Abstract
Drosophila melanogaster is a key experimental system in the study of fat regulation. Numerous techniques currently exist to measure levels of stored fat in Drosophila, but most are expensive and/or laborious and have clear limitations. Here, we present a method to quickly and cheaply determine organismal fat levels in L3 Drosophila larvae. The technique relies on the differences in density between fat and lean tissues and allows for rapid detection of fat and lean phenotypes. We have verified the accuracy of this method by comparison to body fat percentage as determined by neutral lipid extraction and gas chromatography coupled with mass spectrometry (GCMS). We furthermore outline detailed protocols for the collection and synchronization of larvae as well as relevant experimental recipes. The technique presented below overcomes the major shortcomings in the most widely used lipid quantitation methods and provides a powerful way to quickly and sensitively screen L3 larvae for fat regulation phenotypes while maintaining the integrity of the larvae. This assay has wide applications for the study of metabolism and fat regulation using Drosophila.
Keywords: Cellular Biology, Issue 117, lipid quantitation, density, Drosophila melanogaster, larvae, fat regulation, screening technique, buoyancy
Introduction
Drosophila melanogaster has been used for over a century in the study of genetics and other basic biological questions. In the last few decades, it has become clear that Drosophila is a powerful tool in the study of many human diseases. As 70 - 80% of genes associated with human diseases have an identified fly ortholog1-4, Drosophila provides a simplified yet translatable system in which to study complex diseases. Metabolism in particular has benefited from such study. Not only are the genetics of metabolism well conserved between flies and humans, but the relevant organs and cell types are also very similar2,5. For example, the fat body of the fly stores both triacylglycerides (TAG) and glycogen, functions analogous to those performed in mammalian liver and white adipose tissue6. Using Drosophila as a model for human obesity has vastly improved our understanding of lipid metabolism and the genetics of obesity7. The larval stage of development is particularly useful for studying the segregation of nutrients to storage or utilization as it is dedicated to feeding and the storage of energy to be used during pupariation.
Currently, there are many different quantitative methods of determining fat storage levels in Drosophila. The most widely used method is the coupled colorimetric assay (CCA)8,9. CCA was developed for determining TAG levels in human serum and operates on the premise that glycerol liberated from the backbone of triglycerides will undergo several reactions, ultimately resulting in a redox-coupled reaction generating a colored product. Absorbance of specific wavelengths of light is then measured to determine the initial amount of glycerol present. However, glycerol can also be liberated from mono- and diacylglycerides in addition to TAG and therefore may not be an accurate measure of stored body fat9. Furthermore, eye pigment of crushed adult flies can interfere with some absorbance readings and complicate results9,10. Therefore, CCA must be accompanied and validated by thin layer chromatography (TLC), which allows for the separation of most lipid families that can be quantitated by densitometry10,11. However, some lipid classes like sterols cannot be analyzed and must be quantified a different way12. Mass spectrometry (MS) is an accurate way to quantitate all classes of major cellular lipids12,13. However, the lipid extraction procedures necessary to analyze by MS are both time consuming (most taking nearly a full day) and costly. Here we present an alternative method to quickly and cheaply determine organismal fat levels in the L3 larvae of Drosophila melanogaster.
The method presented below exploits the difference in density between fat tissue and lean tissue. Mammalian fat tissue has a density of approximately 0.9 g/ml14 while skeletal muscle as a density of 1.06 g/ml15. This difference means that animals with higher stores of fat will have lower density than animals with lower stores of fat, which will allow them to float better in a solution of fixed density. This property allows for extremely quick screening of a large number of animals while being both inexpensive and non-invasive. Buoyancy-based analysis has been used both to confirm the phenotypes of altering known regulators of fat levels as well as to identify new genetic and neurological regulators of obesity16,17.
Protocol
1. Collect Eggs from Flies with Genotypes of Interest
NOTE: Ideal crosses consist of 150 virgin flies and at least 75 males. Stock collections should consist of at least 200 flies.
