Abstract
Sec20p is an essential endoplasmic reticulum (ER) membrane protein in yeasts, functioning as a tSNARE component in retrograde vesicle traffic. We show that Sec20p in the human fungal pathogen Candida albicans is extensively O mannosylated by protein mannosyltransferases (Pmt proteins). Surprisingly, Sec20p occurs at wild-type levels in a pmt6 mutant but at very low levels in pmt1 and pmt4 mutants and also after replacement of specific Ser/Thr residues in the lumenal domain of Sec20p. Pulse-chase experiments revealed rapid degradation of unmodified Sec20p (38.6 kDa) following its biosynthesis, while the stable O-glycosylated form (50 kDa) was not formed in a pmt1 mutant. These results suggest a novel function of O mannosylation in eukaryotes, in that modification by specific Pmt proteins will prevent degradation of ER-resident membrane proteins via ER-associated degradation or a proteasome-independent pathway.
Secretory proteins in eukaryotes initially traverse the membrane of the endoplasmic reticulum (ER) via the Sec61 pore complex and immediately thereafter fold in the ER lumen to obtain their native three-dimensional structures. It has been established that asparagine-linked (N) glycosyl chains have important roles in folding and degradation of unfolded secretory proteins (reviewed in references 2, 9, and 16). In the ER, unfolded N-glycoproteins are glucosylated, leading to ER retention by resident lectins. After repeated folding attempts, N-glycosyl chains may be trimmed to a Man8 structure, which directs retrograde secretion across the Sec61 pore, followed by protein deglycosylation, ubiquitination, and degradation in proteasomes (ER-associated degradation [ERAD]) (10). During a prolonged stay in the ER, further trimming to a Man7 structure directs proteins to an undefined degradation machinery within the ER, which is independent of proteasomes (3).
In fungi, O glycosylation at serine or threonine residues occurs in the ER (reviewed in references 5 and 21). Mannosyltransferase (Pmt) proteins catalyze O mannosylation of proteins traversing the secretory pore. In Saccharomyces cerevisiae, the PMT family encodes seven isoforms, and the human fungal pathogen Candida albicans has five isoforms (Pmt1, -2, -4, -5, and -6), which modify specific target proteins (7, 14, 17, 18, 23, 24). The close coupling of secretion and O glycosylation in fungi suggested that the latter process, like N glycosylation in higher eukaryotes, is functionally related to folding and/or the stability of secretory glycoproteins. It was shown that in S. cerevisiae a fraction of a model protein, mutant alpha-factor precursor, was partially protected from proteasomal degradation by Pmt2p-mediated O mannosylation (8). In addition, O mannosylation of Axl2p by Pmt4p (18) and of surface sensors Wsc1p, Wsc2p, and Mid2p by Pmt2p/Pmt4p (14, 17) prevents a specific proteolytic cleavage. In C. albicans, Als1p, chitinase, and Kre9p are targets for Pmt1p (23, 24; S. K.-H. Prill, B. Klinkert, C. Timpel, and J. F. Ernst, unpublished data), but protective O mannosylation has not been reported.
Sec20p is an essential type II membrane protein of the ER that functions as part of a complex containing the tSNARE Ufe1p involved in retrograde vesicle traffic in S. cerevisiae and C. albicans (4, 13, 22, 25, 26). In sec20 mutants, retrograde and consequently anterograde traffic of secretory vesicles is blocked; in addition, Golgi glycosylation is defective (19). We show here that C. albicans Sec20p is an O-glycosylated protein, which in its nonglycosylated form, in mutants lacking Pmt1p or Pmt4p isoforms, or in Sec20p variants lacking potential O-glycosylation sites is rapidly degraded. Thus, Sec20p is the first example of an essential component of the eukaryotic secretory apparatus and of an ER membrane protein that is effectively protected from proteolytic degradation in the ER by specific Pmt-mediated O glycosylation.
MATERIALS AND METHODS
Strains and media.
