Abstract
Zebrafish are emerging as a model of dietary lipid processing and metabolic disease. This protocol describes how to feed larval zebrafish a lipid-rich meal, which consists of an emulsion of chicken egg yolk liposomes created by sonicating egg yolk in embryo media. Detailed instructions are provided to screen larvae for egg yolk consumption so that larvae that fail to feed will not confound experimental results. The chicken egg yolk liposomes can be spiked with fluorescent lipid analogs, including fatty acids and cholesterol, enabling both systemic and subcellular visualization of dietary lipid processing. Several methods are described to mount larvae that are conducive to short- and long-term live imaging with both upright and inverted objectives at high and low magnification. Additionally presented is an assay to quantify larval food intake by extracting the lipids of larvae fed fluorescent lipid analogs, spotting the lipids on a thin layer chromatography plate, and quantifying the fluorescence. Finally, critical aspects of the procedures, important controls, options for modifying the protocols to address specific experimental questions, and potential limitations are discussed. These techniques can be applied not only to focused, hypothesis driven inquiries, but also to a variety of screens and live imaging techniques to study dietary lipid metabolism and the control of food intake.
Keywords: Neuroscience, Issue 116, Zebrafish, high-fat feed, food intake, fluorescent labeling, confocal microscopy, thin layer chromatography, fatty acids, cholesterol, live imaging
Introduction
The mechanisms by which the intestine regulates dietary lipid processing, the liver controls complex lipid synthesis and lipoprotein metabolism, and how these organs work with the central nervous system to control food intake are incompletely understood. It is of biomedical interest to elucidate this biology in light of the current epidemics of obesity, cardiovascular disease, diabetes, and non-alcoholic fatty liver disease. Studies in cell culture and mice have provided the majority of our understanding of the mechanistic relationships between dietary lipids and disease, and zebrafish (Danio rerio) are emerging as an ideal model to complement this work.
Zebrafish have similar gastrointestinal (GI) organs, lipid metabolism, and lipoprotein transport to higher vertebrates 1,2, develop rapidly, and are genetically tractable. The optical clarity of the larval zebrafish facilitates in vivo studies, a particular advantage for study of the GI system as its extracellular milieu (i.e., bile, microbiota, endocrine signaling) is virtually impossible to model ex vivo. In accordance, a body of research combining the genetic tractability and conduciveness to live imaging of zebrafish larvae with a variety of dietary manipulations (high-fat3,4, -cholesterol5, and -carbohydrate diets6,7), and models of cardiovascular disease8, diabetes9,10, hepatic steatosis11-13, and obesity14-16, are emerging to provide a host of metabolic insights.
An essential aspect of transitioning the larval zebrafish into metabolic research is the optimization of techniques developed in other model animals to the zebrafish and the development of novel assays that exploit the unique strengths of the zebrafish. This protocol presents techniques developed and optimized to feed larval zebrafish a lipid-rich meal, visualize dietary lipid processing from whole body to subcellular resolution, and measure food intake. Chicken egg yolk was chosen to compose the lipid-rich meal as it contains high levels of fats and cholesterol (lipids compose ~58% of chicken egg yolk, of which ~5% is cholesterol, 60% are triglycerides, and 35% are phospholipids). Chicken egg yolk provides more fat than typical commercial zebrafish micropellet foods (~15% lipids) and the advantage that it is a standardized feed with known percentages of specific fatty acids species, as zebrafish diets and feeding regiments have not been standardized across labs17. Moreover, fluorescent lipid analogs provided in the egg yolk visualize transport and accumulation of dietary lipids18, image cellular components including lipid droplets by acting both as vital dyes3 and through covalent incorporation into complex lipids, investigate metabolism through thin layer chromatography (TLC)19 and high performance liquid chromatography (HPLC) (S.A.F. unpublished data), and provide a quantitative assay for total food intake20.
Protocol
These protocols have been approved by the Carnegie Institution for Science Institutional Animal Care and Use Committee (protocol no. 139).
1. Animal Preparation
Maintain adults and larvae at 28 °C on a 14 hr:10 hr light:dark cycle. Feed adults twice daily with shell free Artemia (decapsulated, non-hatching, starting at 14 dpf) and commercial micropellets.
These protocols are optimized for the use of 6-7 dpf larvae collected by natural spawning of the AB background. Protocols can be modified for larvae of other ages and backgrounds. Do not provide exogenous food prior to 6 dpf.
Anesthetize larvae with Tricaine in embryo media (EM) (4.2 ml of 4 mg/ml Tricaine per 100 ml EM) at room temperature (RT).
