Abstract
Receiver operating characteristic (ROC) analysis is a fundamental tool used for the evaluation and comparison of diagnostic systems that provides estimates of the combinations of sensitivity and specificity that can be achieved with a given technique. Along with critical considerations of practical limitations, such as throughput and time to availability of results, ROC analyses can be applied to provide meaningful assessments and comparisons of available biodosimetry methods. Accordingly, guidance from the Food and Drug Administration to evaluate biodosimetry devices recommends using ROC analysis. However, the existing literature for the numerous biodosimetry methods that have been developed to address the needs for triage either do not contain ROC analyses or present ROC analyses where the dose distributions of the study samples are not representative of the populations to be screened. The use of non-representative sample populations can result in a significant spectrum bias, where estimated performance metrics do not accurately characterize the true performance under real-world conditions. Particularly, in scenarios where a large group of people is screened because they were potentially exposed in a large-scale radiation event, directly measured population data do not exist. However, a number of complex simulations have been performed and reported in the literature that provide estimates of the required dose distributions. Based on these simulations and reported data about the output and uncertainties of biodosimetry assays, we illustrate how ROC curves can be generated that incorporate a realistic representative sample. A technique to generate ROC curves for biodosimetry data is presented along with representative ROC curves, summary statistics and discussion based on published data for triage-ready electron paramagnetic resonance in vivo tooth dosimetry, the dicentric chromosome assay and quantitative polymerase chain reaction assay. We argue that this methodology should be adopted generally to evaluate the performance of radiation biodosimetry screening assays so that they can be compared in the context of their intended use.
INTRODUCTION
In the aftermath of an improvised nuclear device (IND) detonation, screening for life-threatening radiation exposures will be required to enable identifying those who would benefit from effective medical inventions. Emergency planning guidelines call for the application of biodosimetry tools to discriminate between individuals who have received doses above and below a defined threshold, e.g. 2 gray (Gy), where only those with doses above the threshold would receive appropriate medical intervention.(1–6) High sensitivity is required for triage (i.e. low false negatives) so irradiated subjects can be identified and provided appropriate care. High specificity is needed to increase the efficiency of the care so that limited time and resources can be focused on those in need of care, i.e. low false positives. The numbers of people who are falsely identified, either as having received a life-threatening dose and subsequently given scarce treatment resources without benefit or conversely are misidentified as not needing and therefore not given life-saving care, play a critical role in developing effective triage strategies for IND detonation scenarios that depend on accurately sorting those who truly need acute care from those who do not.
Despite the crucial importance of correctly and quickly sorting people in this scenario, conventional precision (or uncertainty) metrics, e.g. confidence intervals, are not in themselves sufficient to characterize the real-world quantitative diagnostic performance of such screening assays. While these metrics can be used to provide confidence intervals for estimates of an individual's absorbed dose, or used to estimate the likelihood that an individual has received a dose above a given threshold, they cannot be used to directly specify the level of precision that a triage decision-maker requires to sort subjects, nor do they provide medical response planners with the information necessary to support effective triage or to compare multiple technologies in their abilities to meet this need.
Recently released guidance from the United States Food and Drug Administration (FDA) for the assessments of radiation biodosimetry medical countermeasure devices(7) includes a statement that receiver operating characteristic (ROC) analysis may be useful to evaluate the diagnostic accuracy of a biodosimetry device. This document cites the Clinical and Laboratory Standards Institute (CLSI) approved guideline as a technical reference for the use of the technique.(8) The CLSI guideline provides a summary of the formation and use of ROC analyses, including definitions, examples and discussion of considerations critical to the proper use of the technique. In the case of radiation biodosimetry for triage following an event where a large population is at risk for developing acute radiation syndrome (ARS), positive and negative populations may be defined based on classifying subjects according to their true exposure status, with positives being those who were exposed to doses greater than or equal to a defined classification threshold (e.g. 2 Gy) and negatives being those who were exposed to doses below this threshold. A biodosimetry assay uses measurements of an associated biomarker and comparison of those measurements to a chosen decision threshold to assign a diagnosis of either positive or negative, noting that this diagnosis may be correct or incorrect.
