Abstract
Carbonic anhydrases (CAs) are zinc metalloenzymes that catalyze the interconversion of CO2 and HCO3− and are ubiquitous in nature. Higher plants contain three evolutionarily distinct CA families, αCAs, βCAs, and γCAs, where each family is represented by multiple isoforms in all species. Alternative splicing of CA transcripts appears common; consequently, the number of functional CA isoforms in a species may exceed the number of genes. CAs are expressed in numerous plant tissues and in different cellular locations. The most prevalent CAs are those in the chloroplast, cytosol, and mitochondria. This diversity in location is paralleled in the many physiological and biochemical roles that CAs play in plants. In this review, the number and types of CAs in C3, C4, and crassulacean acid metabolism (CAM) plants are considered, and the roles of the α and γCAs are briefly discussed. The remainder of the review focuses on plant βCAs and includes the identification of homologs between species using phylogenetic approaches, a consideration of the inter- and intracellular localization of the proteins, along with the evidence for alternative splice forms. Current understanding of βCA tissue-specific expression patterns and what controls them are reviewed, and the physiological roles for which βCAs have been implicated are presented.
Keywords: carbonic anhydrase, regulation, alternative splicing, physiological role
Carbonic anhydrase catalyzes the interconversion of carbon dioxide and bicarbonate, the two forms of inorganic carbon used in multiple metabolic pathways in plants. This review covers the current understanding of the numerous and diverse carbonic anhydrase isoforms found in terrestrial plants. The regulation, alternative splicing, and physiological roles of the different carbonic anhydrase isoforms are discussed.
Introduction
Carbonic Anhydrases Are Essential for Photosynthetic Organisms and Their Intracellular Location Is Critical
Carbonic anhydrases (CAs) play essential roles in all photosynthetic organisms. In cyanobacteria, CAs located in the carboxysome are required for the conversion of accumulated HCO3− to CO2 for fixation by ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) (Espie and Kimber, 2011). Loss of this carboxysomal CA results in abnormal growth of cyanobacteria when they are grown at ambient levels of CO2 (Price and Badger, 1989). In Chlamydomonas reinhardtii, a thylakoidal CA is necessary for the functioning of the CO2 concentrating mechanism (CCM) (Moroney and Ynalvez, 2007), and in the diatom, Pheodactylum tricorutum, a CA in the pyrenoid is required for the CCM of this species (Harada et al., 2005). In these organisms, the conversion of HCO3− to CO2 for Rubisco is needed in a very specific location in the cell. In addition, the CCM of C4 plants requires CA activity specifically in the mesophyll (M) cell cytosol (Gutierrez et al., 1974). While the correct inter- and intracellular location of CAs is essential for efficient physiological functioning of photosynthetic organisms, it is also important that CA activity is not present in certain organelles or cell types. Price et al. (1992) demonstrated this when they transformed cyanobacteria with a gene encoding a human CA. This CA was expressed in the cytoplasm of the Synechocystis cells, effectively short circuiting the CCM (Price et al., 1992). Similarly, a defective C4 CCM resulted when a cytosolic CA was expressed in bundle-sheath (BS) cells of the C4 plant Flaveria bidentis (Ludwig et al., 1998). Recently, a number of research initiatives have been working to improve photosynthesis in plants by introducing CCM components from cyanobacteria, algae, or C4 plants into terrestrial C3 plants. While the introduction of active transporters and enzymes is required for these initiatives to work, it is also necessary to know where the endogenous CAs are active within the recipient plant, as introducing CA activity in the wrong location could short circuit attempts to improve photosynthesis.
This review focus on what is known about the genes encoding CA, and the locations of the CA isoforms in both C3 and C4 plants. Up-to-date research on the physiological roles of the different CA isoforms is also covered, as well as our current understanding of the molecular changes that were responsible for the evolution of the genes encoding C4-associated CAs from their ancestral C3 orthologs.
Plants Have Three Types of Carbonic Anhydrases
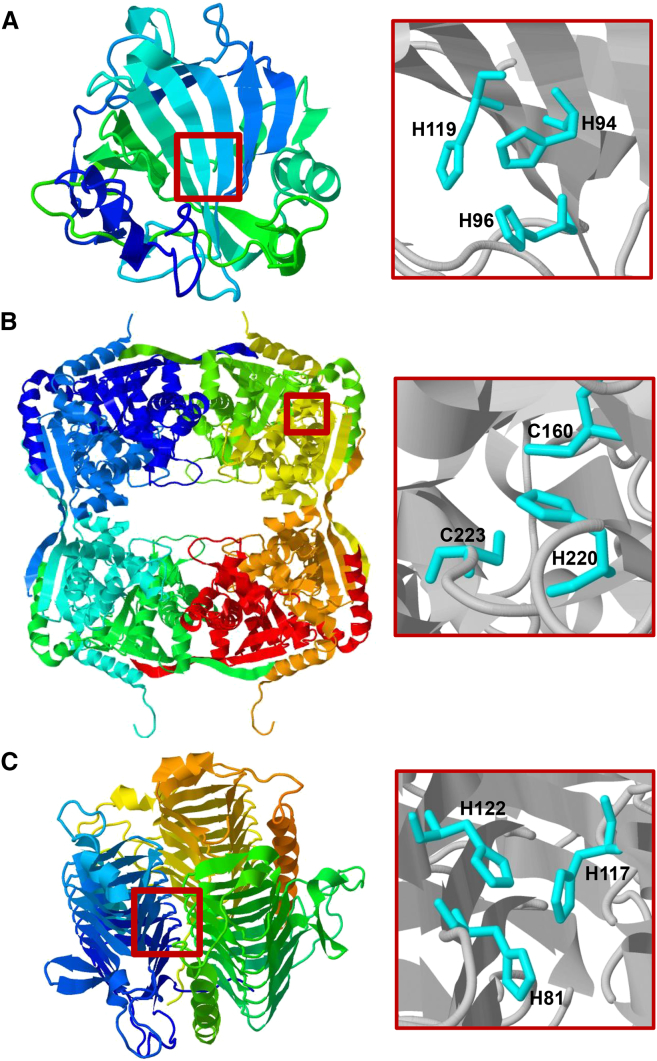
All CAs are zinc metalloenzymes that catalyze the interconversion of CO2 and HCO3−. The enzymes are ubiquitous in nature and are an example of convergent evolution, as multiple, structurally and sequentially distinct families of CA have been discovered (Hewett-Emmett and Tashian, 1996). Plants have three types of CA: α-, β-, and γ-type CAs (Moroney et al., 2001). The α-type CA (αCA) was first found in erythrocytes and was the first CA family discovered (Brinkman et al., 1932, Meldrum and Roughton, 1932). The majority of the enzyme is composed of 10 β strands that create a large central β sheet, which is surrounded by seven α helices on the periphery of the protein (Figure 1A; Liljas et al., 1972). The zinc at the αCA active site is coordinated by three His residues and one water molecule organized in a tetrahedral conformation (Liljas et al., 1972, Eriksson et al., 1988, Håkansson et al., 1992), and is located in the central part of the protein, at the bottom of a cone-shaped crevice (Liljas et al., 1972). While most αCAs are monomers, multimeric αCAs have been discovered as well as αCAs containing extra domains (Ishida et al., 1993, Hilvo et al., 2008). However, even in multimeric αCAs, the zinc ion is always coordinated by His residues from a single polypeptide.
Figure 1.
Structures of α, β, and γ Carbonic Anhydrase Proteins with Their Active Site Architecture.
(A) The human CAII monomer (Mangani and Håkansson, 1992) mostly consists of β strands and contains a single active site with three zinc coordinating histidine residues.
(B) The Pisum sativum βCA octamer (Kimber and Pai, 2000) contains eight active sites where each zinc is coordinated by two cysteines and a histidine.
(C) The Methanosarcina thermophila γCA (Iverson et al., 2000) forms a trimer with three active sites. Although the γCA active site also contains three histidine resides, one monomer provides the H81 and H122 residues, while a second monomer provides the H117 residue to form the γCA active site. Red boxes indicate the enlarged locations of each protein structure to display their active-site architecture. CA protein structures and active-site images were generated using Jmol (http://www.jmol.org/).
The β-type CA (βCA) was first discovered in plants (Burnell et al., 1990, Fawcett et al., 1990, Roeske and Ogren, 1990), and its protein sequence and structure are very different from that of the αCAs. In βCAs, the zinc ion is coordinated by two Cys residues, one His residue, and a water molecule (Figure 1B; Kimber and Pai, 2000). The structure of a βCA monomer is mostly composed of α helices that surround a β sheet consisting of four parallel β strands. There is also a fifth, C-terminal β strand involved in the oligomerization of βCA (Kimber and Pai, 2000). The functional unit of the βCA is a dimer, although the most common βCA oligomerization is a tetramer (Kimber and Pai, 2000, Rowlett, 2010). The βCA dimer is formed via extensive interactions created by two N-terminal α helices of one monomer wrapping around the second monomer and by minor hydrogen bonding between the second β strand of each monomer (Kimber and Pai, 2000). Tetramers are formed by interactions made primarily by the fifth, C-terminal β strand (Kimber and Pai, 2000). In pea, the chloroplastic βCA forms an octamer. For some βCAs, dicots have a unique C-terminal extension of the fifth β strand, whereas monocots do not (Kimber and Pai, 2000, Rowlett, 2010). Octamers are formed via slightly different interactions with these fifth β-strand extensions (Kimber and Pai, 2000, Rowlett, 2010).
The γ-type CA (γCA) was first discovered in archaea (Alber and Ferry, 1994) but has since been found in photosynthetic bacteria (Price et al., 1993, Peña et al., 2010) and in plants (Parisi et al., 2004). The first crystal structure of γCA from Methanosarcina thermophila was reported by Kisker and colleagues in 1996 (Figure 1C). Much like the active site of αCA, the active site of γCA also contains a zinc atom coordinated by three His and a water molecule (Kisker et al., 1996). However, unlike the structure of αCAs, which are monomers, the functional unit of γCA is a trimer, with three active sites spanning the monomer-monomer interfaces. The zinc ion is coordinated by His residues provided by two different subunits (Kisker et al., 1996). A β-strand region dominates the structure of γCA and consists of seven complete turns creating a left-handed β helix (Kisker et al., 1996). Each full turn contains three β strands making the β helix look like an equilateral triangle from the top view (Kisker et al., 1996). In photosynthetic organisms, γCA may contain extra domains as seen in the cyanobacterial CcmM proteins of cyanobacteria that have two or three repeated C-terminal domains with high similarity to the small subunit of Rubisco (Long et al., 2007). In cyanobacteria, CcmM sometimes functions as an active CA, but some CcmM proteins do not have activity (Peña et al., 2010, de Araujo et al., 2014). However, CcmM is thought to organize the packing of Rubisco in the carboxysome even when it does not have CA activity.
Plants Have Multiple Genes Encoding All Three Types of Carbonic Anhydrases
Plants have a large number of genes encoding CA. There are 17 distinct genes in total encoding all α, β, and γ, isoforms in Arabidopsis, including two γ-like CAs (Table 1). A similar number of genes are present in the genomes of other plant species, including mosses, monocots, and dicots (Table 1). Since plants can be polyploids or paleopolyploids, having undergone genome duplication in the past, the number of CA genes may be much higher. For example, in soybean, a diploid plant thought to have undergone genome duplication relatively recently, the total number of genes coding for CA is in excess of 25. Genes encoding CA are expressed in almost all tissues of the plant and CA isoforms can be found in most intracellular compartments, such as chloroplasts, mitochondria, the plasma membrane, and the cytoplasm.
Table 1.
Total Number of α, β, and γ Carbonic Anhydrase Genes in Different Plants.
| Plant type | Species | PS type | Type and number of CA genes |
||
|---|---|---|---|---|---|
| α | β | γ | |||
| Moss | Physcomitella patens | C3 | 5 | 6 | 5 |
| Club moss | Selaginella moellendorffii | C3 | 10 | 5 | 4 |
| Dicots | Arabidopsis thaliana | C3 | 8 | 6 | 5 |
| Medicago truncatula | C3 | 8 | 7 | 4 | |
| Vitis vinifera | C3 | 5 | 6 | 3 | |
| Populus trichocarpa | C3 | 8 | 7 | 5 | |
| Monocots | Brachypodium distachyon | C3 | 6 | 4 | 3 |
| Oryza sativa | C3 | 9 | 3 | 4 | |
| Setaria italica | C4 | 9 | 4 | 3 | |
| Sorghum bicolor | C4 | 9 | 5 | 3 | |
| Ananas comosus | CAM | 4 | 3 | 3 | |
Genes were identified based on multiple sequence alignment in Clustal Omega using Arabidopsis carbonic anhydrase (CA) genes as query. Sequences for Physcomitrella patens, Selaginella moellendorffii, Medicago truncatula, Vitis vinifera, Populus trichocarpa, Brachypodium distachyon, Oryza sativa, Setaria italica, Sorghum bicolor were obtained from Phytozome (https://phytozome.jgi.doe.gov) and NCBI. Sequences for Ananas comosus were obtained from CoGe as described in Ming et al., 2015. γCA gene numbers include γ-like CA genes.