2. Preparation of Lipid-rich Egg Yolk Feed
Prepare and store the chicken egg yolks. Separate the yolk and white of 12 chicken eggs. Pool the yolks, aliquot 1 ml into 1.5 ml tubes, and store at -80 °C up to 1 yr (12 yolks will make ~80 aliquots). Pool the whites, store, and use as a lipid-poor, protein-rich feed.
- Prepare fluorescent lipid analog(s) to be added to the egg yolk feed if desired.
- Suspend the purchased, powdered fluorescent lipid analog in 100% ethanol or 100% chloroform at 0.1 µg/µl (see following note discussing solvents), aliquot in opaque glass tubes, seal with parafilm, and store according to manufacturer's instructions. Due to rapid evaporation of organic solvents do not use aliquot volumes less than ~400 µl for long-term storage. Note: If the lipid analog is suspended in ethanol, large volumes of the lipid analog will be used because it dries faster under N2 than chloroform. If the lipid analog is suspended in ethanol it does not have to be dried and resuspended before adding it to the liposome feed in step 2.2.4, but the total ethanol concentration should not exceed 0.1% in the liposome feed to avoid potentially confounding physiological effects of ethanol.
- For fluorescent fatty acid analogs: Prepare a total volume of 20 ml 5% egg yolk emulsion in EM. From that prepare 5 ml aliquots with a final concentration of 6.4 µM 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene (BODIPY C16) for live confocal imaging and food intake assays or 1 µg/ml BODIPY C12 for stereomicroscope imaging. Transfer the desired amount of the fluorescent lipid analog into a 1.5 ml plastic tube. Dry the lipid under a stream of N2, taking care not to blow the lipid out of the tube.
- Immediately, resuspend in 5 µl 100% ethanol by pipetting a stream down the sides of the tube, add 95 µl EM, and mix well. Note: The mixture should appear homogenous; if colored lipid remains on the sides of the tube or the liquid appears clumpy, the lipid has not fully solubilized and requires more ethanol. Protect the tube from light and store on ice.
- For fluorescent cholesterol analog: prepare a final concentration of 2.4 µg/ml cholesterol in 5 ml 0.5-5% egg yolk emulsion for live imaging studies. Transfer the desired amount of cholesterol into a 1.5 ml plastic tube. Dry the lipid under a stream of N2, taking care not to blow the lipid out of the tube.
- Immediately, resuspend in 15 µl of RT 100% ethanol by pipetting a stream down the sides of the tube and mix with 85 µl of RT 1% fatty acid-free BSA in purified H2O. Protect the tube from light and store on ice.
- Prepare Egg Yolk Liposome Feed.
- Add a thawed aliquot of egg yolk to EM in a 50 ml conical tube. Vortex the diluted egg yolk mixture for 1-2 min; the mixture should appear to be a homogenous emulsion.
- Pour the mixture through a fine mesh strainer to remove protein conglomerates and collect in a new, fresh 50 ml conical tube.
- Pulse sonicate the egg mixture with a one-fourth inch tapered microtip (5 times (5 x 1 sec on, 1 sec off) pausing 5-10 sec between each set; output intensity: 6 W) at RT to create liposomes. Sonicator power settings are instrument dependent and therefore require optimization for each instrument and microtip. Note: Continuously rock liposome mixture at RT to maintain homogeneity until use.
Add the fluorescent lipid analog to the egg yolk liposomes, if desired. Immediately after sonication, pipet the prepared lipids into the liposome mixture and vortex at high speed for 30 sec to incorporate into liposomes. Protect the mixture from light by wrapping the tube in foil and continue rocking at RT.
3. Feeding Paradigm
- Prepare the Larvae for Feeding.
- Prior to beginning the feed, screen larvae for obvious morphological defects that may impede feeding. Transfer larvae to 60 mm x 15 mm Petri dishes or 6- or 12-well plates, reduce EM to a minimum, and replace with 5 ml of the prepared liposome solution.
- If required, transfer unfed control larvae to 5 ml EM and/or 5 ml 5% egg white in EM and treat in parallel.
Gently rock the larvae in an incubated rocker (29-31 °C at 30 rpm) throughout the feed to encourage food intake while continuing to shield from light if fluorescent lipid analogs are present. If larvae need to be exposed to light, minimize the exposure with a tinted glass incubator cover.
At the end of the feeding period, rinse the freely swimming larvae in EM 3 times. Note: Larvae may begin consuming liposomes at any point during the feed. If it is critical that larvae commence feeding at the same time, screen for food intake (see 3.4) after 1 hr of feeding and return only larvae that have commenced feeding to the egg yolk emulsion to complete the feed.