Many biodosimetry techniques apply a dosimetric calibration relationship to convert biomarker measurements to estimates of individuals’ absorbed doses. For the described ROC framework described here, the decision threshold will also have units of absorbed dose.
ROC analysis summarizes the diagnostic performance of a biodosimetry technique by calculating the complete set of paired sensitivity and specificity values that can be achieved with a defined assay, as the decision threshold is varied across the entire range of observed measurements, where sensitivity is defined as the true-positive fraction (TPF) and specificity is defined as the true-negative fraction (TNF). The TPF is equal to the fraction of positives (i.e. individuals with true dose at or above a defined threshold) that are identified relative to the total number of positives. Similarly, TNF is equal to the fraction of negatives (i.e. individuals with true dose below the threshold) that are identified relative to the total number of negatives.
The CLSI document notes several biases that may be generated by inappropriate study design. For a given test and its intended application, spectrum bias is defined as the bias between the estimated test performance and the true test performance when the sample used for evaluating an assay does not properly represent the entire disease spectrum over the target (intended-use) population. As a consequence of such improper sampling of the intended population, estimates of the sensitivity and specificity of the assay are biased, with subsequent impacts on establishment of realistic expectations about the assay or any comparisons with other assays. Accordingly, ROC analyses of biodosimetry assays must be based on population samples that have the same distribution of doses as those expected for the intended ‘real-world’ conditions.
The population screened following an IND event will have a continuous distribution of true doses, likely ranging from a predominance of people with essentially negligible doses to a smaller number of people with doses predicted to lead to imminent death (referred to as expectant casualties). This distribution differs markedly from those that have been used to evaluate biodosimetry technologies thus far. Instead, the populations used in evaluations often consist of equal numbers of samples irradiated to several discrete dose levels between 0 and 10 Gy.
Sensitivity and specificity values computed directly from discrete sample sets do not completely reflect the performance of a biodosimetry technique under real-world conditions and may be misleading. For example, studies of an assay's performance that include a higher proportion of doses in close proximity to the threshold dose will have an increased number of false positives and false negatives. As a consequence of this study design, the reported performance statistics will artificially appear to have poorer sensitivity and specificity compared to a study design using the same assay but which assessed discrete doses further away from the threshold. The use of artificially chosen dose distributions, especially when they are not standardized across studies of different biodosimetry methods, may mislead planners who are trying to select ‘high-performing’ assays unless additional more detailed analyses are performed. Only with additional quantitative consideration of the expected distribution of doses across the affected population can test data, gathered in small controlled studies using limited discrete ‘ordinal’ doses, be given the appropriate real-world context of the population to be screened.
In this article, we illustrate how ROC curves can be generated that incorporate a realistic representative sample. We base our simulations on the expected dose distribution for an IND detonation in New York City (NYC) and reported data about the outputs and uncertainties of several biodosimetry assays.
METHODS
Estimation of true dose distribution
Simulations have been performed to estimate the numbers of people irradiated to various absorbed dose levels to aid in planning for triage and management following a malicious IND detonation. These include Waselenko et al.(9) which estimated the casualties following 1- and 10-kiloton nuclear detonations in a city of 2 million people. Buddemeier and colleagues presented simulation data for the detonation of a 10-kiloton detonation in Washington, DC.(10) More recently, a simulation was presented based on the detonation of a similar device in NYC.(11) This simulation describes a workday population of 4 million people in affected zones. Of 3.3 million immediate survivors, ~2 million are classified as uninjured (including consideration of radiation injury) and the remaining 1.3 million are subdivided based on three levels of injury: expectant, at risk and recoverable. These populations are further divided into injury categories that distinguish between minor versus major trauma and eight midline deep dose levels. Across the moderate damage zone (0.5–1 mile radius), light damage zone (1–3 mile radius) and dangerous fallout region (extending many miles) the ‘at risk’ populations are estimated to include ~250 000, 150 000 and 100 000 people, respectively. Of these 500 000 people, 50% are predicted to succumb to their injuries without medical care, with the potential to save 100 000 with appropriate medical care. In order to care for this population, this at risk population must be identified from within the total number of survivors in the affected zones.