PS, photosynthetic type; C3, C3 photosynthesis; C4, C4 photosynthesis; CAM, crassulacean acid metabolism.
Green Algae Also Contain the Three Types of Carbonic Anhydrase
It is likely that the numbers and types of CA are quite ancient in the plant lineage as the unicellular green alga Chlamydomonas reinhardtii also has multiple genes encoding α-, β- and γCAs. C. reinhardtii has three genes encoding αCA, six encoding βCA, and three encoding γCA and γCA-like proteins (Mitra et al., 2005). The CA isoforms of C. reinhardtii are found throughout the algal cell: in the periplasmic space (cell wall), chloroplast, cytoplasm, and mitochondria. As seen in higher plants, the C. reinhardtii γCAs and γCA-like proteins are mitochondrial with evidence suggesting they are part of Complex I of the mitochondrial electron transport chain, and the βCAs are found in similar intracellular locations as in higher plants, with isoforms in the mitochondria, chloroplast, and cytoplasm. However, two of the C. reinhardtii βCAs have hydrophobic C-terminal extensions (Ynalvez et al., 2008), this is not observed so far in terrestrial plants. In addition, in C. reinhardtii the αCAs seem to play different physiological roles. In terrestrial plant species, only β- and γCAs have been implicated in CCMs, whereas two αCAs play important roles in the CCM of C. reinhardtii: CAH1, which is located in the periplasmic space, and CAH3, which is found in the thylakoid lumen. For more information on Chlamydomonas CAs, see Moroney et al. (2011).
Plant α Carbonic Anhydrases
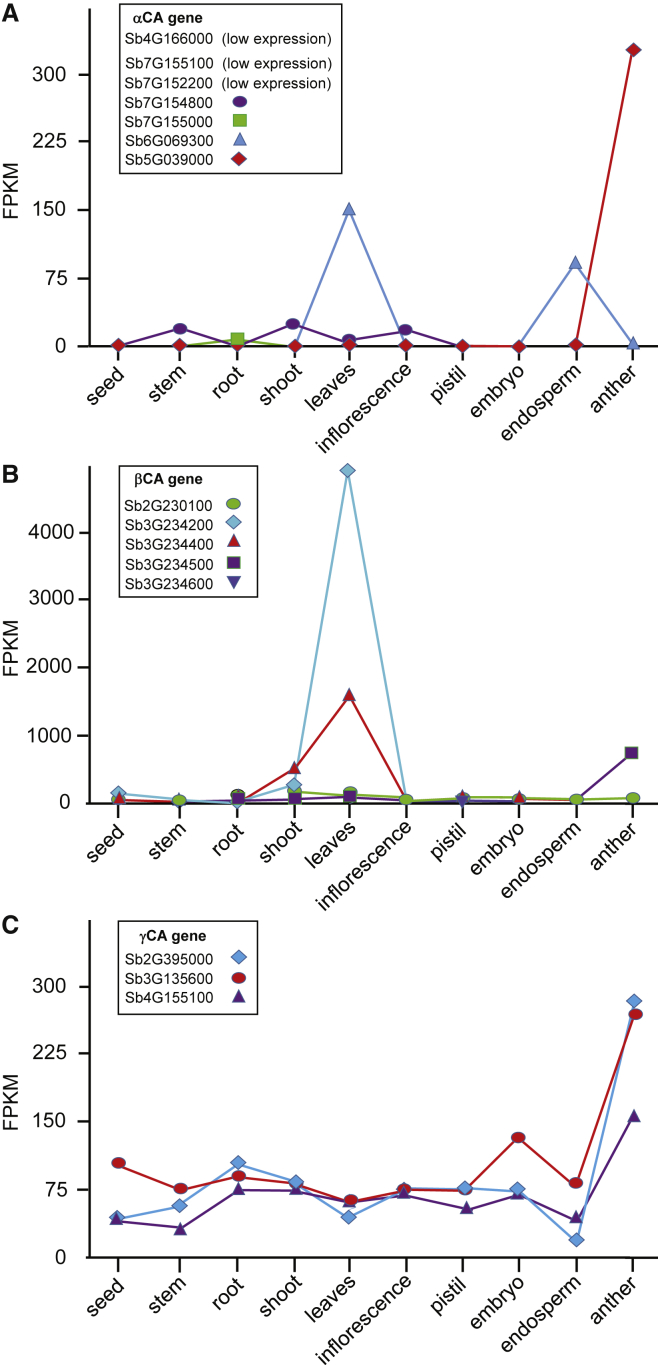
αCAs are the largest CA gene family in most plants, but they are also the least studied. The scarcity of published work on αCAs is most likely because the proteins are not highly abundant in leaves and roots. In Arabidopsis, complete expressed sequence tags (ESTs) exist for only three of the eight αCA genes, and RNA-seq data also only poorly cover the other five annotated genes. Arabidopsis αCA8 is clearly a pseudogene as it encodes in-frame stop codons. Limited expression information for some αCA genes is available through RNA-seq data on genome sites. Interestingly, in sorghum, the αCA Sb5G039000 is expressed specifically in anthers (Figure 2; Makita et al., 2015), while in Medicago trunculata, the αCAs Mt1g059900 and Mt1g059940 are expressed in root nodules (Tang et al., 2014). In Arabidopsis, αCA2 is expressed in trichomes of the leaf. Clearly at least some αCA genes show quite specific organ or tissue expression patterns. To date, there are no reports of plants where one or more αCA genes have been disrupted.
Figure 2.
Relative Expression of Carbonic Anhydrases in Sorghum bicolor Organs.
(A) α, (B) β, and (C) γ carbonic anhydrase expression in different organs of Sorghum bicolour taken from MOROKOSHI - The Sorghum Transcriptome Database (Makita et al., 2015), in fragments per kilobase of transcript per million mapped reads (FPKM). Note that the y axis for the βCA expression is different than that of the αCA or γCA graphs.
There is very little information on the intracellular location of αCA isoforms. Two reports suggest that Arabidopsis αCA1 is a chloroplastic protein (Villarejo et al., 2005, Blanco-Rivero et al., 2012). The targeting of this αCA1 is unusual as it moves through the endoplasmic reticulum and is glycosylated. However, the location of αCA1 in the chloroplast has not yet been confirmed by proteomic studies. Possibly the amount of αCA in leaves is low, or perhaps the glycosylation obscures its detection. There are no reports on the subcellular location or function of αCAs from other plants at this time.
Plant γ Carbonic Anhydrases
Genes encoding γCAs and γCA-like proteins have been found in all plants. In fact, every species appears to have at least two genes encoding γCAs and at least one encoding a γCA-like protein (Table 1). For example, Arabidopsis has three γCA genes and two genes encoding γCA-like proteins (Parisi et al., 2004, Perales et al., 2004). γCAs are well conserved in photosynthetic organisms, from green algae, to mosses, monocots, and dicots. While no higher plant γCA with CA activity has been identified, the proteins have the active-site residues found in γCAs from archaebacteria and cyanobacteria. In contrast, the γCA-like proteins do not have the required Zn coordinating amino acid residues. While γCAs are encoded by the nucleus, they are mitochondrial proteins. They have been shown to be part of the mitochondrial Complex I (NADH-ubiquinone oxidoreductase), and make up an extrinsic domain known as the carbonic anhydrase domain of the oxidoreductase (Sunderhaus et al., 2006), which is composed of three subunits: two γCA subunits and one γCA-like subunit (Klodmann et al., 2010). The γCA and γCA-like proteins are part of nine plant-lineage-specific subunits.
The expression level of genes coding for γCA and γCA-like isoforms is average or above in almost all tissues for which expression data are available (Figure 2). This is not surprising for a subunit of Complex I as the mitochondrial electron transport chain is found in most plant tissues and cell types. If either AtγCA1 or AtγCA2 is knocked out, there is a small reduction in Complex I (Perales et al., 2005); however, if both AtγCA1 and AtγCA2 are knocked out, the plant is profoundly and adversely affected. The γca1γca2 mutants lack Complex I altogether, and do not produce viable seed, having to be maintained using an embryo rescue method, which involves supplying the embryos with sucrose in the growth medium (Fromm et al., 2016a, Fromm et al., 2016b). The double mutants also exhibit high levels of Complexes II and IV (succinate dehydrogenase and cytochrome oxidase, respectively), and the alternative oxidase, and in contrast, reduced levels of photosynthetic proteins (Fromm et al., 2016c).
Plant β Carbonic Anhydrases
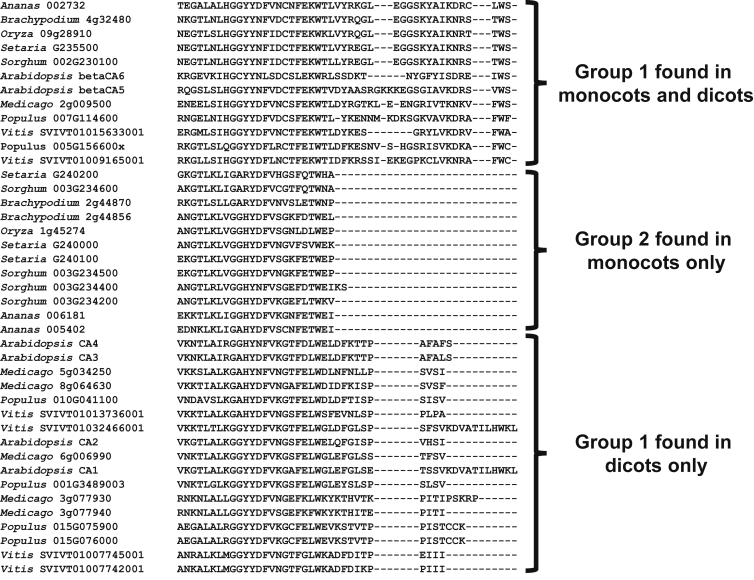
Plants have a moderate number of βCA genes, usually between four and seven (Table 1). There have been a number of studies on the βCAs as they are highly expressed in leaf tissue (Figure 2). βCAs have been found in chloroplasts, mitochondria, the cytosol, and the plasma membrane of Arabidopsis (Table 2), and in the cytosol and chloroplasts of many plants. When the predicted amino acid sequences of βCAs from monocots and dicots are aligned, the isoforms can be divided roughly into three groups (Figure 3). One group was found in all monocots and dicots considered (Figure 3) and is represented by AtβCA5 and AtβCA6, which localize to the chloroplast and mitochondria, respectively. The second group was found only in dicots, and the Arabidopsis AtβCA1, AtβCA2, AtβCA3, and AtβCA4 proteins are in this group. All dicots examined had at least two βCA proteins in this group (Figure 3). The third group contains only monocot CAs, and these proteins are known to localize to the chloroplast and cytosol. The length of the C termini of the proteins was an indicator for the group in which an isoform clusters. The monocot-specific proteins were the shortest, with the dicot-specific isoforms about 10 amino acids longer and the AtβCA5/AtβCA6-related proteins, found in all species examined, about 20 amino acids longer (Figure 3).
Table 2.
Experimentally Derived Subcellular Locations of Plant β Carbonic Anhydrase Isoforms.
Data for Flaveria βCA subcellular locations were obtained using chloroplast import assays, and Arabidopsis and Neurachne βCA subcellular locations were determined using fluorescent protein fusion constructs. Dash indicates that the protein is not targeted to the chloroplast. Arabidopsis βCA sequences are available in TAIR, Flaveria sequences are identified by GenBank accession numbers, and Neurachne sequences are as described in Clayton et al., 2016.
Figure 3.
Multiple Sequence Alignment of C-termini of β Carbonic Anhydrase Proteins from Different Plants.
Sequences were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/; Sievers et al., 2011). Sequences for Physcomitrella patens (Lang et al., 2005;Rensing et al., 2005, Zimmer et al., 2013), Selaginella moellendorffii (Banks et al., 2011), Medicago truncatula (Young et al., 2011, Tang et al., 2014), Vitis vinifera (Jaillon et al., 2007), Populus trichocarpa (Tuskan et al., 2006, Du et al., 2015, Ye and Zhong, 2015), Brachypodium distachyon (Vogel et al., 2010), Oryza sativa (Ouyang et al., 2007), Setaria italic (Bennetzen et al., 2012), Sorghum bicolor (Makita et al., 2015) were obtained from Phytozome (https://phytozome.jgi.doe.gov). Sequences for Arabidopsis thaliana were obtained from TAIR (Lamesch et al., 2011). Sequences for Ananas comosus were obtained from CoGe (https://genomevolution.org; Ming et al., 2015).