Screen the larvae at this point for food intake as a small number may not eat (generally ~0-5%). Examine the intestines of larvae that have been anesthetized or slightly cooled on a metal block on ice to determine if they have fed: larvae that have consumed liposomes will have a darkened intestine (Figure 1).
Image or process larvae for food intake immediately. Alternately, place larvae in fresh EM and incubate at 28 °C to allow metabolic processing to occur.
4. Mounting Larvae for Live Imaging
- Prepare the Mounting Media.
- For 3% methyl cellulose: add 0.75 g methyl cellulose to 25 ml of near boiling EM, vortex, rock at RT 2-3 days until completely dissolved, and store at 4 °C for years at -20 °C. Repeated cycles of freezing and thawing at 4 °C will remove air bubbles. Store 3% methyl cellulose on ice while mounting larvae.
- For 1.2% low-mount agarose: add 0.3 g of low-melt agarose to 25 mL EM and dissolve by bringing to a boil, aliquot into 2 ml tubes, and maintain at 26-28 °C on a heat block. Unused aliquots can be stored at RT or -20 °C and boiled again at the time of use. Be sure that agarose is cool enough to handle before exposing to larvae or they may be over-heated.
For low-magnification, medium-throughput live imaging, place a glass slide on a tissue on a metal block on ice and wait for the slide to cool. Apply a generous amount of 3% methyl cellulose (which has been maintained at 4 °C on ice) onto the slide, transfer a larva onto the 3% methyl cellulose, using a fishing-line poker (0.41 mm diameter fishing line glued to the end of a glass capillary tube) create a troth to drain excess EM (so that methyl cellulose will not be diluted) and position the larvae as desired.
- For short-term (<20 min), high-magnification live imaging with an upright oil emersion objective, mount larvae with the coverslip lean-to method.
- Use cyanoacrylate based glue (Superglue) to attach a a 22 mm x 30 mm coverslip to one end of a glass slide. Use a poker to put a thin line of 3% methyl cellulose on the slide adjacent to the coverslip and transfer larvae to the slide with a wide bore Pasteur pipet. Remove excess EM so it will not dilute the methyl cellulose.
- Using a poker, gently position larvae in the methyl cellulose on their sides (left side up to image more of the liver, right side up to image the gallbladder) with their heads next to the coverslip.
- Spot superglue in the corners of a second coverslip and gently place one edge of the coverslip on the mounted coverslip and the other on the slide, bridging the larvae. Take care not to apply excess pressure during this step or with the objective while imaging so that larvae remain uninjured.
- For long-term, high-magnification imaging with an upright immersion objective, mount larvae in agarose. Add a larva to a small aliquot of 1.2% low melt agarose then transfer the larva in a small volume of agarose to a petri dish.
- Quickly position the larvae with a poker before the agarose solidifies. Allow the agarose to dry for 2-3 min and cover the agarose fully with fresh EM to prevent it from drying.
- For long-term, high-magnification imaging with an inverted objective, mount larvae on a glass-bottom dish in agarose.
- Cool a metal block on ice. Place the dish on the metal block separated by a tissue to prevent excessive cooling and larval freezing. Transfer a larva to the dish and remove the EM by wicking with a tissue.
- Place a drop of 28 °C low-melt agarose (1.2%) on top of the larva and immediately position with a poker (left side down to best image the liver, right side down to image the gallbladder). Remove dish from the cold block, allow to dry for 2-3 min, and add a sufficient volume of fresh EM to cover the agarose (~2-5 ml).
5. Food Intake Assay
At the end of an egg yolk liposome feed containing 6.4 µM BODIPY C16, pool a minimum of 10 washed larvae in a plastic 1.5 or 2 ml tube, remove EM, and snap freeze. The samples may be stored at -80 °C protected from light for several months. Note: If possible, collect multiple replicates. It is important to collect unfed larvae at this step to normalize for background lipid fluorescence (ideally larvae from the same clutch are used).
- Perform a modified Bligh-Dyer lipid extraction3,21.
- Add 100 µl homogenization buffer (20 mM Tris-Cl, 1 mM EDTA) to the sample on ice (alternatively purified H2O can be used). Homogenize on ice with a microtip sonicator (5 sec total: 1 sec on 1 sec off). Check that larvae are totally homogenized; if not, repeat. Transfer to a 13 ml disposable glass culture tube (other glass tubes may be used). Note: Chloroform is hazardous; perform all subsequent lipid extraction steps at RT in a chemical hood.
- For each 100 µl of homogenization buffer used, add 375 µl of 1:2 (chloroform:methanol). Vortex 30-60 sec, incubate 10 min, add 125 µl of chloroform per 100 µl homogenization buffer used, and vortex 30 sec.
