Abstract
Background
The color of berry skin is an important economic trait for grape and is essentially determined by the components and content of anthocyanins. The fruit color of Chinese wild grapes is generally black, and the profile of anthocyanins in Chinese wild grapes is significantly different from that of Vitis vinifera. However, V. davidii is the only species that possesses white berry varieties among Chinese wild grape species. Thus, we performed a transcriptomic analysis to compare the difference of transcriptional level in black and white V. davidii, in order to find some key genes that are related to anthocyanins accumulation in V. davidii.
Results
The results of anthocyanins detection revealed that 3,5-O-diglucoside anthocyanins is the predominant anthocyanins in V. davidii. It showed obvious differences from V. vinifera in the profile of the composition of anthocyanins. The transcriptome sequencing by Illumina mRNA-Seq technology generated an average of 57 million 100-base pair clean reads from each sample. Differential gene expression analysis revealed thousands of differential expression genes (DEGs) in the pairwise comparison of different fruit developmental stages between and within black and white V. davidii. After the analysis of functional category enrichment and differential expression patterns of DEGs, 46 genes were selected as the candidate genes. Some genes have been reported as being related to anthocyanins accumulation, and some genes were newly found in our study as probably being related to anthocyanins accumulation. We inferred that 3AT (VIT_03s0017g00870) played an important role in anthocyanin acylation, GST4 (VIT_04s0079g00690) and AM2 (VIT_16s0050g00910) played important roles in anthocyanins transport in V. davidii. The expression of some selected DEGs was further confirmed by quantitative real-time PCR (qRT-PCR).
Conclusions
The present study investigated the transcriptomic profiles of berry skin from black and white spine grapes at three fruit developmental stages by Illumina mRNA-Seq technology. It revealed the variety specificity of anthocyanins accumulation in V. davidi at the transcriptional level. The data reported here will provide a valuable resource for understanding anthocyanins accumulation in grapes, especially in V. davidii.
Electronic supplementary material
The online version of this article (doi:10.1186/s41065-016-0021-1) contains supplementary material, which is available to authorized users.
Keywords: Vitis davidii, Berry skin, Anthocyanins, Transcriptome analysis, Candidate genes
Background
Grapevine (Vitis L.) is an important economic crop used as table fruit, dried raisins, and for wines or juice. The color of berry skin is an important economic trait for grapes. And it is closely related to the components and content of anthocyanins [1, 2]. In recent years, the profile of anthocyanins in grape berry skin has been widely studied. The anthocyanins are glycosides and acylglycosides of anthocyanidins. The main anthocyanidins found in grapes are pelargonidin, cyanidin, delphinidin, peonidin, petunidin as well as malvidin which is usually the predominant anthocyanidin in most red grapes [3, 4]. However, the kind of anthocyanins exhibits obvious differences in different grape species [5]. In the V. vinifera cultivars, only 3-O-monoglucoside anthocyanins are detected, and just a few cultivars produce pelargonidin-based anthocyanins [6, 7]. In non-V. vinifera species, 3,5-O-disglucoside anthocyanins widely exist, even the pelargonidin-based anthocyanins are also detected [7, 8]. In addition, most V. vinifera cultivars possess acylated forms of the anthocyanins, but some other grape species do not produce acylated forms of anthocyanins, such as V. rotundifolia [9].
Anthocyanins in grape berry skin are synthesized via the flavonoid pathways, which has been extensively studied. In fact, the synthesis of anthocyanins shares the same upstream pathways with proanthocyanidins and flavonol derivatives [10]. Phenylalanine is the precursor of flavonoid, which is used as substrate, phenylalanine ammonia lyase (PAL), cinnamate-4-hydroxylase (C4H) and 4-coumaroyl-CoA synthase (4CL) catalyze a series of reactions to produce 4-coumaroyl-CoA. The catalysis of chalcone synthase (CHS) is the first committed step in the flavonoid biosynthetic pathway, which can catalyze the synthesis of chalcones [10]. Subsequently, after the action of chalcone isomerase (CHI), the basic three rings of the general C6-C3-C6 flavonoid skeleton is produced [11]. The B ring of the naringenin flavanone can be further hydroxylated by flavonoid -3′-hydroxylase(F3′H) or flavonoid-3′5′-hydroxylase (F3′5′H) to form eriodictyol or pentahydroxyflavanone [12, 13]. The naringenin, eriodictyol and pentahydroxyflavanone can be modified by the catalysis of flavanone-3β-hydroxylase (F3H) to form the corresponding dihydrokaempferol, dihydroquercetin and dihydromyricetin, respectively. Besides, the dihydrokaempferol can also be catalyzed by F3′H or F3′5′H to produce other two dihydroflavonols, dihydroquercetin or dihydromyricetin [1]. Then, dihydroflavonol-4-reductase (DFR) catalyzes these dihydroflavonols to form their corresponding leucoanthocyanidins [1]. In the past, leucoanthocyanidin dioxygenase (LDOX) used to be considered as the first key enzyme that could catalyze the formation of anthocyanidins [14, 15] and lead the flavonoid flux into the anthocyanin branch. However, more studies showed that LDOX also play an important role in the biosynthesis of proanthocyanidins [16–19]. And the anthocyanidins which are the production of LDOX can also be catalyzed by anthocyanidin reductase (ANR) to produce the substrates for the proanthocyanidins synthesis [20, 21]. The glycosylation, methylation and acylation of anthocyanidins are very important for their stabilization. Glycosylation of the anthocyanidins is the key step to produce anthocyanins which is catalyzed by the UDP-glucose: anthocyanidin: flavonoid glucosyltransferase (UFGT) [1, 22–25]. In V. vinifera, the anthocyanidins can only be catalyzed by UFGT to glycosylate at C3 position [24]. Thus, it is also called 3GT. The S-adenosyl-L-methionine (SAM) or O-methyltransferase (OMT) can catalyze the methylation of the hydroxyl groups at the C3 positions or both at the C3 and C5 positions on the B rings of the anthocyanins [26, 27]. Acylation can greatly enhance the structural diversity and stability of anthocyanins [28]. It is catalyzed by the action of anthocyanin acyltransferases (AAT) [3, 29]. There are mainly two types of ACTs that are classified based on the acyl group donors: the BAHD family using acyl-CoA and the serine carboxypeptidase-like(SCPL) group using acyl-activated sugars [30, 31]. Anthocyanins are synthesized in the cytoplasm but accumulate in the vacuoles. Recent researches showed that glutathione S-transferase (GST), multidrug resistance-associated protein (MRP) and multidrug and toxic compound extrusion (MATE) were closely related to the transport of anthocyanins [32].
As part of the flavonoid pathway, the synthesis of anthocyanins are regulated by a complex regulation at the transcriptional level [4, 10, 15, 33–35]. Generally, the flavonoid pathway of anthocyanins biosynthesis is under the control of Myb transcriptional factors, basic helix–loop-helix proteins(bHLH) and WD40-like proteins, which also play crucial roles in the regulation of flavonols and proanthocyanidins [10, 17–19, 36–38]. The research showed that the bHLH gene, VvMYC1, was characterized as a component of the transcriptional complex regulating anthocyanins biosynthesis in grapevine [39]. In grapes, a series of R2R3-Myb transcriptional factors are related to the synthesis of anthocyanins [40]. The first Myb transcriptional factor Myba in grape was identified and isolated from V. labrusca hybrids [41], and the results suggested that VlmybA1-1, VlmybA1-2 and VlmybA2 transcriptional factors are involved in the regulation of anthocyanin biosynthesis in the grape by regulating the expression of the UFGT gene [41–43]. In red V. vinifera grapes, the functional VvmybA1 can regulate the expression of the UFGT gene to promote the synthesis of anthocyanin. However, in white V. vinifera grapes, a retrotransposon (Gret1) is inserted in the 5′-flanking region of VvmybA1 gene to form a non-functional VvmybA1a gene, resulting in the transcription factor losing its function [44]. Subsequently, two other VvmybA regulator genes, VvmybA2 and VvmybA3 have been cloned and identified [45]. The VvmybA1 and VvmybA2 are very similar, both of them can regulate the accumulation of anthocyanins in the grape berries, and the white-fruited grapes are caused by the mutation of these two similar and adjacent regulatory genes [46, 47]. More recent researches suggest that variation in anthocyanins content in grapes is involved with the VvmybA gene cluster [48–50].
China is the concentrated distribution area of East Asian Vitis species, which has more than 35 Vitis species [51–53]. In recent years, the research of anthocyanins in Chinese wild grapes has been carried out, and the profile of anthocyanins in Chinese wild grapes is significantly different from V. vinifera [5, 7, 54–56]. Generally, only 3-O-monoglucoside anthocyanins are detected in V. vinifera cultivars, and almost all of the cultivars of V. vinifera are devoid of pelargonidin-based anthocyanins. However, in Chinese wild grapes (such as V. amurensis, V. davidii and V. quinquangularis), the 3,5-O-disglucoside anthocyanins widely exist, and pelargonidin-based anthocyanins are detected in some wild species [5, 7, 55, 56]. The berry color of most Chinese wild grapes is black, except for the white-fruited varieties of V. davidii. The subsequent research found that VvmybA1a gene was detected in white-fruited varieties of V. davidii [57]. The white fruit may be caused by this mutation of mybA1 gene. In order to find the causes that lead to the different profiles of anthocyanins between Chinese wild grapes and V. vinifera, we used the black and white varieties of V. davidii as the materials, and the method of transcriptome analysis to detect the differences at the transcriptional level that might be involved in anthocyanins accumulation of these two varieties.
Results
Anthocyanin composition and content
A total of 24 kinds of anthocyanins were detected at three fruit developmental stages of black and white spine grapes (Table 1). Five categories of anthocyanins were detected: 11 kinds of anthocyanidin diglucosides and 13 kinds of anthocyanidin monoglucosides. And 14 kinds of coumaroylated anthocyanins were detected. The highest content of anthocyanins was detected in the third stage black spine grape (B3, DAF120), it was 1382.127 mg.kg−1. In black spine grape, the major anthocyanin was Malvidin-3,5-O-diglucoside, which accounted for 87% of the total anthocyanins. Besides, we also detected trace amount of anthocyanins in white V. davidii.