As the βCAs have been the most intensely studied family of plant CAs, the remainder of this review focuses on this group.
Evidence of Alternative Splicing of β Carbonic Anhydrase Transcripts
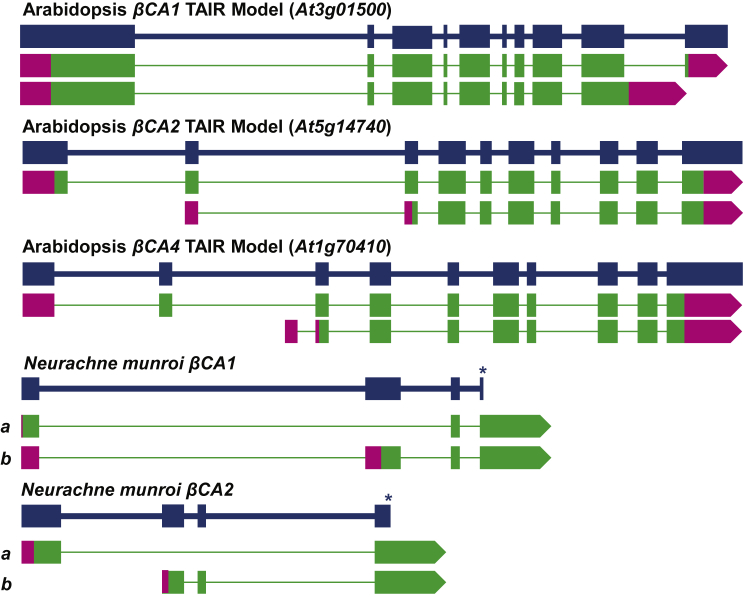
Alternative splicing can result in a single gene coding for multiple proteins that may show tissue-specific expression patterns and/or be targeted to different organelles of the cell. Deposited ESTs in TAIR, as well as RNA-seq data (Oh et al., 2014), indicate that transcription of the AtβCA1 gene may result in two different mRNAs (Figure 4). The RNA variants arise from the splicing of the ninth and tenth exons, where one variant has all 10 exons, and the other has an extended ninth exon, making proteins that differ slightly at their C termini. Two different transcription start sites for AtβCA2 (Figure 4) are predicted to encode two AtβCA2 isoforms with different N termini, resulting in the two proteins having different projected destinations in the plant cell. AtβCA4 is another example of a gene that can produce multiple mRNA forms (Figure 4; Aubry et al., 2014), due to different transcription start sites (Figure 4). The shorter AtβCA4 mRNA lacks the first two exons, encoding a different N terminus relative to the longer form. Interestingly, RNA-seq data for AtβCA4 indicate that the shorter mRNA has a unique first exon that is not present in the longer transcript and is expressed in both roots and leaves (DiMario et al., 2016). In contrast, the longer AtβCA4 mRNA is expressed specifically in leaves of Arabidopsis (DiMario et al., 2016).
Figure 4.
Alternative Splicing of β Carbonic Anhydrase Genes in Arabidopsis thaliana and Neurachne munroi.
Blue lines indicate genomic DNA with larger boxes representing exons. Green and magenta boxes indicate exons present in different splice forms, with green representing open reading frame sequence and magenta representing untranslated regions. Asterisks indicate that the Neurachne munroi CA1 and CA2 genomic DNA sequences are incomplete; for each gene, exon 4 and the downstream exons are present in both splice forms, as represented by the green arrows. Data from DiMario et al. (2016) and Clayton et al. (2016).
In Neurachne munroi leaves, four βCA transcripts are expressed that are derived from two genes by way of alternative splicing (Figure 4; Clayton et al., 2016). For each gene, the alternatively spliced transcripts encode polypeptides that share the same active-site residues but have distinct N termini, thereby influencing the subcellular location of each isoform (Clayton et al., 2016). In N. munroi, the CA1 gene encodes NmCA1a and NmCA1b. Exon 2 is not present in NmCA1a transcripts, such that in CA1a and CA1b, the initiating AUG and N-terminal regions are encoded in different frames (Figure 4; Clayton et al., 2016). In the case of the NmCA2 gene, NmCA2a transcripts do not contain exons 2 and 3, while exon 1 is not present in the NmCA2b transcript, again resulting in each splice form encoding a distinct N-terminal region (Figure 4; Clayton et al., 2016). The same gene-transcript relationship was observed in all other Neurachne species for which transcript and genomic DNA sequences have been obtained (data not shown). While experimental evidence for alternative splicing of other monocot βCA genes has not been reported, parsing of EST databases and comparison with genome sequences suggest that alternative splice forms do exist in other species.
Subcellular Locations of β Carbonic Anhydrase from Experimental Data
Linking CA to photosynthesis has been a major focus in the plant CA field. Studies using reverse transcription (RT)–PCR, microarrays, and RNA-seq have shown that all six βCA genes are expressed in leaves of Arabidopsis (Schmid et al., 2005, Fabre et al., 2007, Winter et al., 2007, Ferreira et al., 2008, Hu et al., 2010, Wang et al., 2014, DiMario et al., 2016); however, subcellular localization studies using green fluorescent protein (GFP) fusion constructs showed that not all of the proteins localize to the chloroplast. AtβCA1 was shown to have a long chloroplast transit peptide over 100 amino acid residues in length (Fett and Coleman, 1994, Kim et al., 1994), and was later confirmed to be located in the chloroplast via GFP studies (Table 2; Fabre et al., 2007, Hu et al., 2015). AtβCA2 and AtβCA3 are cytosolic βCAs (Table 2; Fabre et al., 2007, DiMario et al., 2016) although AtβCA2 is expressed at a much higher level than AtβCA3 in Arabidopsis leaves (Schmid et al., 2005, Winter et al., 2007, Ferreira et al., 2008, Hu et al., 2010, DiMario et al., 2016). The long form of AtβCA4, AtβCA4.1, localizes to the plasma membrane while the short form, AtβCA4.2, is cytosolic, as it no longer has a secretory transit peptide (Table 2; Fabre et al., 2007, DiMario et al., 2016). Studies using GFP fusions also showed that AtβCA5 localizes to the chloroplast (Fabre et al., 2007), and AtβCA6 is a mitochondrial form of the enzyme (Table 2; Fabre et al., 2007, Jiang et al., 2014).
To date, three transcripts encoding distinct βCA isoforms (CA1, CA2, and CA3) have been found in leaves of Flaveria species (Tetu et al., 2007, Tanz et al., 2009). Chloroplast import assays showed in both F. bidentis (a C4 species) and F. pringlei (a C3 species) that CA1 was imported into chloroplasts, while CA2 was not (Table 2; Tetu et al., 2007, Tanz et al., 2009). However, the subcellular location of CA3 was not conserved between the two species; FpCA3 was found to be chloroplast targeted, while FbCA3 was not (Table 2; Tetu et al., 2007, Tanz et al., 2009). Comparison of the predicted amino acid sequences of the CA3 polypeptides revealed that FbCA3 lacks 71 amino acids at the N terminus, including the chloroplast transit peptide, when compared with FpCA3, a situation that was proposed to be important to the molecular evolution of C4 photosynthesis in F. bidentis (Tanz et al., 2009).
The only monocot species so far in which the subcellular location of βCA isoforms has been experimentally determined are from the genus Neurachne (Clayton et al., 2016). GFP fusion constructs indicated that N. munroi CA2b is imported into tobacco chloroplasts or mitochondria, while NmCA1a, NmCA1b, and NmCA2a localize to the cytosol (Table 2; Clayton et al., 2016). As in Flaveria, the location of the isoforms is not conserved between C3 and C4 Neurachne species. While the GFP localization experiments showed that NaCA1b from the C3 species N. alopecuroidea is cytosolic as predicted, NaCA1a is imported into the chloroplasts, in contrast with the cytosolic location of CA1a from N. munroi (Table 2; Clayton et al., 2016). Analyses of the predicted proteins indicated that 11 amino acids present in the N-terminal region of the NaCA1a polypeptide, but absent in NmCA1a, is important for chloroplast targeting (Clayton et al., 2016).
Predicting β Carbonic Anhydrase Subcellular Locations
Numerous algorithms exist for predicting protein subcellular location. Predicted βCA amino acid sequences from various monocot and dicot species were analyzed using four algorithms: Predotar, ChloroP, TargetP, and MultiLoc (Supplemental Table 1; Emanuelsson et al., 1999, Small et al., 2004, Höglund et al., 2006, Emanuelsson et al., 2007). The results indicate that each species contains at least one isoform that is chloroplast targeted and at least one isoform that is likely cytosolic, and that many also contain a mitochondrial βCA (Supplemental Table 1). Comparison with actual experimental results for Arabidopsis, Flaveria, and Neurachne (Table 2) indicates that these predictions are often but not always correct (cf. Table 2 and Supplemental Table 1).
The above results indicate that all predictions must be experimentally tested, keeping in mind that the proteins resulting from different splice forms of a single gene may have different subcellular locations (Table 2; Clayton et al., 2016, DiMario et al., 2016). In addition, predicting βCA subcellular location based on the products of orthologous genes in closely related species may not assist in assigning location since it has been shown that locations are not conserved between closely related C3 and C4 species (Tanz et al., 2009, Clayton et al., 2016).
Organ-, Tissue-, and Cell-Type-Specific Expression of β Carbonic Anhydrases
To date, the specific expression patterns of all identified βCA isoforms have been reported in the scientific literature for only three species: Arabidopsis thaliana (Schmid et al., 2005, Fabre et al., 2007, Winter et al., 2007, Ferreira et al., 2008, Wang et al., 2014, DiMario et al., 2016), F. bidentis (Tetu et al., 2007), and F. pringlei (Tanz et al., 2009). An in-depth CA expression study of Arabidopsis rosette leaves found that AtβCA1, AtβCA2, and AtβCA4 are the most highly expressed CA genes in M cells, and AtβCA1, AtβCA4, and AtβCA6 are the most highly expressed CA genes in guard cells (Hu et al., 2010). Many studies also report CA gene expression in Arabidopsis roots (Schmid et al., 2005, Fabre et al., 2007, Winter et al., 2007, Wang et al., 2014, DiMario et al., 2016), with RNA-seq data showing that AtβCA2 and AtβCA3 are the two lowest expressed βCA genes in Arabidopsis roots, whereas AtβCA4 and AtβCA5 are the two most highly expressed genes (DiMario et al., 2016). Microarray analyses indicated all AtβCA genes are expressed in roots of Arabidopsis, albeit AtβCA1, AtβCA2, and AtβCA3 are expressed at very low levels (Schmid et al., 2005, Winter et al., 2007). Results of an RT–PCR analysis showed that transcripts from most of the AtβCAs genes are found in Arabidopsis roots with the exception of AtβCA2 (Wang et al., 2014), whereas another RT–PCR experiment found that AtβCA3 and AtβCA6 show the highest expression in roots (Fabre et al., 2007). Both RT–PCR studies found that Arabidopsis genes encoding βCA are expressed in stems and floral tissues (Fabre et al., 2007, Wang et al., 2014), and all six AtβCA genes are expressed in Arabidopsis stem and floral tissues according to microarray analyses (Schmid et al., 2005, Winter et al., 2007). Interestingly, the microarray data indicate that all six AtβCA genes are expressed in Arabidopsis seeds although, with the exception of AtβCA5 and AtβCA6, their expression diminishes as the seeds develop (Schmid et al., 2005, Winter et al., 2007).
In F. pringlei, RT–quantitative (q)PCR assays indicated that transcripts encoding FpCA1 and FpCA3 were primarily expressed in leaves, whereas FpCA2 was expressed in leaves, roots, and flowers (Tanz et al., 2009). By contrast, transcripts encoding CA1, CA2, and CA3 were detected in F. bidentis leaves, roots, and flowers, but FbCA1 and FbCA3 transcripts were most abundant in leaves, whereas FbCA2 mRNA levels were consistent among the three tissues (Tetu et al., 2007).
While numerous reports have shown that the majority of CA activity in leaves of C4 species is in the M cells (Gutierrez et al., 1974, Ku and Edwards, 1975, Burnell and Hatch, 1988), the location of multiple βCA isoforms within a leaf has not been comprehensively examined in any species. In F. bidentis, immunocytochemical experiments showed that CA is expressed predominantly in M cells and is undetectable in BS cells (Tetu et al., 2007). Presumably this is the FbCA3isoform previously shown to be essential for C4 photosynthesis (von Caemmerer et al., 2004). The tissue/cell type-specific expression patterns of FbCA1 and FbCA2 were not examined.