- Add 125 µl of 200 mM Tris pH 7.5 per 100 µl homogenization buffer used, vortex 30 sec, and centrifuge (2,000 x g, 5 min, RT, protected from light). Carefully remove the samples from the centrifuge. They will be separated into two phases: an upper phase is aqueous, an interface of larval debris, and a lower organic phase (contains lipids).
- With a clean glass pipette collect the bottom, organic phase and transfer it into a clean 13 ml glass tube. Note: Take care to avoid the aqueous phase and larval debris at the interface; if contamination occurs, repeat centrifugation step and recollect. Discard the upper, aqueous phase; it may be helpful to remove and discard this phase before collecting the organic phase. The sample can be stored at -80 °C for up to 1 month.
- Dry the organic phase under vacuum (0.12 atm) while protecting from light. Do not continue to dry the samples after the liquid has evaporated as it will reduce lipid solubility. Resuspend the lipids in 2:1 (chloroform:methanol) and store on ice. The optimal volume of the chloroform:methanol solution depends on the detection method used; start low and continue to dilute as needed. A general guideline is to use 10 µl for 10 pooled larvae.
- Spot the entire sample volumes in evenly spaced intervals onto a channeled TLC plate and scan the plate with a laser scanner routinely used for biomolecular imaging (e.g., a fluorescent plate reader). For the fluorescent lipid analogs included in the materials list that have an excitation maxima from 500-650 nm and emission maxima from 510-665 nm, use a 488 nm laser and with emission collection at 520 nm.
- Quantify the total fluorescence of each spot with imaging software. Note: Correct for naturally occurring background lipid fluorescence by subtracting the fluorescence of paired, unfed larval samples.
Representative Results
When fed on a rocker at 29-31 °C, the majority of healthy larvae (≥95%) will eat within 1 hr. Upon consuming the egg yolk emulsion, the larval intestine darkens in color. Very dark intestines can be observed at 2 hr (Figure 1). If larvae are unfed or fail to feed, the intestine remains clear. Larvae fed egg white exhibit a distended intestinal lumen that does not darken in color.
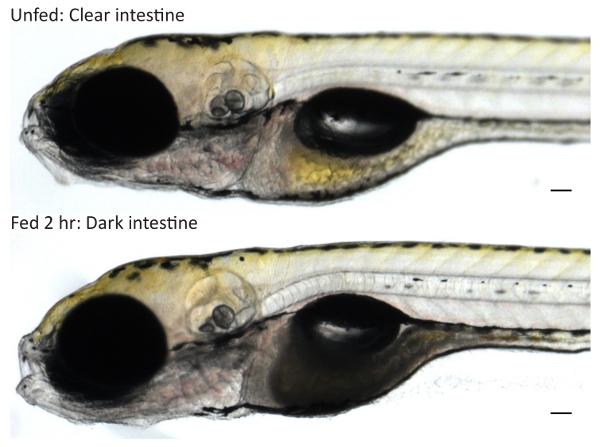 Figure 1: Screening Larvae for Food Intake. Wild type 6-dpf larvae were fed 5% egg yolk in EM for 2 hr or treated in parallel in EM to obtain unfed controls. Unfed larvae have clear intestines while fed larvae that have consumed egg yolk have dark intestines when imaged at 10X on a stereoscope. Scale bars represent 0.1 mm. Please click here to view a larger version of this figure.
Figure 1: Screening Larvae for Food Intake. Wild type 6-dpf larvae were fed 5% egg yolk in EM for 2 hr or treated in parallel in EM to obtain unfed controls. Unfed larvae have clear intestines while fed larvae that have consumed egg yolk have dark intestines when imaged at 10X on a stereoscope. Scale bars represent 0.1 mm. Please click here to view a larger version of this figure.
Feeding with fluorescent lipid analogs permits visualization of systemic transport and subcellular accumulation of dietary lipids. The fluorescent signal is present throughout the digestive organs (intestine, liver, and pancreas) of larvae fed 5% egg yolk with 6.4 µM fluorescent C16 analog for 8 hr mounted with the coverslip lean-to method and imaged with an upright objective (Figure 2)3.