- Prepare grape plates.
- Add 37.5 g agar to 1.5 L distilled water in a 4 L flask.
- Autoclave water/agar mix for 50 min at 121 ºC (250 ºF).
- While water/agar mix is autoclaving, add 50 g sucrose to 500 ml grape juice and stir with a stir bar on a heated stir plate until sucrose is dissolved.
- Turn off the heat and continue stirring to cool the grape juice/sucrose mixture to below 70 ºC. Test the internal temperature with an alcohol thermometer.
- Add 3 g Drosophila anti-fungal agent (e.g., Tegosept) to warm grape juice/sucrose mixture and stir to dissolve.
- When water/agar mixture is out of the autoclave, allow the flask to cool to the touch by swirling occasionally.
- Combine water/agar mixture and grape juice/sucrose mixture and stir until well mixed.
- Use a serological pipette to add 8 - 10 ml of the grape mixture to the lid of a small petri plate (35 x 10 mm style). Allow the mixture to crown higher than the height of the lid walls to form a convex shape.
- Allow grape plates to cool for 2 hr at room temperature and then store in an airtight container at 4 ºC.
- Prepare yeast paste.
- Add 6 ml phosphate buffer solution (PBS) to 4 g live active dried yeast in a 50 ml conical tube and mix with a spatula until smooth.
- Store yeast paste at 4 ºC.
Add a small dollop (approximately 1/8 tsp) of yeast paste to the middle of a grape plate. Smooth down any rough edges with a spatula.
Place grape plates in an incubator until they have reached ambient temperature.
Transfer flies of interest into an egg collection chamber and place grape plate with yeast paste facing inwards on top. Tape grape plate to the egg collection chamber to avoid letting the plate fall out. Make sure to write important genotype and date information on the bottom of the grape plate.
Upend the egg collection chamber to allow the flies to lay eggs on the grape plate.
Place a box or cover over the egg collection chambers and place them in an incubator at 25 ºC.
Allow flies to lay eggs for 4 - 6 hr.
End collection by taking the grape plate out of the egg collection chamber and transferring the flies back into their bottle of food. Use a spatula to wipe off any excess yeast paste from the grape plate. Store grape plate in a larger petri dish in a humidified incubator at 25 ºC for ~ 24 hr.
2. Transfer Larvae to Experimental Food
- Prepare experimental food. NOTE: This food recipe is based on the Bloomington stock center recipe18. Important changes include increased amount of yeast to optimize the nutritional value of the food as assessed by timing of wandering stage (between 112 - 120 hr after egg collection at 25 ºC). While we have found this recipe ideal for the health and development of our experimental larvae, any food recipe is acceptable given calibration with positive and negative controls, examples of which are listed in step 3.10.
- Measure 1,600 ml distilled water.
- Mix 134 g cornmeal, 10.6 g Drosophila Agar Type II, and some of the water (~ 500 ml) in a 1 L beaker. Blend for 2 min with a motorized hand mixer.
- Mix 70 g live active dry yeast, 18.4 g soy flour, 84.8 g malt extract, and some of the water (~ 500 ml) in another 1 L beaker. Blend for 2 min with a motorized hand mixer.
- Swirl each beaker and pour into a 4 L flask. Use remaining water to rinse out the beakers and add into the flask.
- Measure 140 ml light corn syrup and add to the flask, mix by swirling.
- Autoclave for 50 min at 121 ºC (250 ºF) in the liquid setting.
- After autoclaving, pour food into a 4 L beaker and allow to cool. Help the cooling process by blending with a motorized hand mixer.
- Once the flask has cooled enough to touch, add 8.8 ml propionic acid and 16.8 ml Drosophila anti-fungal agent/ethanol solution. Mix well. (Make Drosophila anti-fungal agent solution by adding 380 g Drosophila anti-fungal agent to 1 L 200 proof ethanol and stirring on a warm plate until dissolved, making sure the solution does not exceed 70 ºC.)