Recently, advances in laser-capture microdissection and next-generation sequencing have enabled M and BS cell transcriptomes to be obtained. Comparative transcriptome analyses of leaf M and BS cells of Panicum virgatum (Rao et al., 2016), Setaria viridis (John et al., 2014), Gynandropsis gynandra (formerly Cleome gynandra; Aubry et al., 2014), and Zea mays (Li et al., 2010, Chang et al., 2012) showed that particular βCA transcripts are enriched in M cells. Specifically, Pavir.J08788 and Pavir.J05107 transcripts are approximately three times more abundant in M cells than in BS cells in P. virgatum (Rao et al., 2016), and a similar fold difference is also observed for two transcripts encoding βCA in G. gynandra, GgβCA1 and GgβCA2 (At3g01500 and At5g14740 orthologs; Aubry et al., 2014), one transcript in S. viridis (Si03061m.g ortholog; John et al., 2014), and one in Z. mays (GRMZM2G414528; Li et al., 2010, Chang et al., 2012). However, in the latter two species, there are several βCA transcripts that show much higher M-specific abundance, with at least a 20-fold enrichment compared with BS cells. These include Si003885m.g in S. viridis (John et al., 2014), and GRMZM2G121878, GRMZM2G348512, and GRMZM2G094165 in Z. mays (Li et al., 2010, Chang et al., 2012, Rao et al., 2016). Interestingly, the maize GRMZM2G145101 transcript is the only βCA transcript identified so far that shows higher abundance in BS cells than M cells (Li et al., 2010, Chang et al., 2012, Rao et al., 2016), while no βCA mRNAs from S. viridis, G. gynandra, and P. virgatum show this pattern. Taken together, the current findings suggest that within a C4 species, genes encoding the different βCA isoforms show different tissue- and cell-specific expression patterns, with some isoforms showing preferential expression in the leaf M cells.
Activity of β Carbonic Anhydrase in C3 and C4 Species
Total leaf CA activity within herbaceous dicotyledonous plants ranges from 2- to 10-fold (Everson and Slack, 1968, Atkins et al., 1972, Triolo et al., 1974, Reed and Graham, 1981, Hatch and Burnell, 1990, Gillon and Yakir, 2001, supplementary material), whereas the leaves of some monocotyledons reportedly contain 1000 times more CA activity than other monocot species (Everson and Slack, 1968, Atkins et al., 1972, Triolo et al., 1974, Reed and Graham, 1981, Burnell and Hatch, 1988, Hatch and Burnell, 1990, Gillon and Yakir, 2001, supplementary material; Cousins et al., 2008). Total leaf CA activity in C3 monocots can be 500 times higher than that of C4 monocots (Everson and Slack, 1968, Triolo et al., 1974, Reed and Graham, 1981, Hatch and Burnell, 1990, Gillon and Yakir, 2001, supplementary material), while leaves of herbaceous C4 dicots demonstrate total CA activities that fall within the range of values for C3 dicot leaves (Everson and Slack, 1968, Atkins et al., 1972, Reed and Graham, 1981, Hatch and Burnell, 1990, Gillon and Yakir, 2001, supplementary material).
Very few studies have looked at CA activity in both isolated M and BS cells from C4 plant leaves; however, depending on the comparison being made, this is important. As it is the cytosolic CA in M cells that is associated with the C4 CCM, total leaf CA activity measurements may be misleading. Two forms of CA were isolated from Amaranthus cruentus leaves (Guliev et al., 2003). One form was found associated with the chloroplasts of the BS cells and was responsible for 8% of total leaf CA activity. In contrast, the other form was found in the M cell cytoplasmic fraction, where it represented 62% of the total CA activity in amaranth leaves (Guliev et al., 2003). In another C4 dicot, Flaveria bidentis, BS cell CA activity was found to contribute 0.5% of total leaf CA activity (Ludwig et al., 1998).
Burnell and Hatch (1988) also found low CA activity in BS cells of species representing the three C4 subtypes: NADP-malic enzyme (NADP-ME), NAD-malic enzyme (NAD-ME), and phosphoenolpyruvate carboxykinase (PCK). In the two NADP-ME-subtype species examined, sorghum and maize, BS cell CA activity was 1.8% and 1.6%, respectively, of total leaf CA activity. The activity of CA in the BS of NAD-ME-type (P. miliaceum and Atriplex spongiosa) and PCK-type species (Urochloa panicoides and Chloris gayana) was even lower, representing just 0.5%–0.8% of total leaf CA activity (Burnell and Hatch, 1988). These results suggested low CA activity in the BS was a requisite for efficient functioning of the C4 pathway (Burnell and Hatch, 1988). This idea was later supported by a transgenic approach in which wild-type (WT) plants of the C4 species F. bidentis were transformed with the sequence encoding mature tobacco CA (i.e., no chloroplast transit peptide) that was under the control of a constitutive promoter (Ludwig et al., 1998). This allowed tobacco CA expression in the cytosol of all cells, including leaf BS cells. The transformants showed increased BS leakiness to inorganic carbon (Ci), reduced rates of photosynthesis, and an impaired CCM (Ludwig et al., 1998). Together these results support the idea that in C4 plants demonstrating Kranz leaf anatomy, the strict M and BS cell compartmentalization of CA is essential for the proper functioning of the C4 CCM.
Regulation of Cell-Type-Specific Expression of β Carbonic Anhydrase in C4 Plants
Progress has been made in our understanding of cis elements and chromatin marks that control the preferential accumulation of transcripts encoding βCA in M cells of C4 species; however, the associated trans-acting factors remain elusive. Studies suggest several regulatory mechanisms were already present in ancestral C3 genes coding for βCA but were modified through recruitment of posttranscriptional pathways or the binding of different transcription factors during the evolution of C4 photosynthesis.
In leaves of the C4 species Gynandropsis gynandra, transcripts encoding the homolog of the Arabidopsis plasma-membrane-associated CA (AtβCA4; Fabre et al., 2007, Hu et al., 2010, Kajala et al., 2012), showed abundances similar to the levels of mRNAs coding for other C4-associated proteins (Bräutigam et al., 2011, Kajala et al., 2012). Elements in either the 5′-untranslated region (UTR) or 3′-UTR of the G. gynandra gene encoding βCA4 were found to be sufficient for M-cell-specific expression using GUS fusion constructs (Kajala et al., 2012). Similar sequences in the 5′- and 3′-UTRs of AtβCA4 were also shown to independently direct M-cell-specific expression when they were used to transform G. gynandra.
A more recent study showed the 3′-UTR of a second G. gynandra CA gene, GgCA2, for which high transcript levels are found in leaves, and the homologous region from AtβCA2 also direct preferential accumulation of GUS in M cells (Williams et al., 2016). A common, nine-nucleotide motif in these CA2 3′-UTRs, as well as in the 5′- and 3′-UTRs of AtβCA4 and GgCA4, were identified as sufficient to direct M cell-specific expression and was designated MEM2 for mesophyll expression module 2 (Williams et al., 2016). This study also showed that the MEM2 element does not control the level of GgCA4 gene expression but instead works post-transcriptionally through a mechanism that increases the amount of CA4 protein made in M cells relative to the BS. The high levels of GgCA4 transcripts in G. gynandra M cells appear to result from the loss of elements in the promoter region and introns from the ancestral CA4 gene that repress its expression in C3 species (Williams et al., 2016).
Epigenetic marks have been identified that contribute to M cell-specific expression of genes encoding CA isoforms important in C4 photosynthesis. Trimethylation of the Lys residue at position 4 on histone H3 (H3K4me3) is associated with transcriptionally active genes and is enriched in the 5′-region of the transcribed sequence (Santos-Rosa et al., 2002). Heimann et al. (2013) found the gene encoding one of the C4-associated CAs in maize, GRMZM2G121878, showed a high ratio of H3K4me3 to the dimethylated form (H3K4me2) in M cells. This is consistent with the methylation state of histone H3K4 found at analogous positions in genes coding for the C4-associated forms of PEPC and pyruvate phosphodikinase (PPDK), which also show preferential accumulation of transcripts in M cells (Heimann et al., 2013).
Physiological Roles of β Carbonic Anhydrases
The total number of genes encoding CA is similar in dicots and monocots, and in plants using C3 and C4 photosynthesis (Table 1; Williams et al., 2012), with some of these genes encoding isoforms that likely perform the same function in all species. However, as highlighted above, differences in total CA activity and control of βCA expression patterns have been detected within and between these plant groups, and are responsible for the very specific physiological roles exhibited by some of the enzymes.
C3 Photosynthesis
In the leaves of C3 plants, the majority of CA activity localizes to M cell chloroplasts (Everson and Slack, 1968, Everson, 1970, Poincelot, 1972), where the enzyme can make up 1%–2% of total leaf protein (Okabe et al., 1984, Peltier et al., 2006). Although a major component of the C3 leaf proteome, the actual role of βCAs in C3 photosynthesis remains ambiguous (Figure 5). Initial suggestions included the conversion of HCO3− to CO2 to ensure maximum rates of fixation by Rubisco (Everson, 1970, Poincelot, 1972, Werdan and Heldt, 1972), facilitating the diffusion of CO2 across the chloroplast membranes (Poincelot, 1972), buffering short-term changes in pH in the chloroplast stroma induced by changing light conditions (Jacobson et al., 1975), and the hydration of compounds other than CO2 (Jacobson et al., 1975). However, to date, none of these proposed functions has strong empirical support.
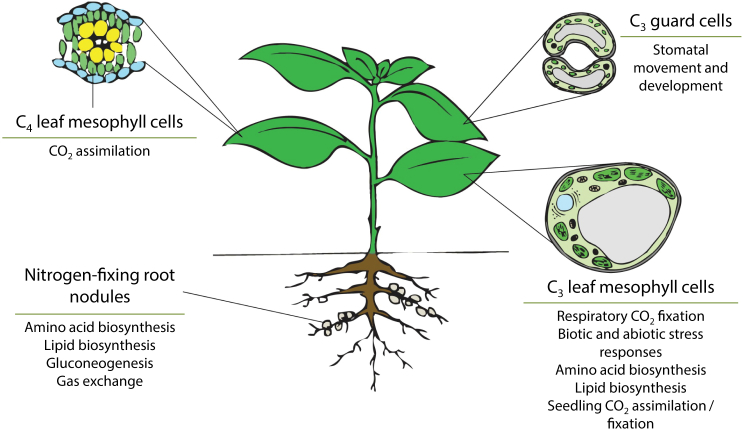
Figure 5.
Schema Illustrating the Physiological Functions of β Carbonic Anhydrases in Plant Cells and Organs.
In leaf mesophyll cells of C4 plants, a cytosolic βCA catalyzes the first step in C4 photosynthesis. βCAs are involved in a CO2 sensing pathway in guard cells and implicated in stomatal development. A number of roles have been attributed to βCAs found in leaf mesophyll cells of C3 plants, including involvement in refixation of respiratory CO2, stress responses, amino acid and lipid biosynthesis, and seedling establishment. In nitrogen-fixing root nodules of legumes, βCAs are implicated in different functions during nodule maturation, including roles in primary metabolism and gas exchange. Note: the roles of βCA in C3 guard cells and C3 leaf mesophyll cells are likely to be performed by homologs in the corresponding cell types of C4 plants. See text for details and references.
To determine the physiological function of C3 plant chloroplast βCA, WT tobacco plants were transformed with antisense constructs directed against transcripts encoding the tobacco chloroplastic βCA isoform (Majeau et al., 1994, Price et al., 1994). Primary transformants with 1%–2% of the CA activity of WT plants showed no significant differences in CO2 assimilation rates, Rubisco activity, and chlorophyll content relative to WT plants.
More recently, Arabidopsis antisense transformants and knockout lines of βCA1 were examined (Ferreira et al., 2008), and these plants did demonstrate an obvious phenotype. Both lines of transformants showed reduced seedling survival that could be rescued by including sucrose in the growth medium, or growing the seedlings in elevated CO2. The cotyledons were found to have compromised CO2 assimilation rates that resulted in the observed reduced seedling establishment before development of the first true leaves (Figure 5). However, when the transformants did survive, the mature plants showed no phenotypic differences from WT plants, strongly suggesting that AtβCA1 plays no direct role in photosynthesis of mature Arabidopsis plants (Ferreira et al., 2008).
C4 Photosynthesis
In contrast to C3 plants, most βCA activity in C4 plants is found in the cytosol of M cells (Gutierrez et al., 1974), where it catalyzes the first reaction in the C4 CCM (Figure 5; Hatch and Burnell, 1990), the conversion of atmospheric CO2 to HCO3−.