In order to estimate a continuous distribution of exposure levels, the data presented by Buddemeier et al. must be normalized to account for the varying ranges of the described dose levels. For the purposes of modeling the dose distribution in this analysis, the minimum and maximum midline deep doses were set to 0.07 and 21 Gy, respectively. Within each dose level, an absorbed dose density function with units of (# exposed)/Gy was calculated by dividing the total number exposed in each level by the magnitude of the dose range for the level (Table 1). These values were assigned to the midpoint dose for each level. This density function, f, as a function of midpoint dose, D, is well modeled by a power function of the form
| (1) |
with an R2 of 0.978. Integration of this function over the full dose range yields a value consistent with the expected 3.3 million survivors, with 13% of this population receiving doses above 2 Gy according to the model.
Table 1.
Estimated distribution of absorbed doses.a
| Midline deep dose (Gy) | Total population affected | Mean #/Gy |
|---|---|---|
| <0.35 | 2 131 847 | 5 329 615 |
| 0.35–0.5 | 179 092 | 895 460 |
| 0.5–0.875 | 241 636 | 439 338 |
| 0.875–2.1 | 259 821 | 148 469 |
| 2.1–3.7 | 152 570 | 66 335 |
| 3.7–5.8 | 112 151 | 37 384 |
| 5.8–10.5 | 150 518 | 22 465 |
| >10.5 | 51 125 | 3408 |
| Sum | 3 278 760 |
aBuddemeier, B.R., Medical Needs in the Aftermath of Nuclear Terrorism (NYC Ed.), Lawrence Livermore National Laboratory, LLNL-PRES-677346 (EPR BioDose 2015).
Numerous biodosimetry techniques have been developed for triage to identify individuals who have received absorbed doses above a defined classification threshold, e.g. 2 Gy. For many methods, an a priori calibration curve is derived to relate the measured quantities to absorbed doses. Examples of measured quantities include electron paramagnetic resonance (EPR) signal amplitude from irradiated teeth, frequency of dicentric chromosomes and gene expression levels. Properly derived calibration curves provide accurate dose estimates when averaged across the population, without gross systematic error, but errors will exist for individual dose estimates due to factors such as measurement uncertainty and unaccounted interpersonal variability. These errors may be interpreted as the precision of dose estimation across the population. As the precision of dose estimates may be variable as a function of the true dose, an accurate assessment of the performance of a biodosimetry technique will depend on both the dose distribution across the affected population and the potentially dose-dependent precision of the assay of interest.
Formation of ROC curves
A ‘perfect’ point-of-care (POC) dosemeter would accurately estimate each individual person's absorbed dose and properly assign a diagnostic category based on comparison to the defined classification threshold. In this case, the density of estimated doses is identical to the true dose density and the division between groups above and below the classification threshold will be discrete, forming vertical edges for the group densities (Figure 1a). The limited precision of a biodosimetry technique will act to blur these group densities, potentially in a dose-dependent manner (Figure 1b). If the precision is not dose dependent, this blurring can be modeled by convolving the true-positive and negative dose densities with a kernel function that characterizes the uncertainty, for example a Gaussian distribution with appropriate variance. If the precision is dose dependent, a similar mathematical process must be carried out where the variance of the kernel depends on dose. Paired sensitivity and specificity values may be calculated from unit-area normalized group density functions as the decision threshold is varied across the full range of measured doses (Figure 1c). A ROC curve is generated by plotting the sensitivity as a function of (1-specificity) (Figure 1d). Note that the decision threshold value used for assigning a diagnostic category does not need to be equal to the defined threshold used to guide triage (e.g. 2 Gy) and different values can be used to optimize the trade-off between sensitivity and specificity for specific triage scenarios.
Figure 1.
