Epileptic encephalopathies are severe childhood-onset epilepsies which often have a genetic aetiology. Using next-generation sequencing, Shen et al. identify six de novo pathogenic variants in GABRG2, the gene that encodes the β2 subunit of the GABAA receptor. Functional analysis confirms that all six mutations impair GABAA receptor biogenesis and/or channel function.
Keywords: epileptic encephalopathy, GABAA receptor, GABRG2, de novo mutation, next generation sequencing
Abstract
Epileptic encephalopathies are a devastating group of severe childhood onset epilepsies with medication-resistant seizures and poor developmental outcomes. Many epileptic encephalopathies have a genetic aetiology and are often associated with de novo mutations in genes mediating synaptic transmission, including GABAA receptor subunit genes. Recently, we performed next generation sequencing on patients with a spectrum of epileptic encephalopathy phenotypes, and we identified five novel (A106T, I107T, P282S, R323W and F343L) and one known (R323Q) de novo GABRG2 pathogenic variants (mutations) in eight patients. To gain insight into the molecular basis for how these mutations contribute to epileptic encephalopathies, we compared the effects of the mutations on the properties of recombinant α1β2γ2L GABAA receptors transiently expressed in HEK293T cells. Using a combination of patch clamp recording, immunoblotting, confocal imaging and structural modelling, we characterized the effects of these GABRG2 mutations on GABAA receptor biogenesis and channel function. Compared with wild-type α1β2γ2L receptors, GABAA receptors containing a mutant γ2 subunit had reduced cell surface expression with altered subunit stoichiometry or decreased GABA-evoked whole-cell current amplitudes, but with different levels of reduction. While a causal role of these mutations cannot be established directly from these results, the functional analysis together with the genetic information suggests that these GABRG2 variants may be major contributors to the epileptic encephalopathy phenotypes. Our study further expands the GABRG2 phenotypic spectrum and supports growing evidence that defects in GABAergic neurotransmission participate in the pathogenesis of genetic epilepsies including epileptic encephalopathies.
Introduction
Epileptic encephalopathies are a devastating group of severe infantile and childhood onset epilepsies, which are clinically and aetiologically heterogeneous and characterized by intractable seizures, neurodevelopmental impairment, and poor prognosis (Berg et al., 2010). Because of the severity of the seizures and the associated intellectual and behavioural disabilities, the children and their families often suffer from substantial economic, social, and emotional burdens (Katsnelson et al., 2014).
Due to developments in massively parallel sequencing technologies, a significant proportion of epileptic encephalopathy patients’ aetiologies have been shown to be genetic in nature. Patients with epileptic encephalopathy usually have limited or no family history of epilepsy and pathogenic variants typically arise de novo (Thomas and Berkovic, 2014). Trio whole exome sequencing, in which the genomes of the individual with epilepsy and both parents are sequenced, is a powerful tool for dissecting the genetic basis of epileptic encephalopathies (Epi PMC, 2015). Use of targeted epilepsy-related gene panels for next generation sequencing is an alternative approach for identifying candidate de novo variants in sporadic cases of epileptic encephalopathy (Carvill et al., 2013). Increased efficiency and reduced cost of these technologies have enabled discovery of numerous new epileptic encephalopathy genes with unprecedented success (McTague et al., 2016). The majority of the genes identified, to date, are involved in regulating synaptic transmission (Euro et al., 2014), which is not surprising given the importance of synaptic function in regulating excitability in the brain.
GABAA receptors mediate the majority of fast inhibitory neurotransmission and control network excitability in the brain. They are heteropentameric GABA-gated chloride ion channels, and the α1β2γ2 receptor is the most abundant GABAA receptor subtype in the CNS (Farrant and Nusser, 2005). The γ2 subunits are abundantly expressed and play important roles in receptor trafficking, clustering, synaptic maintenance (Essrich et al., 1998; Schweizer et al., 2003) and current kinetic properties (Haas and Macdonald, 1999). Hence, dysfunctions of GABAA receptor γ2 subunits have been postulated to be involved in the aetiology of epilepsy. In fact, among currently known epilepsy-associated mutations identified in GABAA receptor subunits, over half of them are found in the GABAA receptor γ2 subunit gene, GABRG2 (Macdonald and Kang, 2009). A substantial number of GABRG2 mutations have been associated with autosomal dominant genetic epilepsies, ranging from relatively benign febrile seizures and childhood absence epilepsy to more severe genetic epilepsy with febrile seizures plus (GEFS+) and Dravet syndrome (Kang et al., 2015). In vitro studies have demonstrated that these GABRG2 mutations exhibited a wide array of functional deficits, including alternation of RNA processing or protein stability, channel kinetic defects, and dominant negative effects (Macdonald and Kang, 2012). Moreover, heterozygous knock-in mice bearing human GABRG2 epilepsy mutations had reduced cortical inhibition and displayed epilepsy phenotypes (Tan et al., 2007; Reid et al., 2013; Kang et al., 2015).
Given the critical role of γ2 subunits and the reported GABRG2 mutations in a broad spectrum of epilepsy syndromes, we wondered whether rare pathogenic GABRG2 variants might also contribute to the aetiology of epileptic encephalopathy. To test this hypothesis, we carried out next-generation sequencing in parent-offspring trios with a wide range of intractable epileptic encephalopathy phenotypes and searched for de novo GABRG2 mutations. Six de novo missense GABRG2 mutations (A106T, I107T, P282S, R323Q, R323W and F343L) were discovered in eight isolated patients. We obtained the patients’ clinical history and investigated functional effects of these de novo GABRG2 mutations on GABAA receptor biogenesis, trafficking and function in vitro. GABAA receptor α1 and β2 subunits were co-expressed with wild-type or mutant γ2 subunits in HEK293T cells. Using this heterologous expression system, we found that all of these de novo GABRG2 mutations impaired GABAA receptor biogenesis and/or channel function, but to different extents. Furthermore, we characterized mutation-induced alternations of secondary and tertiary structures of GABAA receptors based on structural modelling. Our genetic and functional findings provide strong evidence that GABRG2 mutations are a genetic risk factor for the development of epileptic encephalopathy.
Material and methods
Patient phenotypes
Seven patients (six female/one male) were selected for sequencing due to having an intractable early onset epilepsy. The eighth patient (female) was tested for severe intellectual disability, movement disorder and early onset seizures. The patients were collected from multiple sites, four European clinical programs (University of Basel, University of Leipzig, Clinic for Children and Adolescents Munich, Clinic for Children and Adolescents Halle), and three American paediatrics programs (Children’s Hospital of Philadelphia, Boston Children’s Hospital, and Center for Rare Childhood Disorders, TGen). De-identified clinical information was collected and compared across all patients as part of a case series. Five patients were identified on comprehensive epilepsy panels as clinical testing, one by clinical whole exome sequencing, and two by research exome sequencing.
Whole exome sequencing and analysis
Whole exome sequencing was performed for one patient at the Duke University Sequencing core (Duke CHGV) using the Illumina Genome Analyzer IIx massively parallel sequencing system (Illumina, Inc.) as previously published (Poduri et al., 2013). Alignment to the human genome (reference build hg18) was conducted with BWA version 0.5.5. Consensus and variant calls were performed using SAMtools version 0.1.7. Annotation, filtering for quality and removal of potential variants present in dbSNP129 or in 220 individuals from a group non-enriched for neuropsychiatric phenotypes, and prediction of functional effects of potential mutations were performed using Sequent Variant Analyzer (SVA) (http://people.genome.duke.edu). The research laboratory believed the variant was pathogenic.
Whole exome sequencing was performed at the TGen research laboratory in another patient using the following protocol. Libraries were prepared using the Illumina’s TruSeq DNA sample preparation kit and the TruSeq exome enrichment kit following the manufacturer’s protocol. Sequencing was done by 100-bp paired-end sequencing on a Illumina HiSeq2000 instrument. Reads were aligned to the Human Genome (Hg19/GRC37) using Burrows-Wheeler transform alignment (BWA v.0.7.5)1. PCR duplicates were removed using Picard v.1.922, and base quality recalibration, indel realignment and single nucleotide polymorphism (SNP) and indel discovery were performed using the Genome Analysis Toolkit (GATK v.2.5-2)3. Variants were annotated with SnpEff 3.2a and selected (SnpSift) for protein-coding events. Prediction scores were loaded from dbNSFP and used for filtering. This variant was considered probably pathogenic and was validated by GeneDx.
In one patient the GABRG2 variant was found on clinical exome sequencing through GeneDx (XomeDX, Gene DX, Gaithersburg MD) as per their clinical protocol (for details see http://www.genedx.com/test-catalog/xomedx/). GeneDx reported the mutation (c.1027T>C) as variant, likely mutation.
Epilepsy panels
One patient was identified on the GeneDx comprehensive epilepsy panel (Infantile Epilepsy Panel, Gene DX) and reported as a variant of uncertain significance. Subsequent parental testing revealed the mutation to be de novo. The panel was performed as per GeneDx available methodology (http://www.genedx.com/test-catalog/available-tests/infantile-epilepsy-panel/).
Four European patients were identified through the CeGaT epilepsy panel (CeGaT GmbH). All were called pathogenic or likely pathogenic based on the recent guideline from the ACMG (Richards et al., 2015). The panel targeted 119 genes (www.cegat.de/diagnostik/panel-diagnostik/epilepsie-und-migraene/) and was performed as previously described (Lemke et al., 2012). In brief, the sequencing was performed by enriching for coding regions and exon-intron boundaries using Agilent SureSelect technology (Agilent Technologies) and sequencing on an Illumina HiSeq2500 platform (Illumina). Annotation was performed using SAMtools (v0.1.18) and VarScan (v2.3). Variants were selected with a minor allele frequency below 5% (according to 1000 Genomes, dbSNP, EVS and in-house database). More than 98% of targets had at least 30× coverage. Validation of suspicious variants as well as segregation analysis in both parents were performed by post hoc standard Sanger sequencing.
Complementary DNA constructs
The coding sequences of human α1, β2 and γ2L GABAA receptor subunits and EGFP were cloned into pcDNA3.1(+) expression vectors (Invitrogen). Mutant γ2L subunit constructs were generated using the QuikChange site-directed mutagenesis kit (Agilent) and confirmed by DNA sequencing. Due to the lack of a highly specific antibody against the extracellular domain of the γ2 subunit, N-terminal haemagluttinin (HA)-tagged (γ2LHA) subunits were used. The HA epitope was inserted between the fourth and fifth residue of the mature γ2L subunit, a functionally silent position (Connolly et al., 1996). Note that all subunit residues were numbered based on the immature peptide that includes the signal peptide.