Transgenic approaches have been used to test the suggestion that only enough CA is present in the M cytosol of C4 plants to not limit photosynthesis (Hatch and Burnell, 1990). In one study (von Caemmerer et al., 2004), WT plants of the C4 dicot F. bidentis were transformed with an antisense construct directed against transcripts encoding the C4-associated CA3 (von Caemmerer et al., 2004, Tetu et al., 2007). A decrease in CO2 assimilation rates was seen only when the transformants contained less than 20% of WT CA activity, and transformants exhibiting less than 10% of WT activity had very reduced rates of photosynthesis, about 8% of WT plants, and required a high CO2 environment to survive. In addition, this study also showed that the hydration rate of CO2 was about 58 times the photosynthetic rate. Taken together, these results indicate that CA is not limiting photosynthesis in F. bidentis; however, the CO2 response curves of the transformants indicated that a cytosolic CA is essential for an efficient C4 CCM in this dicot species (von Caemmerer et al., 2004).
Maize plants carrying mutations in genes encoding two isoforms of CA that have been correlated with C4 photosynthesis (ZmCa1 and ZmCa2; Studer et al., 2014) demonstrated that CA is not limiting for growth in this C4 monocot species. Unlike the F. bidentis CA3 antisense plants, however, both the ca1 single mutant and the ca1ca2 double mutant, which contained 3% of WT maize CA activity, showed no impairment in CO2 assimilation at ambient levels of CO2. It was not until the concentration of CO2 was sub-ambient that a decrease in CO2 assimilation was detected (Studer et al., 2014). It was concluded from gas exchange and carbon isotope data, and CA and PEPC activities, that the ca1 mutant contains only enough CA activity to supply PEPC with HCO3−, while the activity in the double mutant is below this level, and the plants rely, at least to some extent, on the uncatalyzed conversion of CO2 to HCO3− (Studer et al., 2014). Clearly, while CA in both WT maize and F. bidentis is not rate limiting for photosynthesis, and is necessary for efficient operation of the C4 pathway when CO2 availability to the leaf is limited (Boyd et al., 2015), differences exist between these two C4 species with respect to the levels of CO2 that result in impaired CO2 assimilation. The basis for this discrepancy may be structural, enzymatic, or a combination of mechanisms (Studer et al., 2014, Ludwig, 2016).
A Ubiquitous, Basal Carbon-Concentrating Mechanism in Plants
A mitochondrial βCA along with the γ and γ-like CAs associated with the mitochondrial Complex I are proposed to be part of a mechanism found in all plants that facilitates the fixation of mitochondrial respiratory CO2 in the chloroplasts (Figure 5; Zabaleta et al., 2012). Relative to WT plants, Arabidopsis mutants lacking the mitochondrial AtβCA6 gene show a decrease in leaf area and overall biomass, inhibition of growth at low CO2, and a significant increase in respiration rates (Jiang et al., 2014). In contrast, the overexpression of AtβCA6 in Arabidopsis resulted in larger plants with higher shoot fresh and dry weights, and decreased rates of respiration compared with WT plants (Jiang et al., 2014). Interestingly, there appeared to be no significant difference in photosynthetic rates although the CO2 compensation point of the knockout lines was reportedly increased relative to WT values. From this work, the authors suggested that increasing expression levels of mitochondrial AtβCA6 affect cellular respiration, which impacts positively on biomass production.
Carbonic Anhydrase Activity and Photosystem II
The finding that acetazolamide inhibited photosystem II (PS II) activity (Swader and Jacobson, 1972) raised the possibility that CA activity was associated with PSII. Later, Stemler (1997) presented evidence that included the finding of low levels of CA activity with thylakoid preparations and even core PSII fractions. The discovery of CrCAH3, in the thylakoid lumen of C. reinhardtii (Karlsson et al., 1998), resulted in two different hypotheses as to its physiological role. In one proposal, CAH3 functions in light-driven generation of CO2 from accumulated HCO3−, taking advantage of the low pH of the thylakoid lumen to drive the reaction toward CO2 formation (Raven, 1997, Hanson et al., 2003, Moroney and Ynalvez, 2007). A competing hypothesis was that CrCAH3 was required on the oxidizing side of PSII (Park et al., 1999, Villarejo et al., 2002). Since CrCAH3 is an αCA, this hypothesis presented an attractive physiological role for αCAs in plants. However, evidence over the past 10–15 years strongly argues against a role for CA in PSII, with probably the most persuasive argument being the lack of CA in any of the crystal structures of PSII to date.
Most cyanobacteria do not have an αCA, and for most cyanobacteria, the only CA in the cell is in the carboxysome as part of the CCM. In Arabidopsis, total chloroplast (intact chloroplasts) proteome studies indicate the only CAs in the chloroplasts are βCA1, βCA2, and βCA5 (Friso et al., 2004; Ferro et al., 2010). The other CA reported to be in the chloroplast, αCA1, has not been detected in proteome studies to date, and no other αCA has been found in chloroplasts. None of the Arabidopsis β-type CAs (AtβCA1, AtβCA2 and AtβCA5) have a leader sequence consistent with a thylakoid lumen location, and only the stromal AtβCA1 is present at high levels in chloroplasts of photosynthetically active cells. Finally, Hillier et al. (2006) and McConnell et al. (2007) found no CA activity in highly active PSII preparations using the very sensitive membrane inlet mass spectrometry assay. They convincingly argued that any CA activity associated with PSII was due to contamination. This is not surprising as stromal CA activity is extremely high and even a relatively low level of contamination by this protein could result in measurable activity in enriched PSII preparations (McConnell et al., 2007).
Stomatal Movement and Development
Increased transcript abundances are found for the genes encoding AtβCA1 and AtβCA4 in Arabidopsis guard cells (Hu et al., 2010 and references therein). While single Atβca1 and Atβca4 T-DNA mutants demonstrated no CO2-sensitive phenotype compared with WT plants, the double ca1ca4 mutant showed impaired stomatal conductance in response to changing CO2 concentration as well as higher stomatal numbers and density (Hu et al., 2010). Consequently AtβCA1 and AtβCA4 were implicated in guard cell movement through a role in the early steps of the CO2 signaling pathway, and were suggested to function also in guard cell development (Hu et al., 2010). Recent work using reconstituted systems in Xenopus oocytes has suggested a model in which AtβCA4 functions alongside the aquaporin PIP2;1 at the guard cell plasma membrane, influencing intracellular CO2/HCO3− levels, which when elevated, enhance S-type anion channel activity and stomatal closure (Figure 5; Wang et al., 2016). As yet, no direct interactions between AtβCA4 and PIP2;1 have been reported.
The Arabidopsis ca1ca4 mutants show an inverted response to CO2 relative to WT plants in that, at high CO2, they have increased stomatal numbers in cotyledons and mature leaves (Engineer et al., 2014). Taking these results into account, as well as the earlier characterization of the double mutant (Hu et al., 2010), a preliminary model for the control of stomatal development has been constructed and involves an extracellular signaling pathway mediated by CA (Engineer et al., 2014, Engineer et al., 2016). Not all the components or steps in the model have been identified (Engineer et al., 2014, Engineer et al., 2016); however, it has been proposed that CA activity is necessary for the increased expression of the genes encoding the epidermal patterning factor EPF2, and the CO2-inducible protease that cleaves it, facilitating its binding to the receptor kinase ERECTA, which has been implicated in the regulation of stomatal development (Figure 5; Shpak, 2013).
Biotic and Abiotic Stress Responses
Chloroplastic βCAs from C3 plants are part of a defense mechanism that is induced upon attack by various pathogens (Figure 5; Slaymaker et al., 2002, Restrepo et al., 2005, Jung et al., 2008, Wang et al., 2009, Collins et al., 2010). In tobacco and Arabidopsis, the CAs have been identified as salicylic-acid-binding proteins that function in an antioxidant role during viral infections (Slaymaker et al., 2002, Wang et al., 2009). Recombinant inbred lines of Arabidopsis with resistance to the insect herbivore, Plutella xylostella, had at least a 2-fold increase in abundance of AtβCA1 and AtβCA4 proteins (Collins et al., 2010).
Salinity induces an increase in βCA transcript abundance in maize, and it was suggested that this response paralleled the antioxidant role seen with the biotic stressors described above (Figure 5; Kravchik and Bernstein, 2013). Both salinity and an osmotic stress treatment using polyethylene glycol led to an increase in rice seedling total CA enzyme activity, and the level of mRNA coding for a predicted chloroplastic CA isoform (Yu et al., 2007). Overexpression of the rice CA in Arabidopsis led to improved growth on media containing salt compared with WT Arabidopsis (Yu et al., 2007).
Amino Acid Biosynthesis
Cytosolic CAs have been implicated in affecting amino acid biosynthesis levels (Figure 5; Raven and Newman, 1994). While PEPC, the primary carboxylase of C4 plants, uses HCO3− produced by cytosolic βCA activity to form C4 acids, as part of the C4 CCM, in C3 plants an estimated 50% of the free aspartate pool is created by PEPC activity (Melzer and O'Leary, 1987). Arabidopsis double knockout mutants of the Atβca2 and Atβca4 genes, which code for cytosolic CAs, showed reduced growth rates and chlorosis of the younger leaves relative to WT plants when grown at 200 μL L−1 CO2. This phenotype was ameliorated when the plants were grown under high levels (1000 μL L−1) of CO2 (DiMario et al., 2016). The Atβca2ca4 double mutants also demonstrated reduced levels of aspartate, and a concomitant increase in glycine and serine levels (DiMario et al., 2016). The low CO2 growth phenotype and amino acid profile could be mitigated by complementation of the double mutant with the AtβCA2 gene (DiMario et al., 2016). The elevated amounts of glycine and serine in the double mutant were unanticipated and hints CA activity affecting other biochemical pathways.
Metabolism of Nitrogen-Fixing Root Nodules
The nitrogen-fixing root nodules of numerous legumes contain relatively high CA activity (Atkins, 1974), and transcripts encoding βCAs have been isolated from the nodules of several species (Coba de la Peña et al., 1997, Kavroulakis et al., 2000, Flemetakis et al., 2003). The location of these transcripts and the proteins they encode changes during maturation of the nodules, and this suggests that the role of the enzymes likely varies over the course of nodule development (Kavroulakis et al., 2000, Flemetakis et al., 2003). The functions put forward involve the provision of HCO3− for processes such as amino acid and lipid biosynthesis and gluconeogenesis in the early developmental stages, and the release of CO2 generated from bacteroid respiration to the rhizosphere in mature nodules (Figure 5; Kavroulakis et al., 2000, Flemetakis et al., 2003). However, these have not been supported experimentally. The presence of additional forms of CA in nitrogen-fixing nodules complicates the identification of the precise role(s) of the βCA enzymes (Gálvez et al., 2000, Flemetakis et al., 2003, Yahyaoui et al., 2004; supplemental data; Kalloniati et al., 2009, Tsikou et al., 2011, Tang et al., 2014).
Lipid Biosynthesis
Fatty acid synthesis is a primary metabolic pathway in which acetyl-CoA carboxylase (ACC) uses HCO3− to carboxylate acetyl-CoA to produce malonyl-CoA, the building block of fatty acid chains (Sasaki and Nagano, 2004). In plants, the production of acyl chains takes place in the chloroplast while their utilization occurs in essentially every cellular compartment (Ohlrogge and Jaworski, 1997). Since ACC requires HCO3−, and previous results have shown significant expression and activity of CA in cotton seedlings (Hoang et al., 1999, Hoang and Chapman, 2002a), Hoang and Chapman (2002b) examined the level of radiolabeled acetate incorporation into lipids in cottonseed embryos and tobacco cell suspensions. When embryos and suspension culture cells were incubated with [14C]acetate in the presence of the CA inhibitor, ethoxyzolamide, the rates of lipid synthesis were greatly decreased. Antisense tobacco lines with 5% of WT CA activity (Price et al., 1994) also showed lower levels of radiolabeled lipids (Hoang and Chapman, 2002b), which is consistent with the transgenic plants demonstrating reduced rates of Ci entering the chloroplast (Price et al., 1994). Hoang and Chapman (2002b) suggested that CA activity traps Ci within chloroplasts in the form of HCO3−, which is then used by ACC in fatty acid synthesis (Figure 5).
Conclusions and Perspectives
Plants have many genes encoding α-, β- and γ-type CAs, which are found in most tissues and many intracellular compartments. In addition, alternative splicing and multiple transcription start sites have been shown in a number of βCAs, often leading to different proteins targeted to different organelles. Programs developed to predict protein targeting should be used with caution, particularly when working with monocot CAs or CAs predicted from gene models. The number of CA genes is relatively similar in monocot and dicots, and in plants using C3 or C4 photosynthesis, or CAM. Evidence is building that, during the evolution of the C4 pathway, C3 genes coding for CA were co-opted through changes in cis-regulatory sequences, modification of posttranscriptional controls, and/or recruitment of different transcription factors. CAs, while clearly important in photosynthesis, are also required for other metabolic pathways as well as signaling and developmental pathways.
