Abstract
Cephalopods can undergo rapid and adaptive changes in dermal coloration for sensing, communication, defense, and reproduction purposes. These capabilities are supported in part by the areal expansion and retraction of pigmented organs known as chromatophores. While it is known that the chromatophores contain a tethered network of pigmented granules, their structure-function properties have not been fully detailed. We describe a method for isolating the nanostructured granules in squid Doryteuthis pealeii chromatophores and demonstrate how their associated pigments can be extracted in acidic solvents. To accomplish this, the chromatophore containing dermal layer is first manually isolated using a superficial dissection, and the pigment granules are removed using sonication, centrifugation, and washing cycles. Pigments confined within the purified granules are then extracted via acidic methanol solutions, leaving nanostructures with smaller diameters that are void of visible color. This extraction procedure produces a 58% yield of soluble pigments isolated from granules. Using this method, the composition of the chromatophore pigments can be determined and used to provide insight into the mechanism of adaptive coloration in cephalopods.
Keywords: Biochemistry, Issue 117, Pigment granule, Dissection, Chromatophore, Squid, Doryteuthis pealeii, Nanomaterials, Color, Cephalopods
Introduction
Cephalopods such as squid, cuttlefish, and octopus have the ability to dynamically alter their appearance for camouflaging and signaling.1-6 This capability is supported in part by the selective areal expansion of pigmented organs known as chromatophores.4,7-9 Chromatophores are soft actuators that contain a network of nanostructured pigment granules confined within a cytoelastic sacculus that is anchored radially by muscle fibers.1,3 As they are actuated, chromatophores expand by 500% in presented surface area distributing the granules throughout the organ.3,7,10,11 When this action is concerted over a number of chromatophores, the overall coloration of the animal is changed. While it is known that the pigment granules contribute to this color change, their composition remains unknown. We describe a procedure to isolate and purify chromatophore pigments which may be adapted for future compositional studies.
The isolation of pigment granules involves a multistep extraction, homogenization, and purification procedure.3,12 Chromatophore containing tissue is harvested through careful extirpation from the cephalopod. A digestion and homogenization process is then used to dissociate the surrounding tissue and separate the chromatophore cells. The nanostructured granules are then isolated and purified from the remaining chromatophores using repeated sonication and centrifugation. Upon purification, pigments are extracted from the granules in a process that is adapted from the extraction of visible color from butterfly wings using acidic methanol solutions.13 Scanning electron microscopy (SEM) and spectrophotometry are used to confirm that the chromatophore pigments are successfully extracted using this process.
This method describes the isolation of chromatophore granules which is used to explore the molecular contributions to coloration in cephalopods.12 Small molecule extractions from whole animals can often be a long and tedious process. The aim here is to inform future researchers of an effective and facile protocol for the acquisition of pigments from the nanostructured granules in cephalopods.
Protocol
Invertebrate animal studies conducted herein are not regulated in the United States; therefore the Institutional Animal Care and Use Committee has no authority for review of such protocols. In lieu of these not falling under the jurisdiction of regulation in the United States, the authors hereby state that these studies were conducted with sincere effort towards the ethical use, care, and treatment of these animals, the number of individuals was minimized and these efforts are consistent with the Basel Declaration and the International Council for Laboratory Animal Science (ICLAS) ethics guidelines.
1. Doryteuthis pealeii (D. pealeii) Dissection
- Purchase decapitated squid D. pealeii from the Marine Resources Department at the Marine Biological Laboratory at Woods Hole, MA or from any fresh-fish market. Do not use specimens that have been filleted or previously altered in order to apply this procedure. Specifically, the chromatophore layer must still be intact on the specimen.
- If the animals are not used immediately, store them at -20 °C until use. To defrost the specimens, place them in room temperature water for 1 hr prior to dissection.
Using stainless steel straight fine-point dissection scissors, cut along the posterior side of the animal. Begin at the base of the ventral mantle, and end at the fin region.
Remove all of the internal organs (stomach, gill, ink gland, reproductive organs, systemic heart, and brachial heart) by lifting them with dissecting forceps and severing their connective tissue using dissection scissors. Discard organs in a specimen bag.
Use dissecting T-pins to pin the mantle (fin side up) in a standard 11-1/2" x 7-1/2" x 1-1/2" aluminum dissection pan containing a flexible dissection pad. Submerge the tissue in 250 ml of seawater that has been filtered through a disposable polystyrene vacuum filtration system containing 0.2 µm pore sizes.
Carefully remove the epidermal skin layer (opaque color) with dissecting forceps, keeping the chromatophore containing skin layer intact. Once the epidermal layer is removed, discard it in a specimen bag.