Table 1.
Anthocyanins concentration in three stages of black and white spine grapes
| Compound | Anyhocyanins content/(mg.kg−1) FW of berry skin | |||||
|---|---|---|---|---|---|---|
| B1 | B2 | B3 | W1 | W2 | W3 | |
| Dp-3-glu | 0.03 ± 0.001d | 0.317 ± 0.006a | 0.093 ± 0.006b | 0.019 ± 0.002e | 0.01 ± 0.001e | 0.069 ± 0.003c |
| Dp-3,5-diglu | nd | 2.71 ± 0.243b | 10.813 ± 0.307a | nd | 0.02 ± 0.003d | 1.73 ± 0.098c |
| Cy-3,5-diglu | 0.02 ± 0.003d | 0.347 ± 0.065a | 0.25 ± 0.04b | nd | 0.011 ± 0.001d | 0.133 ± 0.015c |
| Cy-3-glu | 0.66 ± 0.046b | 0.437 ± 0.067c | 1.417 ± 0.095a | 0.074 ± 0.009e | 0.217 ± 0.031d | 0.133 ± 0.012de |
| Cy-3,5-coum-diglu(trans) | nd | nd | 0.039 ± 0.002a | nd | nd | nd |
| Cy-3-coum-glu(cis) | 0.031 ± 0.003b | nd | 0.056 ± 0.005a | nd | nd | nd |
| Cy-3-coum-glu(trans) | 0.117 ± 0.006b | nd | 0.167 ± 0.015a | nd | nd | nd |
| Pt-3-glu | 0.079 ± 0.005b | 0.347 ± 0.045a | 0.39 ± 0.046a | 0.04 ± 0.003b | 0.020 ± 0.004b | 0.036 ± 0.004b |
| Pt-3,5-diglu | nd | 0.717 ± 0.050b | 7.16 ± 0.120a | nd | nd | 0.383 ± 0.051c |
| Pt-3,5-coum-diglu(trans) | nd | 0.079 ± 0.006b | 4.203 ± 0.307a | nd | nd | nd |
| Pt-3-coum-glu(cis) | nd | nd | nd | 0.009 ± 0.002a | nd | nd |
| Pt-3-coum-glu(trans) | 0.13 ± 0.01c | 0.387 ± 0.021b | 3.02 ± 0.193a | 0.030 ± 0.003c | 0.025 ± 0.006c | 0.052 ± 0.009c |
| Pn-3,5-diglu | 0.063 ± 0.006c | 9.263 ± 0.091b | 26.043 ± 0.419a | 0.03 ± 0.006c | 0.04 ± 0.002c | 0.133 ± 0.006c |
| Pn-3-glu | 2.093 ± 0.104a | 0.663 ± 0.0153c | 1.263 ± 0.025b | 0.05 ± 0d | 0.097 ± 0.006d | 0.1 ± 0d |
| Pn-3,5-coum-diglu(cis) | 0.01 ± 0.004c | 0.1 ± 0.005b | 0.763 ± 0.025a | nd | nd | nd |
| Pn-3,5-coum-diglu(trans) | 0.03 ± 0.005c | 2.777 ± 0.119b | 16.24 ± 0.586a | nd | nd | nd |
| Pn-3-coum-glu(cis) | 0.256 ± 0.009b | 0.247 ± 0.012b | 0.316 ± 0.006a | 0.068 ± 0.002e | 0.104 ± 0.001d | 0.181 ± 0.007c |
| Pn-3-coum-glu(trans) | 0.817 ± 0.031b | 0.237 ± 0.006c | 0.88 ± 0.01a | 0.08 ± 0.004d | 0.04 ± 0.001e | 0.05 ± 0.002de |
| Mv-3,5-diglu | 0.113 ± 0.006c | 31.827 ± 1.462b | 1204.697 ± 5.548a | 0.03 ± 0.002c | 0.177 ± 0.006c | 1.043 ± 0.021c |
| Mv-3-glu | 0.46 ± 0.009c | 1.17 ± 0.026b | 6.443 ± 0.170a | 0.02 ± 0.003d | nd | 0.06 ± 0.003d |
| Mv-3,5-coum-diglu(trans) | nd | nd | 0.873 ± 0.021a | nd | nd | nd |
| Mv-3,5-coum-diglu(trans) | nd | 0.24 ± 0.01b | 19.743 ± 0.396a | nd | nd | nd |
| Mv-3-coum-glu(cis) | 0.03 ± 0.002b | 0.03 ± 0.004b | 1.443 ± 0.0351a | nd | nd | nd |
| Mv-3-coum-glu(trans) | 0.147 ± 0.006b | 0.787 ± 0.025b | 75.8 ± 1.899a | 0.03 ± 0.002b | 0.02 ± 0.001b | 0.03 ± 0.001b |
| Total | 5.096 ± 0.127c | 52.690 ± 1.867b | 1382.127 ± 4.633a | 0.490 ± 0.018c | 0.790 ± 0.029c | 4.135 ± 0.064c |
B1, B2 and B3 refer to the berries of black spine grape of 40DAF, 80DAF and 120DAF, respectively. W1, W2 and W3 refer to the berries of white spine grape of 40DAF, 80DAF and 120DAF, respectively
Dp delphinidin, Cy cyanidin, Pt petunidin, Pn peonidin, Mv malvidin, glu glucoside, diglu diglucoside, coum coumaroyl, nd not detected; A significance level of p < 0.01 was applied
mRNA sequencing
A total of 12 cDNA libraries were constructed from the total RNA of black and white spine grape berry skin in three fruit developmental stages (Fig. 1), two biological replicates were made at each stage. These cDNA libraries were subjected to pair-end reading with the Illumina HiSeq 2000 platform, generating from 56 million to 64 million pair-end raw reads of 100 bp in length, respectively. And the raw data have been submitted to SRA database of NCBI (Accession ID: SRP070860; Link: https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP070860). After removing the low-quality reads and trimming the adapter sequences, we obtained from 27 million to 31 million clean reads (Table 2). The subsequent analyses were based on the clean data. Though only two biological replicates were used for RNA-seq, the correlation analysis showed that the R2 of the two replicates of all the samples were greater than 0.95. It fully illustrated the consistency of two biological replicates and the reliability of the RNA-seq results.
Fig. 1.

Berries of black and white spine grapes at different ripening stages. B1, B2 and B3 refer to the berries of black spine grape of 40 DAF, 80 DAF and 120 DAF. W1, W2 andW3 refer to the berries of white spine grape of 40 DAF, 80 DAF and 120 DAF
Table 2.
Summary of the sequencing data
| Sample name | Raw reads | Clean reads | Clean bases | Error rate(%) | Q20(%) | Q30(%) | GC content(%) |
|---|---|---|---|---|---|---|---|
| B1A | 56801854 | 54944048 | 6.86G | 0.03 | 96.90 | 93.75 | 46.02 |
| B1B | 58642586 | 56945838 | 7.12G | 0.03 | 97.00 | 93.94 | 45.84 |
| B2A | 60467584 | 58763160 | 7.34G | 0.03 | 96.4 | 92.73 | 46.45 |
| B2B | 57458296 | 55118356 | 6.88G | 0.03 | 96.23 | 92.42 | 47.42 |
| B3A | 56058442 | 54572136 | 6.82G | 0.03 | 96.25 | 92.43 | 46.49 |
| B3B | 62161910 | 60249422 | 7.54G | 0.03 | 96.32 | 92.57 | 46.41 |
| W1A | 58127846 | 56521424 | 7.06G | 0.03 | 96.30 | 92.47 | 46.14 |
| W1B | 50809432 | 49850444 | 6.24G | 0.03 | 96.40 | 92.59 | 46.59 |
| W2A | 60765156 | 59192440 | 7.40G | 0.03 | 96.47 | 92.84 | 46.10 |
| W2B | 61258710 | 59500464 | 7.44G | 0.03 | 96.54 | 92.96 | 46.53 |
| W3A | 64217138 | 61521590 | 7.70G | 0.03 | 96.19 | 92.28 | 47.07 |
| W3B | 64263360 | 62332092 | 7.80G | 0.03 | 96.23 | 92.35 | 46.76 |
A and B: two biological replicates of each stages; Raw reads: the original number of reads by sequencing; Clean reads: the number of reads after removing the low-quality reads and trimming the adapter sequences from raw reads; Clean bases: number of clean reads multiplied by length of clean reads. Error rate: the average base sequencing error rates; Q20 and Q30: Phred score, refer to the accuracy of sequenced bases were 99 and 99.9%; GC content: the percentage of G and C in total bases
Sequence alignment and mapping to the reference genome
We used the French-Italian Public Consortium for Grapevine Genome Characterization, publically accessible version of the complete V. vinifera genome at 12× coverage (ftp://ftp.ensemblgenomes.org/pub/release-23/plants/fasta/vitis_vinifera/dna/) [58], as the reference genome. After quality control, the reads from black spine grape (64–74%) and white spine grape (63–76%) successfully aligned to the reference genome. Most of the reads from each fruit developmental stage for black and white spine grape aligned to a single position. These uniquely mapped reads account on average for approximately 67 and 71% of the total number of sequenced reads for the black and white spine grape, respectively. The number of reads from each fruit developmental stage for both black and white spine grape mapped to ‘+’ and ‘–’ strand were mostly equal. The number of non-splice reads was approximately twice to splice reads (Table 3).
Table 3.