Cell culture and transfection
HEK293T cells (ATCC, CRL-11268) were cultured at 37°C in humidified 5% CO2 incubator and maintained in Dulbecco’s modified Eagle medium (Invitrogen) supplemented with 10% foetal bovine serum (Life technologies), and 100 IU/ml penicillin streptomycin (Life Technologies). Cells were transfected using polyethylenimine (PEI) reagent (40 kD, Polysciences) at a DNA:transfection reagent ratio of 1:2.5, and harvested 36 h after transfection. To express wild-type and mutant α1β2γ2 receptors, a total of 3 µg of subunit cDNAs were transfected at a ratio of 1:1:1 into 6 cm dishes for most experiments except for whole-cell recording. For the mock-transfected condition, empty pcDNA3.1 vector was added to make a final cDNA transfection amount to 3 μg.
Western blot and surface biotinylation
Transfected HEK293T cells were collected in modified RIPA buffer [50 mM Tris (pH = 7.4), 150 mM NaCl, 1% NP-40, 0.2% sodium deoxycholate, 1 mM EDTA] and 1% protease inhibitor cocktail (Sigma). Collected samples were subjected to gel electrophoresis using 4–12% BisTris NuPAGE precast gels (Invitrogen) and transferred to PVDF-FL membranes (Millipore). Polyclonal anti-γ2 antibodies (Alomone or Millipore) were used to detect GABAA receptor γ2 subunits. Anti-Na+/K+ ATPase antibody (Abcam) was used as a loading control. IRDye® (LI-COR Biosciences) conjugated secondary antibody was used at a 1:10 000 dilution in all cases. Membranes were scanned using the Odyssey Infrared Imaging System (LI-COR Biosciences). The integrated intensity value of bands was determined using the Odyssey Image Studio software (LI-COR Biosciences).
Biotinylation protocols have been described previously (Huang et al., 2014). Briefly, transfected cells were incubated in membrane-impermeable reagent sulf-HNS-SS-biotin (1 mg/ml, Thermo Scientific) at 4°C for 40 min. Cells were lysed after being quenched with 0.1 M glycine. Lysates were cleared by after centrifugation and then incubated overnight with High Binding Capacity NeutrAvidin™ beads (Thermo Scientific Pierce). After incubation, protein was eluted in sampling buffer (Invitrogen) containing 10% beta-mercaptoethanol and subjected to immunoblotting.
Immunocytochemistry and confocal microscopy
For immunofluorescence, coverslip-grown HEK293T cells were washed with phosphate-buffered saline (PBS) and fixed with Prefer (Anatech) to stain surface proteins or permeabilized with 0.5% TritonTM X-100 to stain total proteins. The fixed/permeabilized cells were blocked for 2 h with 5% bovine serum albumin in PBS, and then stained with primary antibodies overnight, followed by incubation in Alexa 488-conjugated donkey anti-rabbit IgG antibodies and Cy3-conjugated donkey anti-mouse IgG antibodies. Primary antibodies used were as the follows: rabbit monoclonal HA antibody (Cell Signaling), mouse monoclonal α1 subunit antibody (Millipore), mouse monoclonal anti-calnexin antibody (Abcam). Coverslips were mounted with ProLong Gold® anti-fade reagent (Thermo Fisher Scientific Inc.).
Confocal images were obtained from HEK293T cells using a Zeiss LSM 710 Meta inverted confocal microscope. Stained HEK293T cells were excited with the 488 nm laser for the Alexa 488 fluorophore signal and the 543 nm laser for the Cy3 fluorophore signal. Images were taken with 8 bit, 1024 × 1024 pixel resolution. Pinholes were adjusted so that the sample thickness was 0.9 μm. An average of four scans was taken to decrease the background noise. Confocal experiments were performed in part using the VUMC Cell Imaging Shared Resource.
Co-localization analysis was performed using the Coloc2 plugin in the open source image processing program Fiji (Schindelin et al., 2012). Microscopic image files were imported, and the two channels (green and red) were separated. The two channels being compared were assigned to Channel 1 (green) and Channel 2 (red) in a manner consistent across all samples. A region of interest surrounding individual cells was selected in the green channel, and its location was set in the Coloc2 panel. Both Pearson’s correlation coefficient (R) and Manders’ co-localization coefficient (MCC) were calculated.
Electrophysiology
Whole-cell recordings of wild-type and mutant GABAA receptor currents were obtained at room temperature from lifted HEK293T cells (Hernandez et al., 2011). The external solution was composed of (in mM): 142 NaCl, 8 KCl, 10 D(+)-glucose, 10 HEPES, 6 MgCl2.6H2O, and 1 CaCl2 (pH 7.4, ∼326 mOsm). The internal solution consisted of (in mM): 153 KCl, 10 HEPES, 5 EGTA 2 Mg-ATP, and 1 MgCl2.6H2O (pH 7.3, ∼300 mOsm). The Cl− reversal potential was near 0 mV, and cells were voltage clamped at −20 mV. GABA (1 mM) was applied for 4 s for measurements of current amplitude and zinc inhibition. Zinc (10 µM) was pre-applied for 10 s followed by co-application with GABA. GABAA receptor current concentration–response curves were fitted using GraphPad Prism version 6.07 for Windows (GraphPad Software, La Jolla, CA). GABA was applied using a four-barrel square glass pipette connected to a SF-77B Perfusion Fast-Step system (Warner Instruments Corporations). The solution exchange time across the open electrode tip was ∼200–400 μs, and the exchange around lifted cells (∼8–10 pF) occurred within 800 μs, which was sufficiently fast for these experiments (Bianchi and Macdonald, 2002) and guaranteed rapid solution exchanges and accurate measure of the kinetic properties of the receptor. All experiments were performed at room temperature (22–23°C). Whole cell currents were amplified and low-pass filtered at 2 kHz using an Axopatch 200B amplifier, digitized at 10 kHz using Digidata 1550, and saved using pCLAMP 10.4 (Axon Instruments). Data were analysed offline using Clampfit 10.4 (Axon Instruments). Activation onset and deactivation weight time constants (τ) were measured from currents obtained by application of 1 mM GABA for 10 ms, while peak current amplitude was measured from currents obtained by application of 1 mM GABA for 4 s. Activation and deactivation time constants (τ) were fitted using the Levenberg–Marquardt least squares method with up to four component exponential functions of the form ∑anexp(–t/τn) + C, where n is the number of the exponential components, t is time, a is the relative amplitude, τn is the time constant, and C is the residual current at the end of GABA application. Additional components were accepted only if they significantly improved the fit, as determined by an F test on the sum of squared residuals. The multi-exponential time course of deactivation was presented as a weighted time constant, defined by the following expression: ∑anτn/∑an
Structural modelling and simulation
GABAA receptor α1, β2 and γ2 subunit raw sequences in FASTA format were individually loaded into Swiss-PdbViewer 4.10 for template search against the ExPDB database. The structure of the Caenorhabditis elegans glutamate-gated chloride channel (GluCl; PDB: 3RHW) was identified as a template using DeepView/Swiss-PdbViewer 4.02 (Schwede et al., 2003). The long cytoplasmic regions of GABAA receptor subunits were excluded from modelling as they were absent in the solved GluCl structure and separate alignments were generated for the TM4 domains. Full-length multiple alignments were submitted for automated comparative protein modelling incorporated in SWISS-MODEL program suite. The resulting subunit models were energy-optimized using GROMOS96 of the Swiss-PdbViewer. To generate pentameric GABAA receptor 3D models, α1, β2 and γ2 subunit structural models were assembled in a counter-clockwise β2-α1-β2-α1-γ2 order by superposition onto the C. elegans GluCl channel. Neighbourhood structural variability on the 3D GABAA receptor predicted by the γ2 subunit mutations were implemented using Rosetta 3.1 (Smith and Kortemme, 2008) (https://kortemmelab.ucsf.edu/backrub/cgi-bin/rosettaweb.py). Up to 20 of the best-scoring structures were generated for each mutation by choosing parameters recommended by the application. We measured mutation-induced structural differences by analysing the root mean squared (RMS) deviation between the initial (wild-type) structures and superimposed simulated (mutated) structures. RMS deviation provides carbon-α/carbon-α comparisons between two structurally aligned models; the larger the RMS deviation, the more the mutant structure deviates from the wild-type structure. For each mutation, the average RMS deviation over 10 lowest energy structures was computed. We prepared the figures using Chimera 1.7 (Pettersen et al., 2004).
Statistical analysis
Numerical data were reported as mean ± standard error of the mean (SEM). Statistical analysis was performed using GraphPad Prism version 6.07 for Windows (GraphPad Software, La Jolla, CA). Statistically significant differences were taken as P < 0.05 using one-way ANOVA followed by Dunnett’s multiple comparison test.
Results
Mutation screening and de novo GABRG2 variants
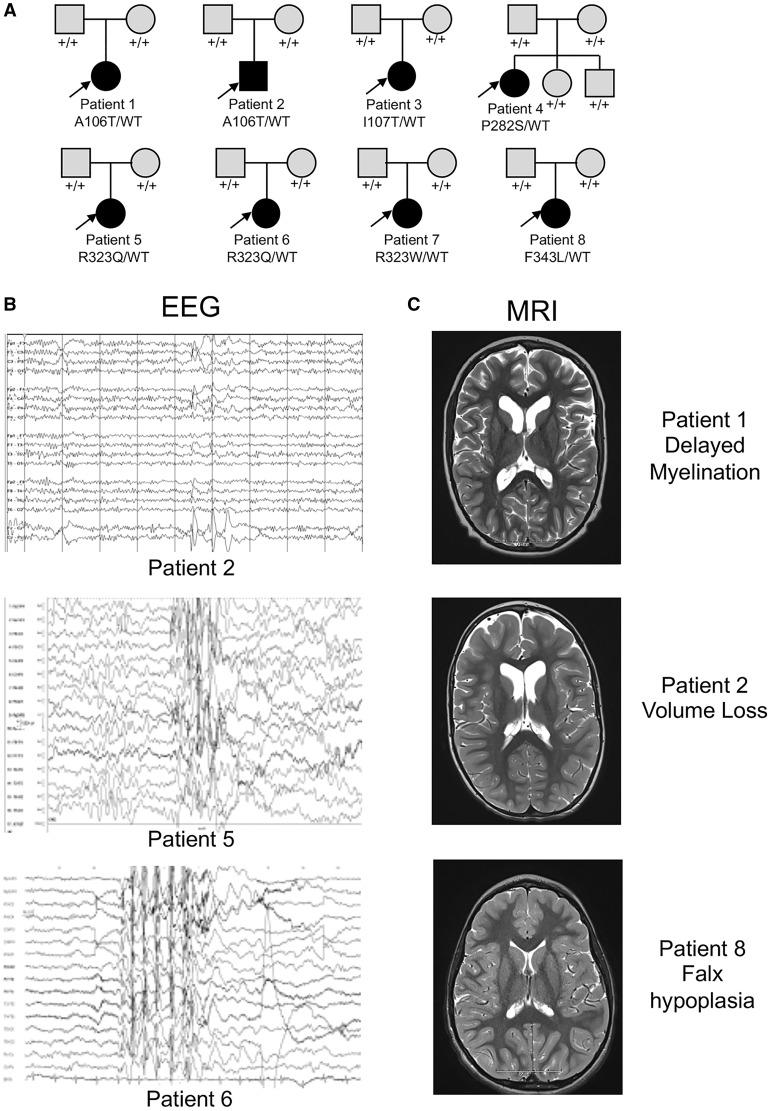
All eight patients (seven female/one male) were selected for sequencing due to having an intractable early onset epilepsy. The testing was done at a mean age of 6.4 years (range 3 years to 10 years old). For six patients, sequencing was done as part of clinical evaluation using either epilepsy panels (n = 5) or whole exome sequencing (n = 1) at GeneDx or CeGaT. For the others, research whole exome sequencing was performed. In all eight patients, the variant was found to occur de novo in the child after testing the parental DNA (Fig. 1A).