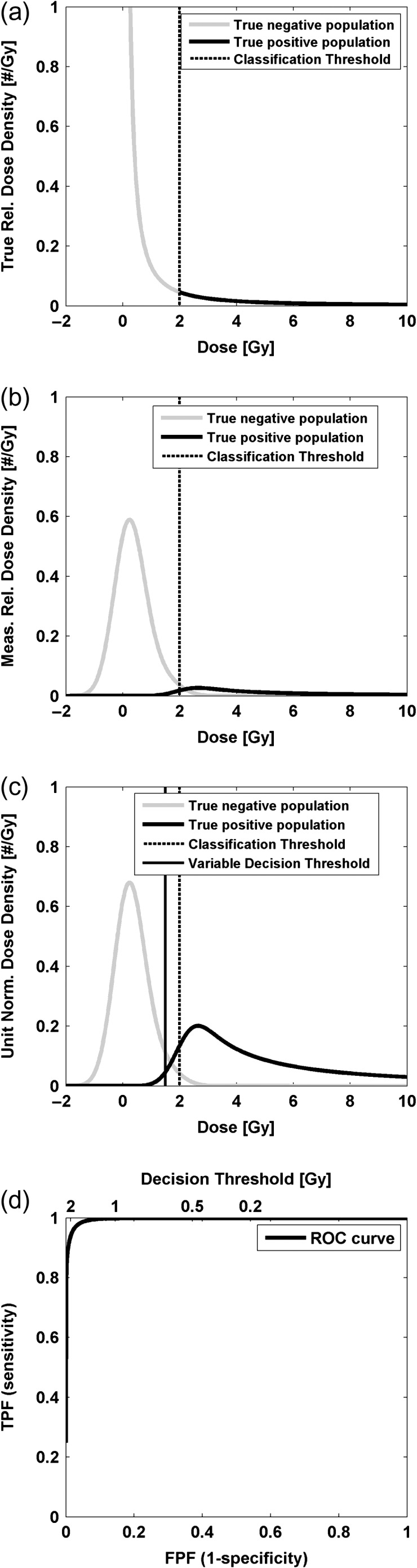
The modeled population dose density distributions for subjects classified as positive and negative (a) is blurred to reflect the limited precision of an assay (b). Unit normalized measured dose density functions (c) are used to estimate sensitivity and specificity pairs based on a variable decision threshold to form an ROC curve (d). FPF, false-positive fraction.
Applications
The described methodology for estimating ROC curves to characterize the diagnostic performance of biodosimetry techniques can be applied generally using the dose density function described above, or an alternate density appropriate for the scenario of interest, along with specification of the precision of dose estimation for the assay across all doses. Although published studies have reported quantitative characterizations of the precision of dose estimation for many of the biodosimetry techniques in development,(12–18) none has presented ROC analyses based on pertinent dose distributions for the intended population.
In order to demonstrate the application of the proposed ROC analysis methodology, we present three case studies based on use of Buddemeier's NYC IND simulation data and published uncertainty data for each biodosimetry method. Case studies are developed based on published data for EPR tooth dosimetry,(17, 18) the dicentric chromosome assay (DCA),(13) and a genetic assay based on quantitative polymerase chain reaction (qPCR).(16) The data included in these reports provide estimates of assay performance at the respective times of publication and may not be representative of fully optimized performance. The case studies presented here are not intended as definitive assessments or comparisons. A classification threshold of 2 Gy is used, where ‘positive’ is defined as a true dose of greater than or equal to 2 Gy and ‘negative’ is defined as a true dose below 2 Gy.
