEHS or the National Institutes of Health.
REFERENCES
- 1.Pellmar T. C. and Rockwell S.. Priority list of research areas for radiological nuclear threat countermeasures. Radiat. Res. 163, 115–123 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Salerno A., et al. Design considerations for a minimally invasive high-throughput automation system for radiation biodosimetry. In: IEEE International Conference on Automation Science and Engineering, CASE2007, Scottsdale, AZ (2007).
- 3.Garty G., et al. The RABIT: a rapid automated biodosimetry tool for radiological triage. Health Phys. 98(2), 209–217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garty G., et al. The RABiT: A Rapid Automated BIodosimetry Tool for radiological triage II. Technological developments. Int. J. Radiat. Biol. 87(8), 776–790 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., et al. Design and Preliminary Validation of a Rapid Automated Biodosimetry Tool for High Througput Radiological Triage. In: Proceedings of the ASME Design Engineering Technical Conferences ASME Design Engineering Technical Conferences 3, 61–67 (2009). [DOI] [PMC free article] [PubMed]
- 6.Chen Y., et al. Development of a robotically-based automated biodosimetry tool for high-throughput radiological triage. Int. J. Biomechatron. Biomed. Rob. 1(2), 115–125 (2010). [Google Scholar]
- 7.Chen Y., Wang H., Zhang J., Garty G., Simaan N., Lawrence Yao Y. and Brenner D. J.. Automated recognition of robotic manipulation failures in high-throughput biodosimetry tool. Exp. Syst. Appl. 39(10), 9602–9611 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayata I., Tabuchi H., Furukawa A., Okabe N., Yamamoto M. and Sato K.. Robot system for preparing lymphocyte chromosome. J. Radiat. Res. 33(Suppl), 231–241 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Schunck C., Johannes T., Varga D., Lorch T. and Plesch A.. New developments in automated cytogenetic imaging: unattended scoring of dicentric chromosomes, micronuclei, single cell gel electrophoresis, and fluorescence signals. Cytogenet Genome Res. 104(1–4), 383–389 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Martin P. R., Berdychevski R. E., Subramanian U., Blakely W. F. and Prasanna P. G. S.. Sample tracking in an automated cytogenetic biodosimetry laboratory for radiation mass casualties. Radiat. Meas. 42(6–7), 1119–1124 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garty G., Karam P. A. and Brenner D. J.. Infrastructure to support ultra high throughput biodosimetry screening after a radiological event. Int. J. Radiat. Biol. 87(8), 754–765 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner H. C., et al. Adapting the γ-H2AX assay for automated processing in human lymphocytes. 1. Technological aspects. Radiat. Res. 175(3), 282–290 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura A., Sedelnikova O. A., Redon C., Pilch D. R., Sinogeeva N. I., Shroff R., Lichten M. and Bonner W. M.. Techniques for gamma-H2AX detection. Methods Enzymol. 409, 236–50 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Lue S. W., Repin M., Mahnke R. and Brenner D. J.. Development of a high-throughput and miniaturized cytokinesis-block micronucleus assay for use as a biological dosimetry population triage tool. Radiat. Res. 184(2), 134–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyulko O. V., Garty G., Randers-Pehrson G., Turner H. C., Szolc B. and Brenner D. J.. Fast image analysis for the micronucleus assay in a fully automated high-throughput biodosimetry system. Radiat. Res. 181(2), 146–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenech M., Chang W. P., Kirsch-Volders M., Holland N., Bonassi S. and Zeiger E.. HUMN project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat. Res. 534(1-2), 65–75 (2003). [DOI] [PubMed] [Google Scholar]
- 17.IAEA.. Cytogenetic Analysis for Radiation Dose Assessment: A Manual (International Atomic Energy Agency; ) (2001) ISBN 9201021011 IAEA technical report #405: http://www-pub.iaea.org/MTCD/publications/PDF/TRS405_scr.pdf [Google Scholar]