Figure 1.
De novo GABRG2 variants were identified in eight individuals with epileptic encephalopathy. (A) Pedigrees and segregation analysis of the six GABRG2 missense variants identified in eight patients. Arrows indicate probands. (B) Three representative EEGs are presented. The top EEG demonstrates excessive beta activity and focal discharges over the vertex head region. The lower two EEG traces show more diffuse background slowing and irregularly generalized (middle) and generalized (bottom) epileptiform discharges. (C) Most of the brain MRIs were normal (not shown), but three patients had non-specific findings and are presented (top MRI: delayed myelination of the frontal lobes; middle MRI: hypoplasia of the falx; bottom MRI: enlarged ventricles and extra-axial CSF spaces for age). All MRIs are presented at level of head of caudate.
Patient phenotypes
The clinical features of the eight patients with GABRG2 variants were summarized in Table 1, and their representative EEG and brain MRI images were shown in Fig. 1. The age of onset of epilepsy was within the first year of life in all eight patients (range Day of life 1 to 1 year of age). Seizure semiology at onset was described as tonic-clonic seizures in two patients, tonic seizures in three patients, partial seizures with secondary generalization in one patient, and febrile seizures in combination with myoclonic seizures in two patients. The epilepsy in these patients progressed in all except one patient (Patient 8) with development of additional seizure types, which included atonic, generalized tonic clonic, absence and focal seizures. As epileptic encephalopathies are a spectrum of disorders that include a number of named syndromes, we asked if any patient fit criteria for a specific electroclinical syndrome diagnosis (i.e. infantile spasms syndrome). No patients were given a diagnosis early, but three patients eventually had features of Lennox-Gastaut syndrome. Epilepsy outcome was variable, with two patients eventually becoming seizure-free (Patients 1 and 7), whereas the six other patients’ seizures remained intractable as of last follow-up despite combination therapy with antiepileptic drugs. Physical and neurological examinations were remarkable for the presence of hypotonia in six patients, abnormal eye movements in four patients, and choreiform movements in four patients. There were no dysmorphic or other pathognomonic features on exam, and two patients (Patients 5 and 7) were described as having normal physical and neurological examinations. Developmentally, all eight individuals had severe intellectual disability, were non-verbal, and had severe motor disabilities.
Table 1.
Clinical features of all individuals with GABRG2 variants identified in this study.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Patient 7 | Patient 8 | |
|---|---|---|---|---|---|---|---|---|
| Variant | c.316G>A | c.316G>A | c.320T>C | c.844C>T | c.968G>A | c.968G>A | c.967C>T | c.1027 T>C |
| Origin | De novo | De novo | De novo | De novo | De novo | De novo | De novo | De novo |
| Protein change | p.A106T | p.A106T | p.I107T | p.P282S | p.R323Q | p.R323Q | p.R323W | p.F343L |
| Sex | Female | Male | Female | Female | Female | Female | Female | Female |
| Age at inclusion | 7 years | 9 years | 3 years | 10 years | 4 years | 3 2/12 years | 9 years | 6 years |
| Age at seizure onset | Day of life 1 | 3 months | 1.5 months | 1 year | 10 months | 1 year | 11 months | 1 year |
| Seizure type at onset | GTCS | Tonic | Tonic | Secondary generalized | FS, GTCS, myoclonic | FS, myoclonic | GTCS | Tonic |
| Seizure frequency at onset | Daily | Unknown | Daily | Unknown | Sporadic GTCS | Daily | Weekly | Daily |
| Further seizure types | Tonic, CPS | CPS, secondary generalized, atonic | Infantile spasms, tonic | Atypical absences | Myoclonic, absences, GTCS, CPS | Atonic, myoclonias during sleep, atypical absences, GTCS | Absences | None |
| AED responses | Seizure-free for 2 years on LEV | No clear response | No clear response | Slight improvement with LTG | No clear response | No clear response | VPA and TPM best combination | Some improvement on LEV |
| Seizure outcome | Seizure-free for 3 years (1 year seizure free off AED) | Remains intractable | Remains intractable | Remains intractable | Remains intractable | Remains intractable | Seizure-free for 3 years | Seizures Persist |
| EEG at onset | Normal | Normal | High voltage, slowing of background, sharp transients on the right side | Generalized and multifocal spikes | Normal | Generalized spikes, irregular spike-wave- complexes | Normal background, rare generalized spike waves | Excess diffuse beta and intermittent left temporal slowing. |
| Other EEG | No epileptiform activity seen | Diffuse xs beta, multifocal sharps | Background slowing, rare sharp transients right more than left | Generalized spike-wave nearly continuous | Generalized irregular spike wave | Generalized spikes, irregular spike-wave-complexes | Normal background frequencies, rare single generalized or hemispheric accentuated spike waves | poor organization, diffuse xs beta, frequent sharps maximal at the central vertex |
| Development | Severe global delay | Severe global delay | Severe global delay | Severe global delay | Severe global delay | Severe global delay | Severe global delay | Severe global delay |
| Language | Non-verbal | Non-verbal | Non-verbal | Non-verbal | Non-verbal | Non-verbal | Non-verbal | Non-verbal |
| Neurological exam | Hypotonia, nystagmus | Hypotonia, nystagmus, hyperkinetic movements with some choreoathetotic components | Hypotonia, nystagmus, hand stereotypies, choreoathetosis | Hypotonia, roving eye movements | Normal | Hypotonia, mild ataxia | Normal | Hypotonia, intermittent hand posturing |
| MRI findings | Delayed myelination | Volume loss | Normal | Normal | Normal | Normal | Normal | Falx hypoplasia |
AED = antiepileptic drug; CPS = complex partial seizures; GTCS = generalized tonic-clonic seizures; FS = febrile seizures; LEV = levetiracetam; LTG = lamotrigine; TPM = topiramate; VPA = valproic acid; xs = excess.
Additional studies of EEG and MRI in this cohort were also variable with no consistent findings. The initial EEG was normal in three patients (Patients 1, 2 and 5), but over time all became abnormal. A variety of EEG abnormalities was found in patients with GABRG2 variants (Fig. 1B) including seven of eight with either focal (n = 3) or generalized (n = 4) interictal epileptiform discharges. Brain MRIs were normal in five patients and showed mild non-specific findings in three patients (delayed myelination, volume loss, and falx hypoplasia in one patient each) (Fig. 1C). These data suggest that GABRG2 can lead to variable neurodevelopmental outcomes, including epileptic encephalopathy and abnormal motor development.
De novo GABRG2 variants were located in different structural domains of GABAA receptor γ2 subunits
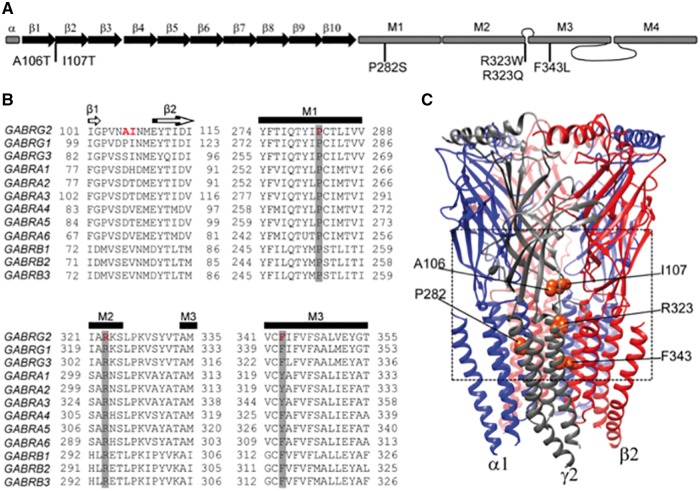
Individual GABAA receptor γ2 subunits are composed of a large extracellular N-terminal domain, followed by four transmembrane domains (M1-M4) as well as extracellular (M2-M3) and two intracellular (M1-M2; M3-M4) loops (Macdonald and Olsen, 1994; Miller and Aricescu, 2014), and the six variants identified here were located in functionally important regions of the receptor channel (Fig. 2A). By analysing the sequence alignment among the GABRs, we found that P282 and R323 were invariant residues across all GABAA receptor subunits, and F343 was a highly conserved residue (Fig. 2B). Consistently, in silico analysis using PolyPhen-2 (Adzhubei et al., 2010) and SIFT (Kumar et al., 2009) predicted that the substitutions P282S, R323Q, R323W and F343L would not be tolerated and might damage protein structure. In contrast, the variants A106T and I107T, which were located in the non-conserved residue (Fig. 2B), were predicted to be tolerated.
Figure 2.