- Pour food into vials until the food is approximately 1 inch deep and allow to cool at RT for 2 - 3 hr. Plug vials and store in an incubator at 25 ºC. Use within a week.
Plunge a spatula several times into the top layer of the vial of food to score it. This will help the larvae to burrow and feed.
22 - 24 hr after egg collection has ended, use a small paintbrush to collect 50 first instar (L1) larvae of the same approximate size. Place larvae in the experimental food. Clean the paintbrush on a laboratory wipe and ensure that there are no remaining larvae on the paintbrush before continuing to another genotype.
Place vial of larvae in an incubator at 25 ºC and allow to develop.
3. Determine Fat Levels in Larvae
- Prepare solutions.
- Prepare 1 L 10x PBS stock.
- Add 80 g NaCl, 2 g KCl, 14.4 g Na2HPO4, and 2.4 g KH2PO4 to 800 ml water in a 2 L beaker containing a stir bar. Stir with no heat until all salts are incorporated. Bring pH to 7.4. Increase volume to 1 L with water.
- Prepare 2 L 1x PBS. Add 200 ml 10x PBS stock solution to 1,800 ml water.
- Use this stock of 1x PBS to make 20% w/v sucrose.
Remove the vial of larvae from the incubator once there are 20 - 40 larvae wandering up the sides of the vial (generally on the fifth day after egg collections). Record the date and time of the assay as well as the number of wandering larvae.
Prepare experimental solution by adding 11.5 ml PBS and 9 ml 20% sucrose to a 50 ml conical tube.
Add 20% sucrose to the vial until it is 0.5 - 1 inch from the top of the vial.
Use a spatula to gently stir up the top layer of food to free larvae that are still burrowing in the food. All larvae will rise to the surface of the sucrose solution.
Use forceps to gently transfer the larvae into the initial sucrose concentration from step 3.3.
Use a spatula to gently stir the larvae in this solution.
Cap the conical tube and upend several times to thoroughly mix the solution.
Uncap the conical tube and swirl to create a gentle vortex.
Allow the larvae to settle, either floating to the top of the solution or sinking to the bottom. Wait 2 - 5 min for larvae to settle. NOTE: If there are still larvae that are not definitely either floating or sinking, pick a line to use as a cutoff point to label the larvae as either a floater or a sinker. Apply this same criterion to all genotypes. We use the angle at the bottom of the conical tube as our boundary. adp or sir217 mutant larvae may be used as a positive control and lsd219 mutants may be used as a negative control for calibration of the assay.
Record the number of floating larvae and the specific concentration tested.
Add 1 ml 20% sucrose to the experimental solution to increase its density.
Repeat steps 3.7 to 3.10. Record the number of floating larvae and this new concentration.
Continue steps 3.12 and 3.13 until at least 95% of the larvae are floating.
Use forceps to collect all larvae into a dissection dish filled with PBS.
Observe larvae under a microscope and note if there are any small larvae, pre-pupae, pupae, or any other differences between genotypes. NOTE: Ideally, all larvae should appear homogeneous and at the same developmental stage. We have only observed consistent fat measurements under these conditions.
Record the total number of larvae.
Using forceps, transfer 10 larvae to a laboratory wipe to dry. Label a microcentrifuge tube and place 10 larvae in the tube.
Flash freeze the larvae in liquid nitrogen. Store the tube at -20 ºC.
Representative Results
Figure 1 represents an example of how the buoyancy-based assay can detect both higher and lower fat stores in genetically manipulated larvae. Figure 1A shows that excitation or silencing of a certain subset of neurons (E347) in the larval brain induces lower and higher fat stores respectively. The same genetic background control was used in both cases. Figure 1B provides an example of how this phenotype obtained by the density assay is corroborated by neutral lipid extraction and quantification by GCMS.