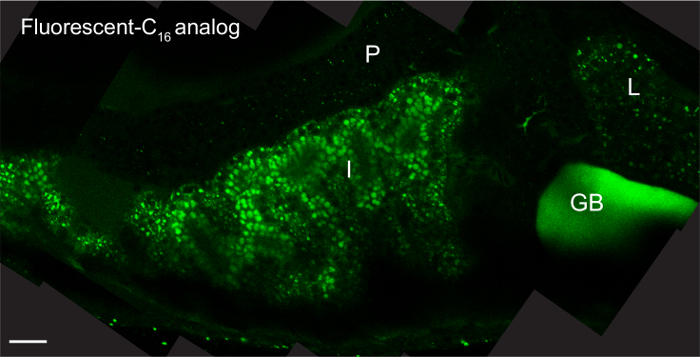 Figure 2: Fluorescent Lipid Analogs Visualize Dietary Lipid Transport. Representative composite image of a 6-dpf larva (head facing right) fed 5% egg yolk in EM with 6.4 µM BODIPY FL C16 for 8 hr. The larvae was mounted in 3% methyl cellulose by the coverslip lean-to method and imaged with an upright 63X oil immersion objective on a single photon confocal microscope with an argon laser. Scale bars represent 20 μm. Liver, L; intestine, I; pancreas, P, gallbladder, GB; reprinted with permission from Dev Bio3. Please click here to view a larger version of this figure.
Figure 2: Fluorescent Lipid Analogs Visualize Dietary Lipid Transport. Representative composite image of a 6-dpf larva (head facing right) fed 5% egg yolk in EM with 6.4 µM BODIPY FL C16 for 8 hr. The larvae was mounted in 3% methyl cellulose by the coverslip lean-to method and imaged with an upright 63X oil immersion objective on a single photon confocal microscope with an argon laser. Scale bars represent 20 μm. Liver, L; intestine, I; pancreas, P, gallbladder, GB; reprinted with permission from Dev Bio3. Please click here to view a larger version of this figure.
Various lipid analogs allow visualization of unique sets of cellular structures since they are differentially metabolized into complex lipids (i.e., triglycerides, cholesterol esters, phospholipids). Following 4 - 8 hr feeds, fluorescent C16 and C12 analogs label lipid droplets, fluorescent FL C5 labels lipid droplets, hepatic and pancreatic ducts, cellular membranes, and arterial networks, and fluorescent C2 analog labels hepatic and pancreatic ducts and cellular membranes (Figure 3)3.
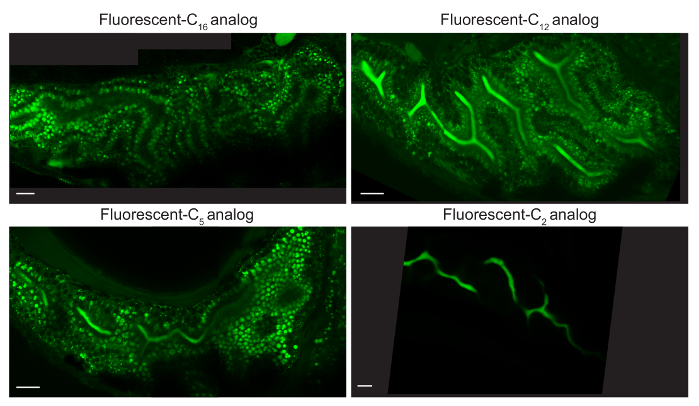 Figure 3: Different Fluorescent Lipid Analogs Label Unique Cellular Organs. Representative composite images of larvae fed 5% egg yolk in EM with 6.4 µM BODIPY FL C16, C12, C5, orC2 for 4-8 hr (6 dpf). C16 analog is observed in lipid droplets (LD) of enterocytes, C12 and C5 analogs in enterocyte LD and the intestinal lumen, and C2 analog in the intestinal lumen. Larvae were mounted in 3% methyl cellulose by the coverslip lean-to method and imaged with an upright 63X oil immersion objective on a single photon confocal microscope with an argon laser. Scale bars represent 20 microns. Reprinted with permission from Drug Discov Today Dis Models22.
Please click here to view a larger version of this figure.
Figure 3: Different Fluorescent Lipid Analogs Label Unique Cellular Organs. Representative composite images of larvae fed 5% egg yolk in EM with 6.4 µM BODIPY FL C16, C12, C5, orC2 for 4-8 hr (6 dpf). C16 analog is observed in lipid droplets (LD) of enterocytes, C12 and C5 analogs in enterocyte LD and the intestinal lumen, and C2 analog in the intestinal lumen. Larvae were mounted in 3% methyl cellulose by the coverslip lean-to method and imaged with an upright 63X oil immersion objective on a single photon confocal microscope with an argon laser. Scale bars represent 20 microns. Reprinted with permission from Drug Discov Today Dis Models22.
Please click here to view a larger version of this figure.