The clean reads mapped to the reference genome
| Sample name | Total reads | Total mapped | Multiple mapped | Uniquely mapped | Reads map to ‘ + ’ | Reads map to ‘-’ | Non-splice reads | Splice reads |
|---|---|---|---|---|---|---|---|---|
| B1A | 54944048 | 40808925 (74.27%) | 794409 (1.45%) | 40014516 (72.83%) | 20002451 (36.41%) | 20012065 (36.42%) | 24600401 (44.77%) | 15414115 (28.05%) |
| B1B | 56945838 | 42479460 (74.6%) | 859482 (1.51%) | 41619978 (73.09%) | 20805711 (36.54%) | 20814267 (36.55%) | 25634857 (45.02%) | 15985121 (28.07%) |
| B2A | 58763160 | 37649622 (64.07%) | 748838 (1.27%) | 36900784 (62.8%) | 18441742 (31.38%) | 18459042 (31.41%) | 23233578 (39.54%) | 13667206 (23.26%) |
| B2B | 55118356 | 36141693 (65.57%) | 728542 (1.32%) | 35413151 (64.25%) | 17717304 (32.14%) | 17695847 (32.11%) | 22963769 (41.66%) | 12449382 (22.59%) |
| B3A | 54572136 | 36055433 (66.07%) | 800693 (1.47%) | 35254740 (64.6%) | 17628599 (32.3%) | 17626141 (32.3%) | 22458722 (41.15%) | 12796018 (23.45%) |
| B3B | 60249422 | 40232538 (66.78%) | 961911 (1.6%) | 39270627 (65.18%) | 19632707 (32.59%) | 19637920 (32.59%) | 24890373 (41.31%) | 14380254 (23.87%) |
| W1A | 56521424 | 43356467 (76.71%) | 856353 (1.52%) | 42500114 (75.19%) | 21230181 (37.56%) | 21269933 (37.63%) | 26214684 (46.38%) | 16285430 (28.81%) |
| W1B | 49850444 | 38208017 (76.65%) | 751623 (1.51%) | 37456394 (75.14%) | 18713704 (37.54%) | 18742690 (37.6%) | 22835496 (45.81%) | 14620898 (29.33%) |
| W2A | 59192440 | 44324521 (74.88%) | 750136 (1.27%) | 43574385 (73.61%) | 21759103 (36.76%) | 21815282 (36.85%) | 26852753 (45.37%) | 16721632 (28.25%) |
| W2B | 59500464 | 45258909 (76.06%) | 808909 (1.36%) | 44450000 (74.71%) | 22202737 (37.32%) | 22247263 (37.39%) | 27353360 (45.97%) | 17096640 (28.73%) |
| W3A | 61521590 | 39182938 (63.69%) | 793374 (1.29%) | 38389564 (62.4%) | 19130918 (31.1%) | 19258646 (31.3%) | 24966612 (40.58%) | 13422952 (21.82%) |
| W3B | 62332092 | 42273326 (67.82%) | 830849 (1.33%) | 41442477 (66.49%) | 20661523 (33.15%) | 20780954 (33.34%) | 26550475 (42.6%) | 14892002 (23.89%) |
Approximately 80% of the reads from each fruit developmental stage for both black and white spine grape mapped to exons, 20% mapped to intergenic regions, and 1% mapped to introns (Fig. 2). The reads mapped to introns resulted from the residual of pre-mRNA and intron retention in the process of alternative splicing. However, the incomplete genome annotation led to the reads mapped to intergenic regions.
Fig. 2.
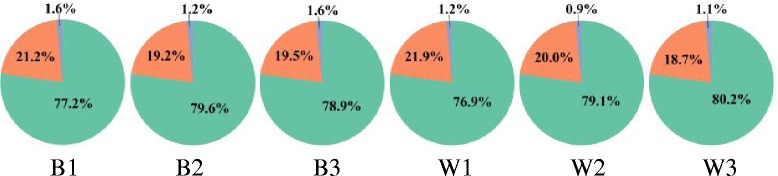
Percent of reads mapped to genome regions. The green part refers to percentage of reads mapped to exons; the orange part refer to percentage of reads mapped to intergenic regions; the blue part refer to percentage of reads mapped to introns
Differential expression analysis
We used the RPKM value to show the gene expression level (Table 4). About 45–50% of the genes fall into the RPKM range of 0–1, 9–10% of the genes fall into the RPKM range of 1–3, 18–20% of the genes fall into the RPKM range of 3–15, 14–17% of the gens fall into the RPKM range of 15–60, and approximately 7% of the genes that RPKM value is greater than 60.
Table 4.
The statistic of different gene expression level interval number
| RPKM Interval | B1 | B2 | B3 | W1 | W2 | W3 |
|---|---|---|---|---|---|---|
| 0 ~ 1 | 14793(45.10%) | 16496(50.29%) | 16410(50.03%) | 15195(46.33%) | 15234(46.45%) | 16526(50.38%) |
| 1 ~ 3 | 3220(9.82%) | 2944(8.98%) | 3061(9.33%) | 3221(9.82%) | 3253(9.92%) | 2978(9.08%) |
| 3 ~ 15 | 6771(20.64%) | 5977(18.22%) | 6174(18.82%) | 6509(19.84%) | 6347(19.35%) | 6177(18.83%) |
| 15 ~ 60 | 5668(17.28%) | 5033(15.34%) | 4961(15.12%) | 5494(16.75%) | 5389(16.43%) | 4900(14.94%) |
| >60 | 2348(7.16%) | 2350(7.16%) | 2194(6.69%) | 2381(7.26%) | 2577(7.86%) | 2219(6.77%) |
Numbers of transcripts from the V. vinifera RefSeq dataset detected at various levels of abundance at each time-point, as calculated by reads per kilobase of exon per million reads (RPKM)
There were thousands of differential expression genes in three fruit developmental stages. But, we focused on the genes which were related to the change of berry skin color in fruit development. Thus, we compared the expression of genes in different stages, such as B2vsB1, B3vsB2, B3vsB1, B1vsW1, B2vsW2 and B3vsW3. Then, we used the differential expression genes which were gained from the comparison to make venn diagrams (Fig. 3). In the venn diagram of B2vsB1, B3vsB2 and B3vsB1, 1440 differential expression genes were found. And in the venn diagram of B1vsW1, B2vsW2 and B3vsW3, it was 1006. Accordingly, the scope of selecting the candidate differential expression genes is effectively narrowed.
Fig. 3.
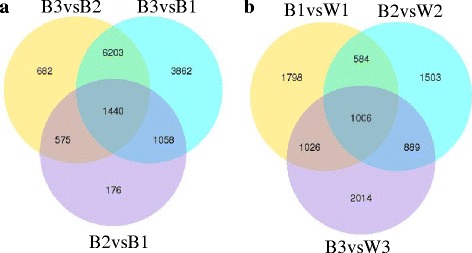
The venn diagrams of differential expression genes. Venn diagrams indicate the overlap of differential expression genes between B2vsB1, B3vsB1 and B3vsB2 (a), and between B1vsW1, B2vsW2 and B3vsW3 (b)
Selection of candidate genes
Functional enrichment analysis
Gene Ontology (GO, http://www.geneontology.org/) is an international standardized gene function classification system which can be used to classify the function of the predicted genes. GO include three parts: molecular function, biological process and cellular component. We used the differential expression genes between B2 and W2 to make GO enrichment analysis. Thirty significant terms were found, which belonged to biological process and molecular function (Fig. 4). The color of the berry skin is determined by the pigment, and the anthocyanins are the main pigment in grapes [1, 2]. The anthocyanins were synthesized in the endoplasmic reticulum, and transported into vacuole by a serious of anthocyanins transporters [32]. Thus, in these 30 terms, transmembrane transport (GO:0055085) and pigment catabolic process(GO:0046149), which possibly contained the candidate genes, came into our attention. The transmembrane transport term contained 107 up-regulated expression genes and 110 down-regulated expression genes. The pigment catabolic process contained 6 up-regulated expression genes and 14 down-regulated expression genes (Additional file 1: Table S1).
Fig. 4.
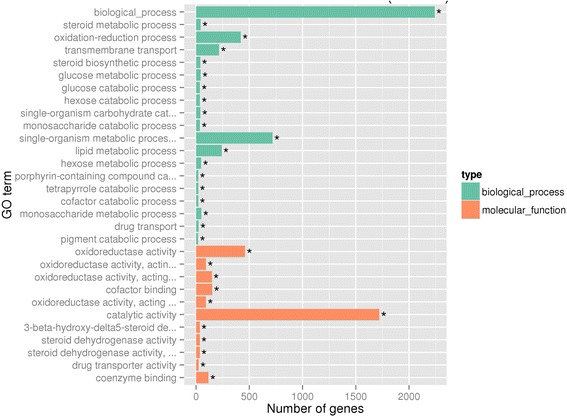
The most enriched GO terms (B2vsW2). Thirty most enriched GO terms were displayed, included 19 biological processes (green) and 11 molecular function (orange)
To identify the biological pathways activated in grape fruit, pathway enrichment was quantified in the KEGG database [59]. In grapes, anthocyanins synthesis is via the flavonoid pathways [10], which were significantly enriched in our study (Fig. 5). There were 33 background genes in the flavonoid biosynthesis pathway, 27 genes showed expressional differences (Table 5). In these 27 genes, VIT_00s0361g00040 was annotated as anthocyanidin reductase (ANR), VIT_01s0011g02960 and VIT_17s0000g04150 were annotated as leucoanthocyanidin reductase(LAR), which were related to the synthesis of proanthocyanidins [20, 21]. VIT_18s0001g03470 was annotated as flavonol synthase (FLS), which was related to the synthesis of flavonol aglycones [60, 61]. VIT_02s0033g00410 was annotated as transcription factor MYBA1, which can effectively regulate the anthocyanins biosynthesis [41–43]. VIT_02s0025g04720 and VIT_16s0039g02230 were respectively annotated as LDOX and UFGT, which were directly related to anthocyanins biosynthesis [14, 15, 22–25]. VIT_03s0063g00140, VIT_07s0031g00350, VIT_11s0016g02610 and VIT_12s0028g03110 were annotated as OMT, which participated in the methylation modification of anthocyanins [26, 27]. Other genes were related to the upstream pathways of the flavonoid biosynthesis.
Fig. 5.