Dissect small (1 cm x 1 cm) sections of the chromatophore layer with forceps and dissection scissors. Collect samples in 1.5 ml micro-centrifuge tubes (fill one-half of each empty tube with the dissected tissue).
Add 500 µl of filtered seawater to the tissue containing tubes. The samples can be stored overnight at 4 °C.
2. Isolating Chromatophore Pigment Granules
- Tissue Digestion
- Preparation of Papain/collagenase
- Weigh out 30 mg of collagenase and 5 mg of papain using a 4" x 4" low nitrogen weigh paper. Transfer these into a 50 ml polypropylene centrifuge tube with a screw cap.
- Measure 10 ml of deionized water using a graduated pipette. Add directly to vial containing papain/collagenase. Vortex on the highest setting until the solution is clear.
- Removing Extracellular Tissue
- Centrifuge the collected chromatophore tissue from step 1.7 for 5 min at 14,000 x g. Discard the top layer of sea water into a plastic waste bucket.
- Add 500 µl of the papain/collagenase solution to the tissue using a P1000 variable volume single channel micropipette. Vortex each sample for 1-2 min on the highest setting.
- Sonicate the samples for 5 min at a frequency of 40 kHz to dissociate cells from the surrounding tissue.
- Centrifuge for 5 min at 14,000 x g. Discard the supernatant into a plastic waste bucket.
- Centrifuge the collected chromatophore tissue for 5 min at 14,000 x g. Discard the top layer of sea water into a plastic waste bucket. Repeat once again.
- Manually remove any remaining large tissue sections that did not digest in this process with forceps before proceeding to the next step.
- Isolating the Pigment Granules
- Preparation of Homogenization Buffer
- Weigh 2.38 g of (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) (HEPES, 100 mM), 0.095 g of magnesium chloride (10 mM), 0.856 g of potassium-aspartate (50 mM), and 0.015 g of dithiothreitol (1 mM). Transfer into a 150 ml polystyrene container with a screw cap. Add one commercial mini tablet protease inhibitor.
- Measure 100 ml of deionized water using a graduated cylinder. Add directly to the container containing the homogenization salts. Vortex until the solution is clear.
- Homogenization of Pigment Granules
- Centrifuge each sample from step 2.1 at 14,000 x g for 5 min. Discard the supernatant into a plastic waste bucket. This sample should be void of extracellular tissue.
- Add 500 µl of homogenization buffer to each tube and vortex for 1-2 min each to thoroughly mix the samples. Sonicate the samples for 30 min (~40 kHz) and follow with 5 min of centrifugation at 14,000 x g.
- Discard the supernatant into a plastic waste bucket, and repeat the previous step 2-3 times to ensure the complete digestion of the chromatophore sacculus and protein tethers. The remaining solution should contain a light yellow supernatant and a red-colored pellet containing the isolated pigment granules.
3. Pigment Extraction
- Preparation of Acidic Methanol (HCl-MeOH)
- Carefully add 25 µl of concentrated (36.5-38 % (w/w)) hydrochloric acid (HCl) to 5 ml of methanol (MeOH). Store the HCl-MeOH solution in a screw cap glass sample vial. Swirl gently to uniformly mix the solution. NOTE: The more acidic the solution, the more pigment will be extracted during the first extraction.
- Extraction of Pigment
- Centrifuge the pigment granule suspension (from 2.2.2) for 5 min at 14,000 x g and discard the supernatant into a plastic waste bucket.
- Add 500 µl of the HCl-MeOH solution and vortex each sample for ~3-4 min. Sonicate the samples for 10 min (~40 kHz), then centrifuge for 5 min at 14,000 x g.
- Using a transfer pipette, collect the supernatant in a 1 dram screw-top glass vial. The supernatant should be dark red in color.
- Repeat the extraction step 3.2.2 until no further color is extracted (~4-5 washes). NOTE: Each extraction will yield ~1.5 ml of pigment. The remaining colorless pellet is the pigment extracted granules. NOTE: Both colorless granules and extracted pigments can be stored in water at 4 °C until further use.
4. Characterization throughout Extraction Process
- Scanning Electron Microscopy
- Image the granules (unreacted from step 2.2.1 and pigment-extracted from step 3.2.3) using scanning electron microscopy (SEM) to verify the purity of the extraction process.
- To prepare granule samples for imaging, first centrifuge both unreacted granules and pigment extracted granules for 5 min at 14,000 x g, and discard the supernatant into a plastic waste bucket. Re-suspend these in 1 ml of ethanol, and deposit 2-3 drops of the granule suspension onto aluminum SEM stubs using a transfer pipette.