The larval food intake assay was developed to investigate how genetic mutations, transgenic gene overexpression, and/or exogenous treatment affect larval food intake. Larvae are fed a lipid-rich meal spiked with a fluorescent lipid analog to indicate the amount of food consumed. Unfed larvae are collected in parallel to normalize for the amount of background fluorescence present in larvae, increasing the sensitivity at which the fluorescent lipid analog can be detected. This assay was used to show that overexpression of Apolipoprotein A-IVb.1 (apoA-IVb.1) decreases food intake. Previously unfed Tg(hsp70:apoA-IVb.1:mCherry) larvae fed 10% egg yolk with 6.4 µM fluorescent C16 analog for 4 hr at 7 dpf consumed approximately 30% fewer lipids than wild type larvae (Figure 4)20.
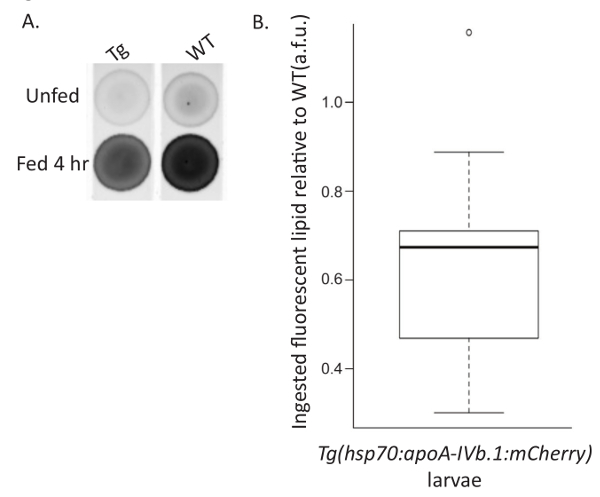 Figure 4: Representative Food Intake Assay. Wild type (WT) and Tg(hsp70:apoA-IVb.1:mCherry) (Tg) larvae were fed 10% chicken egg yolk with 6.4 µM BODIPY FL C16 for 4 hr (Fed) or treated in parallel in EM (Unfed). (A) Larvae were pooled (n= 10), lipids were extracted, and extracts were spotted on a TLC plate. (B) Total fluorescence was quantified, expressed in arbitrary fluorescent units (a.f.u.), normalized for background fluorescence in unfed siblings by subtraction, and expressed relative to WT. Tg larvae overexpressing ApoA-IVb.1 ingest fewer fluorescently labeled lipid analogs as shown by comparison to WT (paired Student's t-test, p <0.001; n= 9, 20 larvae per experiment). The box represents the 25-75th percentiles, and the median is indicated. The whiskers show the 10-90th percentiles. Reprinted with permission from Dis Model Mech20. Please click here to view a larger version of this figure.
Figure 4: Representative Food Intake Assay. Wild type (WT) and Tg(hsp70:apoA-IVb.1:mCherry) (Tg) larvae were fed 10% chicken egg yolk with 6.4 µM BODIPY FL C16 for 4 hr (Fed) or treated in parallel in EM (Unfed). (A) Larvae were pooled (n= 10), lipids were extracted, and extracts were spotted on a TLC plate. (B) Total fluorescence was quantified, expressed in arbitrary fluorescent units (a.f.u.), normalized for background fluorescence in unfed siblings by subtraction, and expressed relative to WT. Tg larvae overexpressing ApoA-IVb.1 ingest fewer fluorescently labeled lipid analogs as shown by comparison to WT (paired Student's t-test, p <0.001; n= 9, 20 larvae per experiment). The box represents the 25-75th percentiles, and the median is indicated. The whiskers show the 10-90th percentiles. Reprinted with permission from Dis Model Mech20. Please click here to view a larger version of this figure.
Discussion
The techniques described here allow researchers to treat larval zebrafish with a lipid-rich feed, visualize dietary lipid processing in live larvae, and quantify larval food intake. To ensure success, special attention should be given to several critical steps. Commercial chicken eggs vary; to minimize potential variability we perform all assays on organic eggs from cage-free chickens that have not been enriched for omega-3 fatty acids. Lower feeding rates may be observed in fish younger than 6 dpf with remaining endogenous yolk energy supplies, unhealthy fish, or fish with developmental mutations that impede food intake (i.e., jaw or intestine malformation, inability to swim properly). The Farber laboratory generally performs feeding studies at 6-7 days post fertilization (dpf) because larvae do not require exogenous food until their endogenous yolk stores are exhausted and yolk depletion allows for better visualization of the digestive system. Feeding at RT also greatly reduces the number of larvae that eat. Failure to screen larvae for food intake and unintentional inclusion of unfed larvae in subsequent experiments may confound results. Additionally, it is important to protect the fluorescent lipid analogs, and all larval samples fed with the analogs, from light when possible to maintain maximum fluorescence, and thus assay sensitivity. Finally, if food intake will be measured, do not allow the feed and chase (post-feed metabolism period) to exceed a total of 4 hr, since the meal will begin to be excreted. Currently, the rate at which the meal and associated fluorescent lipid analogs are excreted is unknown, but fluorescence can persist throughout the larval body for at least 2 days post feed (J.P.O. and S.A.F. unpublished data).