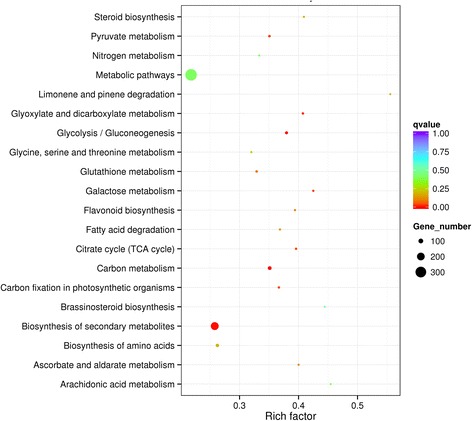
Statistics of KEGG pathway enrichment. The vertical axis refers to pathways, horizontal axis refers to rich factor. The sizes of the dots represent the numbers of differential expression genes in this pathway. The colors of the dots correspond to the scopes of Q-value
Table 5.
The differential expression genes in the KEGG pathway of flavonoid biosynthesis
| Gene_ID | RPKM value | Annotation | |||||
|---|---|---|---|---|---|---|---|
| B1 | B2 | B3 | W1 | W2 | W3 | ||
| VIT_02s0033g00410 | 0.04 | 140.49 | 127.82 | 0.00 | 0.00 | 7.97 | MYBA1 |
| VIT_00s0361g00040 | 2.79 | 1.66 | 0.63 | 2.92 | 1.88 | 0.67 | ANR |
| VIT_01s0011g02960 | 5.94 | 1.75 | 0.42 | 1.85 | 0.70 | 0.78 | LAR1 |
| VIT_02s0025g04720 | 38.96 | 1659.75 | 1030.34 | 26.99 | 36.62 | 87.43 | LDOX |
| VIT_03s0063g00140 | 0.09 | 0.30 | 7.61 | 0.44 | 1.01 | 4.66 | OMT |
| VIT_04s0023g03370 | 6.98 | 561.89 | 230.96 | 3.71 | 2.35 | 6.48 | F3H |
| VIT_05s0136g00260 | 8.42 | 4686.98 | 3990.83 | 5.62 | 4.05 | 41.12 | CHS |
| VIT_06s0004g08150 | 23.71 | 109.70 | 79.78 | 29.92 | 18.20 | 42.08 | Trans-cinnamate 4-monooxygenase |
| VIT_06s0009g02830 | 6.79 | 237.73 | 281.50 | 0.13 | 0.20 | 0.12 | F3′5′H |
| VIT_06s0009g02860 | 0.05 | 6.73 | 1.89 | 0 | 0 | 0 | F3′5′H |
| VIT_06s0009g03050 | 0 | 0.04 | 0.57 | 0 | 0 | 0 | F3′5′H |
| VIT_07s0031g00350 | 1.15 | 448.44 | 1191.02 | 16.33 | 50.62 | 648.28 | OMT |
| VIT_08s0040g00780 | 11.03 | 40.57 | 48.50 | 8.59 | 8.74 | 29.68 | Cytochrome P450 |
| VIT_11s0016g02610 | 1.04 | 1.64 | 1.03 | 0.07 | 0 | 0 | OMT |
| VIT_11s0065g00350 | 0.02 | 0.57 | 0.60 | 0.05 | 0.82 | 0.10 | Trans-cinnamate 4-monooxygenase |
| VIT_12s0028g03110 | 2.76 | 0.12 | 0.04 | 1.62 | 0.59 | 0.18 | OMT |
| VIT_13s0067g03820 | 48.72 | 700.54 | 545.88 | 33.78 | 26.95 | 91.33 | CHI |
| VIT_14s0068g00920 | 2.25 | 490.58 | 278.42 | 3.15 | 7.42 | 13.49 | CHS |
| VIT_14s0068g00930 | 0.20 | 2.97 | 2.04 | 2.17 | 6.63 | 1.69 | CHS |
| VIT_16s0039g02350 | 2.31 | 3.48 | 13.25 | 0.00 | 0.17 | 1.02 | DFR |
| VIT_17s0000g04150 | 14.12 | 10.12 | 20.50 | 9.55 | 11.13 | 9.64 | LAR2 |
| VIT_17s0000g07200 | 6.74 | 47.12 | 57.77 | 12.97 | 17.55 | 53.45 | F3′H |
| VIT_17s0000g07210 | 0.60 | 3.35 | 3.37 | 0.53 | 0.79 | 2.40 | F3′H |
| VIT_18s0001g03470 | 0.02 | 0.00 | 0.03 | 0.07 | 10.20 | 0.39 | FLS |
| VIT_18s0001g12800 | 4.86 | 35.81 | 24.90 | 7.04 | 7.24 | 9.41 | DFR |
| VIT_16s0039g02230 | 0.04 | 1214.43 | 1009.98 | 0.08 | 0.04 | 0.16 | UFGT |
| VIT_18s0001g14310 | 45.39 | 120.46 | 67.93 | 46.73 | 66.97 | 124.79 | Flavanone 3-dioxygenase |
Expression levels are shown in RPKM for each sample. The annotation was the results of the gene blast in Swiss-prot. ANR anthocyanidin reductase, LAR leucoanthocyanidin reductase, LDOX leucoanthocyanidin dioxygenase, OMT O-methyltransferase, F3H flavanone-3β-hydroxylase, CHS chalcone synthase, F3′5′H flavonoid-3′5′-hydroxylase, CHI chalcone isomerase, DFR dihydroflavonol 4-reductase, F3′H flavonoid-3′-hydroxylase, FLS flavonol synthase, UFGT UDP-glucose:flavonoid-3-O-glucosyltransferase;
Expression pattern analysis
We also used differential expression gene clustering methodology to find the genes that are related to the biosynthesis of anthocyanins in spine grape. The genes that showed same expression pattern were clustered together, and they may obtain the same function or belong to the same biological pathway. We used all of the differential expression genes to draw a heatmap (Fig. 6a). The genes that had the same or similar expression patterns were effectively clustered together. However, so many clusters were produced, and it was difficult to find the clusters that contained the genes closely related to the color of the berry skin. But, the previous studies have suggested that UFGT(VIT_16s0039g02230) [1, 24, 25] and MybA1(VIT_02s0033g00410) [41] were the key genes to regulate the synthesis of the anthocyanins. Thus, we selected the genes whose expression patterns were similar to these two genes. Then, we obtained 100 genes and used them to make a heatmap (Fig. 6b). The genes were divided into two groups that contained 68 and 32 genes respectively. And the expression pattern of the group that contained 68 genes was very similar to UFGT and MybA1. Then, we can select the genes that were related to the biosynthesis and transport of anthocyanins.
Fig. 6.
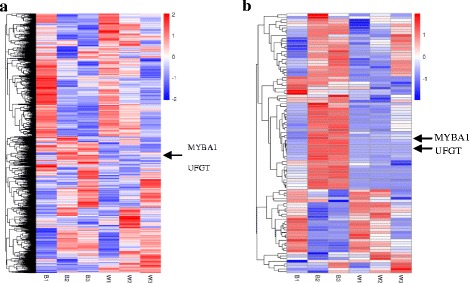
The heatmap of the differential expression genes. a The heatmap of all the differential expression genes. b The heatmap of the selected 100 differential expression genes. Using log10(RPKM + 1) value to cluster, the red color referred to high expressed genes, the blue color refer to low expressed genes. From blue to red, refer to the log10(RPKM + 1) value gradually rised up. The vertical axis refer to the genes showed similar expressed pattern were clustered in a group
The candidate genes
After these analyses, we finally obtained 41 differential expression genes, which were considered to be related to the biosynthesis and transport of anthocyanins in the berry skin of spine grape (Table 6). The expression patterns of these genes were very similar, mainly expressed in B2 and B3 (Fig. 7). In these candidate genes, just one gene (VIT_14s0128g00600) was not annotated. Yet its expression pattern was very similar to the other candidate genes, and we inferred that it was probably related to the anthocyanins accumulation.
Table 6.