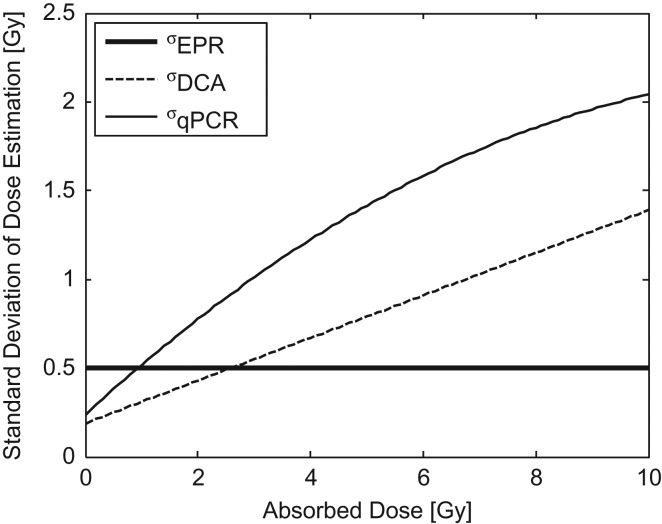
For EPR tooth dosimetry, published in vitro data using whole intact teeth indicate that individual doses can be estimated with an uncertainty of 0.43–0.5 Gy.(17, 18) Recent in vivo data demonstrate a comparable uncertainty of 0.66 Gy.(18) Both in vitro and in vivo uncertainty estimates are based on the use of settings appropriate for triage with total times for measurement of ~5 and 10 min, respectively. Dose estimates for EPR tooth dosimetry are linear across a dose range from 0 to at least 10 Gy.(17) For the present case study, an uncertainty of 0.5 Gy is applied.
For the DCA analysis, uncertainty data are based on triage dose estimates of manual scoring of 50 metaphase spreads or 30 dicentrics reported across 6 institutions for blood samples taken from a single healthy male and irradiated to discrete doses between 0.1 and 6.4 Gy.(13) The uncertainty was linearly dependent on dose across the reported range with a slope of 0.12 ± 0.02 Gy/Gy and an intercept of 0.19 ± 0.05 Gy.
For the qPCR analysis, based on interpretation of data presented for Gene Set B 1 d after irradiation of human peripheral blood from 60 adult donors, the uncertainty is non-linearly dependent on dose and well characterized by a quadratic relationship with coefficients of −0.011, 0.29 and 0.24 for the quadratic, linear and constant terms, respectively.(16)
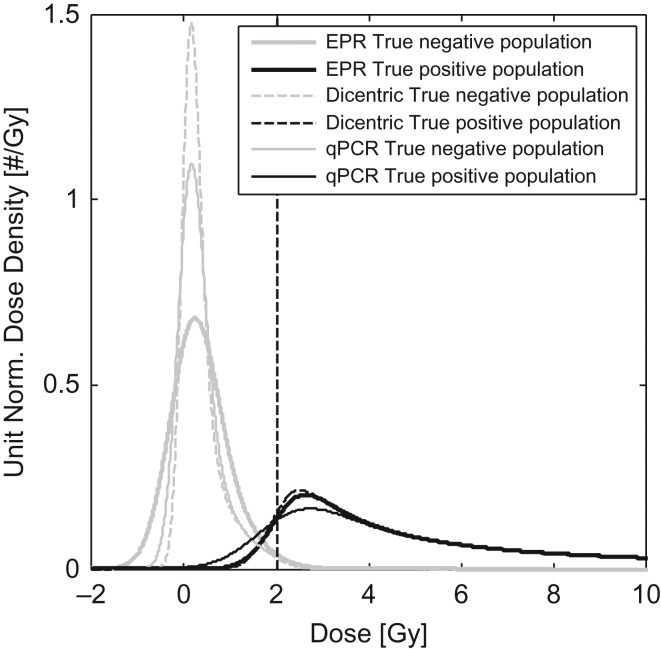
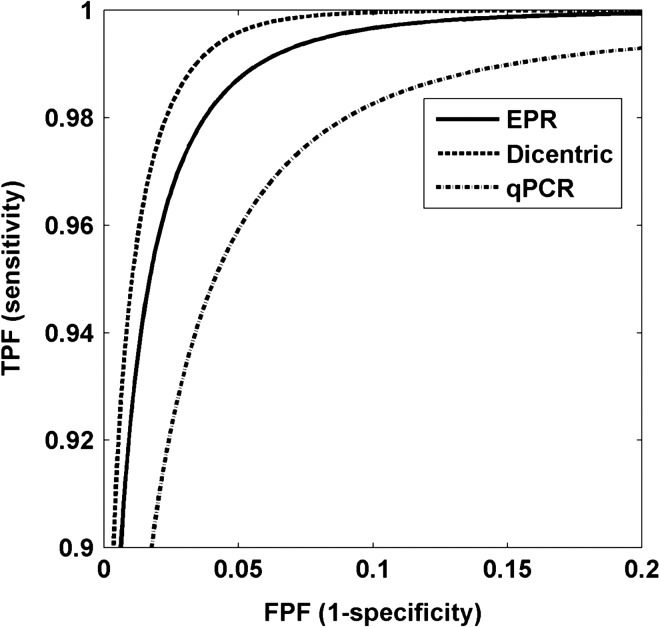
For each of these assays, the uncertainty in dose estimation, characterized by the standard deviation, is shown as a function of true dose in Figure 2 and the resulting distributions of estimated doses for the positive and negative populations are shown in Figure 3. ROC curves for each of the three assays are shown in Figure 4. Note that the sensitivity range has been limited to show only values ≥90% and (1-specificity) limited to show only values ≤20% to focus on high sensitivity and specificity operational conditions that would be suitable for the POC triage application.
Figure 2.
Uncertainty (standard deviation) of dose estimation based on published data for EPR, DCA and qPCR.
Figure 3.
Estimated measured dose density function for the three assays, demonstrating variable degrees of overlap between the measured positive and negative densities and relation to the dashed classification threshold.
Figure 4.
Estimated ROC curves. All techniques can operate at levels where both sensitivity and specificity are >95%. Higher diagnostic accuracy is afforded by curves closer to the upper right corner.
RESULTS