The C. albicans strains used were strain CAI4 (Δura3::imm434 Δura3::imm434) (6) and its mutated derivatives, including the pmt1 mutant CAP1-3121 (pmt1Δ::hisG pmt1Δ::hisG) (21), the pmt4 mutant CAP4-2164 (Prill et al., unpublished), the pmt6 mutant CAP2-1341 (pmt4Δ::hisG pmt4Δ::hisG) (22), and the mnt1 mutant NGY24 (mnt4Δ::hisG mnt4Δ::hisG) (1). Strains were grown in YPD medium or on supplemented SD minimal medium at 30°C (20). Transformation of C. albicans strains was carried out by the spheroplast method (20). The expression plasmids pYW7, encoding a PCK1p-SEC20 fusion, and pYW69, encoding a PCK1p-SEC20-myc fusion, have been described (24). The PCK1 promoter in transformants was repressed in S4D medium (SD medium with 4% glucose) and induced in SCAA medium (0.67% yeast nitrogen base, 2% Casamino Acids) or SLac medium (0.67% yeast nitrogen base, 2% sodium lactate) (12) supplemented by a mix of amino acids (20) but lacking methionine and cysteine.
SEC20 mutagenesis.
The SEC20 expression vector pYW69 was mutated by specific oligonucleotides, using the QuikChange site-directed mutagenesis kit (Stratagene). Plasmids pYW96 (S268A), pYW98 (S272A, S274A), pYW94 (S279A, S280A), pYW92 (T285A, T286A), pYW97 (T294A, T296A), pYW95 (T300A, S302A), pYW93 (T321A, T322A, S323A, S324A), pYW91 (N201A), pYWΔK245 (K245A), pYWΔK253 (K253A), pYWΔK268 (K268A), and pYWΔK294 (K294A) were constructed.
Preparation of crude extracts and immunoblottings.
Twenty milliliters of cells were grown in SCAA medium to an optical density at 600 nm of 1 to 4 and harvested by centrifugation (5 min; 2,150 × g). The cell pellet was washed twice with water and resuspended in 500 μl of RE buffer (50 mM HEPES-KOH; 150 mM NaCl; 5 mM EDTA; 1% Triton X-100; 1 μg of protease inhibitors antipain, pepstatin A, and leupeptin [Sigma]/ml; pH 7.5). An equal volume of glass beads (diameter, 0.25 to 0.5 mm) was added, and the cells were shaken on a Vibrax VX 2E (Janke and Kunkel) at maximum speed at 4°C. The cell debris and beads were pelleted by centrifugation (3 min; 2,150 × g) and discarded, while the supernatant was considered the crude extract. Protein concentrations were determined by the Bradford assay using a commercial kit (Bio-Rad).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were carried out as described previously (26). An anti-c-myc antibody (9E10; Babco) diluted 1:500 was used as the primary antibody, while polyclonal goat anti-mouse antibody coupled to peroxidase (Jackson Research Laboratory, Inc.), diluted 1:60,000, was used as the secondary antibody. Staining was done with Super-SignalULTRA chemoluminescent substrate (Pierce). Anti-ubiquitin antibody (Babco) was used at a dilution of 1:200.
Binding to ConA.
According to a previous protocol (22), crude extracts were prepared from 10 ml of cells as described above, but using concanavalin A (ConA) buffer (1.6% Triton X-100, 0.1% SDS, 0.5 M NaCl, 20 mM sodium phosphate buffer, pH 7.6) during cell breakage. Crude extract (120 μl; 2.5 μg of protein/μl) was split into two 60-μl portions that were treated differently. One portion was incubated with 100 μl of ConA-Sepharose and 900 μl of ConA buffer for 4 h at 4°C and then centrifuged (45 s at 16,060 × g). The resulting supernatant containing mostly unglycosylated Sec20p was designated S1, while the ConA-Sepharose pellet was resuspended again in 1 ml of ConA buffer and treated for 45 min at room temperature before being centrifuged as before. The resulting ConA-Sepharose pellet was resuspended in Laemmli buffer and labeled fraction P1, while the corresponding supernatant was designated S2. The second portion of the crude extract was treated similarly, except that 10% methyl α-d-mannopyranoside was present in the second incubation and after centrifugation, pellet P3 and a supernatant fraction, S3, were obtained.
Pulse-chase experiments.