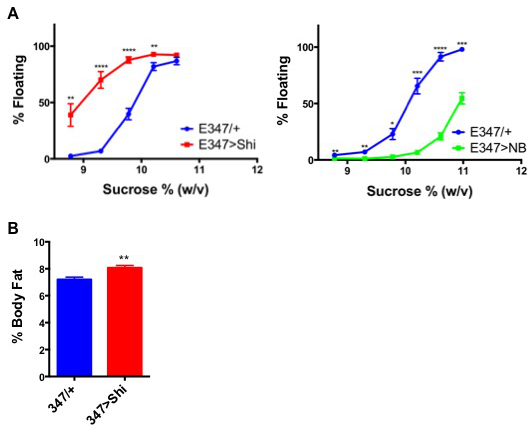 Figure 1.Neuronal Function in Specific Regions of the Larval Brain is Necessary and Sufficient for Body Fat Regulation. Neuronal activity in the indicated E347-GAL4 line was silenced by overexpression of ShiDN, or activated by overexpression of NB, as indicated in the panels, and effects on body fat were assessed. (A) Percent of floating larvae at equilibrium in different density solutions. Fifty larvae were examined per biological replicate, n= 7 - 9 biological replicates per genotype. (B) Percent body fat (total neutral lipids including triacylglycerides and phospholipids divided by body weight) as measured by GCMS17,20 (n= 8). Blue lines or bars represent GAL4-only (no UAS) animals crossed to w1118 to control for effects of genetic background. P values represent results from two-tailed t tests. **** P < 0.0001, *** P = 0.0001 to 0.001, ** P = 0.001 to 0.01, * P = 0.01 to 0.05. Error bars show SEM. This figure has been modified16. Please click here to view a larger version of this figure.
Figure 1.Neuronal Function in Specific Regions of the Larval Brain is Necessary and Sufficient for Body Fat Regulation. Neuronal activity in the indicated E347-GAL4 line was silenced by overexpression of ShiDN, or activated by overexpression of NB, as indicated in the panels, and effects on body fat were assessed. (A) Percent of floating larvae at equilibrium in different density solutions. Fifty larvae were examined per biological replicate, n= 7 - 9 biological replicates per genotype. (B) Percent body fat (total neutral lipids including triacylglycerides and phospholipids divided by body weight) as measured by GCMS17,20 (n= 8). Blue lines or bars represent GAL4-only (no UAS) animals crossed to w1118 to control for effects of genetic background. P values represent results from two-tailed t tests. **** P < 0.0001, *** P = 0.0001 to 0.001, ** P = 0.001 to 0.01, * P = 0.01 to 0.05. Error bars show SEM. This figure has been modified16. Please click here to view a larger version of this figure.
Discussion
There are a multitude of techniques that have been developed to measure lipid levels8-10. However, each of these methods comes with some major drawbacks that are addressed by the buoyancy-based assay outlined above. First, this assay is extremely quick. Testing a full concentration gradient takes no more than 30 - 60 min. This is a huge improvement on most of the techniques currently in use. For example, lipid quantitation by MS takes 7 - 9 hr to isolate the lipids and another several hours to analyze them. This is a strong deterrent when performing a screen on a large number of animals. By measuring a quick buoyancy phenotype, one can quickly focus on the genotypes that have the phenotype of interest. Second, the density assay requires only common reagents such as salts (for PBS) and sucrose. This makes it much easier and cheaper to screen a large number of larvae or quickly test an interesting genotype. Finally, this protocol is non-invasive and the larvae are still alive at the end. This allows further experimentation to be performed on the animals such as specific lipid quantitation or transcript or protein analysis. Additionally, recovered larvae may be allowed to continue growth to adulthood.
An additional benefit of this technique over others is increased sensitivity to small changes in fat levels in a population. By performing a whole concentration gradient on a population of 50 larvae, slight shifts in the population fat levels will be identified by a change in the intermediate concentrations, although the concentration at which the larvae begin floating and are totally floated may not be different than controls. This small shift in the population may not always be identified by other fat quantitation methods as they require a large group of larvae to analyze. The density assay on the other hand interrogates individuals within the population and can consequently identify these small changes or shifts in the overall population distribution.