Through these ROC analyses, i.e. by employing an estimate of the expected distribution of doses in the intended-use population and published uncertainties in dose estimation, there are several observations of note. First focusing on measures of sensitivity and specificity as an indicator of performance, all three assays are capable of performing well, i.e. at a level where both sensitivity and specificity are >95%. Second, the three assays can be compared using another widely used metric of accuracy: the area under the ROC curve (AUC), which (among other interpretations) is equivalent to the probability that a randomly chosen positive subject will be evaluated to be at higher risk than a randomly chosen negative subject. Accordingly, an AUC of 0.5 is equivalent to random chance and an AUC of 1.0 denotes a perfect test. For all three assays, the AUC was above 0.99 (Table 2), with EPR and the dicentric assay ≥0.997.
Table 2.
ROC and screening descriptors with discrimination cut-off equal to classification threshold of 2 Gy.
| EPR | Dicentric | qPCR | |
|---|---|---|---|
| AUC | 0.997 | 0.998 | 0.991 |
| Sens.|2Gy | 0.942 | 0.943 | 0.899 |
| Spec.|2Gy | 0.986 | 0.991 | 0.983 |
| % incorrectly identified as positive | 1.25 | 0.77 | 1.50 |
| % incorrectly identified as negative | 0.77 | 0.76 | 1.35 |
| Total % incorrectly identified | 2.02 | 1.53 | 2.85 |
DISCUSSION
While these results might seem to imply equivalency of performance across the three biodosimetry methods in this case, even small differences in sensitivity and specificity (<1%) can lead to significant differences in the absolute numbers of misidentified subjects. For example, if these assays were applied with the discrimination cut-off set equal to the classification threshold of 2 Gy (Table 2), the numbers of subjects incorrectly diagnosed as positive vary by approximately a factor of 2 across these assays (0.77–1.50%), which may have considerable impacts on the allocation and use of limited resources when very large populations are to be screened. Similarly, between 0.76 and 1.35% of subjects are incorrectly diagnosed as negative, and potentially denied critical medical care.
In the case of screening for ARS risk, especially as first-stage triage, it would be appropriate to place a high priority on maintaining a very high sensitivity value to minimize the number of people who could benefit from care who are erroneously denied entry into the medical system. As demonstrated in the ROC curves, the cost of increased sensitivity is a concomitant loss of specificity, but to varying degrees dependent on the performance of the assay.