To maximally induce the PCK1p-SEC20-myc fusion, cells were grown twice in SLac medium before being harvested by centrifugation. Twenty units of cells at an optical density at 600 nm of 1 in the exponential growth phase were pelleted and resuspended in 2.5 ml of SLac medium. Following preincubation at 30°C for 20 min, 100 μCi of [35S]methionine (TRAN35S-LABEL; ICN) was added, and the cells were incubated for 15, 20, or 45 min (pulse). Thereafter, 1/50 volume of chase solution (0.3% cysteine, 0.4% methionine) was added, and incubation was continued for various times (chase). At each time point, an aliquot of 500 μl was removed, and the cells were pelleted by brief centrifugation and resuspended in 1 ml of ice-cold 10 mM NaN3. After another centrifugation step, the cell pellet was resuspended in 110 μl of lysis buffer (0.3 M sorbitol, 50 mM HEPES, 10 mM NaN3, pH 7.5), transferred to a microcentrifuge tube containing 250 mg of glass beads (diameter, 0.25 to 0.5 mm), and shaken at maximum speed on a Vibrax VX 2E for 7 min. One hundred microliters of 2× Laemmli sample buffer was added, and proteins were denatured at 95°C for 10 min.
For immunoprecipitation, 800 μl of IP dilution buffer (1.25% Triton X-100, 6 mM EDTA, 60 mM Tris-HCl, pH 7.6) was added to 200 μl of the denatured proteins, and insoluble cell material was pelleted by centrifugation at 16,060 × g for 2 min. Five microliters of anti-c-myc antibody (9E10; Babco) was added, and the solution was incubated overnight at 4°C; 50 μl of a 20% solution of protein A-Sepharose in IP buffer was added, and incubation was continued for 4 h at 4°C. Following centrifugation (16,060 × g; 20 s), a pellet was obtained, which was washed three times with washing buffer (1% Triton X-100, 0.2% SDS, 150 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl, pH 7.6), and then 50 μl of IP buffer, 50 μl of 2× Laemmli buffer, and 100 mM dithiothreitol was added, and the proteins were denatured at 95°C for 10 min. Following SDS-PAGE (11.5% acrylamide), the gel was bathed first in fixing solution (20% methanol, 7% acetic acid) for 30 min and then for 30 min in Amplify Fluorgraphic Reagent (Amersham). Proteins were detected by autoradiography.
RESULTS
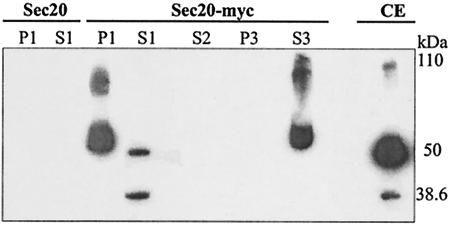
Sec20p in C. albicans is O glycosylated. We previously reported a plasmid (pYW69) directing the biosynthesis of Myc-tagged Sec20p in C. albicans (expected molecular mass, 38.645 kDa) (26). To determine if any of the three Sec20p electrophoretic forms (major 50-kDa form and minor 38.6- and 110-kDa forms) (Fig. 1, CE) is mannosylated, we allowed crude extracts of a pYW69 transformant to react with concanavalin A-Sepharose beads. Beads were found to bind the 50- and 110-kDa forms (P1), while the nonbound fraction contained mainly the 38.6-kDa nonglycosylated form and little of the 50-kDa form (S1). If beads retaining the 50- and 110-kDa forms were treated with methyl α-d-mannopyranoside, both forms were released (S3). In control experiments with transformants synthesizing untagged Sec20p (encoded by plasmid pYW7), no significant signals were detected by immunoblotting (Fig. 1).
FIG. 1.
Binding of Sec20p to concanavalin A. Crude extracts of C. albicans strain CAI4(pYW7) expressing a PCK1p-SEC20 fusion (Sec20) or strain CAI4(pYW69) expressing a PCK1p-SEC20-myc fusion (Sec20-myc) were allowed to react with concanavalin A-Sepharose for 4 h at 4°C. Centrifugation yielded a supernatant (S1) and a pellet, which was incubated with buffer for 45 min at room temperature and then centrifuged to yield pellet P1 and supernatant S2. If, instead of buffer, 10% methyl α-d-mannopyranoside was used for incubation, pellet P3 and S3 were obtained. Fractions were separated by SDS-PAGE (10% acrylamide) and immunoblotted using a mouse anti-c-myc antibody. In parallel, a crude extract of strain CAI4(pYW69) was tested by immunoblotting (CE). The migrations of the unglycosylated 38.6-kDa form and the 50- and 110-kDa glycosylated forms are indicated.