De novo γ2 subunit variants were located between the interface of the N-terminal and transmembrane domains of the GABAAreceptor. (A) A cartoon representation of the linearized secondary structure of a γ2 subunit was made displaying putative locations of the substitutions identified in this study. β-strands were represented as black arrows and α-helices as grey rectangles. (B) Alignment of human γ(1-3), α(1-6), and β(1-3) subunits from the GABAA receptor subunit family were performed. Sites of de novo variants in the γ2 subunit are shown in red. A106 and I107 residues were not conserved (shown in red in γ2 subunit only). Across all sequences, P282 and R323 residues were identical (highlighted in dark grey), and the residue F343 was conserved (highlighted in light grey). Secondary structures such as β-strands (β1 and β2) or transmembrane domains (M1, M2 and M3) are also represented across subunits above the alignments. (C) A 3D structural model of the GABAA receptor was constructed. GABRG2 de novo variants were mapped onto the γ2 subunit in orange. The dashed box highlights the observation that the variants were closely connected with the structural domains between the interface of the N-terminal (β1 -β2 loop, Cys-loop, loop F) and transmembrane domains (M2-M3 loop, M1, M2, M3). See extended details in Fig. 7.
GABAA receptors are hetero-pentameric proteins assembled with γ-β-α-β-α stoichiometry (Fig. 2C). Remarkably, γ2 subunit variants were mapped to locations that were closely connected among structural domains between the interface of the N-terminal (β1-β2 loop) and transmembrane domains (M1, M2 and M3). In the N-terminal domain, γ2(A106T) and γ2(I107T) variants occurred in the β1-β2 loop, whereas γ2(P282S), γ2(R323W), γ2(R323Q) and γ2(F343L) variants occurred in the transmembrane domains M1, M2 and M3 delineating the pore region of the receptor (Fig. 2B and C).
De novo GABRG2 variants decreased GABA-evoked currents to different extents and altered their Zn2+ sensitivity
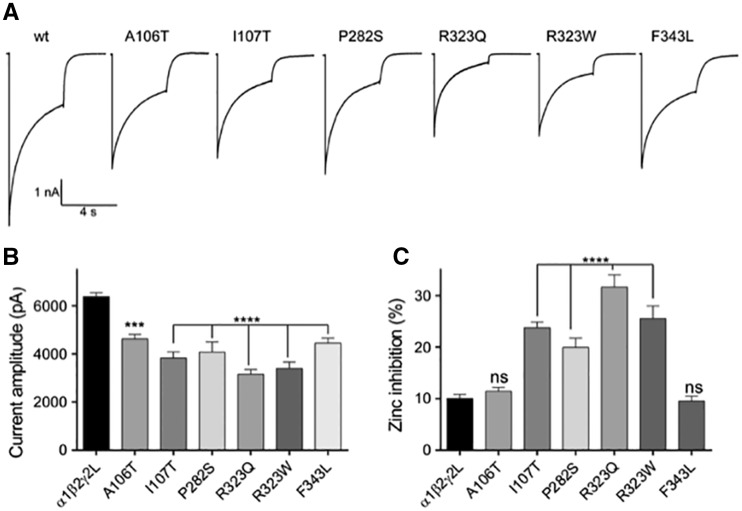
We determined the functional consequences of epileptic encephalopathy-associated γ2 subunit variants by measuring macroscopic GABA-evoked currents in transfected HEK293T cells (Fig. 3). All γ2 subunit variants decreased GABAA currents, but to different extents. While γ2L(A106T), γ2L(I107T), γ2L(P282S), and γ2L(F343L) variants located at the N-terminal and M1 and M3 domains, decreased currents by ∼30% (Supplementary Table 1), γ2L(R323W) and γ2L(R323Q) variants located in the pore forming M2 domain decreased channel current ∼50%, relative to wild-type currents (Supplementary Table 1 and Fig. 3A and B). In addition, GABAA receptors containing γ2L(I107T), γ2L(P282S), γ2L(R323W) and γ2L(R323Q) variants increased by ∼25% the fractional Zn2+ inhibition of currents (Supplementary Table 1) of the wild-type receptor (10 ± 1%, n = 51) (Fig. 3C). No changes in Zn2+ sensitivity were found for GABAA receptors containing γ2L(A106T) and γ2L(F343L) variants (Supplementary Table 1).
Figure 3.
Mutant α1β2γ2L receptors showed decreased GABA-evoked whole-cell currents and increased zinc sensitivity. (A) Representative GABA current traces are shown that were obtained following rapid application of 1 mM GABA for 4 s to lifted HEK293T cells voltage clamped at −20 mV. (B and C) Bar graphs showing average peak current and zinc inhibition from cells co-expressing α1β2 subunits with wild-type (wt) or mutant γ2 subunits. Values are expressed as mean ± SEM (Supplementary Table 1). One-way ANOVA with Dunnett’s post-test was used to determine significance compared to the wild-type condition. ****P < 0.0001, ***P < 0.001, and nsP > 0.05, respectively.
Decrease of current amplitudes can be produced by impaired biogenesis of receptors leading to decreased or altered expression of surface receptors or to mutation-induced alteration of surface receptor channel gating. Increased sensitivity of GABAA receptors to Zn2+ inhibition may be the result of the variant itself or of alterations in the subunit composition of receptors expressed on the cell surface such as formation of surface αβ receptors.
Mutant γ2 subunits were stable in transfected HEK293T cells, but with different total levels
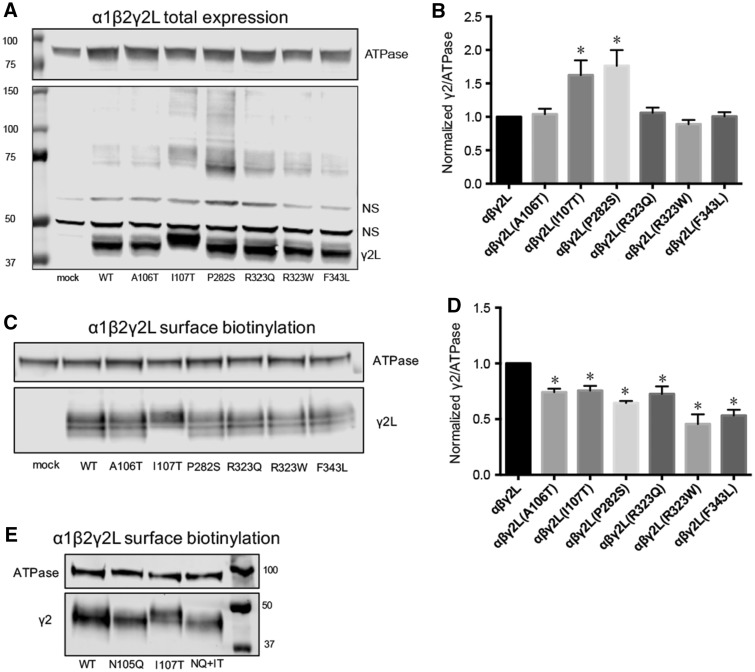
The GABRG2 epileptic encephalopathy-associated variants all decreased GABAA receptor currents due to impaired biogenesis or channel gating. To determine if these variants affected biogenesis of γ2 subunits, we expressed wild-type and mutant γ2L subunits with α1 and β2 subunits in HEK293T cells. Whole-cell lysates were analysed by western blot and immunoblotted using polyclonal γ2 subunit antibodies (Fig. 4).
Figure 4.
Immunoblotting studies obtained for mutant γ2L subunits. Wild-type or mutant γ2L subunits were cotransfected with α1β2 subunits into HEK293T cells. (A) Total cell lysates were collected, analysed by SDS-PAGE and blotted by anti-γ2 and anti-ATPase antibodies. In this representative western blot, NS = non-specific control. (B) Band intensity of γ2L subunits was normalized to the ATPase signal (n = 4, mean ± SEM). Both the lower and higher molecular mass bands were included. (C) Surface protein samples were collected through biotinylation and probed by anti-γ2 and anti-ATPase antibodies. A representative western blot is presented. (D) Band intensities of γ2L subunits were normalized to the ATPase signal (n ≥ 4, mean ± SEM). One-way ANOVA followed by Dunnett’s multiple comparison test were used to determine significance. *P < 0.05, compared with wild-type condition. (E) Surface proteins of HEK293T cells co-expressing α1β2γ2L, α1β2γ2L(N105Q), α1β2γ2L(I107T) or α1β2γ2L(N105Q/I107T) subunits were collected and probed with anti-γ2 and anti-ATPase antibodies. A representative western blot is presented. WT = wild-type.
Wild-type γ2L and mutant γ2L(A106T), γ2L(P282S), γ2L(R323Q), γ2L(R323W), and γ2L(F343L) subunits all migrated at the same molecular mass, predicted to be ∼45 kD (Fig. 4A). The variant γ2L(I107T) introduced a new amino acid threonine two amino acids after asparagine 105 (N105), thus creating a new fourth potential glycosylation motif (NXS/T) in the extracellular domain. Unsurprisingly, in cells co-transfected with mutant γ2L(I107T) subunits, a main band with a shift in molecular mass compared with wild-type γ2L was detected, consistent with the increased glycosylation of the mutant protein (Fig. 4A). Interestingly and unexpectedly, mutant γ2L(P282S) and γ2L(I107T) subunits also formed substantial amounts of protein complexes that migrated at a high molecular mass (∼75–150 kD). It is possible that these high molecular mass protein complexes are oligomers formed by mutant γ2 subunits as observed in γ2(Q390X) subunits (Kang et al., 2010, 2015).
We then quantified the γ2 subunit band intensity of each lane, normalized to the ATPase band intensity of the same lane, and compared the normalized γ2/ATPase ratio among wild-type and mutant subunits (Fig. 4B). Total levels of mutant γ2L(A106T), γ2L(R323Q), γ2L(R323W), and γ2L(F343L) subunits did not differ from those of wild-type γ2L subunits (1.00, n = 4). In contrast, the total amount of mutant γ2L(I107T) and γ2L(P282S) subunits were increased to 1.62 ± 0.22 (P < 0.05, n = 4) and 1.76 ± 0.23 (P < 0.05, n = 4), respectively, suggesting that mutant γ2L(I107T) and γ2L(P282S) subunits were more stable than wild-type subunits and/or were retained in the endoplasmic reticulum (Fig. 4B).