Funding
This work was supported by the University of Illinois as part of the Bill & Melinda Gates Foundation-funded Realizing Increased Photosynthetic Efficiency (RIPE) consortium, by NSF grant IOS-1146597 to J.V.M. and the Australian Research Council Discovery Projects DP130102243 and DP150101037 to M.L.
Author Contributions
R.J.D., H.C., A.M., M.L., and J.V.M. all contributed to the writing of the original draft and to the reviewing and editing of the manuscript.
Acknowledgments
No conflict of interest declared.
Published: September 16, 2016
Footnotes
Published by the Molecular Plant Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, SIBS, CAS.
Supplemental Information is available at Molecular Plant Online.
Supplemental Information
References
- Alber B.E., Ferry J.G. A carbonic anhydrase from the archaeon Methanosarcina thermophila. Proc. Natl. Acad. Sci. USA. 1994;91:6909–6913. doi: 10.1073/pnas.91.15.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins C.A. Occurrence and some properties of carbonic anhydrases from legume root nodules. Phytochemistry. 1974;13:93–98. [Google Scholar]
- Atkins C.A., Patterson B.D., Graham D. Plant carbonic anhydrases I. Distribution of types among species. Plant Physiol. 1972;50:214–217. doi: 10.1104/pp.50.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S., Smith-Unna R.D., Boursnell C.M., Kopriva S., Hibberd J.M. Transcript residency on ribosomes reveals a key role for the Arabidopsis thaliana bundle sheath in sulfur and glucosinolate metabolism. Plant J. 2014;78:659–673. doi: 10.1111/tpj.12502. [DOI] [PubMed] [Google Scholar]
- Banks J.A., Nishiyama T., Hasebe M., Bowman J.M., Gribskov M., dePamphilis C., Albert V.A., Aono N., Aoyama T., Ambrose B.A. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L., Schmutz J., Wang H., Percifield R., Hawkins J., Pontaroli A.C., Estep M., Feng L., Vaughn J.N., Grimwood J. Reference genome sequence of the model plant Setaria. Nat. Biotechnol. 2012;30:555–561. doi: 10.1038/nbt.2196. [DOI] [PubMed] [Google Scholar]
- Blanco-Rivero A., Shutova T., Román M.J., Villarejo A., Martinez F. Phosphorylation controls the localization and activation of the lumenal carbonic anhydrase in Chlamydomonas reinhardtii. PLoS One. 2012;7:e49063. doi: 10.1371/journal.pone.0049063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd R.A., Gandin A., Cousins A.B. Temperature responses of C4 photosynthesis: biochemical analysis of Rubisco, phosphoenolpyruvte carboxylase, and carbonic anhydrase in Setaria viridis. Plant Physiol. 2015;169:1850–1861. doi: 10.1104/pp.15.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., Kajala K., Wullenweber J., Sommer M., Gagneul D., Weber K.L., Carr K.M., Gowik U., Maß J., Lercher M.J. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 2011;155:142–156. doi: 10.1104/pp.110.159442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman R., Margaria R., Meldrum N., Roughton F. The CO2 catalyst present in blood. J. Physiol. 1932;75:3–4. [Google Scholar]
- Burnell J.N., Hatch M.D. Low bundle sheath carbonic anhydrase is apparently essential for effective C4 pathway operation. Plant Physiol. 1988;86:1252–1256. doi: 10.1104/pp.86.4.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnell J.N., Gibbs M.J., Mason J.G. Spinach chloroplastic carbonic anhydrase nucleotide sequence analysis of cDNA. Plant Physiol. 1990;92:37–40. doi: 10.1104/pp.92.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.M., Liu W.Y., Shih A.C.C., Shen M.N., Lu C.H., Lu M.Y.J., Yang H.W., Wang T.Y., Chen S.C.C., Chen S.M. Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 2012;160:165–177. doi: 10.1104/pp.112.203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton H., Saladié M., Rolland V., Sharwood R., Macfarlane T., Ludwig M. CoGe Comparative Genomics Research; 2016. Carbonic Anhydrase Evolution in Neurachne: Alternative Splicing and C4-associated N-terminal Changes.https://genomevolution.org [Google Scholar]
- Coba de la Peña T., Frugier F., McKhann H.I., Bauer P., Brown S., Kondorosi A., Crespi M. A carbonic anhydrase gene is induced in the nodule primordium and its cell-specific expression is controlled by the presence of Rhizobium during development. Plant J. 1997;11:407–420. doi: 10.1046/j.1365-313x.1997.11030407.x. [DOI] [PubMed] [Google Scholar]
- Collins R.M., Afzal M., Ward D.A., Prescott M.C., Sait S.M., Rees H.H., Tomsett A.B. Differential proteomic analysis of Arabidopsis thaliana genotypes exhibiting resistance or susceptibility to the insect herbivore, Plutella xylostella. PLoS One. 2010;5:e10103. doi: 10.1371/journal.pone.0010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins A.B., Badger M.R., von Caemmerer S. C4 photosynthetic isotope exchange in NAD-ME- and NADP-ME-type grasses. J. Exp. Bot. 2008;59:1695–1703. doi: 10.1093/jxb/ern001. [DOI] [PubMed] [Google Scholar]
- de Araujo C., Arefeen D., Tadesse Y., Long B.M., Price G.D., Rowlett R.S., Kimber M.S., Espie G.S. Identification and characterization of a carboxysomal γ-carbonic anhydrase from the cyanobacterium Nostoc sp. PCC 7120. Photosynth. Res. 2014;121:135–150. doi: 10.1007/s11120-014-0018-4. [DOI] [PubMed] [Google Scholar]
- DiMario R.J., Quebedeaux J.C., Longstreth D.J., Dassanayake M., Hartman M.M., Moroney J.V. The cytoplasmic carbonic anhydrases βCA2 and βCA4 are required for optimal plant growth at low CO2. Plant Physiol. 2016;171:280–293. doi: 10.1104/pp.15.01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q., Wang L., Yang X., Gong C., Zhang D. Populus endo-β-1,4-glucanases gene family: genomic organization, phylogenetic analysis, expression profiles and association mapping. Planta. 2015;241:1417–1434. doi: 10.1007/s00425-015-2271-y. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Engineer C.B., Ghassemian M., Anderson J.C., Peck S.C., Hu H., Schroeder J.I. Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature. 2014;513:246–250. doi: 10.1038/nature13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engineer C.B., Hashimoto-Sugimoto M., Negi J., Israelsson-Nordström M., Azoulay-Shemer T., Rappel W.-J., Iba K., Schroeder J.I. CO2 sensing and CO2 regulation of stomatal conductance: advances and open questions. Trends Plant Sci. 2016;21:16–30. doi: 10.1016/j.tplants.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson A.E., Jones T.A., Liljas A. Refined structure of human carbonic anhydrase II at 2.0 Å resolution. Proteins. 1988;4:274–282. doi: 10.1002/prot.340040406. [DOI] [PubMed] [Google Scholar]
- Espie G.S., Kimber M.S. Carboxysomes: cyanobacterial Rubisco comes in small packages. Photosynth. Res. 2011;109:7–20. doi: 10.1007/s11120-011-9656-y. [DOI] [PubMed] [Google Scholar]
- Everson R.G. Carbonic anhydrase and CO2 fixation in isolated chloroplasts. Phytochemistry. 1970;9:25–32. [Google Scholar]
- Everson R.G., Slack C.R. Distribution of carbonic anhydrase in relation to the C4 pathway of photosynthesis. Phytochemistry. 1968;7:581–584. [Google Scholar]
- Fabre N., Reiter I.M., Becuwe-Linka N., Genty B., Rumeau D. Characterization and expression analysis of genes encoding alpha and beta carbonic anhydrases in Arabidopsis. Plant Cell Environ. 2007;30:617–629. doi: 10.1111/j.1365-3040.2007.01651.x. [DOI] [PubMed] [Google Scholar]
- Fawcett T., Browse J., Volokita M., Bartlett S. .Spinach carbonic anhydrase primary structure deduced from the sequence of a cDNA clone. J. Biol. Chem. 1990;265:5414–5417. [PubMed] [Google Scholar]
- Ferreira F.J., Guo C., Coleman J.R. Reduction of plastid-localized carbonic anhydrase results in reduced Arabidopsis seedling survivorship. Plant Physiol. 2008;147:585–594. doi: 10.1104/pp.108.118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M., Brugière S., Salvi D., Seigneurin-Berny D., Moyet L., Ramus C., Miras S., Mellal M., Le Gall S., Kieffer-Jaquinod S. AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol. Cell. Proteomics. 2010;9:1063–1084. doi: 10.1074/mcp.M900325-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett J.P., Coleman J.R. Characterization and expression of two cDNAs encoding carbonic anhydrase in Arabidopsis thaliana. Plant Physiol. 1994;105:707–713. doi: 10.1104/pp.105.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemetakis E., Dimou M., Cotzur D., Aivalakis G., Efrose R.C., Kenoutis C., Udvardi M., Katinakis P. A Lotus japonicus β-type carbonic anhydrase gene expression pattern suggests distinct physiological roles during nodule development. Biochim. Biophys. Acta. 2003;1628:186–194. doi: 10.1016/s0167-4781(03)00142-8. [DOI] [PubMed] [Google Scholar]
- Friso G., Giacomelli L., Ytterberg A.J., Peltier J.-B., Rudella A., Sun Q., van Wijk K.J. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell. 2004;16:478–499. doi: 10.1105/tpc.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm S., Braun H.P., Peterhänsel C. Mitochondrial gamma carbonic anhydrases are required for complex I assembly and plant reproductive development. New Phytol. 2016;211:194–207. doi: 10.1111/nph.13886. [DOI] [PubMed] [Google Scholar]
- Fromm S., Göing J., Lorenz C., Peterhänsel C., Braun H.-P. Depletion of the “gamma-type carbonic anhydrase-like” subunits of complex I affects central mitochondrial metabolism in Arabidopsis thaliana. Biochim. Biophys. Acta. 2016;1857:60–71. doi: 10.1016/j.bbabio.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Fromm S., Senkler J., Eubel H., Peterhänsel C., Braun H.P. Life without complex I: proteome analyses of an Arabidopsis mutant lacking the mitochondrial NADH dehydrogenase complex. J. Exp. Bot. 2016;67:3079–3093. doi: 10.1093/jxb/erw165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez S., Hirsch A.M., Wycoff K.L., Hunt S., Layzell D.B., Kondorosi A., Crespi M. Oxygen regulation of a nodule-located carbonic anhydrase in alfalfa. Plant Physiol. 2000;124:1059–1068. doi: 10.1104/pp.124.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillon J., Yakir D. Influence of carbonic anhydrase activity in terrestrial vegetation on the 18O content of atmospheric CO2. Science. 2001;291:2584–2587. doi: 10.1126/science.1056374. [DOI] [PubMed] [Google Scholar]
- Guliev N.M., Babaev G.G., Bairamov Sh.M., Aliev D.A. Purification, properties, and localization of two carbonic anhydrases from Amaranthus cruentus leaves. Russ. J. Plant Physiol. 2003;50:213–219. [Google Scholar]
- Gutierrez M., Huber S.C., Ku M.S.B., Kanai R., Edwards G.E. Intracellular localization of carbon metabolism in mesophyll cells of C4 plants. In: Avron M., editor. Proceedings of the Third International Congress on Photosynthesis. Elsevier; Amsterdam: 1974. pp. 1219–1230. [Google Scholar]
- Håkansson K., Carlsson M., Svensson L.A., Liljas A. Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes. J. Mol. Biol. 1992;227:1192–1204. doi: 10.1016/0022-2836(92)90531-n. [DOI] [PubMed] [Google Scholar]
- Hanson D.T., Franklin L.A., Samuelsson G., Badger M.R. The Chlamydomonas reinhardtii cia3 mutant lacking a thylakoid lumen-localized carbonic anhydrase is limited by CO2 supply to rubisco and not photosystem II function in vivo. Plant Physiol. 2003;132:2267–2275. doi: 10.1104/pp.103.023481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Nakatsuma D., Ishida M., Matsuda Y. Regulation of the expression of intracellular β-carbonic anhydrase in response to CO2 and light in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 2005;139:1041–1050. doi: 10.1104/pp.105.065185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M.D., Burnell J.N. Carbonic anhydrase activity in leaves and its role in the first step of C4 photosynthesis. Plant Physiol. 1990;93:825–828. doi: 10.1104/pp.93.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimann L., Horst I., Perduns R., Dreesen B., Offermann S., Peterhansel C. A common histone modification code on C4 genes in maize and its conservation in sorghum and Setaria italica. Plant Physiol. 2013;162:456–469. doi: 10.1104/pp.113.216721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett-Emmett D., Tashian R.E. Functional diversity, conservation, and convergence in the evolution of the α-, β-, and γ-carbonic anhydrase gene families. Mol. Phylogenet. Evol. 1996;5:50–77. doi: 10.1006/mpev.1996.0006. [DOI] [PubMed] [Google Scholar]
- Hillier W., McConnell I., Badger M.R., Boussac A., Klimov V.V., Dismukes G.C., Wydrzynski T. Quantitative assessment of intrinsic carbonic anhydrase activity and the capacity for bicarbonate oxidation in photosystem II. Biochemistry. 2006;21:2094–2102. doi: 10.1021/bi051892o. [DOI] [PubMed] [Google Scholar]
- Hilvo M., Baranauskiene L., Salzano A.M., Scaloni A., Matulis D., Innocenti A., Scozzafava A., Monti S.M., Di Fiore A., De Simone G. Biochemical characterization of CA IX, one of the most active carbonic anhydrase isozymes. J. Biol. Chem. 2008;283:27799–27809. doi: 10.1074/jbc.M800938200. [DOI] [PubMed] [Google Scholar]