In further experiments, we treated the crude extracts with glycopeptidase F or we produced a Sec20p variant lacking the single possible N-glycosylation site (see below); in both experiments, the Sec20p forms were not altered. We conclude that Sec20p in C. albicans, similar to its homologue in S. cerevisiae (22), is partially O mannosylated to generate the 50- and 110-kDa forms.
Sec20p is not detectable in pmt1 and pmt4 mutants.
To examine Sec20p in mutants lacking known components of O glycosylation in C. albicans, we transformed pYW69 into a pmt1 mutant (23), a pmt4 mutant (Prill et al., unpublished), a pmt6 mutant (24), and an mnt1 mutant lacking the transferase extending monomannosylated proteins (1).
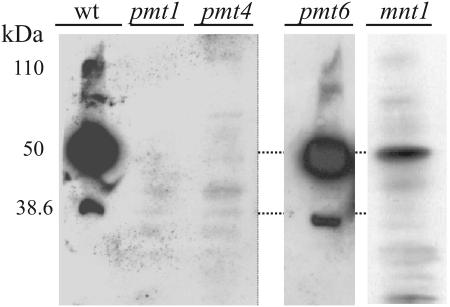
Following SDS-PAGE, the three electrophoretic forms of Sec20-Myc were observed in the wild-type strain and the pmt6 mutant but not in pmt1 and pmt4 mutants (Fig. 2). Minor bands detected in pmt mutants appear to be due to proteins in the extract that cross-react with the anti-Myc antibody, because they were also seen repeatedly in extracts of a control strain producing untagged Sec20p (data not shown and Fig. 3). The mnt1 strain contained the 50-kDa major form but none or little of the 38.6- and 110-kDa forms. We conclude from these results (i) that to obtain wild-type levels of Sec20p, specific Pmt isoforms, Pmt1p and Pmt4p, are required; (ii) that the major 50-kDa form of Sec20p is monomannosylated at multiple O-glycosylation sites; and (iii) that the 110-kDa form either contains extended O-glycosyl chains or consists of a stable dimer of the 50-kDa form.
FIG. 2.
Influence of O glycosylation mutants on Sec20p. Crude extracts (30 μg of protein) of the control strain CAI4(pYW69) (wt) and mutants CAP1-3121(pYW69) (pmt1), CAP4-2164(pYW69) (pmt4), CAP2-1341(pYW69) (pmt6), and NGY24(pYW69) (mnt1) were separated by SDS-PAGE, and immunoblots were tested using a monoclonal anti-c-myc antibody.
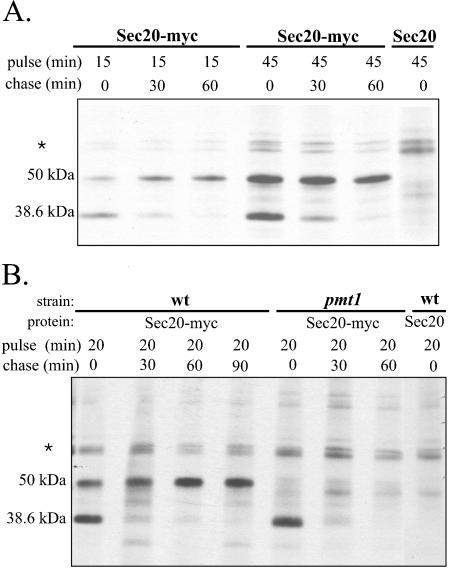
FIG. 3.
Stability measurements of Sec20p. (A) Pulse-chase experiments were carried out using strain CAI4(pYW69) (Sec20-myc) and, as a control, strain CAI4(pYW7) (Sec20). Cells were labeled with TRAN35S-LABEL (ICN) for 15 or 45 min (pulse), followed by an excess of methionine-cysteine and further incubation (chase). At 30 or 60 min of chase, samples were immunoprecipitated, followed by SDS-PAGE and autoradiography. (B) Strains CAI4(pYW69) (wt) and CAP1-3121(pYW69) (pmt1) were examined by pulse-chase measurements as for panel A; strain CAI4(pYW7) (Sec20) was used as the control. The migration of a nonspecifically immunoprecipitated protein is labeled by the asterisk.
Stability of Sec20 p in a pmt1 mutant.