Expression profiles of anthocyanin biosynthesis and transport candidate genes in spine grape
| Gene_ID | RPKM value | Annotation | |||||
|---|---|---|---|---|---|---|---|
| B1 | B2 | B3 | W1 | W2 | W3 | ||
| VIT_02s0033g00410 | 0.04 | 140.49 | 127.82 | 0.00 | 0.00 | 7.97 | MYBA1 |
| VIT_02s0033g00390 | 0.03 | 128.67 | 322.83 | 0.00 | 0.16 | 248.31 | MYBA2 |
| VIT_02s0033g00450 | 0.00 | 358.15 | 409.69 | 1.21 | 2.49 | 222.68 | MYBA3 |
| VIT_01s0011g04760 | 97.59 | 124.81 | 117.63 | 67.22 | 61.35 | 46.54 | MYB4 |
| VIT_14s0060g01010 | 24.39 | 38.55 | 27.01 | 3.48 | 17.65 | 20.80 | bHLH |
| VIT_14s0108g01070 | 3.26 | 11.77 | 29.28 | 0.91 | 2.80 | 11.50 | NAC |
| VIT_06s0004g02620 | 0.93 | 202.54 | 219.54 | 0.40 | 0.88 | 5.22 | PAL |
| VIT_13s0019g04460 | 4.30 | 232.02 | 340.47 | 8.63 | 10.27 | 89.01 | PAL2 |
| VIT_16s0039g02040 | 9.04 | 30.83 | 9.78 | 12.76 | 14.09 | 10.35 | 4CL |
| VIT_16s0050g00390 | 9.67 | 45.72 | 129.44 | 15.09 | 20.51 | 45.27 | 4CL |
| VIT_05s0136g00260 | 8.42 | 4686.98 | 3990.83 | 5.62 | 4.05 | 41.12 | CHS3 |
| VIT_14s0068g00920 | 2.25 | 490.58 | 278.42 | 3.15 | 7.42 | 13.49 | CHS2 |
| VIT_13s0067g03820 | 48.72 | 700.54 | 545.88 | 33.78 | 26.95 | 91.33 | CHI1 |
| VIT_13s0067g02870 | 132.01 | 654.60 | 660.10 | 99.54 | 89.00 | 184.20 | CHI2 |
| VIT_04s0023g03370 | 6.98 | 561.89 | 230.96 | 3.71 | 2.35 | 6.48 | F3H1 |
| VIT_11s0016g01020 | 62.59 | 74.70 | 15.97 | 11.77 | 3.55 | 0.21 | F3′H |
| VIT_17s0000g07200 | 6.74 | 47.12 | 57.77 | 12.97 | 17.55 | 53.45 | F3′H |
| VIT_06s0009g02830 | 6.79 | 237.73 | 281.50 | 0.13 | 0.20 | 0.12 | F3′5′H |
| VIT_06s0009g03010 | 2.84 | 262.91 | 424.92 | 0.23 | 0.32 | 0.26 | F3′5′H |
| VIT_06s0009g02810 | 1.28 | 45.46 | 55.50 | 0.00 | 0.10 | 0.04 | F3′5′H |
| VIT_06s0009g02920 | 2.99 | 75.91 | 133.90 | 0.06 | 0.09 | 0.04 | F3′5′H |
| VIT_18s0001g12800 | 4.86 | 35.81 | 24.90 | 7.04 | 7.24 | 9.41 | DFR |
| VIT_02s0025g04720 | 38.96 | 1659.75 | 1030.34 | 26.99 | 36.62 | 87.43 | LDOX |
| VIT_16s0039g02230 | 0.04 | 1214.43 | 1009.98 | 0.08 | 0.04 | 0.16 | UFGT |
| VIT_09s0002g06590 | 0.55 | 511.11 | 202.65 | 0.68 | 0.00 | 0.10 | 5GT |
| VIT_07s0031g00350 | 1.15 | 448.44 | 1191.02 | 16.33 | 50.62 | 648.28 | OMT |
| VIT_01s0010g03510 | 9.97 | 736.92 | 512.06 | 0.87 | 0.12 | 0.03 | OMT1 |
| VIT_01s0010g03490 | 7.95 | 79.79 | 44.64 | 0.56 | 0.24 | 0.00 | OMT2 |
| VIT_14s0068g01440 | 21.78 | 11.03 | 4.82 | 2.91 | 0.93 | 0.39 | AT |
| VIT_03s0017g00870 | 0.00 | 246.75 | 227.53 | 0.02 | 0.00 | 0.02 | 3AT |
| VIT_12s0028g00920 | 48.17 | 23.64 | 20.90 | 19.40 | 18.62 | 16.04 | GST3 |
| VIT_04s0079g00690 | 0.03 | 3274.70 | 3166.24 | 0.22 | 0.06 | 0.14 | GST4 |
| VIT_19s0015g02690 | 8.25 | 238.49 | 58.89 | 3.48 | 5.57 | 26.74 | GSTu25 |
| VIT_19s0015g02730 | 5.81 | 84.17 | 96.10 | 1.79 | 4.27 | 27.43 | GST5 |
| VIT_19s0015g02880 | 21.32 | 192.30 | 211.83 | 27.78 | 28.43 | 70.32 | GST |
| VIT_16s0050g00910 | 0.72 | 494.01 | 746.08 | 13.13 | 3.05 | 4.36 | AM2 |
| VIT_16s0050g02480 | 79.44 | 76.26 | 99.26 | 40.72 | 39.48 | 79.18 | ABCC1 |
| VIT_10s0003g04390 | 97.89 | 82.88 | 145.60 | 78.68 | 66.86 | 91.25 | ABCC2 |
| VIT_09s0002g02430 | 34.16 | 36.74 | 49.76 | 16.68 | 6.89 | 20.86 | ABCC8 |
| VIT_03s0017g01290 | 10.84 | 42.46 | 222.45 | 3.05 | 4.87 | 153.61 | ABCG11 |
| VIT_14s0128g00600 | 0.00 | 118.71 | 290.45 | 0.07 | 0.00 | 6.32 | Unkown |
Expression values are shown in RPKM for each sample. The annotation was the results of the gene blast in swiss-prot. bHLH basic helix–loop-helix proteins, PAL phenylalanine ammonia lyase, 4CL 4-coumaroyl-coA synthase, CHS chalcone synthase, CHI chalcone isomerase, F3H flavanone-3β-hydroxylase, F3′H flavonoid-3′-hydroxylase, F3′5′H flavonoid-3′5′-hydroxylase, DFR dihydroflavonol 4-reductase, LDOX leucoanthocyanidin dioxygenase, UFGT UDP-glucose:flavonoid-3-O-glucosyltransferase, 5GT UDP-glucose: anthocyanin 5-O-glucosyltransferase, OMT O-methyltransferase, AT anthocyanin acyltransferases, GST glutathione S-transferase, AM anthoMATEs, ABCC ATP binding cassette
Fig. 7.

The heatmap of the 41 candidate genes. The denotations are the same as in Fig. 5. The corresponding gene IDs were noted on the right side of the picture
Selected candidate UGT genes in white spine grape
Trace amount of anthocyanins were detected in white spine grape (Table 1). Thus, we inferred that some other UGTs are probably related to anthocyanins accumulation in white spine grape. We compared the third stage of white spine grape with the first stage, 62 significant differential expression genes were annotated as UGTs (Additional file 2: Table S2), containing 20 up-regulated and 42 down-regulated genes. From the 20 up-regulated genes, we selected the most 5 significant differential expression genes as the candidate genes (Table 7). All of these five candidate genes were above 4.5-fold higher in W3 than in W1. Especially, VIT_00s0324g00050 showed highly significant differential expression (P = 3.79E-129).
Table 7.
The candidate UGT genes selected from comparison between W3 and W1
| Gene_ID | Readcount_W3 | Readcount_W1 | Log2FoldChange | P-value | Blast swiss prot |
|---|---|---|---|---|---|
| VIT_17s0000g08100 | 214.94 | 0.00 | - | 1.41E-06 | UDP-glycosyltransferase 90A1 |
| VIT_00s0324g00070 | 27.64 | 0.43 | 5.9911 | 0.0001105 | UDP-glycosyltransferase 85A2 |
| VIT_00s0324g00050 | 9505.73 | 172.13 | 5.7873 | 3.79E-129 | UDP-glycosyltransferase 85A2 |
| VIT_06s0004g05780 | 45.61 | 0.87 | 5.714 | 4.10E-09 | UDP-glycosyltransferase 76E2 |
| VIT_08s0007g04570 | 382.63 | 15.45 | 4.6299 | 2.18E-46 | UDP-glycosyltransferase 73D1 |
Log2FoldChange: log2 (Readcount_W3/ Readcount_W1); P-value was adjusted; Predicted amino acid sequence was used to blast in swiss prot, showing a high homology with Arabidopsis thaliana UGTs
qRT-PCR analysis
To confirm the results obtained from RNA-seq, relative expression profiles of 30 genes were analyzed by real-time RT-PCR at the three fruit developmental stages in black and white spine grapes. These 30 genes related to the biosynthesis and transport of anthocyanins were chosen for qRT-PCR analysis. The black spine grape showed much higher gene expression of the selected genes than that of the white spine grape. For most of the genes, the qRT-PCR results were consistent with those obtained from the expression profile determined from RNA-Seq data (Fig. 8).
Fig. 8.
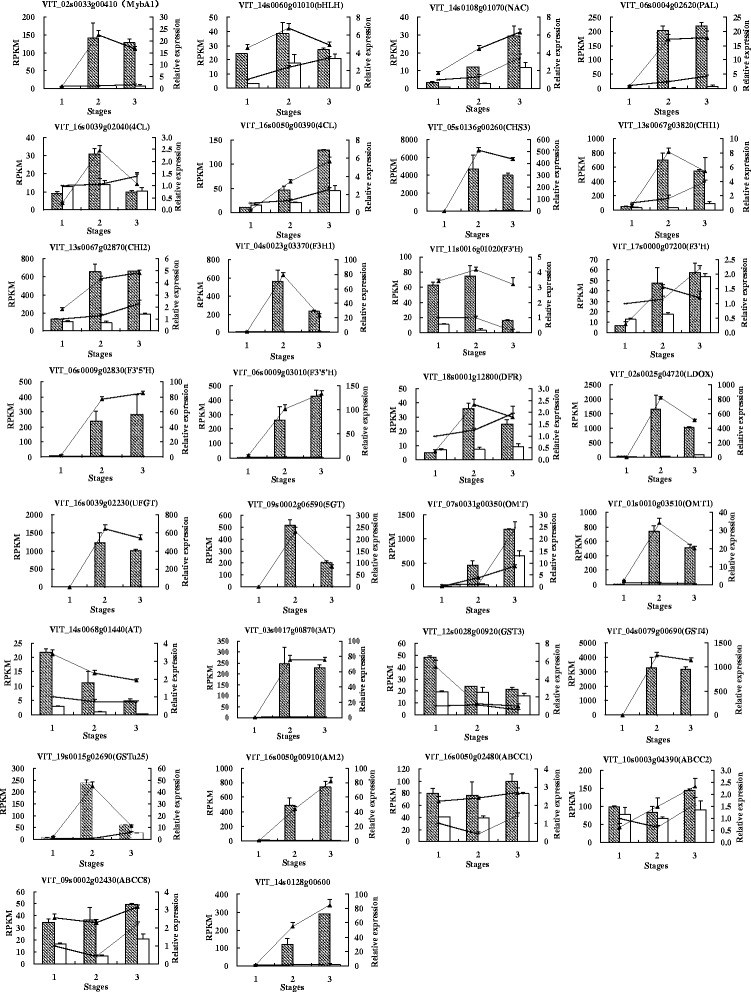
Quantitative real-time PCR validation of RNA-Seq data. Relative expression profiles of 30 genes showed the expression fold changes (FC) in comparison between the three fruit developmental stages in the black and white spine grapes. Histograms represent expression levels as assessed by RNA-Seq, data are reported as means ± SE of two biological replicates (left axis), the columns with diagonal lines or blank represent the RPKM of black or white spine grape, respectively. The line charts represent expression fold changes as assessed by qRT-PCR, data are reported as means ± SE of three replicates (right axis), the lines with black triangle or ‘×’ mark represent black or white spine grape, respectively
Discussion
Anthocyanin composition and content in black and white V. davidii
In V. vinifera, only 3-O-monoglucoside anthocyanins were detected [6]. However, in our research, 3,5-O-diglucoside anthocyanins was the predominant anthocyanins in V. davidii. The studies have shown that, the 3,5-O-diglucoside anthocyanins were usually detected in other Chinese wild grapes [5, 7, 55, 56]. Therefore, the V. davidii was closer to the other Chinese wild grapes from the perspective of the composition of anthocyanins. We inferred that the high 3-5-O-disglucoside anthocyanins concentration in black spine grape was closely related to the expression of VIT_09s0002g06590 (5GT).