- Hoang C.V., Chapman K.D. Regulation of carbonic anhydrase gene expression in cotyledons of cotton (Gossypium hirsutum L.) seedlings during post-germinative growth. Plant Mol. Biol. 2002;49:449–458. doi: 10.1023/a:1015554024633. [DOI] [PubMed] [Google Scholar]
- Hoang C.V., Chapman K.D. Biochemical and molecular inhibition of plastidial carbonic anhydrase reduces the incorporation of acetate into lipids in cotton embryos and tobacco cell suspensions and leaves. Plant Physiol. 2002;128:1417–1427. doi: 10.1104/pp.010879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang C.V., Wessler H.G., Local A., Turley R.B., Benjamin R.C., Chapman K.D. Identification and expression of cotton (Gossypium hirsutum L.) plastidial carbonic anhydrase. Plant Cell Physiol. 1999;40:1262–1270. doi: 10.1093/oxfordjournals.pcp.a029514. [DOI] [PubMed] [Google Scholar]
- Höglund A., Dönnes P., Blum T., Adolph H.-W., Kohlbacher O. MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics. 2006;22:1158–1165. doi: 10.1093/bioinformatics/btl002. [DOI] [PubMed] [Google Scholar]
- Hu H., Boisson-Dernier A., Israelsson-Nordström M., Böhmer M., Xue S., Ries A., Godoski J., Kuhn J.M., Schroeder J.I. Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat. Cell Biol. 2010;12:87–93. doi: 10.1038/ncb2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H.H., Rappel W.J., Occhipinti R., Ries A., Bohmer M., You L., Xiao C.L., Engineer C.B., Boron W.F., Schroeder J.I. Distinct cellular locations of carbonic anhydrases mediate carbon dioxide control of stomatal movements. Plant Physiol. 2015;169:1168–1178. doi: 10.1104/pp.15.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida S., Muto S., Miyachi S. Structural analysis of periplasmic carbonic anhydrase 1 of Chlamydomonas reinhardtii. Eur. J. Biochem. 1993;214:9–16. doi: 10.1111/j.1432-1033.1993.tb17890.x. [DOI] [PubMed] [Google Scholar]
- Iverson T.M., Alber B.E., Kisker C., Ferry J.G., Rees D.C. A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila. Biochemistry. 2000;39:9222–9231. doi: 10.1021/bi000204s. [DOI] [PubMed] [Google Scholar]
- Jacobson B.S., Fong F., Heath R.L. Carbonic anhydrase of spinach: studies on its location, inhibition, and physiological function. Plant Physiol. 1975;55:468–474. doi: 10.1104/pp.55.3.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Aury J., Noel B., Policriti A., Clepet C., Casagrande A., Choisne N., Aubourg S., Vitulo N., Jubin C. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jiang C.Y., Tholen D., Xu J.M., Xin C.P., Zhang H., Zhu X.G., Zhao Y.X. Increased expression of mitochondria-localized carbonic anhydrase activity resulted in an increased biomass accumulation in Arabidopsis thaliana. J. Plant Biol. 2014;57:366–374. [Google Scholar]
- John C.R., Smith-Unna R.D., Woodfield H., Covshoff S., Hibberd J.M. Evolutionary convergence of cell-specific gene expression in independent lineages of C4 grasses. Plant Physiol. 2014;165:62–75. doi: 10.1104/pp.114.238667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H.W., Lim C.W., Lee S.C., Choi H.W., Hwang C.H., Hwang B.K. Distinct roles of the pepper hypersensitive induced reaction protein gene CaHIR1 in disease and osmotic stress, as determined by comparative transcriptome and proteome analyses. Planta. 2008;227:409–425. doi: 10.1007/s00425-007-0628-6. [DOI] [PubMed] [Google Scholar]
- Kajala K., Brown N.J., Williams B.P., Borrill P., Taylor L.E., Hibberd J.M. Multiple Arabidopsis genes primed for recruitment into C4 photosynthesis. Plant J. 2012;69:47–56. doi: 10.1111/j.1365-313X.2011.04769.x. [DOI] [PubMed] [Google Scholar]
- Kalloniati C., Tsikou D., Lampiri V., Fotelli M.N., Rennenberg H., Chatzipavlidis I., Fasseas C., Katinakis P., Flemetakis E. Characterization of a Mesorhizobium loti α-type carbonic anhydrase and its role in symbiotic nitrogen fixation. J. Bacteriol. 2009;191:2593–2600. doi: 10.1128/JB.01456-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson J., Clarke A.K., Chen Z.Y., Hugghins S.Y., Park Y.I., Husic H.D., Moroney J.V., Samuelsson G. A novel α-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J. 1998;17:1208–1216. doi: 10.1093/emboj/17.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavroulakis N., Flemetakis E., Aivalakis G., Katinakis P. Carbon metabolism in developing soybean root nodules: the role of carbonic anhydrase. Mol. Plant Microbe. 2000;13:14–22. doi: 10.1094/MPMI.2000.13.1.14. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Bracey M.H., Bartlett S.G. Nucleotide-sequence of a gene encoding carbonic-anhydrase in Arabidopsis thaliana. Plant Physiol. 1994;105:449. doi: 10.1104/pp.105.1.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimber M.S., Pai E.F. The active site architecture of Pisum sativum beta-carbonic anhydrase is a mirror image of that of alpha-carbonic anhydrases. EMBO J. 2000;19:1407–1418. doi: 10.1093/emboj/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisker C., Schindelin H., Alber B.E., Ferry J.G., Rees D.C. A left-handed beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J. 1996;15:2323–2330. [PMC free article] [PubMed] [Google Scholar]
- Klodmann J., Sunderhaus S., Nimtz M., Jänsch L., Braun H.P. Internal architecture of mitochondrial complex I from Arabidopsis thaliana. Plant Cell. 2010;22:797–810. doi: 10.1105/tpc.109.073726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchik M., Bernstein N. Effects of salinity on the transcriptome of growing maize leaf cells point at cell-age specificity in the involvement of the antioxidative response in cell growth restriction. BMC Genomics. 2013;14:24. doi: 10.1186/1471-2164-14-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku S.B., Edwards G.E. Photosynthesis in mesophyll protoplasts and bundle sheath-cells of various types of C4 plants: 4. Enzymes of respiratory metabolism and energy utilizing enzymes of photosynthetic pathways. Z. Pflanzenphysiol. 1975;77:16–32. [Google Scholar]
- Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2011;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D., Eisinger J., Reski R., Rensing S.A. Representation and high-quality annotation of the Physcomitrella patens transcriptome demonstrates a high proportion of proteins involved in metabolism in mosses. Plant Biol. 2005;7:238–250. doi: 10.1055/s-2005-837578. [DOI] [PubMed] [Google Scholar]
- Li P.H., Ponnala L., Gandotra N., Wang L., Si Y.Q., Tausta S.L., Kebrom T.H., Provart N., Patel R., Myers C.R. The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 2010;42:1060–1067. doi: 10.1038/ng.703. [DOI] [PubMed] [Google Scholar]
- Liljas A., Kannan K.K., Bergstén P.C., Waara I., Fridborg K., Strandberg B., Carlbom U., Jrup L., Lvgren S., Petef M. Crystal-structure of human carbonic anhydrase-C. Nat. New Biol. 1972;235:131–137. doi: 10.1038/newbio235131a0. [DOI] [PubMed] [Google Scholar]
- Long B.M., Badger M.R., Whitney S.M., Price G.D. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple rubisco complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 2007;282:29323–29335. doi: 10.1074/jbc.M703896200. [DOI] [PubMed] [Google Scholar]
- Ludwig M. Evolution of carbonic anhydrase in C4 plants. Curr. Opin. Plant Biol. 2016;31:16–22. doi: 10.1016/j.pbi.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Ludwig M., von Caemmerer S., Price G.D., Badger M.R., Furbank R.T. Expression of tobacco carbonic anhydrase in the C4 dicot Flaveria bidentis leads to increased leakiness of the bundle sheath and a defective CO2-concentrating mechanism. Plant Physiol. 1998;117:1071–1081. doi: 10.1104/pp.117.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeau N., Arnoldo M., Coleman J.R. Modification of carbonic anhydrase activity by antisense and over-expression constructs in transgenic tobacco. Plant Mol. Biol. 1994;25:377–385. doi: 10.1007/BF00043867. [DOI] [PubMed] [Google Scholar]
- Makita Y., Shimada S., Kawashima M., Kondou-Kuriyama T., Tetsuro Toyoda T., Matsui M. MOROKOSHI: transcriptome database in Sorghum bicolor. Plant Cell Physiol. 2015;56:e6. doi: 10.1093/pcp/pcu187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangani S., Håkansson K. Crystallographic studies of the binding of protonated and unprotonated inhibitors to carbonic anhydrase using hydrogen sulphide and nitrate anions. Eur. J. Biochem. 1992;210:867–871. doi: 10.1111/j.1432-1033.1992.tb17490.x. [DOI] [PubMed] [Google Scholar]
- McConnell I.L., Badger M.R., Wydrzynski T., Hillier W. A quantitative assessment of the carbonic anhydrase activity in photosystem II. Biochim. Biophys. Acta. 2007;1767:639–647. doi: 10.1016/j.bbabio.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Meldrum N., Roughton F. Some properties of carbonic anhydrase, the CO2 enzyme present in blood. J. Physiol. 1932;75:15. [Google Scholar]
- Melzer E., O'Leary M.H. Anapleurotic CO2 fixation by phosphoenolpyruvate carboxylase in C3 plants. Plant Physiol. 1987;84:58–60. doi: 10.1104/pp.84.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R., VanBuren R., Wai C.M., Tang H., Schatz M.C., Bowers J.E., Lyons E., Wang M., Chen J., Biggers E. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015;47:1435–1442. doi: 10.1038/ng.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra M., Mason C.B., Xiao Y., Ynalvez R.A., Lato S.M., Moroney J.V. The carbonic anhydrase gene families of Chlamydomonas reinhardtii. Can. J. Bot. 2005;83:780–795. [Google Scholar]
- Moroney J.V., Ynalvez R.A. Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Euk. Cell. 2007;6:1251–1259. doi: 10.1128/EC.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney J.V., Bartlett S.G., Samuelsson G. Carbonic anhydrases in plants and algae. Plant Cell Environ. 2001;24:141–153. [Google Scholar]
- Moroney J.V., Ma Y., Frey W.D., Fusilier K., Pham T.T., Simms T.A., DiMario R.J., Yang J., Mukherjee B. The carbonic anhydrase isoforms of Chlamydomonas reinhardtii: expression, intracellular location and physiological roles. Photosynth. Res. 2011;109:133–149. doi: 10.1007/s11120-011-9635-3. [DOI] [PubMed] [Google Scholar]
- Oh D.-H., Hong H., Lee S.Y., Yun D.-J., Bohnert H.J., Dassanayake M. Genome structures and transcriptomes signify niche adaptation for the multiple-ion-tolerant extremophyte Schrenkiella parvula. Plant Physiol. 2014;164:2123–2138. doi: 10.1104/pp.113.233551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J.B., Jaworski J.G. Regulation of fatty acid synthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997;48:109–136. doi: 10.1146/annurev.arplant.48.1.109. [DOI] [PubMed] [Google Scholar]
- Okabe K., Yang S.-Y., Tsuzuki M., Miyachi S. Carbonic anhydrase: its content in spinach leaves and its taxonomic diversity studied with anti-spinach leaf carbonic anhydrase antibody. Plant Sci. Lett. 1984;33:145–153. [Google Scholar]
- Ouyang S., Zhu W., Hamilton J., Lin H., Campbell M.L.K., Childs K., Thibaud-Nissen F., Malek R.L., Lee Y., Zheng L. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Res. 2007;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi G., Perales M., Fornasari M.S., Colaneri A., Gonzalez-Shain N., Gomez-Casati D., Zimmermann S., Brennicke A., Araya A., Ferry J.G. Gamma carbonic anhydrases in plant mitochondria. Plant Mol. Biol. 2004;55:193–207. doi: 10.1007/s11103-004-0149-7. [DOI] [PubMed] [Google Scholar]
- Park I., Karlsson J., Rojdestvenski I., Pronina N., Klimov V., Oquist G., Samuelsson G. Role of a novel photosystem II-associated carbonic anhydrase in photosynthetic carbon assimilation in Chlamydomonas reinhardtii. FEBS Lett. 1999;444:102–105. doi: 10.1016/s0014-5793(99)00037-x. [DOI] [PubMed] [Google Scholar]
- Peltier J.-B., Cai Y., Sun Q., Zabrouskov V., Giacomelli L., Rudella A., Ytterberg A.J., Rutschow H., van Wijk K.J. The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics. 2006;5:114–133. doi: 10.1074/mcp.M500180-MCP200. [DOI] [PubMed] [Google Scholar]
- Peña K.L., Castel S.E., de Araujo C., Espie G.S., Kimber M.S. Structural basis of the oxidative activation of the carboxysomal γ-carbonic anhydrase, CcmM. Proc. Natl. Acad. Sci. USA. 2010;107:2455–2460. doi: 10.1073/pnas.0910866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M., Parisi G., Fornasari M.S., Colaneri A., Villarreal F., González-Schain N., Echave J., Gómez-Casati D., Braun H.P., Araya A. Gamma carbonic anhydrase like complex interact with plant mitochondrial complex I. Plant Mol. Biol. 2004;56:947–957. doi: 10.1007/s11103-004-6324-z. [DOI] [PubMed] [Google Scholar]
- Perales M., Eubel H., Heinemeyer J., Colaneri A., Zabaleta E., Braun H.P. Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I+III2 levels and alters mitochondrial physiology in Arabidopsis. J. Mol. Biol. 2005;350:263–277. doi: 10.1016/j.jmb.2005.04.062. [DOI] [PubMed] [Google Scholar]
- Phytozome v11.0 https://phytozome.jgi.doe.gov.