The absence of Sec20p in the pmt1 and pmt4 mutants was conceivably caused by defective SEC20 expression, by the lack of biosynthesis, or by degradation of Sec20p. To decide among these alternatives, we performed pulse-chase experiments to monitor the kinetics of Sec20p biosynthesis and degradation.
We first established the parameters of Sec20p labeling in the PMT1 wild-type genetic background of strain CAI4. After pulse-labeling, the 38.6-kDa unglycosylated and 50-kDa O-glycosylated forms could be immunoprecipitated, while the 110-kDa form found in immunoblots was absent (Fig. 3A), suggesting that it is synthesized only after extended times (>100 min). Labeling of a control strain, producing untagged Sec20p, revealed only a doublet of nonspecific proteins. By pulse-labeling for 15 min and subsequent chase, we detected a clear precursor-product relationship between the 38.6-kDa form that was synthesized first and the 50-kDa form, which arose during the chase. From autoradiographies, we deduce a half-time of ∼15 min for the disappearance of the 38.6-kDa form and the appearance of the 50-kDa form; once produced, the 50-kDa form is stable for extended times (>100 min).
In the pmt1 genetic background, Sec20-Myc was initially synthesized as in the control strain (Fig. 3B). However, the 50-kDa form did not occur in the pmt1 mutant, and only minor bands, also found in the untagged Sec20p control, were detected in the 50-kDa range. Instead, the 38.6-kDa form disappeared with a half-time of ∼15 min, indicating that it was degraded. This result showed that the low level of Sec20p production in the pmt1 mutant was not caused by lack of SEC20 expression or defective Sec20p biosynthesis but was due to an inability to produce the stable O-glycosylated 50-kDa form.
In additional experiments, we also tested whether a 200 μM concentration of the proteasomal inhibitor lactacystin would lead to stabilization of Sec20-Myc in a pmt1 strain during a pulse-chase experiment. However, no significant stabilization was achieved (data not shown), and the possibility that lactacystin is unable to enter C. albicans, like S. cerevisiae, cannot be excluded (11). Because ubiquitination is a prerequisite for proteasomal degradation, we also tested whether Sec20-myc could be coimmunoprecipitated with ubiquitin (detected with a monoclonal anti-yeast ubiquitin antibody) in a wild-type or a pmt1 strain (data not shown). No ubiquitination of Sec20p was observed in this experiment, which is consistent with (but does not positively prove) the notion that Sec20p is degraded independently of the proteasome (3).
Biosynthesis of Sec20p in mutants lacking potential glycosylation and ubiquitination sites.
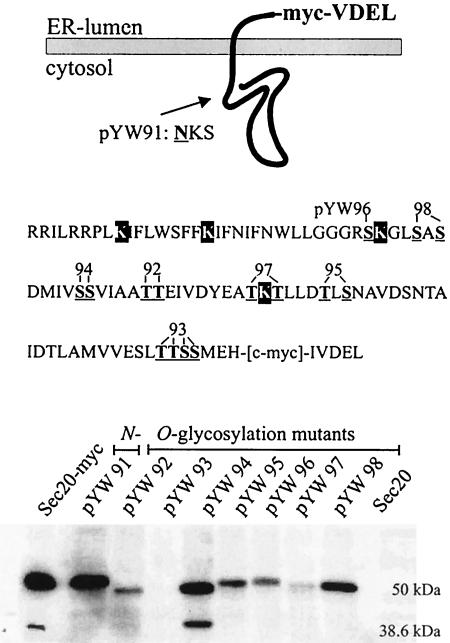
Sec20p is a type II membrane protein that in C. albicans directs a portion of ∼100 C-terminal residues oriented toward the lumen of the ER. Because it is known that Pmt proteins O mannosylate proteins at the lumenal side of the ER, we examined Sec20-Myc variants mutated in the lumenal domain to delete potential O glycosylation sites. A schematic view of the Sec20p lumenal domain, which is rich in Ser and Thr residues, and the locations of serine- or threonine-to-alanine replacements of the variants are shown in Fig. 4, top.
FIG. 4.