- After drying the samples overnight, sputter coat with gold-platinum coating for 3 min to achieve a 15 nm coating.
- Image using a SEM with a Schottky emitter and a secondary electron detector. Use a 5-7 kV beam voltage and a working distance between 8 and 9 mm to image these materials.
- Ultraviolet-visible (UV-vis) Spectrophotometric Measurements
- Monitor the absorbance profiles for all 3 conditions (unreacted granule from 2.2.2, extracted pigment from 3.2.3, and pigment extracted granules from 3.2.4) using a dual beam spectrophotometer from 200-800 nm.
- Collect a blank measurement using deionized water in a 1 ml cuvette for the unreacted granules and the pigment extracted granules. Collect UV-Vis absorption spectra for both samples.
- Collect a blank measurement using the acidic methanol solution in a 1 ml cuvette for the extracted pigment. Collect UV-Vis absorption spectrum for sample.
- Determine maximum wavelength of each sample by identifying the wavelength at which the absorbance peak reaches its maximum relative height.
Representative Results
Chromatophores are dissected from the D. pealeii dorsal mantle (Figure 1A, 1B). Once they are removed, chromatophores are lysed and purified using centrifugation and washing cycles to isolate out the pigmented granules (Figures 2A, 2B). Acidic methanol solutions (HCl-MeOH) are used to extract the pigment from the granules (Figure 2C), yielding a soluble pigment extract and insoluble, colorless pigment-extracted granules.
The extraction process reduces the average diameter of the granules. This is observed using scanning electron microscopy (SEM). Before the granules are exposed to HCl-MeOH, they have an average diameter of (527.3 ± 91.8) nm (N = 100, error is standard deviation; Figure 3A). When the pigment is extracted from the granules, the size decreases to (202.6 ± 32.2) nm (N = 100, error is standard deviation; Figure 3B). This size reduction is hypothesized to be due to the extraction of pigment from the granule when treated with HCl-MeOH.
To test this hypothesis, the differences in visible color pre- and post- extraction are monitored using light microscopy and spectrophotometry. Upon addition of HCl-MeOH, the color associated with the unreacted granules visibly reduces (Figure 4A). This is further characterized using UV-vis spectrophotometry (Figure 4B), where a broad absorbance profile (~280-800 nm) is observed for the pre-extracted granules and post-extracted granules, albeit at a much lower intensity; while, the soluble pigment extract exhibits a lambda max centered at ~500 nm, with a shoulder at ~380 nm. Collectively, these data suggest a method to successfully extract the visible color from the granules.
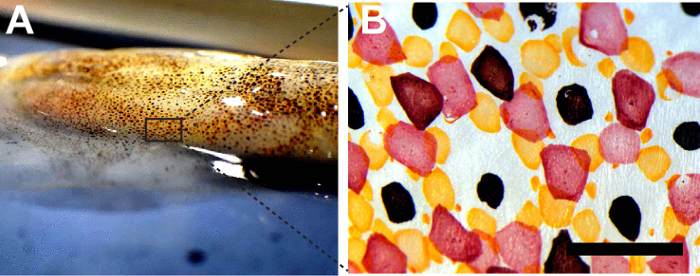 Figure 1:Chromatophores from squid D. pealeii. (A) Squid D. pealeii dorsal mantle. (B) Bright field image of red, yellow, and brown chromatophores. Scale bar = 1 mm. Please click here to view a larger version of this figure.
Figure 1:Chromatophores from squid D. pealeii. (A) Squid D. pealeii dorsal mantle. (B) Bright field image of red, yellow, and brown chromatophores. Scale bar = 1 mm. Please click here to view a larger version of this figure.
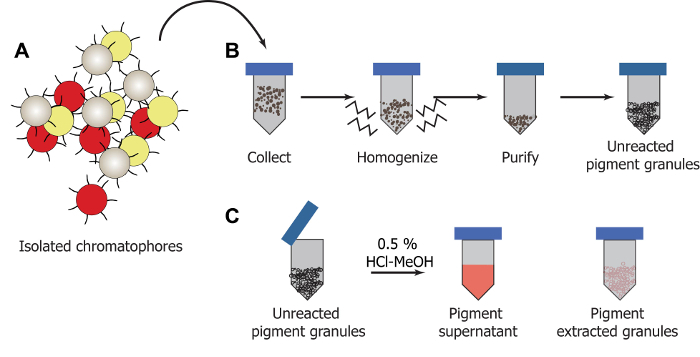 Figure 2:Illustration of extraction process. (A) Chromatophores are dissected. (B) Pigment granules are removed from homogenized chromatophore tissue and purified using a series of centrifugation and washing cycles. (C) The pigment can be extracted from the granules using HCl-MeOH yielding a soluble pigment supernatant which is separated from the colorless pigment extracted granules. Please click here to view a larger version of this figure.