Table 3 provides estimated specificity values for the three assays based on using discrimination cut-offs chosen to provide sensitivities of 99%, resulting in only 0.13% of the tested population being erroneously diagnosed as negative. Note that the discrimination cut-off values required to achieve this level of sensitivity are not equal to the classification threshold of 2 Gy and vary across the assays. For example, in order to achieve 99% sensitivity based on the qPCR assay data all subjects with estimated doses >0.8 Gy would be identified as positive. At this discrimination threshold, 99% sensitivity is achieved with 84.5% specificity. In this case, the relatively low specificity results in 13.4% of the tested population being incorrectly identified as positive, placing elevated demands on medical resources and limiting efficiency of the system. Conversely, 99% sensitivity can be achieved based on the dicentric assay data with specificity of 96.5%, 3% misidentification as positive and use of a 1.5 Gy discrimination threshold. Performance based on the tooth dosimetry data is more similar to that of the dicentric assay, with specificity of 94.2%, 5% misidentification as positive and a 1.4 Gy discrimination threshold. This example demonstrates how ROC analysis can be applied to enable medical response planners to specify an appropriate discrimination threshold, for a given assay, to achieve a desired level of sensitivity (or specificity if resources are a primary limitation).
Table 3.
ROC and screening descriptors at 99% sensitivity.
| EPR | Dicentric | qPCR | |
|---|---|---|---|
| Spec.|99% Sens. | 0.942 | 0.965 | 0.845 |
| % incorrectly identified as positive | 5.04 | 2.99 | 13.40 |
| % incorrectly identified as negative | 0.13 | 0.13 | 0.13 |
| % incorrectly identified | 5.2 | 3.1 | 13.5 |
| Discrimination cut-off (Gy) | 1.4 | 1.5 | 0.8 |
CONCLUSION
The rational design and implementation of plans for triage of populations affected by large-scale radiation events, including choices of applied biodosimetry technologies, necessarily includes consideration of sensitivity and specificity as estimated using ROC analyses with representative sample populations. The proposed methodology combines an estimate of the true distribution of absorbed doses across the affected population with knowledge of the uncertainty in dose estimation for an assay of interest to derive ROC curves that reflect ‘real-world’ conditions. The estimated sensitivity and specificity metrics can be used by planners, in concert with considerations of additional details for implementation, to optimize large-scale IND emergency response.
DISCLOSURES
ABF and HMS are owners of Clin-EPR, LLC, which manufacturers clinical EPR devices for investigational uses.
ACKNOWLEDGEMENTS
The authors are grateful to Brooke Buddemeier, Associate Program Leader (acting), Risk & Consequence Management Division, Lawrence Livermore National Laboratory for supplying improvised nuclear device simulation data and for valuable discussions regarding these data. B.B.W. would like to thank Darrin Edwards for an additional internal review of the manuscript.
FUNDING
This research was supported by the National Institute of Allergy and Infectious Diseases of the US DHSS’ National Institutes of Health (NIH) under Award Number U19-AI091173 and contract HHSO100201100024C with the Biomedical Advanced Research and Development Authority (BARDA), within the Office of the Assistant Secretary for Preparedness and Response, US Department of Health and Human Services.
REFERENCES
- 1.Preparedness and response for a nuclear or radiological emergency: general safety requirements. (Vienna: International Atomic Energy Agency) (IAEA safety standards series, ISSN 1020–525X; no. GSR part 7). (2015).
- 2.Generic procedures for medical response during a nuclear radiological emergency. (Vienna: IAEA) (2005).
- 3.National Security Staff Interagency Policy Coordination Subcommittee for Preparedness and Response to Radiological and Nuclear Events. Planning Guidance for Reaction to a Nuclear Detonation, 2nd ed. Washington, DC: US Executive Office of the President; 2010. Available at: http://www.epa.gov/radiation/docs/er/planning-guidance-for-response-to-nuclear-detonation-2-edition-final.pdf (Accessed 20 April 2016).