The variants all decreased surface levels of γ2 subunits, but to different extents
We asked if mutant γ2 subunits could assemble with α1 and β2 subunits and traffic to cell membranes as functional receptors. To assess surface trafficking of mutant γ2 subunits, we co-transfected HEK293T cells with α1, β2, and wild-type or mutant γ2L subunits at a 1:1:1 α1:β2:γ2 subunit ratio and evaluated surface levels of wild-type and mutant γ2L subunits by surface biotinylation (Fig. 4C). Compared to co-expressed wild-type γ2L subunits (1.00, n = 6), surface levels of co-expressed mutant γ2L subunits were reduced to 0.74 ± 0.03 (P < 0.05, n = 6) for A106T, 0.76 ± 0.06 (P < 0.05, n = 6) for I107T, 0.65 ± 0.02 (P < 0.05, n = 4) for P282S, 0.73 ± 0.07 (P < 0.05, n = 5) for R323Q, 0.46 ± 0.09 (P < 0.05, n = 6) for R323W and 0.53 ± 0.05 (P < 0.05, n = 6) for F343L, respectively. These results demonstrated that A106T, I107T, P282S, R323Q, R323W and F343L substitutions all reduced surface levels of γ2L subunits, but to different extents (24–54%). The reductions in surface levels of γ2 subunits (Fig. 4D) were similar to the reductions in whole cell currents produced by these γ2 subunit variants (Fig. 3), suggesting that the variants may reduce biogenesis of GABAA receptors. All of these GABRG2 variants were de novo and their pathogenicity was confirmed by our functional characterization. Thus, we will refer to them as mutations instead of variants.
The γ2(I107T) subunit mutation introduced a novel glycosylation site
To this point, we observed two principle effects of the γ2(I107T) mutation. First, it added a fourth glycosylation site to γ2(I107T) subunits, and second, there was decreased γ2(I107T) subunit surface expression. However, it remained unclear whether there was a causal relationship between these two phenomena. We therefore mutated the N-glycosylation site N105 to glutamine in wild-type γ2 and mutant γ2(I107T) subunits, thereby creating glycosylation-defective subunits. The double mutant construct γ2(N105Q/I107T) disrupted the novel glycosylation sequence, although it retained the I107T mutation. We then co-expressed α1 and β2 subunits with wild-type γ2LHA, wild-type/glycosylation-deficient γ2L(N105Q)HA, mutant γ2L(I107T)HA and mutant/glycosylation-deficient γ2L(N105Q/I107T)HA subunits, and measured surface levels of γ2LHA subunits in each condition using flow cytometry (Supplementary Fig. 1). With α1β2γ2L(N105Q)HA, α1β2γ2L(I107T)HA, and α1β2γ2L(N105Q/I107T)HA subunit co-expression, surface HA levels were significantly reduced to 0.60 ± 0.06 (P < 0.001, n = 3), 0.48 ± 0.03 (P < 0.001, n = 11), and 0.69 ± 0.05 (P < 0.001, n = 4), respectively, compared with the wild-type condition. Immunoblotting for γ2 subunit surface protein yielded similar results (Fig. 4E). The molecular mass of the double mutant protein γ2(N105Q/I107T) returned to the size of wild-type level, as would be expected as the added glycosylation site was eliminated. Importantly, γ2L(N105Q), γ2L(I107T) and γ2L(N105Q/I107T) subunits all had lower surface expression level relative to wild-type γ2L subunit. Taken together, these results suggested that the I107T mutation itself and not the new glycosylation site at N105 was the mechanism by which the I107T mutation impaired γ2L subunit surface incorporation and, GABAA receptor function.
Mutant γ2 subunits had different surface and intracellular distribution
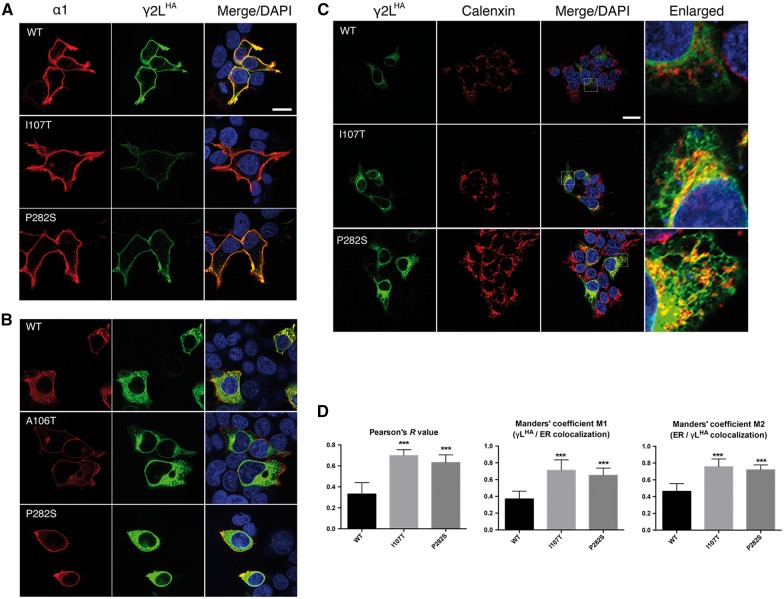
We next extended our study to determine and compare the cellular locations of mutant and wild-type γ2 subunits in HEK293T cells using confocal microscopy (Fig. 5 and Supplementary Fig. 2). Wild-type and mutant γ2LHA subunits were co-expressed in HEK293T cells with α1 and β2 subunits at a 1:1:1 cDNA ratio. We co-labelled cells with anti-α1 subunit (red) and anti-HA (green) antibodies. Without cell permeabilization, wild-type γ2LHA subunit signals were present on the surface and were co-localized well with α1 subunit signals, consistent with co-assembly with α1 and β2 subunits into receptors that were trafficked to the cell surface (Fig 5A, yellow florescence is co-localization). In contrast, γ2L(A106T)HA, γ2L(I107T)HA, γ2L(P282S)HA, γ2L(R323Q)HA, γ2L(R323W)HA and γ2L(F343L)HA subunits all had reduced surface HA signals (lack or reduction of yellow fluorescence in Fig. 5A and Supplementary Fig. 2A).
Figure 5.
γ2L(I107T)HA and γ2L(P282S)HA subunits are retained intracellularly. Wild-type or mutant γ2L(I107T)HA and γ2L(P282S)HA subunits were co-expressed with α1 and β2 subunits in HEK293T cells. Surface (A) and total (B) staining patterns were revealed by confocal microscopy. Both permeabilized and unpermeablized cells were stained with antibodies against the α1 subunit (red) and the HA tag (green). Also shown is DAPI nuclear counterstaining (blue), and a merged image. Scale bars = 10 μm. (C) The transfected cells were permeabilized, and γ2LHA subunits were labelled with anti-HA antibody (green). The endoplasmic reticulum was visualized with anti-calnexin antibody (red). White boxes on the merged images depict the enlarged area shown in the images to the right. Scale bars = 20 μm. (D) Statistical analyses of wild-type or mutant γ2LHA subunits and endoplasmic reticulum co-localization was performed using Pearson’s correlation coefficient (R) and Manders’ co-occurrence coefficient (M1 and M2). Results shown are the mean ± SEM of 15 cells in three independent experiments. (***P < 0.001). WT = wild-type.
With cell permeabilization and co-expression with α1 and β2 subunits, wild-type γ2LHA subunits were well distributed intracellularly (Fig. 5B). Co-expressed γ2L(I107T)HA and γ2L(P282S)HA subunits had more prominent intracellular HA signalling than wild-type γ2LHA subunits. This was consistent with the higher total amount of mutant γ2L(I107T) and γ2L(P282S) subunits in whole-cell lysates. However, the total expression of γ2L(A106T)HA, γ2L(R323Q)HA, γ2L(R323W)HA and γ2L(F343L)HA subunits was indistinguishable from that of wild-type γ2LHA subunits (Supplementary Fig. 2B).
We observed that wild-type γ2LHA subunits were localized primarily to the plasma membrane. In contrast, mutant γ2L(I107T)HA and γ2L(P282S)HA subunits accumulated in cells and had impaired trafficking to the cell surface when co-expressed with α1 and β2 subunits. Given the previous results, we hypothesized that I107T and P282S mutations resulted in the retention of the mutated γ2LHA subunits in the endoplasmic reticulum. This was confirmed by co-labelling permeabilized cells with anti-HA and anti-calnexin antibodies (Fig. 5C). Calnexin, a well-established endoplasmic reticulum marker, exhibits a typical perinuclear and reticular distribution suggestive of its endoplasmic reticulum distribution. Wild-type γ2LHA subunits spread outside the endoplasmic reticulum, which presumably represented the newly synthesized subunits that were in transit to the cell surface. In contrast, mutant γ2L(I107T)HA and γ2L(P282S)HA subunits were found to be predominantly localized to the endoplasmic reticulum as evidenced by their co-localization with calnexin.
Quantification of the co-localization of mutant γ2L(I107T)HA and γ2L(P282S)HA subunits with the endoplasmic reticulum in HEK293T cells was shown in Fig. 5D. Correlation between the signal intensities of γ2LHA subunits and the endoplasmic reticulum was significantly stronger for both I107T (R = 0.70 ± 0.02) and P282S (R = 0.63 ± 0.02) mutations relative to the wild-type condition (R = 0.33 ± 0.06), as measured by Pearson’s correlation analysis. We also determined the interaction between γ2L subunits and the endoplasmic reticulum by quantifying the Manders’ co-localization coefficient (MCC), which measures co-occurrence of two proteins independent of signal proportionality (Manders et al., 1993; Costes et al., 2004). The Manders’ coefficient M1 indicated the fraction of γ2L subunits that co-localized with the endoplasmic reticulum, whereas the Manders’ coefficient M2 indicated the fraction of endoplasmic reticulum that co-localized with the γ2L subunits. In both cases, we observed that mutant γ2L(I107T)HA and γ2L(P282S)HA subunits had significantly increased co-localization with the endoplasmic reticulum (M1 of 0.71 ± 0.03 and 0.65 ± 0.02 in I107T and P282S, respectively, M2 of 0.75 ± 0.02 and 0.72 ± 0.01 in I107T and P282S, respectively), in comparison with wild-type γ2L subunits (M1 of 0.37 ± 0.02 and M2 of 0.46 ± 0.02).