Figure 2:Illustration of extraction process. (A) Chromatophores are dissected. (B) Pigment granules are removed from homogenized chromatophore tissue and purified using a series of centrifugation and washing cycles. (C) The pigment can be extracted from the granules using HCl-MeOH yielding a soluble pigment supernatant which is separated from the colorless pigment extracted granules. Please click here to view a larger version of this figure.
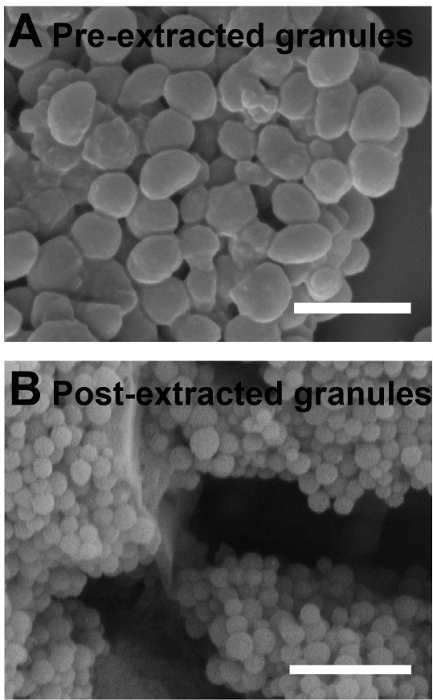 Figure 3:HCl-MeOH extraction alters diameters of the nanostructured granules. Scanning electron micrographs of (A) pre-acidic methanol extracted pigment granules and (B) post-acidic methanol extracted pigment granules. Scale bar = 1 µm. Please click here to view a larger version of this figure.
Figure 3:HCl-MeOH extraction alters diameters of the nanostructured granules. Scanning electron micrographs of (A) pre-acidic methanol extracted pigment granules and (B) post-acidic methanol extracted pigment granules. Scale bar = 1 µm. Please click here to view a larger version of this figure.
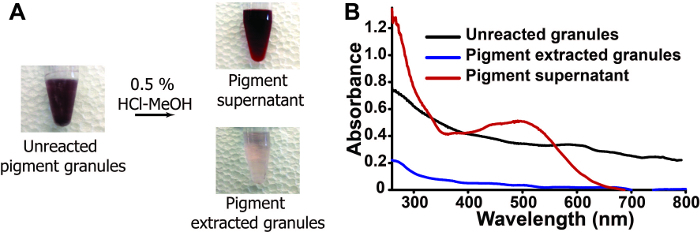 Figure 4:HCl-MeOH extraction alters visible color of nanostructured granules. (A) Bright field images of pigment granules throughout the extraction process. (B) Absorbance spectra of pre (black)- and post (blue)- extraction along with the pigment supernatant (red). Please click here to view a larger version of this figure.
Figure 4:HCl-MeOH extraction alters visible color of nanostructured granules. (A) Bright field images of pigment granules throughout the extraction process. (B) Absorbance spectra of pre (black)- and post (blue)- extraction along with the pigment supernatant (red). Please click here to view a larger version of this figure.
Discussion
We have demonstrated a method to extract pigments from squid chromatophore granules. By specifically targeting the granules, our goal is to determine their role in mediating adaptive coloration. This method differs from previous reports designed to characterize cephalopod pigments using bulk tissue samples14 or freeze-dried skin15.
While this protocol is effective at extracting chromatophore pigments, it is limited to small reaction scales. Dissecting one squid yields ~11 mg of purified granules. Approximately 58% of this mass contains the soluble pigment, which is extracted using the HCl-MeOH solution.12 The purity of the starting chromatophore tissue directly influences the product yield from one dissection. If the samples appear contaminated by excess epidermal or iridophore tissue collected during the dissection, it is best to separate out the sample's contents, resuspend them in additional buffer, and begin the homogenization steps over again. Isolating a pure population of pigment granules from chromatophore tissue may take as many as eight, 4 hr sonication cycles that could span across 4 days. It is important to remain patient in the isolation to ensure a pure population of pigment granules.