As this technique relies on the density of the solution to produce reliable results, it is extremely important that the PBS and sucrose solutions are made consistently. We have found that different PBS recipes produce differences in solution density, leading to varying results. We use the recipe outlined above for consistent results. Furthermore, making the 20% sucrose from the same batch of 1x PBS is important as it results in the same background solution density. If the two solutions were made separately, slight variations in the PBS density could bring in additional density variable beyond the addition of sucrose to the experimental conical vial. Lastly, evaporation can be a problem, because over time the solutions will become more concentrated with the evaporation of water. It is therefore important to cap the bottles tightly and to remake the solution every week so that the experimental solutions remain consistent.
Among the steps outlined above, there are several critical steps that if altered, could result in inaccurate results by the buoyancy assay. First, it is important that the larvae transferred into the experimental food vial be the same age. Transferring larvae that vary in size will result in larvae at different developmental stages at the time the buoyancy assay is performed. This protocol has been developed to determine changes in fat levels in wandering L3 larvae and will not work accurately for early L3 or pre-pupae or later. Early third instar larvae have a tendency to float independently of fat levels while pre-pupae and pupae sink even at the highest concentrations used. For this reason, the age of the L1 larvae transferred is critical. Similarly, the point at which the vial is taken to perform the buoyancy assay is important. For the same developmental timing reasons stated above, the vials should be taken only when 20 - 40 larvae are wandering to ensure accurate density measurements. Additionally, the number of L1 transferred can affect results as well. The wide vials used in this protocol allow 50 larvae to eat without competition during development. A much larger number of larvae transferred than this will result in competition for food and may affect the amount of fat stored independent of genotype.
There may be genotypes with such extremely high fat levels that most or all of the larvae will float at the first concentration. In this case, the protocol may be modified by decreasing the concentration of the starting solution in order to obtain a full spectrum of the population density distribution. On the other hand, a specific genotype may be so lean that no larva floats in the initial sucrose. In this case, the initial steps may be omitted and a higher starting concentration used. This assay is very flexible and can be modified to better fit a wide range of fat levels. We advise the use of controls such as appropriate wild type animals (controlled for genetic background), positive controls (established fat mutants, such as adp or Sir217), and negative controls (published lean mutants such as lsd219). Appropriate use of this protocol will allow future screening of collections to determine new regulators of metabolism and obesity predisposition. Furthermore, this protocol provides an easy way for researchers in any field to test whether their genetic manipulation of interest causes a change in fat levels, without the need for complex protocols or expensive equipment. This protocol will help to elucidate the complex relationship between genetics and obesity.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
K.E.H. was supported by the Training Grant in Molecular Biology NIH-T32-GM08730. This work was supported by NIH, NIDDK Grant 5K01DK095932 and AHA Award 12BGIA11930014 to T.R.