Biosynthesis of mutated Sec20p variants. (Upper panel) SEC20 in plasmid pYW69 was mutated to encode variant versions of Sec20p, in which serine or threonine residues of the Sec20p ER lumenal portion (underlined boldface residues in the sequence shown) were replaced by alanine. Plasmids pYW96 (S267A), pYW98 (S271A, S273A), pYW94 (S278A, S279A), pYW92 (T284A, T285A), pYW97 (T293A, T295A), pYW95 (T299A, S301A), and pYW93 (T321A, T322A, S323A, S324A) were tested; in addition, a sequence of a potential N-glycosylation site was mutated in pYW91 (N201A). Variants with alanine replacements for the indicated lysine residues (black boxes) were also constructed. (Lower panel) Crude extracts of CAI4 transformants carrying pYW69 (Sec20-myc), its derivatives lacking potential glycosylation sites, or a control plasmid pYW7 (Sec20) were tested by immunoblots, using anti-c-myc antibody.
Sec20p levels were found to differ greatly among CAI4 transformants expressing mutated SEC20 genes, with the lowest levels detected for variants encoded by pYW93 and pYW97 (Fig. 4, bottom). This finding suggests that Ser/Thr residues 321 to 324 and 293 and/or 295 are needed to stabilize Sec20p, very likely by their ability to receive Pmt-dependent mannosylation. The electrophoretic migration of other variants was increased slightly compared to the unaltered protein (variants encoded by pYW92 and pYW94), also indicating that the respective residues 278, 279, 284, and 285 were modified. Overall, these results indicated that most, if not all, potential Ser or Thr residues in the lumenal domain of Sec20p are mannosylated, cumulatively resulting in the increase in molecular mass from 38.6 to 50 kDa. Because in the mnt1 background the 50-kDa form had an identical molecular mass (see above), single mannoses appear to be attached at each site.
Because during ERAD, ubiquitin may get attached to Sec20p, we also tested the biosynthesis of variants, in which lysine residues within the ER lumenal domain, some situated adjacent to potential serine or threonine mannosylation sites, were changed to alanine (Fig. 4). However, such variants were not stabilized in pmt1 mutants (data not shown), suggesting that there is more than a single site for ubiquitination in the lumenal domain, that ubiquitination occurs elsewhere, or that Sec20p is degraded via a ubiquitination- and proteasome-independent pathway.
DISCUSSION
We show here that Sec20p in C. albicans is O glycosylated, and very likely this modification occurs within the C-terminal 92 residues in the ER lumen, based on the topology of Sec20p in S. cerevisiae (22). Because the major 50-kDa form of Sec20p has a similar electrophoretic mobility in an mnt1 host, in which O-chain extension is blocked (1), or after exchanges of individual Thr/Ser residues in the ER lumenal domain, it appears that the 38.6-kDa unmodified form gets monomannosylated at multiple sites. We found that the 38.6-kDa form is unstable, while the 50-kDa form is stable (half-time, >100 min), indicating that O mannosylation protects Sec20p from proteolytic degradation. We also discovered that Sec20p is unstable in pmt1 and pmt4 mutants but stable in a pmt6 mutant, suggesting that specific Pmt proteins, Pmt1p and Pmt4p, are responsible for modification and consequently for protection of Sec20p. Sec20p in S. cerevisiae, which has considerably diverged in structure and sequence from its C. albicans homologue (25), is also O mannosylated (22), but the roles of Pmt isoforms in modification and proteolytic degradation have not yet been studied. Specific mannosylation targets for individual Pmt proteins have been described in S. cerevisiae: chitinase, Bar1p protease, Hsp150p, α-agglutinin, and Kre1/9p are targets of Pmt1p and Pmt2p; Kex2p, Gas1p, Fus1p, and Axl2p are targets of Pmt4p (reviewed in reference 21); while Mid2p and Wsc1p are targets of Pmt2p and Pmt4p (14, 17). Some of these proteins, namely, Axl2p, Wsc1p, Wsc2p, and Mid2p, are prevented by Pmt2/4p-mediated O glycosylation from undergoing specific proteolytic cleavage (14, 17, 18). The stabilization of Sec20p in C. albicans described here represents the first case of the stabilization of an ER membrane protein by Pmt-mediated O mannosylation, which may be a paradigm for eukaryotic cells, including mammalian cells, which contain homologues of Pmt proteins that are active in O mannosylation (15, 27). It will be of interest to determine if among species, homologous proteins are subject to the same principle of protective O mannosylation or if this phenomenon is protein and species specific.