The purity of the starting granule solution also influences the pigment extraction steps with HCl-MeOH. If the granules are occluded by undigested tissue, simply vortex and sonicate the samples up to 1 hr and homogenize the sample before beginning the extraction again. Often 4-5 washes using 500 µl of HCl-MeOH per sample are required to extract the pigment from the granules. If done correctly, this procedure will yield between 2-3 ml of soluble pigment per sample. When additional purification is employed via thin layer chromatography, the isolated pigments separate into four unique fractions containing varying concentrations of xanthommatin and decarboxylated xanthommatin.12
This method can be utilized by chemists, biologists, or engineers to better understand the role of pigments in color modulation. The presence of the pigment within the granules suggests a hierarchical mechanism for coloration in cephalopods which originates with the molecular components contained within nanostructured chromatophore pigment granules. These findings can be used to inform engineered systems designed to mimic the dynamic range of adaptive color displayed in cephalopods using pre-packaged pigmented nanoparticles.
Disclosures
We have nothing to disclose.
Acknowledgments
The authors gratefully acknowledge the use of facilities at the University of New Hampshire including the University Instrumentation Center. This work was supported by the University of New Hampshire, Department of Chemistry.
References
- Bell GRR, et al. Chromatophore radial muscle fibers anchor in flexible squid skin. Invertebr Biol. 2013;132(2):120–132. [Google Scholar]
- Crookes WJ, et al. Reflectins: The unusual proteins of squid reflective tissues. Science. 2004;303(5655):235–238. doi: 10.1126/science.1091288. [DOI] [PubMed] [Google Scholar]
- Deravi LF, et al. The structure-function relationships of a natural nanoscale photonic device in cuttlefish chromatophores. J. R. Soc. Interface. 2014;11(93):1–9. doi: 10.1098/rsif.2013.0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäthger LM, Denton EJ, Marshall NJ, Hanlon RT. Mechanisms and behavioural functions of structural coloration in cephalopods. J. R. Soc. Interface. 2009;6(2):149–163. doi: 10.1098/rsif.2008.0366.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäthger LM, Hanlon RT. Anatomical basis for camouflaged polarized light communication in squid. Biol. Lett. 2006;2(4):494–496. doi: 10.1098/rsbl.2006.0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäthger LM, Hanlon RT. Malleable skin coloration in cephalopods: selective reflectance, transmission and absorbance of light by chromatophores and iridophores. Cell Tissue Res. 2007;329(1):179–186. doi: 10.1007/s00441-007-0384-8. [DOI] [PubMed] [Google Scholar]
- Cloney RA, Brocco SL. Chromatophore Organs, Reflector Cells, Iridocytes and Leucophores in Cephalopods. Amer. Zool. 1983;23(3):581–592. [Google Scholar]
- Hanlon RT, Messenger JB. Adaptive coloration in young cutttlefish (Sepia officinalis L)- The morphology and development of body patterns and their relation to behaviour. Philos Trans R Soc Lond B Biol Sci. 1988;320(1200):437–487. [Google Scholar]
- Sutherland RL, Mäthger LM, Hanlon RT, Urbas AM, Stone MO. Cephalopod coloration model. II. Multiple layer skin effects. J. Opt. Soc. Am. 2008;25(8):2044–2054. doi: 10.1364/josaa.25.002044. [DOI] [PubMed] [Google Scholar]
- Florey E. Ultrastructure and function of cephalopod chromatophores. Amer. Zool. 1969;9(2):429–442. doi: 10.1093/icb/9.2.429. [DOI] [PubMed] [Google Scholar]
- Florey E, Kriebel ME. Electrical and mechanical responses of chromatophore muscle fibers of squid, Loligo opalescens, to nerve stimulation and drugs. Z. Vergl. Physiol. 1969;65(1):98–130. [Google Scholar]
- Williams TL, et al. Contributions of Phenoxazone-Based Pigments to the Structure and Function of Nanostructured Granules in Squid Chromatophores. Langmuir. 2016. [DOI] [PubMed]
- Nijhout HF. Ommochrome pigmentation of the linea and rosa seasonal forms of Precis coenia (Lepidoptera: Nymphalidae) Arch. Insect. Biochem. Physiol. 1997;36(3):215–222. [Google Scholar]
- Van Den Branden C, Decleir D. A Study of the Chromatophore Pigments in the Skin of the Cephalopod Sepia Officinalis. L. Biol. Jb. Dodonaea. 1976;44(2):345–352. [Google Scholar]
- Aubourg S, Torres-Arreola W, Trigo M, Ezquerra-Brauer J. Characterization of Jumbo Squid Skin Pigment Extract and its Antioxidant Potential ina Marine Oil System. Eur. J. Lipid Sci. Technol. 2016;118(0) [Google Scholar]