- Poincelot R.P. Intracellular distribution of carbonic anhydrase in spinach leaves. Biochim. Biophys. Acta. 1972;258:637–642. doi: 10.1016/0005-2744(72)90255-0. [DOI] [PubMed] [Google Scholar]
- Price G.D., Badger M.R. Isolation and characterization of high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1989;91:514–525. doi: 10.1104/pp.91.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price G.D., Coleman J.R., Badger M.R. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1992;100:784–793. doi: 10.1104/pp.100.2.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price G., Howitt S., Harrison K., Badger M. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J. Bacteriol. 1993;175:2871–2879. doi: 10.1128/jb.175.10.2871-2879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price G.D., von Caemmerer S., Evans J.R., Yu J.W., Lloyd J., Oja V., Kell P., Harrison K., Gallagher A., Badger M.R. Specific reduction of chloroplast carbonic anhydrase activity by antisense RNA in transgenic tobacco plants has a minor effect on photosynthesis. Planta. 1994;193:331–340. [Google Scholar]
- Rao X.L., Lu N., Li G.F., Nakashima J., Tang Y.H., Dixon R.A. Comparative cell-specific transcriptomics reveals differentiation of C4 photosynthesis pathways in switchgrass and other C4 lineages. J. Exp. Bot. 2016;67:1649–1662. doi: 10.1093/jxb/erv553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven J. CO2-concentrating mechanisms: a direct role for thylakoid lumen acidification? Plant Cell Environ. 1997;20:147–154. [Google Scholar]
- Raven J., Newman J. Requirement for carbonic anhydrase activity in processes other than photosynthetic inorganic carbon assimilation. Plant Cell Environ. 1994;17:123–130. [Google Scholar]
- Reed M.L., Graham D. Carbonic anhydrase in plants: distribution, properties and possible physiological roles. In: Reinhold L., Harborne J.B., Swain T., editors. Progress in Phytochemistry. Pergammon Press; Oxford, U.K.: 1981. pp. 47–94. [Google Scholar]
- Rensing S.A., Fritzowsky D., Lang D., Reski R. Protein encoding genes in an ancient plant: analysis of codon usage, retained genes and splice sites in a moss, Physcomitrella patens. BMC Genomics. 2005;6:43. doi: 10.1186/1471-2164-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo S., Myers K.L., del Pozo O., Martin G.B., Hart A.L., Buell C.R., Fry W.E., Smart C.D. Gene profiling of a compatible interaction between Phytophthora infestans and Solanum tuberosum suggests a role for carbonic anhydrase. Mol. Plant Microbe Interact. 2005;18:913–922. doi: 10.1094/MPMI-18-0913. [DOI] [PubMed] [Google Scholar]
- Roeske C.A., Ogren W.L. Nucleotide sequence of pea cDNA encoding chloroplast carbonic anhydrase. Nucleic Acids Res. 1990;18:3413. doi: 10.1093/nar/18.11.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett R.S. Structure and catalytic mechanism of the β-carbonic anhydrases. Biochim. Biophys. Acta. 2010;1804:362–373. doi: 10.1016/j.bbapap.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C.T., Schreiber S.L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Nagano Y. Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 2004;68:1175–1184. doi: 10.1271/bbb.68.1175. [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Shpak E.D. Diverse roles of ERECTA family genes in plant development. J. Integr. Plant Biol. 2013;55:1238–1250. doi: 10.1111/jipb.12108. [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker D.H., Navarre D.A., Clark D., del Pozo O., Martin G.B., Klessig D.F. The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc. Natl. Acad. Sci. USA. 2002;99:11640–11645. doi: 10.1073/pnas.182427699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I., Peeters N., Legeai F., Lurin C. Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- Stemler A. The case for chloroplast thylakoid carbonic anhydrase. Physiol. Plant. 1997;99:348–353. [Google Scholar]
- Studer A.J., Gandin A., Kolbe A.R., Wang L., Cousins A.B., Brutnell T.P. A limited role for carbonic anhydrase in C4 photosynthesis is revealed by a ca1ca2 double mutant in maize. Plant Physiol. 2014;165:608–617. doi: 10.1104/pp.114.237602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderhaus S., Dudkina N.V., Jänsch L., Klodmann J., Heinemeyer J., Perales M., Zabaleta E., Boekema E.J., Braun H.P. Carbonic anhydrase subunits form a matrix-exposed domain attached to the membrane arm of mitochondrial complex I in plants. J. Biol. Chem. 2006;281:6482–6488. doi: 10.1074/jbc.M511542200. [DOI] [PubMed] [Google Scholar]
- Swader J.A., Jacobson B.S. Acetozolamide inhibition of photosystem II in isolated spinach chloroplasts. Plant Physiol. 1972;11:65–70. [Google Scholar]
- Tang H., Krishnakumar V., Bidwell S., Rosen B., Chan A., Zhou S., Gentzbittel L., Childs K.L., Yandell M., Gundlach H. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics. 2014;15:312. doi: 10.1186/1471-2164-15-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz S.K., Tetu S.G., Vella N.G.F., Ludwig M. Loss of the transit peptide and an increase in gene expression of an ancestral chloroplastic carbonic anhydrase were instrumental in the evolution of the cytosolic C4 carbonic anhydrase in Flaveria. Plant Physiol. 2009;150:1515–1529. doi: 10.1104/pp.109.137513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetu S.G., Tanz S.K., Vella N., Burnell J.N., Ludwig M. The Flaveria bidentis β-carbonic anhydrase gene family encodes cytosolic and chloroplastic isoforms demonstrating distinct organ-specific expression patterns. Plant Physiol. 2007;144:1316–1327. doi: 10.1104/pp.107.098152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triolo L., Bagnara D., Anselmi L., Bassanelli C. Carbonic anhydrase activity and localization in some plant species. Physiol. Plant. 1974;31:86–89. [Google Scholar]
- Tsikou D., Stedel C., Kouri D.C., Udvardi M.K., Wang T.L., Katinakis P., Labrou N.E., Flemetakis E. Characterization of two novel nodule-enhanced α-type carbonic anhydrases from Lotus japonicus. Biochim. Biophys. Acta. 2011;1814:496–504. doi: 10.1016/j.bbapap.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Tuskan G.A., Difazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., Putnam N., Ralph S., Rombauts S., Salamov A. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Villarejo A., Shutova T., Moskvin O., Forssén M., Klimov V.V., Samuelsson G. A photosystem II-associated carbonic anhydrase regulates the efficiency of photosynthetic oxygen evolution. EMBO J. 2002;21:1930–1938. doi: 10.1093/emboj/21.8.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A., Buren S., Larsson S., Dejardin A., Monne M., Rudhe C., Karlsson J., Jansson S., Lerouge P., Rolland N. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 2005;7:1224–1231. doi: 10.1038/ncb1330. [DOI] [PubMed] [Google Scholar]
- Vogel J.P., Garvin D.F., Mockler T.C., Schmutz J., Rokhsar D., Bevan M.W., Barry K., Lucas S., Harmon-Smith M., Lail K. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- von Caemmerer S., Quinn V., Hancock N.C., Price G.D., Furbank R.T., Ludwig M. Carbonic anhydrase and C4 photosynthesis: a transgenic analysis. Plant Cell Environ. 2004;27:697–703. [Google Scholar]
- Wang Y.-Q., Feechan A., Yun B.-W., Shafiei R., Hofmann A., Taylor P., Xue P., Yang F.-Q., Xie Z.-S., Pallas J.A. S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 2009;284:2131–2137. doi: 10.1074/jbc.M806782200. [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang Q., Liu F.C., Xie W.F., Wang G.D., Wang J., Gao Q.H., Duan K. Family-wide expression characterization of Arabidopsis beta-carbonic anhydrase genes using qRT-PCR and Promoter::GUS fusions. Biochimie. 2014;97:219–227. doi: 10.1016/j.biochi.2013.10.020. [DOI] [PubMed] [Google Scholar]
- Wang C., Hu H., Qin X., Zeise B., Xu D., Rappel W.-J., Boron W.F., Schroeder J.I. Reconstitution of CO2 regulation of SLAC1 anion channel and function of CO2-permeable PIP2:1 aquaporin as CARBONIC ANHYDRASE4 interactor. Plant Cell. 2016;28:568–582. doi: 10.1105/tpc.15.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werdan K., Heldt H.W. Accumulation of bicarbonate in intact chloroplasts following a pH gradient. Biochim. Biophys. Acta. 1972;283:430–441. doi: 10.1016/0005-2728(72)90260-5. [DOI] [PubMed] [Google Scholar]
- Williams B.P., Aubry S., Hibberd J.M. Molecular evolution of genes recruited into C4 photosynthesis. Trends Plant Sci. 2012;17:213–220. doi: 10.1016/j.tplants.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Williams B.P., Burgess S.J., Reyna-Llorens I., Knerova J., Aubry S., Stanley S., Hibberd J.M. An untranslated cis-element regulates the accumulation of multiple C4 enzymes in Gynandropsis gynandra mesophyll cells. Plant Cell. 2016;28:454–465. doi: 10.1105/tpc.15.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar V., Nahal H., Ammar R., Wilson G., Provart N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahyaoui F.E., Küster H., Amor B.B., Hohnjec N., Pühler A., Becker A., Gouzy J., Vernié T., Gough C., Niebel A. Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 2004;136:3160–3176. doi: 10.1104/pp.104.043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z., Zhong R. Molecular control of wood formation in trees. J. Exp. Bot. 2015;66:4119–4131. doi: 10.1093/jxb/erv081. [DOI] [PubMed] [Google Scholar]
- Ynalvez R.A., Xiao Y., Ward A.S., Cunnusamy K., Moroney J.V. Isolation and characterization of two closely related β-carbonic anhydrases from Chlamydomonas reinhardtii. Physiol. Plant. 2008;133:15–26. doi: 10.1111/j.1399-3054.2007.01043.x. [DOI] [PubMed] [Google Scholar]
- Young N.D., Debellé F., Oldroyd G.E.D., Geurts R., Cannon S.B., Udvardi M.K., Benedito V.A., Mayer K.F.X., Gouzy J., Schoof H. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Zhang X., Guan Q., Takano T., Liu S. Expression of a carbonic anhydrase gene is induced by environmental stresses in rice (Oryza sativa L.) Biotechnol. Lett. 2007;29:89–94. doi: 10.1007/s10529-006-9199-z. [DOI] [PubMed] [Google Scholar]
- Zabaleta E., Martin M.V., Braun H.-P. A basal carbon concentrating mechanism in plants? Plant Sci. 2012;187:97–104. doi: 10.1016/j.plantsci.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Zimmer A.D., Lang D., Buchta K., Rombauts S., Nishiyama T., Hasebe M., Van de Peer Y., Rensing S.A., Reski R. Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genomics. 2013;14:498. doi: 10.1186/1471-2164-14-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.