The molecular mechanism by which Pmt1/4p-mediated O mannosylation protects Sec20p from degradation is not known. In contrast to the known cases of protective O mannosylation (14, 17, 18), Sec20p appears to be completely degraded immediately after its synthesis in a pmt1 mutant (no cleavage products were detected); furthermore, Sec20p is not transiently but permanently associated with the ER, performing an essential function in retrograde vesicle traffic. It is possible that a specific lectin in the ER lumen recognizes monomannosylated proteins to positively retain them in the ER. In a negative model, mannosylation could also prevent recognition by a specific component of the ERAD or by an ER-resident degradation machinery (3). Such mechanisms could be especially important for ER-resident soluble or membrane proteins containing lumenal domains permanently or transiently in an open, noncompact conformation, which would otherwise be constantly and nonproductively removed from the ER by proteolysis. For fungi, protection by O mannosylation may be especially relevant, because protein translocation across the Sec61p translocon is closely coupled to O modification. It has in fact been shown that a mutated version of pre-pro-α-factor in S. cerevisiae is O glycosylated by Pmt2p within the ER and that in a pmt2 mutant its halftime is shortened from 12 to 10 min (8). Although this result supports our general conclusion about stabilization by Pmt-mediated mannosylation, we note important differences in that (i) unlike Sec20p, pre-pro-α-factor is not normally retained in the ER but secreted into the medium after processing; (ii) the rate of stabilization of Sec20p by O mannosylation is significantly greater than in the case of mutant pre-pro-α-factor; (iii) Pmt1p/Pmt4p instead of Pmt2p are involved in the stabilization of Sec20p; and (iv) we did not obtain evidence for an ERAD pathway of degradation, which involves retrograde export across the Sec61 pore and ubiquitination.
Some combinations of mutations in at least three PMT genes prevent the growth of S. cerevisiae (7), while in C. albicans, even a pmt single mutant (pmt2) and a pmt double mutant (pmt1 pmt4) are not viable (Prill et al., unpublished). On the other hand, Sec20p performs an essential function in the secretory pathway (25), suggesting that lack of Sec20p O mannosylation may be one reason why the pmt1 pmt4 double mutant is nonviable. In support of this notion, we found that moderate lowering of SEC20 expression generated a similar phenotype of antifungal supersensitivity (25) compared to pmt1 and pmt4 single mutants (23; Prill et al., unpublished). We assume that in both single mutants, residual mannosylation of Sec20p still occurs, allowing the presence of residual Sec20p in the ER and thereby allowing survival; on the other hand, the lack of both Pmt1p and Pmt4p would lower Sec20p levels below a threshold level not compatible with growth. Concepts for future strategies to combat fungal infections may include antifungals acting on Pmt proteins but could also focus on specific and essential Pmt targets, including Sec20p, for which no close homologue exists in human cells.
Acknowledgments
We thank N. Gow for sending the mnt1 mutant, and we acknowledge the excellent technical assistance of M. Gerads.
This study was supported by the Deutsche Forschungsgemeinschaft and by EU grant QLK2-CT-2001-02377 (Combating MDR in Pathogens).