There are several ways to modify and troubleshoot the protocols to answer the experimental questions of interest. The lipid-rich feed can be performed for various periods of time: 1 hr will provide a small meal, while 4 hr will fill the intestine with a large amount of lipid. However, feeding the larvae longer than 6 hr is not recommended as the yolk and EM are not prepared in sterile conditions and bacterial overgrowth in the egg yolk/white emulsion may confound results (i.e., increased inflammation, altered intestinal microbiota profile). Larvae can be examined immediately after feeding to investigate acute effects of the feed Larvae immobilized by brief cooling or anesthesia rapidly begin to fed again after rewarming, but cooling is the preferable method of immobilization as larvae may exhibit emesis upon anesthesia (S.A.F. unpublished observation). Alternatively, chase periods can be carried out after the feed to allow for transport and metabolism of the dietary lipids prior to study. Although feeds are usually carried out in 5 ml liposome solution, feeds have been successfully conducted in volumes ranging from 1 to 20 ml. Various concentrations of egg yolk can be used for larval feeds; the Farber laboratory routinely performs experiments with 5% egg yolk (1 ml egg yolk added to 19 ml EM), 0.5% egg yolk, and 10% egg yolk.
There are potential limitations of fluorescent lipid analogs that must be considered. When choosing a fluorescent lipid analog it is important to consider how the lipid is physiologically processed and where the fluorescent moiety is on the chemical structure of the lipid when interpreting results. For example, triglycerides are broken down into free fatty acids and glycerol, and cholesterol esters into free cholesterol and fatty acids, in the intestinal lumen prior to absorption. Therefore, if a fluorescent triglyceride or cholesterol ester analog is provided in the diet the fluorescence observed in the body will track the fatty acid, free cholesterol, or glycerol that the fluorescent moiety is attached to, not the original triglyceride or cholesterol ester. Different fluorescent lipid analogs label unique cellular structures, and this fact should be considered during selection3. Also of note, compared to the metabolism of native fatty acids, the addition of a BODIPY moiety is roughly equivalent to adding ~2-3 carbons to the fatty acid chain length (S.A.F. unpublished data).
The techniques described here represent significant advancements over previous assays described in the field. Prior to development of this chicken egg yolk feed, two manuscripts detailed studies in which larvae were fed powered hard-boiled egg yolk4,23. The technique described here presents the advantage that the egg yolk does not need to be hard boiled and powdered prior to use and that powdered egg yolk has a very short half-life due to its rapid oxidation. Liquid egg yolk can also be prepared as liposomes that readily accept fluorescent lipid analogs for efficient dietary delivery. Alternatively, radiolabeled lipids can be used to investigate dietary lipid metabolism and measure food intake, but fluorescent lipid analogs do not present the safety hazards associated with radioactivity. However if researchers wish to use radiolabeled lipids, the Farber laboratory has performed studies to verify that radiolabeled lipids can be incorporated into, and successfully fed with, liposomes19. The fluorescent lipid analogs described here were chosen because they show highly similar metabolism to endogenous and radiolabeled lipids, and were sufficiently bright for low- and high-power imaging.
Preceding the development of this technique to feed fluorescent lipid analogs to larvae, no other methods were available to visualize dietary lipid transport and accumulation in vivo. Direct visualization of fluorescent lipid analogs can provide an advantage over using indirect measures of physiology, such as serum lipids. Accurate measurement of larval zebrafish food intake was also a long-standing challenge in the field. The only alternative was to culture paramecia, label them with the fluorescent lipophilic tracer 4-(4-(didecylamino)styryl)-N-methylpyridinium iodide (4-Di-10-ASP), allow larvae to feed, and measure the fluorescence of the intra-abdominal area with a plate reader24. Finally, these techniques have the advantage of being applicable to both medium-throughput screens (i.e., forward genetic screens of dietary lipid processing) as well as focused high-magnification imaging studies (i.e., reverse genetic, hypothesis driven studies). In the future, these techniques can be applied to phenotype and screen diverse models of metabolic and GI disease in zebrafish.
Disclosures
The authors have nothing to disclose.