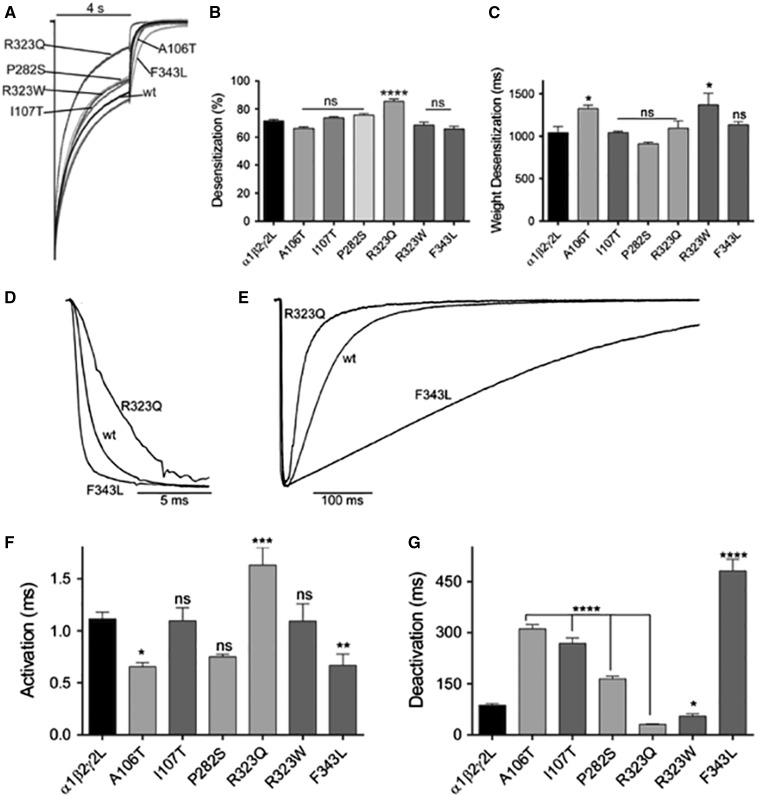
De novo GABRG2 mutations altered the kinetic properties of GABAA receptor currents
Assembly of mutant subunits into surface GABAA receptors may impair channel gating by causing macroscopic kinetic changes of GABA-evoked currents. To address this possibility, we determined whether the epileptic encephalopathy-associated mutations altered the kinetic properties of functional GABAA receptors. Thus, we measured current desensitization, activation and deactivation rates of wild-type and mutant receptor currents. GABAA receptor desensitization during 4 s GABA (1 mM) application was variably affected by γ2 subunit mutations (Fig. 6A). Thus, only the γ2L(R323Q) mutation significantly increased the extent of current desensitization (Supplementary Table 1), whereas γ2L(A106T) and γ2L(R323W) mutations slowed desensitization (Fig. 6B and C). It is noteworthy that all of these mutations occur at the interface between the N-terminal domain and the pore region of the receptor (Figs 2C and 6A and B).
Figure 6.
Mutant γ2 subunits altered the kinetic properties of GABAA receptor currents. (A) Superimposed representative traces show the desensitization of GABA-evoked currents produced by 4-s applications of 1 mM GABA to wild-type (wt) and mutant receptors. Traces were normalized to wild-type currents for clarity. (B) Bar graphs show average extent of desensitization measured at the end of the application of GABA, and (C) the weighted desensitization time constant during 4-s applications of GABA were determined. Representative current traces show activation (D) and deactivation (E) of currents produced by 10 ms GABA (1 mM) applications to wild-type and mutant receptors containing the γ2(R323Q) and γ2(F343L) subunits. Traces are normalized for clarity. Bar graphs show (F) average activation time constant and (G) weighted deactivation time constant from the cells co-expressing α1β2 subunits with mutant or wild-type γ2L subunits. Values are expressed as mean ± SEM (Supplementary Table 1). One-way ANOVA with Dunnett’s post-test was used to determine significance. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, and nsP > 0.05, respectively, relative to wild-type condition.
We determined current activation and deactivation by measuring the current time constant (τ) at current onset (Fig. 6D) and at current offset (Fig. 6E) during and following the 10 ms GABA (1 mM) application. While most of the γ2 subunit mutations accelerated (A106T, F343L) or did not affect (I107T, P282S, R323W) receptor activation, the γ2L(R323Q) mutation significantly slowed it (Supplementary Table 1) (Fig. 6F). The deactivation of the receptor was also affected but in opposite directions. Most of the γ2 subunit mutations slowed deactivation (A106T, I107T, P282S, F343L) (Fig. 6G). The γ2L(F343L) subunit mutation caused the greatest effect about five times the value of the wild-type condition (Supplementary Table 1). Only γ2L(R323W) and γ2L(R323Q) subunit mutations accelerated deactivation (Fig. 6G).
Taken together, these results demonstrate that the primary effect of the γ2 subunit mutations was to reduce receptor biogenesis but the mutations also have variable, subunit-dependent effects on the kinetic properties that appeared to be correlated with the structural domain of the receptor where the mutation occurs. As a result, γ2 subunit mutations located near the interface between N-terminal domain and channel pore (A106T, I107T, P282S, and F343L) mainly accelerated activation and prolonged deactivation, and those in the pore (R323W and R323Q) accelerated deactivation of the receptor and decreased channel function by ∼50%.
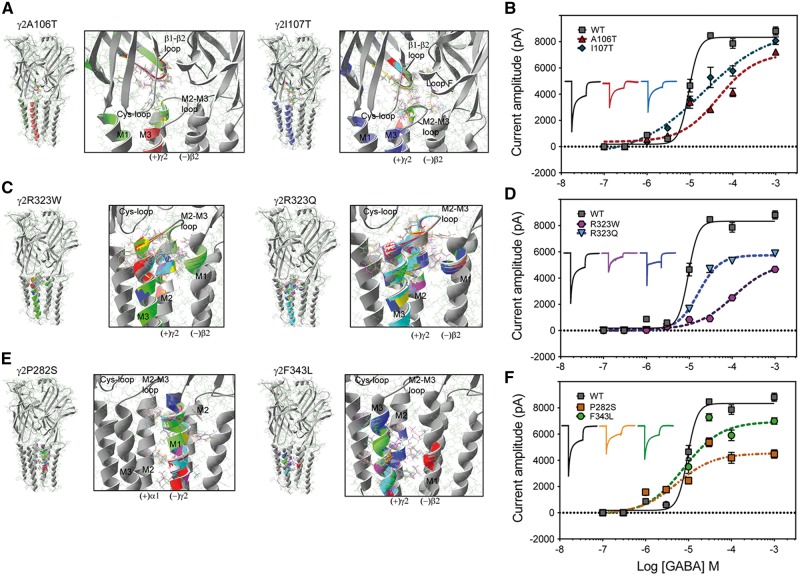
De novo γ2 subunit mutations decreased GABA potency by disrupting structural domains important for GABAA receptor function
Changes in GABAA receptor potency appeared to be correlated with well-defined structural domains, which have been described as essential to receptor function (Klausberger et al., 2000; Bianchi et al., 2001; Bianchi and Macdonald, 2002; Sarto et al., 2002; Venkatachalan and Czajkowski, 2012; Althoff et al., 2014; Lo et al., 2014). To determine whether predicted changes in channel structure caused by the γ2 subunit mutations are related to changes in the GABAA receptor potency, we first generated wild-type and mutant pentameric αβγ GABAA receptor simulations (Fig. 7) using solved structures of the C. elegans GluCl channel as template (Althoff et al., 2014). We computed rearrangements of the subunit’s secondary structure by computing the RMS deviation between the wild-type and mutant structural simulations (Janve et al., 2016) (Fig. 7A, C and E). When the perturbations of the secondary structure and side chain residues had RMS deviation values ≥0.5 Å, they were shown in rainbow colours on the simulation. Our simulations revealed that more than one subunit was affected. Structural changes were propagated through the entire structure, perturbing the Cys-loop, β1 -β2 loop, pre-M1 region, and M2-M3 loop at the extracellular junction between the N-terminal domain and transmembrane domain, which participate in the activation of the receptor. Subsequently, we measured the effects of the γ2 subunit mutations on GABA concentration-response curves (Fig. 7B, D and F). Peak GABAA receptor peak currents were obtained by applying various concentrations of GABA for 4 s to wild-type α1β2γ2L and mutant GABAA receptors. For wild-type α1β2γ2L GABAA receptors, the EC50 for current stimulation was 8.27 ± 1.16 µM, and the maximal current was 8922 ± 216 pA (n = 5–6). Therefore, we clustered the mutations by their structural location when assessing the disturbances that the mutation caused on the receptor structure and the measurable functional changes in GABAA receptor potency (see below).
Figure 7.
De novo γ2 subunit mutations decreased GABA potency by disrupting structural domains important for GABAA receptor function. (A, C and E) On the right are represented two neighbouring subunits where the mutations are located in relation to the γ+/β− and α+/γ− interfaces. Enlarged views of structural domains showing structural rearrangements caused by the γ2(A106T) and γ2(I107T) (A), γ2(R323W) and γ2(R323Q) (C), and γ2(P282S) and γ2(F343L) (E) mutations are shown in black boxes. The structural perturbations in the secondary structure and side chain residues that differed among the wild-type (in grey) and the mutant simulation (RMS deviation ≥0.5 Å) are indicated in a different colour from the wild-type simulation. The principal (+) and complementary (−) interfaces of each subunit are shown, and structural domains along the interface of the N-terminal (C loop, β1-β2 loop, Cys-loop, loop F) and transmembrane domains (M2-M3 loop, M1, M2, M3) are indicated. (B, D and F) GABA concentration-response curves for receptors containing γ2(A106T) and γ2(I107T) (B), γ2(R323W) and γ2(R323Q) (D), and γ2(P282S) and γ2(F343L) (F) mutant subunits (dashed lines) and for wild-type (wt) receptors (solid lines) were obtained. Inside the panels, representative peak currents evoked by a 4-s application of GABA (100 µM) are shown. The colour of the traces indicate the experimental condition as represented in the GABA-concentration response curves. The peak current traces obtained from receptors containing mutant γ2 subunits were normalized with respect to wild-type receptors for comparison. Values are expressed as mean ± SEM (n = 5–6 cells for each experimental condition). The data represents the summary of 37 cells with comparable capacitances (8–12 pF) recorded from three independent transfections.
γ2(A106T) and γ2(I107T) subunit mutations
These mutations are located in the β1-β2 inner loop in the N-terminal domain, at the interface between the principal (+) side of the γ2 subunit and the complementary (−) side of the β2 subunit, which delimits the γ+/β− interface (Fig. 7A). Mainly the structural perturbations were restricted to the γ2 subunit in the Cys-loop, β1–β2 loop and the M2–M3 loop on the mutant γ2(A106T) subunit model and were propagated to loop F of the neighbouring β2 subunit on the mutant γ2(I107T) subunit model. It is noteworthy that when γ2L(A106T) and γ2L(I107T) subunits were co-expressed with wild-type α1 and γ2L subunits (Fig. 7B), the EC50 was shifted 6- to 2-fold to the right, respectively (46.3 ± 1.22 µM; 14.3 ± 1.19 µM), with a reduction of 80–85% in the maximal response to GABA (7117 ± 296 pA and 7585 ± 233 pA, respectively, n = 5–6).