References
- Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- Liu Z, Huang X. Lipid metabolism in Drosophila development and disease Using Drosophila System to Study Lipid Metabolism Lipids Function in Drosophila Early Development. Acta Biochim Biophys Sin. 2013;45(1):44–50. doi: 10.1093/abbs/gms105. [DOI] [PubMed] [Google Scholar]
- Ruden DM, Lu X. Evolutionary conservation of metabolism explains howDrosophila nutrigenomics can help us understand human nutrigenomics. Genes Nutr. 2006;1(2):75–84. doi: 10.1007/BF02829949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf MJ, Rockman HA. Drosophila, genetic screens, and cardiac function. Circ. Res. 2011;109:794–806. doi: 10.1161/CIRCRESAHA.111.244897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh I, Boulianne GL. Modeling obesity and its associated disorders in Drosophila. Physiology (Bethesda) 2013;28(2):117–124. doi: 10.1152/physiol.00025.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azeez OI, Meintjes R, Chamunorwa JP. Fat body, fat pad and adipose tissues in invertebrates and vertebrates: the nexus. Lipids Health Dis. 2014;13:71. doi: 10.1186/1476-511X-13-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dourlen P, Sujkowski A, Wessells R, Mollereau B. Fatty acid transport proteins in disease: New insights from invertebrate models. Prog. Lipid Res. 2015;60:30–40. doi: 10.1016/j.plipres.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Hildebrandt A, Bickmeyer I, Kühnlein RP. Reliable Drosophila body fat quantification by a coupled colorimetric assay. PLoS One. 2011;6(9):e23796. doi: 10.1371/journal.pone.0023796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JM, Barry WE, Cox J, Thummel CS. Methods for studying metabolism in Drosophila. Methods. 2014;68:105–115. doi: 10.1016/j.ymeth.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Anzi B, Zinn K. Colorimetric measurement of triglycerides cannot provide an accurate measure of stored fat content in Drosophila. PLoS One. 2010;5(8):e12353. doi: 10.1371/journal.pone.0012353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh NTK, Stevenson G, Obatoml D, Bach P. Determination of Lipids in Animal Tissues by High-Performance Thin-Layer Chromatography with Densitometry. J. Planar Chromatogr. 2000;13:375–381. [Google Scholar]
- Borrull A, Lopez-Martinez G, Poblet M, Cordero-Otero R, Rozes N. A simple method for the separation and quantification of neutral lipid species using GC-MS. Eur. J. Lipid Sci. Technol. 2015;117:274–280. [Google Scholar]
- Shui G, et al. Toward one step analysis of cellular lipidomes using liquid chromatography coupled with mass spectrometry: application to Saccharomyces cerevisiae and Schizosaccharomyces pombe lipidomics. Mol. Biosyst. 2010;6:1008–1017. doi: 10.1039/b913353d. [DOI] [PubMed] [Google Scholar]
- Farvid MS, Ng TWK, Chan DC, Barrett PHR, Watts GF. Association of adiponectin and resistin with adipose tissue compartments, insulin resistance and dyslipidaemia. Diabetes. Obes. Metab. 2005;7(4):406–413. doi: 10.1111/j.1463-1326.2004.00410.x. [DOI] [PubMed] [Google Scholar]
- Urbanchek MG, Picken EB, Kalliainen LK, Kuzon WM. Specific force deficit in skeletal muscles of old rats is partially explained by the existence of denervated muscle fibers. J. Gerontol. A. Biol. Sci. Med. Sci. 2001;56A(5):B191–B197. doi: 10.1093/gerona/56.5.b191. [DOI] [PubMed] [Google Scholar]
- Mosher J, et al. Coordination between Drosophila Arc1 and a specific population of brain neurons regulates organismal fat. Dev. Biol. 2015;405(2):280–290. doi: 10.1016/j.ydbio.2015.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis T, van Gilst MR, Hariharan IK. A buoyancy-based screen of drosophila larvae for fat- storage mutants reveals a role for Sir2 in coupling fat Storage to Nutrient Availability. PLoS Genet. 2010;6(11):e1001206. doi: 10.1371/journal.pgen.1001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Current Bloomington Recipe for Drosophila Medium. Bloomington Drosophila Stock Center at Indiana University; 2014. Available from: http://flystocks.bio.indiana.edu/Fly_Work/media-recipes/bloomfood.htm. [Google Scholar]
- Teixeira L, Rabouille C, Rørth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech. Dev. 2003;120(9):1071–1081. doi: 10.1016/s0925-4773(03)00158-8. [DOI] [PubMed] [Google Scholar]
- Perez CL, Van Gilst MR. A 13C Isotope Labeling Strategy Reveals the Influence of Insulin Signaling on Lipogenesis in C. elegans. Cell Metab. 2008;8:266–274. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]