Acknowledgments
The authors thank Meng-Chieh Shen for images, Jennifer Anderson for providing helpful comments on the manuscript, and members of the Farber laboratory for their contributions in developing these techniques. This study was funded by NIDDK-NIH award RO1DK093399 (S.A.F.), RO1GM63904 (The Zebrafish Functional Genomics Consortium: PI Stephen Ekker and Co-PI S.A.F), and F32DK096786 (J.P.O.). This content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. Additional support was provided by the G. Harold and Leila Y. Mathers Charitable Foundation to the laboratory of S.A.F and the Carnegie Institution for Science endowment.
References
- Carten JD, Farber SA. A new model system swims into focus: using the zebrafish to visualize intestinal metabolism in vivo. Clin Lipidol. 2009;4(4):501. doi: 10.2217/clp.09.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babin PJ, Vernier JM. Plasma lipoproteins in fish. J Lipid Res. 1989;30(4):467–489. [PubMed] [Google Scholar]
- Carten JD, Bradford MK, Farber SA. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev Biol. 2011;360(2):276–285. doi: 10.1016/j.ydbio.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marza E, et al. Developmental expression and nutritional regulation of a zebrafish gene homologous to mammalian microsomal triglyceride transfer protein large subunit. Dev Dyn. 2005;232(2):506–518. doi: 10.1002/dvdy.20251. [DOI] [PubMed] [Google Scholar]
- Stoletov K, et al. Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ Res. 2009;104(8):952–960. doi: 10.1161/CIRCRESAHA.108.189803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, et al. Programming effects of high-carbohydrate feeding of larvae on adult glucose metabolism in zebrafish, Danio rerio. Br J Nutr. 2014;111(5):808–818. doi: 10.1017/S0007114513003243. [DOI] [PubMed] [Google Scholar]
- Wang Z, Mao Y, Cui T, Tang D, Wang XL. Impact of a combined high cholesterol diet and high glucose environment on vasculature. PLoS One. 2013;8(12):81485. doi: 10.1371/journal.pone.0081485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, et al. In vivo visualization and attenuation of oxidized lipid accumulation in hypercholesterolemic zebrafish. J Clin Invest. 2011;121(12):4861–4869. doi: 10.1172/JCI57755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curado S, et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236(4):1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124(3):218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passeri MJ, Cinaroglu A, Gao C, Sadler KC. Hepatic steatosis in response to acute alcohol exposure in zebrafish requires sterol regulatory element binding protein activation. Hepatology. 2009;49(2):443–452. doi: 10.1002/hep.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132(15):3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- Matthews RP, et al. TNFalpha-dependent hepatic steatosis and liver degeneration caused by mutation of zebrafish S-adenosylhomocysteine hydrolase. Development. 2009;136(5):865–875. doi: 10.1242/dev.027565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, et al. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 2010;10:21. doi: 10.1186/1472-6793-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, et al. Overexpression of Akt1 enhances adipogenesis and leads to lipoma formation in zebrafish. PLoS One. 2012;7(5):36474. doi: 10.1371/journal.pone.0036474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Cone RD. Creation of a genetic model of obesity in a teleost. FASEB J. 2007;21(9):2042–2049. doi: 10.1096/fj.06-7503com. [DOI] [PubMed] [Google Scholar]
- Watts SA, Powell M, D'Abramo LR. Fundamental approaches to the study of zebrafish nutrition. ILAR J. 2012;53(2):144–160. doi: 10.1093/ilar.53.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber SA, et al. Genetic analysis of digestive physiology using fluorescent phospholipid reporters. Science. 2001;292(5520):1385–1388. doi: 10.1126/science.1060418. [DOI] [PubMed] [Google Scholar]
- Miyares RL, de Rezende VB, Farber SA. Zebrafish yolk lipid processing: a tractable tool for the study of vertebrate lipid transport and metabolism. Dis Model Mech. 2014;7(7):915–927. doi: 10.1242/dmm.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otis JP, et al. Zebrafish as a model for apolipoprotein biology: comprehensive expression analysis and a role for ApoA-IV in regulating food intake. Dis Model Mech. 2015;8(3):295–309. doi: 10.1242/dmm.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh E, Dyer W. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–918. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Otis JP, Farber SA. Imaging vertebrate digestive function and lipid metabolism. Drug Discov Today Dis Models. 2013;10(1) doi: 10.1016/j.ddmod.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre M, et al. Intestinal fatty acid binding protein gene expression reveals the cephalocaudal patterning during zebrafish gut morphogenesis. Int J Dev Biol. 2000;44(2):249–252. [PubMed] [Google Scholar]
- Shimada Y, Hirano M, Nishimura Y, Tanaka T. A high-throughput fluorescence-based assay system for appetite-regulating gene and drug screening. PLoS One. 2012;7(12):52549. doi: 10.1371/journal.pone.0052549. [DOI] [PMC free article] [PubMed] [Google Scholar]