γ2(R323W) and γ2(R323Q) subunit mutations
These mutations are located at the extracellular interface of the transmembrane α-helices M2 of the γ2 subunit, in the outermost portion of the pore of the receptor at the γ+/β− subunit interface (Fig. 7C). These mutations caused mainly rearrangements at α-helices M2, M3 and M2-M3 loop of the γ2 subunit towards the γ+/β− subunit interface of the receptor, and propagated to the α-helix pre-M1 of the neighbouring β2 subunit at the extracellular junction between the N-terminal and the transmembrane domains of the receptor. In contrast to wild-type receptors (Fig. 7D), the GABA concentration-response curves of γ2L(R323W) and γ2L(R323Q) subunits was shifted considerably, with EC50 right-shifted 13- to 3-fold (108 ± 1.13 µM; 20.2 ± 1.13 µM, respectively) and had substantial reduction of 58–69% in the maximal response to GABA (5154 ± 165 pA and 6187 ± 129 pA, respectively, n = 5–6).
γ2(P282S) and γ2(F343L) subunit mutations
These mutations are located at the deeper portion of the transmembrane α-helices M1 and M3 of the γ2 subunit, towards the α+/γ− and γ+/β− subunit interfaces, respectively (Fig. 7E). While on the γ2(P282S) subunit simulation, structural perturbations occurred mainly at the α-helix M1 of the γ2 subunit, on the γ2(F343L) subunit simulation, structural perturbations occurred in the α-helices M2 and M3 of the γ2 subunit, and in the deeper region of the α-helix M1 of the neighbouring β2 subunit. Distinct from the aforementioned γ2 subunit mutations that are located at the at the extracellular junction of the receptor, GABAA receptors expressing γ2L(R323W) and γ2L(R323Q) subunits had EC50s similar to those of wild-type α1β2γ2L GABAA receptors (5.35 ± 1.33 µM and 7.82 ± 1.23 µM, respectively). In contrast, receptors with these mutant subunits displayed a similar reduction of 51–78% in the maximal response to GABA (4538 ± 198 pA and 6950 ± 220 pA, respectively, n = 5–6) (Fig. 7F). Remarkably, these findings clearly demonstrated that mutations that disrupt the structure of the coupling interface of GABAA receptors decreased GABA potency.
Discussion
Mutations in GABRG2 have been most frequently associated with genetic epilepsies among all the GABRs (Kang and Macdonald, 2016). However, clinical evidence implicating GABRG2 mutations in epileptic encephalopathies is still lacking. Here we present both genetic information and functional analysis that for the first time provides strong evidence that mutations in GABRG2 may contribute to early onset epileptic encephalopathy.
GABRG2 mutations are associated with early onset epileptic encephalopathy
There were a few consistent clinical features of this cohort, including infantile onset seizures (<1 year) and severe intellectual disability without prominent brain MRI findings. None of these patients were originally diagnosed with a named infantile epilepsy syndrome [Ohtahara syndrome or infantile spasms syndrome (West syndrome)], but three patients did progress to be diagnosed with the electroclinical pattern of Lennox-Gastaut syndrome. From this cohort, there was no distinguishing pathognomonic clinical feature associated with GABRG2 mutations. This level of phenotypic pleiotropy is increasingly recognized across many epilepsy syndromes, and using the broader diagnosis of early onset epileptic encephalopathy is appropriate. While the clinical features may not point to a specific pattern of disease, the genetic data, all patients carrying de novo changes with two recurrent variants, provides strong genetic evidence for the importance of GABRG2 as an epileptic encephalopathy gene.
Pathophysiological mechanisms of epileptic encephalopathy-associated GABRG2 mutations
Our electrophysiological experiments showed significant reductions in current amplitudes for all of these mutations, thus demonstrating directly a clear impairment of GABAA receptor function. Disease severity might be related to the extent of mutation-induced functional channel impairment, but this cannot be definitively established with this small cohort of patients. In addition, we demonstrated that these mutations reduced channel function to different extents and by diverse mechanisms including impaired surface expression, endoplasmic reticulum retention, and gating defects (overview in Supplementary Table 2).
All of these γ2 subunit mutations produced significant, but variable, impairment of γ2 subunit surface expression, which is a common abnormality for GABRG2 missense mutations (Huang et al., 2014; Todd et al., 2014). A106T, R323Q, R323W and F343L mutations did not affect the total expression levels of γ2 subunits. In contrast, mutant γ2(I107T) and γ2(P282S) subunits were more stable than wild-type subunits and were retained predominantly within the endoplasmic reticulum, which is the location where immature GABAA receptor subunits reside once synthesized. The presence of endoplasmic reticulum-retained trafficking-deficient γ2 subunits has been demonstrated to produce endoplasmic reticulum stress (Kang et al., 2013). The sustained endoplasmic reticulum stress could lead to neurodegeneration, as evidenced by increased caspase 3 activation in older Gabrg2+/Q390X mice (Kang et al., 2015). Thus, it is possible that the misfolded mutant γ2(I107T) and γ2(P282S) subunit proteins could progressively accumulate and form aggregates inside neurons, which could affect function and survival of neurons in vivo.
Our finding that surface expression of γ2 subunits was reduced by the R323Q substitution was contrary to a previous study that reported that surface expression of γ2(R323Q) subunits was at the wild-type level (Reinthaler et al., 2015). This conflict may have been due to the different γ2 subunit cDNAs used for transfection. In contrast to the pHluorin-tagged mouse γ2 subunit construct used in their study, we used HA-tagged or untagged human γ2 subunits.
Epileptic encephalopathy-associated GABRG2 mutations altered structural domains that decreased GABA potency
Our results demonstrated a structure-dysfunction correlation with the location of the mutation in the receptor. The substitutions A106T and I107T were located next to each other in the β1 -β2 inner loop of the N-terminal extracellular domain that contributes to the γ+/β− subunit interface at the junction of the transmembrane domain that couples the opening of the receptor. The occurrence of these mutations demonstrates the importance of this domain in transducing GABA-binding-coupling once they caused a significant decrease in GABA sensitivity. In addition, the occurrence of the R323W and R323Q mutations in the outermost region of the transmembrane M2 facing the extracellular junction, also substantially decreased the sensitivity for GABA. Thus, the epileptic encephalopathy-associated GABRG2 mutations located in the outermost region of the pore-forming domain of the receptor, which is the outer ring region between the N-terminal extracellular domain and the pore, directly altered GABAA receptor activation (Bianchi et al., 2001; Bianchi and Macdonald, 2002; Althoff et al., 2014), and may contribute to the pathophysiological mechanism of the disease. In contrast, the P282S and F343L mutations, located in the transmembrane M1 and M3 of the γ2 subunit, respectively, seemed not to contribute directly to the activation of the receptor due to lack of altered sensitivity to GABA. Nevertheless, they produced a significant decrease in the maximum response to GABA that might be accounted for by the altered expression and receptor kinetics. Similar decreases in the maximal response to GABA were found for mutations located in the transmembrane M2 with decreased surface expression. No mutations in transmembrane domains M1 and M3 of the γ2 subunit have ever been reported in epilepsy patients. Recently, three de novo mutations in the M1 domain of GABRA1 were identified in patients with Ohtahara and West syndromes (Kodera et al., 2016), one of those in a homologous position of the γ2 subunit, supporting the important role of the M1 domain in GABAA receptor function (Bianchi et al., 2001).
GABRG2 mutations in genetic epilepsies and phenotype/genotype correlations
The first two genetic epilepsy-associated GABRG2 mutations (K328M and R82Q) were reported in a family with GEFS+ (Baulac et al., 2001) and a family with childhood absence epilepsy and febrile seizures (Wallace et al., 2001). Up to now, 19 GABRG2 epilepsy mutations have been identified in patients with simple febrile seizures and several different epilepsy syndromes (Boillot et al., 2015). Before the present study, it has been generally accepted that missense GABRG2 mutations are associated with mild phenotypes including childhood absence epilepsy and febrile seizures (Baulac et al., 2001; Wallace et al., 2001; Audenaert et al., 2006; Shi et al., 2010), while nonsense GABRG2 mutations lead to more severe phenotypes ranging from GEFS+ to Dravet syndromes (Harkin et al., 2002; Huang et al., 2012; Kang et al., 2013; Johnston et al., 2014).
The current data demonstrated that missense GABRG2 mutations could also lead to severe epilepsy phenotypes. Patients in our cohort showed a broad epilepsy phenotypic spectrum including Lennox-Gastaut syndrome and unclassified epileptic encephalopathies. There were loose correlations between mutation type and disease severity. For example, among these GABRG2 mutations, the I107T mutation had the most striking effect on GABAA receptor macroscopic current properties and cellular localization (Supplementary Table 2). With respect to age of onset, motor development and epilepsy outcomes, the most severe disease course was also seen in Patient 3 with the I107T mutation. However, as there are only a small number of patients with GABRG2 mutations and only the cohort in this report with epileptic encephalopathy, we cannot make a definitive statement about effect of mutation on channel function and epileptic encephalopathy severity. What is likely is that the clinical and biophysical effects of GABRG2 mutations can be modified by the genetic background of the individual as evidenced by the difference in epilepsy phenotypes of knock-in mice with different genetic backgrounds (Reid et al., 2013) and that variants can be found in both an inherited and de novo pattern.
Conclusions
Collectively, our study used a combination of massively parallel sequencing and in vitro functional assays and established that mutations of the GABRG2 gene are genetic risk factors for epileptic encephalopathies. This complemented the prevailing GABAergic channelopathy paradigm in epilepsy and broadened the phenotype of epileptic encephalopathies associated with GABRG2. Our findings are of clinical significance, as GABAA receptors are known to be targets for epilepsy treatment (Braat and Kooy, 2015). Identification of additional GABRG2 mutations will no doubt guide further studies of the precise role of γ2 subunits in epileptogenesis and provide new insights into the targeted treatment for epileptic encephalopathies. Our present results do not cover the full spectrum of possible mutation-induced channel dysfunction, and the precise mechanisms by which mutations cause epileptic encephalopathy in humans remain to be clarified. Future studies in cultured neurons or in animal models will be required to study the downstream effects of these mutations in detail and solidify genotype-phenotype relationships in GABRG2-epileptic encephalopathy.
Supplementary Material
Acknowledgements
The authors would like to thank Rebecca Kamens for clinical data entry and Dr Erin Heinzen for access to the Duke Sequencing core.
Funding
This work was supported by the National Institutes of Health RO1 NS 33300 grant to R.L.M.