REFERENCES
- 1.Buurman, E. T., C. Westwater, B. Hube, A. J. P. Brown, F. C. Odds, and N. A. R. Gow. 1998. Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 95:7670-7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabral, C. M., Y. Liu, and R. N. Sifers. 2001. Dissecting glycoprotein quality control in the secretory pathway. Trends Biochem. Sci. 26:619-624. [DOI] [PubMed] [Google Scholar]
- 3.Cabral, C. M., Y. Liu., K. W. Moremen, and R. N. Sifers. 2002. Organizational diversity among distinct glycoprotein endoplasmic reticulum-associated degradation programs. Mol. Biol. Cell. 13:2639-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dilcher, M., B. Veith, S. Chidambaram, E. Hartmann, H. D. Schmitt, and G. Fischer von Mollard. 2003. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 22:3664-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ernst, J. F., and S. K.-H. Prill. 2001. O-glycosylation. Med. Mycol. 39(Suppl. I):67-74. [PubMed] [Google Scholar]
- 6.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentzsch, M., and W. Tanner. 1996. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 15:5752-5759. [PMC free article] [PubMed] [Google Scholar]
- 8.Harty, C., S. Strahl, and K. Römisch. 2001. O-mannosylation protects mutant alpha-factor precursor from endoplasmic reticulum-associated degradation. Mol. Biol. Cell 12:1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helenius, A., and M. Aebi. 2001. Intracellular functions of N-linked glycans. Science 291:2364-2369. [DOI] [PubMed] [Google Scholar]
- 10.Jakob, C. A., P. Burda, J. Roth, and M. Aebi. 1998. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol. 142:1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, D. H., and A. L. Goldberg. 1996. Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271:27280-27284. [DOI] [PubMed] [Google Scholar]
- 12.Leuker, C. E., A. Sonneborn, S. Delbrück, and J. F. Ernst. 1997. Sequence and regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene 192:235-240. [DOI] [PubMed] [Google Scholar]
- 13.Lewis, M. J., J. C. Rayner, and H. R. B. Pelham. 1997. A novel SNARE complex implicated in vesicle fusion with the endoplasmic reticulum. EMBO J. 16:3017-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lommel, M., M. Bagnat, and S. Strahl. 2004. Abberant processing of the WSC family and Mid2p cell surface sensors results in cell death of Saccharomyces cerevisiae O-mannosylation mutants. Mol. Cell. Biol. 24:46-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manya, H., A. Chiba, A. Yoshida, X. Wang, Y. Chiba, Y. Jigami, R. U. Margolis, and T. Endo. 2004. Demonstration of mammalian protein O-mannosyltransferase activity: coexpression of PMT1 and PMT2 required for enzymatic activity. Proc. Natl. Acad. Sci. USA 101:500-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parodi, A. J. 2000. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem. J. 348:1-13. [PMC free article] [PubMed] [Google Scholar]
- 17.Philip, B., and D. E. Levin. 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders, S. L., M. Gentzsch, W. Tanner, and I. Herskowitz. 1999. O-glycosylation of Axl2/Bud10p by Pmt4p is required for its stability, localization and function in daughter cells. J. Cell Biol. 145:1177-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schleip, I., E. Heiβ, and L. Lehle. 2001. The yeast SEC20 gene is required for N- and O-glycosylation in the Golgi. J. Biol. Chem. 276:28751-28758. [DOI] [PubMed] [Google Scholar]
- 20.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Strahl-Bolsinger, S., M. Gentzsch, and W. Tanner. 1999. Protein O-mannosylation. Biochim. Biophys. Acta 1426:279-307. [DOI] [PubMed] [Google Scholar]
- 22.Sweet, D. J., and H. R. B. Pelham. 1992. The Saccharomyces cerevisiae SEC20 gene encodes a membrane glycoprotein which is sorted by the HDEL retrieval system. EMBO J. 11:423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timpel, C., S. Strahl-Bolsinger, K. Ziegelbauer, and J. F. Ernst. 1998. Multiple functions of Pmt1p-mediated protein O-mannosylation in the fungal pathogen Candida albicans. J. Biol. Chem. 273:20837-20846. [DOI] [PubMed] [Google Scholar]
- 24.Timpel, C., S. Zink, S. Strahl-Bolsinger, K. Schröppel, and J. Ernst. 2000. Morphogenesis, adhesive properties, and antifungal resistance depend on the Pmt6 protein mannosyltransferase in the fungal pathogen Candida albicans. J. Bacteriol. 182:3063-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber, Y., U. J. Santore, J. F. Ernst, and R. K. Swoboda. 2001. Divergence of eukaryotic secretory components: the Candida albicans homolog of the Saccharomyces cerevisiae Sec20 protein is N terminally truncated, and its levels determine antifungal drug resistance and growth. J. Bacteriol. 183:46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber, Y., R. K. Swoboda, and J. F. Ernst. 2002. Sec20p-interacting proteins (Tip20p, Ufe1p) in the retrograde secretory pathway of the fungal pathogen Candida albicans. Mol. Gen. Genom. 268:468-476. [DOI] [PubMed] [Google Scholar]
- 27.Willer, T., M. C. Valero, W. Tanner, J. Cruces, and S. Strahl. 2003. O-Mannosyl glycans: from yeast to novel associations with human disease. Curr. Opin. Struct. Biol. 13:621-630. [DOI] [PubMed] [Google Scholar]