- 18.Thierens H., De Ruyck K., Vral A., de Gelder V., Whitehouse C. A., Tawn E. J. and Boesman I.. Cytogenetic biodosimetry of an accidental exposure of a radiological worker using multiple assays. Radiat. Prot. Dosim. 113(4), 408–414 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Bian D., Tsui J. C., Repin M., Garty G., Turner H., Yao Y. L. and Brenner D. J.. Platform-dependent liquid handling in high-throughput biodosimetry tool. J. Med. Dev. (2016). doi:10.1115/1.4033600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertucci A., Smilenov L. B., Turner H. C., Amundson S. A. and Brenner D. J.. In vitro RABiT measurement of dose rate effects on radiation induction of micronuclei in human peripheral blood lymphocytes. Radiat. Environ. Biophys. 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y., Randers-Pehrson G., Turner H. C., Marino S. A., Geard C. R., Brenner D. J. and Garty G.. Accelerator-based biological irradiation facility simulating neutron exposure from an improvised nuclear device. Radiat. Res. 184(4), 404–410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner H. C., Shuryak I., Weber W., Doyle-Eisele M., Melo D., Guilmette R., Amundson S. A. and Brenner D. J.. Gamma-H2AX kinetic profile in mouse lymphocytes exposed to the internal emitters Cesium-137 and Strontium-90. PLoS One 10(11), e0143815 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkins R. C., Romm H., Oestreicher U., Marro L., Yoshida M. A., Suto Y. and Prasanna P. G. S.. Biological dosimetry by the triage dicentric chromosome assay – further validation of international networking. Radiat. Meas. 46(9), 923–928 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell C. R., Azizova T. V., Hande M. P., Burak L. E., Tsakok J. M., Khokhryakov V. F., Geard C. R. and Brenner D. J.. Stable intrachromosomal biomarkers of past exposure to densely ionizing radiation in several chromosomes of exposed individuals. Radiat. Res. 162(3), 257–263 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Chudoba I., Hickmann G., Friedrich T., Jauch A., Kozlowski P. and Senger G.. mBAND: a high resolution multicolor banding technique for the detection of complex intrachromosomal aberrations. Cytogenet Genome Res. 104(1–4), 390–393 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Fowler B., Liu C., Mims S., Balicki J., Li W., Do H., Appelbaum J. and Vu P., editors. A 5.5Mpixel 100 frames/sec wide dynamic range low noise CMOS image sensor for scientific applications. Proc. SPIE 7536, 753607 (2010).
- 27.Garty G., Bigelow A. W., Repin M., Turner H. C., Bian D., Balajee A. S., Lyulko O. V., Taveras M., Yao Y. L. and Brenner D. J.. An automated imaging system for radiation biodosimetry. Microsc. Res. Tech. 78(7), 587–598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma P. M., Ponnaiya B., Taveras M., Shuryak I., Turner H. and Brenner D. J.. High throughput measurement of γ-H2AX DSB repair kinetics in a healthy human population. PLOS One 10(3), e0121083 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner H. C., Sharma P., Perrier J. R., Bertucci A., Smilenov L., Johnson G., Taveras M., Brenner D. J. and Garty G.. The RABiT: high-throughput technology for assessing global DSB repair. Radiat. Environ. Biophys. 53(2), 265–272 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dove A. High-throughput screening goes to school. Nat. Methods 4(6), 523–532 (2007). [Google Scholar]
- 31.Michael S. A robotic platform for quantitative high-throughput screening. Assay Drug Dev. Technol. 6(5), 637–657 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janzen W. P. Screening technologies for small molecule discovery: the state of the art. Chem. Biol. 21(9), 1162–1170 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Macarron R., et al. Impact of high-throughput screening in biomedical research. Nat. Rev. Drug Discov. 10(3), 188–195 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Repin M., Turner H. C., Garty G. and Brenner D. J.. Next generation platforms for high-throughput biodosimetry. Radiat. Protect. Dosim. 159(1–4), 105–110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]