Interestingly, we also detected trace amount of anthocyanins in white V. davidii. It was generally accepted that anthocyanins were present only in red grapes, and it was used as a standard to define the difference between red grape and white grape [46, 62]. But, a very recent research suggested that some white grapes contained measurable traces of anthocyanins [63]. Previous researches showed that UFGT gene was not expressed in white grape berries, and the expression of UFGT was regulated by two very similar regulatory genes, VvMYBA1 and VvMYBA2, which were not transcribed in white grape berries [46, 62]. The present study revealed that UFGT (VIT_16s0039g02230) did not express in white V. davidii. However, why did we detect anthocyanins in white grape berries? A previous study showed that there were as many as 181 putative UDP-glycosyltranferases (UGTs) found in the genome of V. vinifera [64]. Arapitsas et al. [65] detected measurable trace amounts of anthocyanins in some white grape varieties, and they inferred that the other UGTs expressed in berry skin accepts anthocyanidin as substrate and is therefore involved in the synthesis of trace amount anthocyanins detected in the berry skin of white grape. In our study, 5 candidate UGT genes were above 4.5-fold higher in W3 than in W1, especially VIT_00s0324g00050 showed highly significant differential expression (P = 3.79E-129). These genes are probably related to anthocyanins accumulation in white V. davidii.
The candidate genes related to anthocyanins biosynthesis
The synthesis of anthocyanins via the phenylpropanoid and flavonoid pathway, and it is catalyzed by a serious of enzymes (Fig. 9) [1, 10, 37]. However, most of the genes coding for these enzymes are multi-copied in the grape genome [65]. Phenylalanine ammonia lyase (PAL) catalyzes the first step of the phenylpropanoid pathway. In our study, two differential expression genes (VIT_06s0004g02620 and VIT_13s0019g04460) were annotated as PAL. VIT_06s0004g02620 was mainly expressed in B2 and B3, but just had a trace amount expression in W3. VIT_13s0019g04460 was also mainly expressed in B2 and B3, and had a high expression in W3. Thus, we inferred that VIT_06s0004g02620 played a major role in the pathway. Under the action of C4H and 4CL, 4-Coumaroyl-CoA was produced. We did not find any obvious differential expression genes annotated as C4H. However, VIT_16s0039g02040 and VIT_16s0050g00390 were annotated as 4CL. Chalcone synthase (CHS) is the first committed enzyme of the flavonoid pathway. Previous studies suggested that 3 CHS genes (CHS1, CHS2 and CHS3) played a role in this pathway. Of the CHSs, the mRNAs of CHS1and CHS2 were detected in both leaves and berry skins of white and red grape cultivars, whereas the mRNA of CHS3 was mainly accumulated in the berry skin of red cultivars during coloration [65–67]. In our study, only CHS2 (VIT_14s0068g00920) and CHS3 (VIT_05s0136g00260) showed significant expression differences between black and white spine grapes. Especially, CHS3 (VIT_05s0136g00260) was detected as having a very high expressional level in B2 and B3, and significantly higher than that in white spine grape. It was consistent with the previous studies [65–67]. Two CHI genes (VIT_13s0067g03820 and VIT_13s0067g02870) showed obvious differential expression levels between black and white spine grapes. These two CHI genes (CHI1 and CHI2) have been reported by previous researches [65, 67, 68]. The researches suggested that two F3H genes (F3H1 and F3H2) played a role in the flavonoid pathway, and the mRNA of F3H2 was detected at a high level in grape berry skin during coloration [65, 67]. However, in our study, only the expression of F3H1 (VIT_04s0023g03370) in black spine grape showed significantly higher than white spine grape. Maybe F3Hs play different roles in the flavonoid pathway between V. vinifera and V. davidii. F3′H and F3′5′H belong to the cytochrome P450 super family, and catalyze hydroxylation at the 3′ and 3′,5′ positions of the B-ring of the flavonoid to produce the precursors for cyanidin-based anthocyanins and delphinidin-based anthocyanins, respectively. Thus, components of anthocyanins in grape berry skins are closely related to the expression of F3′H and F3′5′H [62, 69]. The cDNAs of F3′H and F3′5′H were first isolated from petunia [70, 71]. The previous studies showed that grapevines contain two copies of F3′H and sixteen copies of F3′5′H, both of the F3′Hs are located on chr17, fifteen of F3′5′Hs located on chr6, and one of F3′5′Hs located on chr8 [72]. The previous study showed that, the mRNA levels of F3′H and F3′5′H were high in grape berry skins at the harvest stage [73]. In our study, two (VIT_11s0016g01020 and VIT_17s0000g07200) and four (VIT_06s0009g02830, VIT_06s0009g03010, VIT_06s0009g02810 and VIT_06s0009g02920) differential expression genes were annotated as F3′H and F3′5′H, respectively. Of the two F3′Hs, one F3′H gene (VIT_17s0000g07200) was located on chr17. However, the other F3′H gene (VIT_11s0016g01020) was located on chr11. In addition, VIT_11s0016g01020 showed more significantly differential expression level than VIT_17s0000g07200 between black and white grapes. We suggested that VIT_11s0016g01020 (F3′H) played a major role in anthocyanins accumulation in black spine grape. All of the four F3′5′H genes showed high expression level in B2 and B3, especially VIT_06s0009g02830 and VIT_06s0009g03010. But F3′5′H genes were almost not expressed at any of the three stages in white spine grape. Besides, we could see that the expression level of F3′5′H was obviously higher than F3′H in black spine grape. This led to more pentahydroxy-flavone and leucodelphinidin production, which were substrates of malvidin-based anthocyanins. Thus, we detected malvidin-based anthocyanins as the predominant kind of anthocyanins in black spine grape. The dihydroflavonols were catalyzed by DFR and LDOX to produce anthocyanidin. We found the differential expression genes (VIT_18s0001g12800 and VIT_02s0025g04720) annotated as DFR and LDOX, which was consistent with previous studies [67, 68, 74, 75].
Fig. 9.
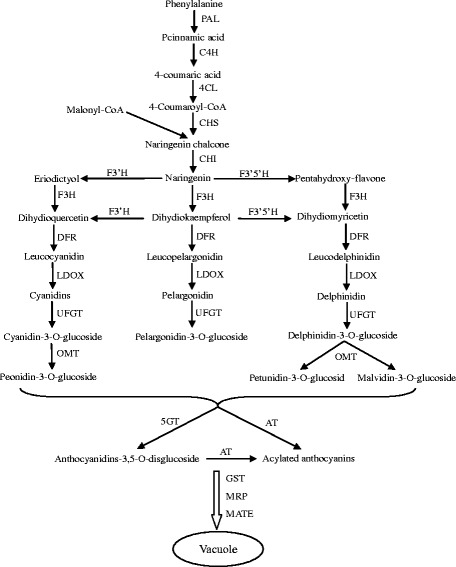
The diagram of the anthocyanins biosynthetic pathway. Abbreviations for the enzymes: PAL phenylalanine ammonia lyase, C4H cinnamate-4-hydroxylase, 4CL 4-coumaroyl-coA synthase, CHS chalcone synthase, CHI chalcone isomerase, F3′H flavonoid-3′-hydroxylase, F3′5′H flavonoid-3′5′-hydroxylase, F3H flavanone-3β-hydroxylase, DFR dihydroflavonol 4-reductase, LDOX leucoanthocyanidin dioxygenase, UFGT UDP-glucose:flavonoid-3-O-glucosyltransferase, OMT O-methyltransferase, 5GT UDP-glucose:anthocyanin-5-O- glucosyltransferase; MRP multidrug resistance-associated protein, MATE toxic compound extrusion, AT anthocyanin acyltransferases, GST glutathione S-transferase
Regulation of anthocyanins synthesis
It has been suggested that there was a mybA gene cluster on chromosome 2 of the grape, which contained MYBA1, MYBA2 and MYBA3 [48]. The expression of MYBA1 and MYBA2 could promote the synthesis of anthocyanins through regulating the expression of UFGT [41, 46, 47]. In our study, MYBA1 (VIT_02s0033g00410) showed high expression levels in B2 and B3, but it did not express in white spine grape. MYBA2 (VIT_02s0033g00390) and MYBA3 (VIT_02s0033g00450) showed high expression levels in B2, B3 and W3. Jiao et al. [57] detected Gret1 in white-fruited varieties of V. davidii. Thus, it was reasonable that MYBA1 did not express in white spine grape. MYBA2 could also promote the expression of UFGT [41]. Though MYBA2 showed high expression level in W3, UFGT (VIT_16s0039g02230) almost did not express and just trace amount anthocyanins were detected in white spine grape. We could infer from this observation that MYBA1 may play a major role in regulating the synthesis of anthocyanins in spine grapes. Besides, another R2R3-MYB gene VIT_01s0011g04760 was annotated as MYB4 expressed both in black and white spine grapes. But it showed significantly differential expression between them. The expression level of VIT_01s0011g04760 in black spine grape was obviously higher than in white spine grape. Thus, it can be regarded as a new candidate gene and needs further studies to confirm whether this gene regulates the biosynthesis of anthocyanins in grapes.