Supplementary material
Supplementary material is available at Brain online.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althoff T, Hibbs RE, Banerjee S, Gouaux E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature 2014; 512: 333–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audenaert D, Schwartz E, Claeys KG, Claes L, Deprez L, Suls A, et al. A novel GABRG2 mutation associated with febrile seizures. Neurology 2006; 67: 687–90. [DOI] [PubMed] [Google Scholar]
- Baulac S, Huberfeld G, Gourfinkel-An I, Mitropoulou G, Beranger A, Prud'homme JF, et al. First genetic evidence of GABA(A) receptor dysfunction in epilepsy: a mutation in the gamma2-subunit gene. Nat Genet 2001; 28: 46–8. [DOI] [PubMed] [Google Scholar]
- Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, van Emde Boas W, et al. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005-2009. Epilepsia 2010; 51: 676–85. [DOI] [PubMed] [Google Scholar]
- Bianchi MT, Haas KF, Macdonald RL. Structural determinants of fast desensitization and desensitization-deactivation coupling in GABAa receptors. J Neurosci 2001; 21: 1127–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi MT, Macdonald RL. Slow phases of GABA(A) receptor desensitization: structural determinants and possible relevance for synaptic function. J Physiol 2002; 544(Pt 1): 3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boillot M, Morin-Brureau M, Picard F, Weckhuysen S, Lambrecq V, Minetti C, et al. Novel GABRG2 mutations cause familial febrile seizures. Neurol Genet 2015; 1: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braat S, Kooy RF. The GABAA Receptor as a Therapeutic Target for Neurodevelopmental Disorders. Neuron 2015; 86: 1119–30. [DOI] [PubMed] [Google Scholar]
- Carvill GL, Heavin SB, Yendle SC, McMahon JM, O'Roak BJ, Cook J, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet 2013; 45: 825–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly CN, Krishek BJ, McDonald BJ, Smart TG, Moss SJ. Assembly and cell surface expression of heteromeric and homomeric-aminobutyric acid type A receptors. J Biol Chem 1996; 271: 89–96. [DOI] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 2004; 86: 3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi PMC. A roadmap for precision medicine in the epilepsies. Lancet Neurol 2015; 14: 1219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essrich C, Lorez M, Benson JA, Fritschy JM, Luscher B. Postsynaptic clustering of major GABAA receptor subtypes requires the gamma 2 subunit and gephyrin. Nat Neurosci 1998; 1: 563–71. [DOI] [PubMed] [Google Scholar]
- Euro E-RESC, Epilepsy Phenome/Genome P, Epi KC. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet 2014; 95: 360–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrant M, Nusser Z. Variations on an inhibitory theme: phasic and tonic activation of GABA(A) receptors. Nat Rev Neurosci 2005; 6: 215–29. [DOI] [PubMed] [Google Scholar]
- Haas KF, Macdonald RL. GABAA receptor subunit gamma2 and delta subtypes confer unique kinetic properties on recombinant GABAA receptor currents in mouse fibroblasts. J Physiol 1999; 514 (Pt 1): 27–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin LA, Bowser DN, Dibbens LM, Singh R, Phillips F, Wallace RH, et al. Truncation of the GABA(A)-receptor gamma2 subunit in a family with generalized epilepsy with febrile seizures plus. Am J Hum Genet 2002; 70: 530–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez CC, Gurba KN, Hu N, Macdonald RL. The GABRA6 mutation, R46W, associated with childhood absence epilepsy, alters 6beta22 and 6beta2 GABA(A) receptor channel gating and expression. J Physiol 2011; 589(Pt 23): 5857–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Hernandez CC, Hu N, Macdonald RL. Three epilepsy-associated GABRG2 missense mutations at the gamma+/beta- interface disrupt GABAA receptor assembly and trafficking by similar mechanisms but to different extents. Neurobiol Dis 2014; 68: 167–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Tian M, Hernandez CC, Hu N, Macdonald RL. The GABRG2 nonsense mutation, Q40X, associated with Dravet syndrome activated NMD and generated a truncated subunit that was partially rescued by aminoglycoside-induced stop codon read-through. Neurobiol Dis 2012; 48: 115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janve VS, Hernandez CC, Verdier KM, Hu N, Macdonald RL. Epileptic encephalopathy de novo GABRB mutations impair GABA receptor function. Ann Neurol 2016; 79: 806–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Kang JQ, Shen W, Pickrell WO, Cushion TD, Davies JS, et al. A novel GABRG2 mutation, p.R136*, in a family with GEFS+ and extended phenotypes. Neurobiol Dis 2014; 64: 131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JQ, Macdonald RL. Molecular pathogenic basis for GABRG2 mutations associated with a spectrum of epilepsy syndromes, from generalized absence epilepsy to dravet syndrome. JAMA Neurol 2016; 73: 1009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JQ, Shen W, Lee M, Gallagher MJ, Macdonald RL. Slow degradation and aggregation in vitro of mutant GABAA receptor gamma2(Q351X) subunits associated with epilepsy. J Neurosci 2010; 30: 13895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JQ, Shen W, Macdonald RL. Trafficking-deficient mutant GABRG2 subunit amount may modify epilepsy phenotype. Ann Neurol 2013; 74: 547–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JQ, Shen W, Zhou C, Xu D, Macdonald RL. The human epilepsy mutation GABRG2(Q390X) causes chronic subunit accumulation and neurodegeneration. Nat Neurosci 2015; 18: 988–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsnelson A, Buzsaki G, Swann JW. Catastrophic childhood epilepsy: a recent convergence of basic and clinical neuroscience. Sci Transl Med 2014; 6: 262ps13. [DOI] [PubMed] [Google Scholar]
- Klausberger T, Fuchs K, Mayer B, Ehya N, Sieghart W. GABA(A) receptor assembly. Identification and structure of gamma(2) sequences forming the intersubunit contacts with alpha(1) and beta(3) subunits. J Biol Chem 2000; 275: 8921–8. [DOI] [PubMed] [Google Scholar]
- Kodera H, Ohba C, Kato M, Maeda T, Araki K, Tajima D, et al. De novo GABRA1 mutations in Ohtahara and West syndromes. Epilepsia 2016; 57: 566–73. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–81. [DOI] [PubMed] [Google Scholar]
- Lemke JR, Riesch E, Scheurenbrand T, Schubach M, Wilhelm C, Steiner I, et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia 2012; 53: 1387–98. [DOI] [PubMed] [Google Scholar]
- Lo WY, Lagrange AH, Hernandez CC, Gurba KN, Macdonald RL. Co-expression of gamma2 subunits hinders processing of N-linked glycans attached to the N104 glycosylation sites of GABAA receptor beta2 subunits. Neurochem Res 2014; 39: 1088–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald RL, Kang JQ. Molecular pathology of genetic epilepsies associated with GABAA receptor subunit mutations. Epilepsy Curr 2009; 9: 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald RL, Kang JQ. mRNA surveillance and endoplasmic reticulum quality control processes alter biogenesis of mutant GABAA receptor subunits associated with genetic epilepsies. Epilepsia 2012; 53Suppl 9: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald RL, Olsen RW. GABAA receptor channels. Annu Rev Neurosci 1994; 17: 569–602. [DOI] [PubMed] [Google Scholar]
- Manders E, Verbeek F, Aten J. Measurement of co-localization of objects in dual-colour confocal images. Journal of microscopy 1993; 169: 375–82. [DOI] [PubMed] [Google Scholar]
- McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol 2016; 15: 304–16. [DOI] [PubMed] [Google Scholar]
- Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature 2014; 512: 270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 2004; 25: 1605–12. [DOI] [PubMed] [Google Scholar]
- Poduri A, Heinzen EL, Chitsazzadeh V, Lasorsa FM, Elhosary PC, LaCoursiere CM, et al. SLC25A22 is a novel gene for migrating partial seizures in infancy. Ann Neurol 2013; 74: 873–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid CA, Kim T, Phillips AM, Low J, Berkovic SF, Luscher B, et al. Multiple molecular mechanisms for a single GABAA mutation in epilepsy. Neurology 2013; 80: 1003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinthaler EM, Dejanovic B, Lal D, Semtner M, Merkler Y, Reinhold A, et al. Rare variants in gamma-aminobutyric acid type A receptor genes in rolandic epilepsy and related syndromes. Ann Neurol 2015; 77: 972–86. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarto I, Wabnegger L, Dogl E, Sieghart W. Homologous sites of GABA(A) receptor alpha(1), beta(3) and gamma(2) subunits are important for assembly. Neuropharmacology 2002; 43: 482–91. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012; 9: 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 2003; 31: 3381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer C, Balsiger S, Bluethmann H, Mansuy IM, Fritschy JM, Mohler H, et al. The gamma 2 subunit of GABA(A) receptors is required for maintenance of receptors at mature synapses. Mol Cell Neurosci 2003; 24: 442–50. [DOI] [PubMed] [Google Scholar]
- Shi X, Huang MC, Ishii A, Yoshida S, Okada M, Morita K, et al. Mutational analysis of GABRG2 in a Japanese cohort with childhood epilepsies. J Hum Genet 2010; 55: 375–8. [DOI] [PubMed] [Google Scholar]
- Smith CA, Kortemme T. Backrub-like backbone simulation recapitulates natural protein conformational variability and improves mutant side-chain prediction. J Mol Biol 2008; 380: 742–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HO, Reid CA, Single FN, Davies PJ, Chiu C, Murphy S, et al. Reduced cortical inhibition in a mouse model of familial childhood absence epilepsy. Proc Natl Acad Sci USA 2007; 104: 17536–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RH, Berkovic SF. The hidden genetics of epilepsy-a clinically important new paradigm. Nat Rev Neurol 2014; 10: 283–92. [DOI] [PubMed] [Google Scholar]
- Todd E, Gurba KN, Botzolakis EJ, Stanic AK, Macdonald RL. GABAA receptor biogenesis is impaired by the gamma2 subunit febrile seizure-associated mutation, GABRG2(R177G). Neurobiol Dis 2014; 69: 215–24. [DOI] [PubMed] [Google Scholar]
- Venkatachalan SP, Czajkowski C. Structural link between gamma-aminobutyric acid type A (GABAA) receptor agonist binding site and inner beta-sheet governs channel activation and allosteric drug modulation. J Biol Chem 2012; 287: 6714–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RH, Marini C, Petrou S, Harkin LA, Bowser DN, Panchal RG, et al. Mutant GABA(A) receptor gamma2-subunit in childhood absence epilepsy and febrile seizures. Nat Genet 2001; 28: 49–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.