- 4.Grace M. B., Moyer B. R., Prasher J., Cliffer K. D., Ramakrishnan N., Kaminski J., Coleman C. N., Manning R. G., Maidment B. W. and Hatchett R. Rapid radiation dose assessment for radiological public health emergencies: roles of NIAID and BARDA. Health. Phys. 98(2), 172–178 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Sullivan J. M., Prasanna P. G. S., Grace M. B., Wathen L. K., Wallace R. L., Koerner J. F. and Coleman C. N.. Assessment of biodosimetry methods for a mass casualty radiological incident: medical response and management considerations. Health. Phys. 105, 540–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.NCRP Report No. 165, Bethesda, MD, 202 pp. (soft cover). ISBN: 978-0-9823843-3-6. (2010).
- 7.The Center for Devices and Radiological Health. Food and Drug Administration (FDA). Radiation biodosimetry medical countermeasure devices: Guidance for industry and Food and Drug Administration Staff. Document 1400045 issued April 18, 2016. http://www.fda.gov/MedicalDevices/DeviceRegulationandGuidance/GuidanceDocuments/ucm418448.htm (Accessed 20 April 2016).
- 8.Clinical and Laboratory Standards Institute (CLSI). Assessment of the Diagnostic Accuracy of Laboratory Tests Using Receiver Operating Characteristic Curves; Approved Guideline – Second Edition. CLSI document EP24-A2. (Wayne, PA: CLSI) (2011).
- 9.Waselenko J. K., MacVittie T. J., Blakely W. F., Pesik N., Wiley A. L., Dickerson W. E., Tsu H., Confer D. L., Coleman C. N. and Seed T.. Medical management of the acute radiation syndrome: recommendations of the strategic national stockpile radiation working group. Ann. Intern. Med. 140(12), 1037–1051 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Buddemeier B. R., Valentine J.E., Millage K.K., Brandt L.D. National Capital Region: Key response planning factors for the aftermath of nuclear terrorism. LLNL-TR-512111. (November 2011) fas.org/irp/agency/dhs/fema/ncr.pdf (Accessed 20 April 2016).
- 11.Buddemeier B. Medical Needs in the Aftermath of Nuclear Terrorism, LLNL-PRES-677346 (Presented at EPR BioDose 2015, Hanover, NH, October 4–8, 2015).
- 12.Badie C., et al. Laboratory intercomparison of gene expression assays. Radiat. Res. 180(2), 138–148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beinke C., et al. Laboratory intercomparison of the dicentric chromosome analysis assay. Radiat. Res. 180(2), 129–137 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Romm H., et al. Laboratory intercomparison of the cytokinesis-block micronucleus assay. Radiat. Res. 180(2), 120–128 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Rothkamm K., et al. Comparison of established and emerging biodosimetry assays. Radiat. Res. 180(2), 111–119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tucker J. D., Joiner M. C., Thomas R. A., Grever W. E., Bakhmutsky M. V., Chinkhota C. N., Smolinski J. M., Divine G. W. and Auner G. W.. Accurate gene expression-based biodosimetry using a minimal set of human gene transcripts. Int. J. Radiat. Oncol. Biol. Phys. 88(4), 933–939 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Williams B. B., Flood A. B., Salikhov I., Kobayashi K., Dong R., Rychert K., Du G., Schreiber W. and Swartz H. M.. In vivo EPR tooth dosimetry for triage after a radiation event involving large populations. Radiat. Environ. Biophys. 53, 335–346 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flood A. B., Williams B. B., Schreiber W., Du G., Wood V. A., Kmiec M. M., Petryakov S. V., Demidenko E. and Swartz H. M., the EPR Center Tooth Dosimetry Project Team . Advances in in vivo EPR tooth biodosimetry: meeting the targets for initial triage following a large-scale radiation event. Radiat. Prot. Dosim. (same issue). [DOI] [PMC free article] [PubMed] [Google Scholar]