The flavonoid biosynthesis pathway is under the control of Myb transcriptional factors, basic helix–loop-helix proteins (bHLH) and WD40-like proteins [7, 10, 17–19, 36, 38]. The first bHLH was submitted as VvMYCA1 (accession number EF193002; gene ID VIT_15s0046g02560) [76, 77], which was considered to regulate the expression of UFGT. Subsequently, the second bHLH was called VvMYC1 (accession number EU447172; gene ID VIT_07s0104g00090) [39], which was characterized as a component of the transcriptional complex regulating anthocyanin biosynthesis in grapevine. However, both of these two genes did not show obvious differences at the transcriptional level between black and white spine grapes in our study. But, another gene (VIT_14s0060g01010) annotated as bHLH exhibited differential expression between black and white spine grapes. It was shown that WDR1 (Genbank accession number DQ517914) contributed positively to the accumulation of anthocyanins [77]. However, we did not find any WD40 gene having obvious differential expression. In recent years, researches showed that NAC TFs were involved in the regulation of anthocyanins accumulation. A NAC TF has been proposed to be involved in the regulation of anthocyanins accumulation during the response of blood orange to cold exposure [78]. In addition, PpNAC1 can activate the transcription of PpMYB10.1, resulting in anthocyanins pigmentation in blood-fleshed peach [79]. In grape, there was no report that the NAC TFs were related to the anthocyanins accumulation. In our study, a candidate gene (VIT_14s0108g01070) was annotated as NAC, which may be related to anthocyanins accumulation in spine grapes.
Modification of anthocyanins
Anthocyanidins need to be modified by glycosylation, methylation and acylation to form stabilized anthocyanins. In V. vinifera, the glycosylation was catalyzed by UDP-glucose: anthocyanidin: flavonoid glucosyltransferase (UFGT) at C3 position, and the gene encoding UFGT had been cloned [24, 80]. The action of UFGT was crucial for anthocyanin accumulation in grape berry skin [1, 22, 23, 25, 50, 67, 81, 82]. In our study, a differential expression gene (VIT_16s0039g02230) was annotated as UFGT, which had a high expression level in B2 and B3, and did not express in white spine grape. However, in non-V. vinifera species, the 3-5-O-disglucoside anthocyanins are widely present [7, 8]. There must be another UDP-glucose: anthocyanidin: flavonoid glucosyltransferase to catalyze glycosylation at C5 position. In recent years, a few studies reported the isolation of 5GT genes in grapes [83, 84]. Jánváry et al. [83] cloned functional Cha5GT and nonfunctional Dia5GT from the heterozygous hybrid cultivar ‘Regent’, a cross of V. vinifera cv. ‘Diana’ and the interspecific hybrid cv. ‘Chambourcin’. The functional analysis of Cha5GT and Dia5GT suggested that two mutations in the 5GT gene eliminated its enzymatic activity. Because of the absence of active 5GT, dis-glucosidic anthocyanins could not be produced in V. vinifera red grapes. He et al. [84] cloned the full-length cDNA of UDP-glucose: anthocyanin 5-O-glucosyltransferase (Va5GT) from V. amurensis Rupr. cv. ‘Zuoshanyi’. The results suggested that Va5GT was a key enzyme in the biosynthesis of dis-glucosidic anthocyanins in V. amurensis grape berries. In our study, a differential expression gene (VIT_09s0002g06590) showed a high expressional level just in B2 and B3, which was annotated as UGT. It has high homology with Va5GT. Thus, we inferred that the expression of VIT_09s0002g06590 (5GT) led to a high 3-5-O-disglucoside anthocyanins concentration in black spine grape.
In plants, methylated anthocyanidins accounted for a large proportion of the total reported anthocyanidins [3]. Especially, three methylated anthocyanidins, peonidin, petunidin and malvidin, were commonly present in grape berry skin. The methylation was catalyzed by S-adenosyl-L-methionine (SAM) or O-methyltransferase (OMT) at the C3 positions or both at the C3 and C5 positions on the B rings of the anthocyanins in grape [26, 27]. And the cDNAs of several OMT had been cloned in grapes [85, 86]. Subsequently, a QTL for anthocyanins methylation variation was identified that was colocalized with a cluster of three putative OMT genes: VIT_01s0010g03470 (OMT3), VIT_01s0010g03490 (OMT2) and VIT_01s0010g03510 (OMT1) [87, 88]. Fournier-Level et al. [87] reported that OMT2 gene presented two SNPs associated with methylation level. It probably led to a structural change of the OMT2 protein, thus significantly affected the enzyme specific catalytic efficiency for the 3′-O-methylation of delphinidin-3-glucoside. In this study, three differential expression genes (VIT_07s0031g00350, VIT_01s0010g03510 and VIT_01s0010g03490) were annotated as OMTs. Of the three genes, VIT_01s0010g03510 (OMT1) and VIT_01s0010g03490 (OMT2) expressions were consistent with the previous researches [87, 88], which did not express in white spine grape. It is interesting that OMT1 showed higher activity than OMT2 in black spine grape. In addition, a new candidate gene (VIT_07s0031g00350) was also annotated as OMT, and showed high expressional level in B2 and B3. It probably played an important role in the methylation of anthocyanins in spine grape berry skin.
Acylation was a common modification of anthocyanins in grapes. It not only increased diversity of anthocyanins, but also improved the color stabilization and intensity for the anthocyanins [28]. Two enzyme families (BAHD-ATs and SCPL-ATs) have been reported as related to the acylation of anthocyanins [30, 31]. A recent study identified a number of QTLs associated with variation in acylated anthocyanin levels in F1 progeny from a ‘Syrah’x‘Pinot Noir’ cross. The strongest candidate genes within these QTLs included those belonging to the BAHD and SCPL acyltransferase family [88]. But, no QTL was found to cause the presence/absence of acylation in berries. In our study, two differential expression genes (VIT_14s0068g01440 and VIT_03s0017g00870) were annotated as acyltransferase (AT). Especially VIT_03s0017g00870 did not express in white spine grape and at the first stage of black spine grape. Yet it showed high expressional level in B2 and B3. Simultaneously, we detected acylated anthocyanins in black spine grape. Thus, this candidate gene can be very possibly related with the acylation of anthocyanins. It is known that V. vinifera cv. Pinot Noir does not synthesise acylated anthocyanins [89]. After SNP analysis of the candidate genes, at the position 36 of the putative coding region of VIT_03s0017g00870, A is replaced by G, compared with the PN40024 grapevine genome [58], but it does not alter the predicted amino acid sequence. Rinaldo et al. [90] identified a gene, anthocyanin 3-O-glucoside-6-O-acyltransferase (Vv3AT), which encoded a BAHD acyltransferase protein. This protein can promote the synthesis of acylated anthocyanins, which was transcriptionally regulated by VvMYBA. This is consistent with our study.
Anthocyanins transport
Anthocyanins are synthesized in the endoplasmic reticulum and then transported into the vacuoles. Three kinds of anthocyanin transporters: glutathione S-transferase (GST), multidrug resistance-associated protein (MRP) and multidrug and toxic compound extrusion (MATE), were reported as related to the anthocyanins transport [32, 91]. In grapes, a few candidate anthocyanin transporters have been reported.
Conn et al. [92] cloned five GSTs (VvGST1,VvGST2,VvGST3,VvGST4 and VvGST5) from V. vinifera. cv. Gamay Fréaux and the study showed that VvGST1 and VvGST4 coded for the enzymes which had the function of anthocyanins transport. However, in our study, GST1 (VIT_19s0093g00320) was not detected, GST4 (VIT_04s0079g00690) and GST5 (VIT_19s0015g02730) showed significantly differential expression between black and white spine grape. Especially, GST4 just express in B2 and B3 (Fig. 10). GST2 (VIT_07s0005g00030) showed the highest expressional level in the first stage, and it possessed higher transcriptional level in white spine grape than black one. GST3 (VIT_12s0028g00920) also showed the highest expressional level at the first stage, but did not show obvious differential expression between black and white spine grapes. Thus, we inferred that GST4 (VIT_04s0079g00690) and GST5 (VIT_19s0015g02730) play an important role in anthocyanins transport in spine grapes. Besides, another two candidate genes (VIT_19s0015g02690 and VIT_19s0015g02880) were annotated as GSTs, which showed higher expressional level in black spine grape than in white one (Fig. 10). Hence, these two candidate genes are also probably related to anthocyanins transport in spine grapes.
Fig. 10.
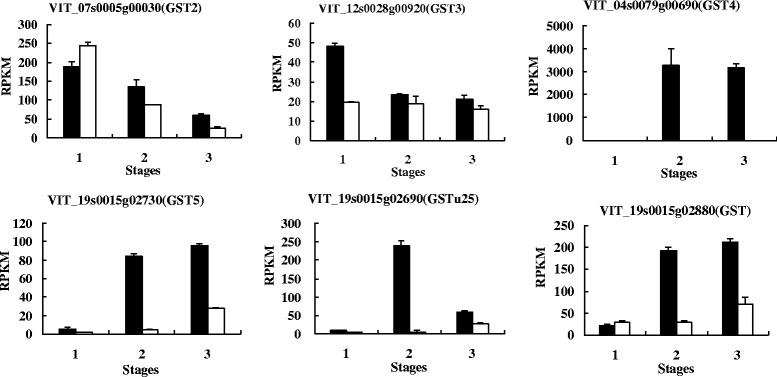
Expressional levels of six GSTs in black and white spine grapes. The vertical axes refer to expression levels as assessed by RNA-Seq, horizontal axes refer to three berry developmental stages. The black columns represent black spine grape, the white columns represent white spine grape
Gomez et al. [93] identified three multidrug and toxic compound extrusion (MATE) genes as candidate anthoMATEs(AMs). In their study, just AM1 and AM3 were cloned from V. vinifera L. cv. Syrah, AM2 was not successfully cloned. They inferred that AM2 was probably not expressed at detectable levels in mature berry or was a pseudogene. Accordingly, their results revealed that AM1 and AM3 just could transport acylated anthocyanins in the presence of Mg ATP [93]. Yet in our study, AM1 (VIT_16s0050g00900) was hardly expressed in spine grapes, the expressional level of AM3 (VIT_16s0050g00930) in white spine grape was higher than in black spine grape. However, AM2 (VIT_16s0050g00910) was mainly expressed in B2 and B3 (Fig. 11). Thus, we infer that AM2 plays an important role in spine grape anthocyanins transportation.
Fig. 11.
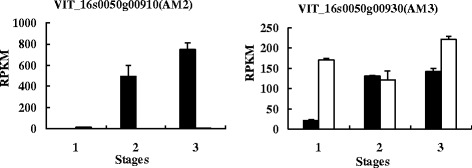
Expressional levels of AM2 and AM3 in black and white spine grapes. The denotations are the same as in Fig. 10
The plant ATP binding cassette (ABC) transporters, in particular from the ABCC subfamily (formerly named multidrug resistance proteins [MRPs]) had been reported as related to the accumulation of flavonoid in vacuoles [94]. In grapes, an ABC transporter, ABCC1 was identified, which localizes to the tonoplast and is involved in the transport of glucosylated anthocyanidins [95]. In our study, four candidate genes were annotated as ABC transporters: VIT_16s0050g02480 (ABCC1), VIT_10s0003g04390 (ABCC2), VIT_09s0002g02430 (ABCC8) and VIT_03s0017g01290 (ABCG11). Francisco et al. [95] were unsuccessful to clone the ABCC2 from the exocarp of grape, and inferred it might not be expressed in detectable amounts during the ripening stage. However, our study showed that both ABCC1 and ABCC2 were expressed in black and white spine grape berry skins. All of the four candidate ABCs were higher in expression in black spine grape than in white spine grape, but the differences were not very significant (Fig. 12). Thus, more studies are needed to confirm the key ABC transporters in grapes.
Fig. 12.
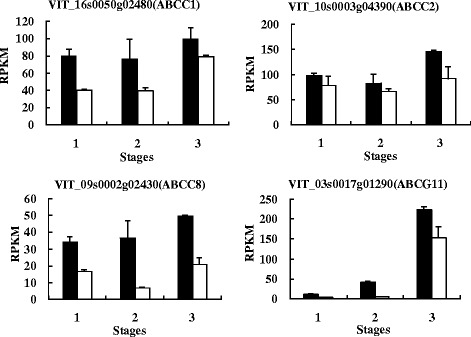
The expressional level of the ABCs in black and white spine grapes. The denotations are the same as in Fig. 10
Conclusions
V. davidii is the only Chinese wild grape species which possesses white berry varieties. High levels of 3,5-O-diglucoside anthocyanins were detected in the black berry skin of V. davidii. The present study investigated the transcriptome profiles of the berry skin from black and white spine grapes at three fruit developmental stages by Illumina mRNA-Seq technology. The examination of absolute expression count for every gene has enabled us to carry out a global investigation of gene expression at these three key time-points in black and white spine grapes. The transcriptome analysis presented thousands of DEGs. We used gene clustering and the enrichment of GO and KEGG to describe the transcriptional patterns of genes involved in anthocyanins accumulation. We found 41 differentially expressed genes probably related to anthocyanins accumulation in V. davidii, including the genes that encode enzymes, transcription factors and transporters involved in anthocyanins biosynthesis, regulation and transport. Some genes were consistent with the previous studies in other grape species, some were newly found in our study. Trace amounts of anthocyanins were detected in berry skins of white V. davidii. Five candidate UGT genes were probably related to the biosynthesis of anthocyanins in white V. davidii. In conclusion, the present study provides new insights into the understanding of anthocyanins accumulation in grapes.
Methods
Sample collection
Berries of black and white spine grapes (V. davidii) were used in this study. Both of the black and white spine grapes were collected from the wild in Hunan, China. The plants were grown in China National Germplasm Resources Repository of Grape, Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences. Skins of grape berries were collected at three different fruit developmental stages: before veraison (Veraison refers to the stage that the berries begin to color and be soft) (40 days after flowering [DAF]), at veraison (80 DAF) and fruit ripe period (120 DAF) (Fig. 1). Two biological replicates were made at each stage. At each stage, we harvested 6–8 clusters at each sampling date, and 5 berries were collected from each cluster. Thus, 30–40 berries were used to collect the skin at each sampling date. After collection, samples were flash-frozen in liquid nitrogen and stored at −80 °C until further processing.
The extraction and determination of anthocyanins
Extraction of total anthocyanins was as described by He et al. [7] with some modifications. The peels were put into mortars and ground in liquid nitrogen. Aliquots of 0.5 g ground powder were added into 10 ml centrifuge tubes with 8 ml 2% formic acid-methanol solution. After ultrasonic oscillation for 10 min, the extracts were put on a shaker in the dark in the table concentrator at 25 °C, 200 rpm for 30 min, followed by centrifugation at 4 °C, 12000 rpm for 10 min. The supernatants were transferred into 50 ml centrifuge tubes. The residues were re-extracted 3 times. The organic fractions were pooled, evaporated by a vacuum rotary evaporator (BUCH, USA) at 40 °C. The residual parts were poured into activated solid phase extraction cartridges. The solid phase extraction cartridges were washed with 5 ml water for 2 times. After removing the leacheate, the solid phase extraction cartridges were eluted with 10 ml methanol for 2 times. The filtrates were collected and evaporated to dryness, then re-dissolved in 5 mL 0.5% hydrochloric acid- methanol solution. Finally, the solutions were filtered through a 0.22 μm Millipore filter for analysis.
The anthocyanins content was determined by an ACQUITY Ultra Performance Liquid Chromatography (UPLC) system (Waters, Milford, MA, USA) linked to both a PDAeλ photo diode array detector (Waters, Milford, MA, USA) and a Micromass Quattro microTM API benchtop triple quadrupole mass spectrometer (Waters MS Technologies, Manchester, UK), with a electrospray ionization (ESI) source operating in multiple reaction monitoring (MRM) mode. Sample solutions were injected into a ACQUITY UPLC®HSS T3 column (2.1 × 150 mm i.d,with 1.8 μm particle size, Waters, Milford, MA, USA), which was maintained at 30 °C. The mass spectrometric acquisition parameters were as follows: ESI source temperature 150 °C, desolvation gas temperature 400 °C, desolvation gas flow rate 800 L/h, cone gas flow rate 50 L/h, collision gas (high purity argon gas) flow rate 0.14 mL/min. The mobile phase had acetonitrile as solvent A, and 0.5% hydrochloric acid solution as solvent B. The gradient profile began with 5–10% A for 1 min, 10–25% A for 16 min, 25–40% A for 18 min, 40–100% A for 19 min, and then returned to initial conditions for 20 min and for 5 min. The flow rate was 1.0 ml min−1 and the column temperature was set at 40 °C. The injection volume was 2.0 μl. The detection wavelength was 520 nm.
Total RNA extraction and qualification
Total RNA was extracted using TIANGEN RNAprep Pure Plant Kit (Tiangen Biotech Beijing, China). RNA degradation and contamination was monitored on 1% agarose gels. RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA). RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
cDNA library construction and transcriptome sequencing
A total amount of 3 μg RNA per sample was used as the input material. Sequencing libraries were generated using NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNAs were purified from the total RNAs using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNaseH−). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. In the reaction buffer, dNTPs with dTTP were replaced by dUTP. The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated for hybridization. In order to select cDNA fragments of preferentially 150–200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 μl USER Enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNAs at 37 °C for 15 min followed by 95 °C for 5 min before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, universal PCR primers and Index (X) Primer. The PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced by an Illumina Hiseq 2000 platform and 100-base paired-end reads were generated.
Sequencing data analysis
Raw data (raw reads) of fastq format were first processed through in-house PERL scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing ploy-N and low quality reads from raw data. At the same time, Q20, Q30 and GC content of the clean data were calculated. All the downstream analyses were based on the clean data with high quality. Reference genome and gene model annotation files were downloaded from genome website directly (ftp://ftp.ensemblgenomes.org/pub/release-23/plants/fasta/vitis_vinifera/ dna/). Index of the reference genome was built using Bowtie v2.0.6 and base paired-end clean reads were aligned to the reference genome using TopHat v2.0.9. We selected TopHat as the mapping tool since TopHat can generate a database of splice junctions based on the gene model annotation file and thus a better mapping result than other non-splice mapping tools.
HTSeq v0.6.1 was used to count the reads numbers mapped to each gene. And then RPKM of each gene was calculated based on the length of the gene and reads count mapped to this gene. RPKM (Reads Per Kilobase of exon model per Million mapped reads) was used for the effect of sequencing depth and gene length for the reads count at the same time, and is currently the most commonly used method for estimating gene expression levels [96].
Differential expression analysis
Differential expression analysis of two conditions/groups (two biological replicates per condition) was performed using the DESeq R package (1.10.1). DESeq provided statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 selected by DESeq were assigned as differentially expressed.
Selection of candidate genes
We used Gene Ontology (GO) and KEGG enrichment analysis of differentially expressed genes to select candidate genes. GO enrichment analysis of differentially expressed genes was implemented by the GOseq R package, in which gene length bias was corrected. GO terms with corrected P-value less than 0.05 were considered significantly enriched by differential expressed genes. KEGG is a database resource for understanding high-level functions and utilities of the biological system, such as the cell, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-through put experimental technologies (http://www.genome.jp/kegg/). We used KOBAS software to test the statistical enrichment of differential expression genes in KEGG pathways. We also used the differentially expressed genes clustering methodology to find the candidate genes. The clustering analysis was made based on the expression pattern of the differentially expressed genes. We selected the genes that the expression patterns were very similar to the genes closely related to the anthocyanins biosynthesis as the candidate genes.
qRT-PCR analysis
Thirty genes were chosen for validation using qRT-PCR. Specific primer pairs for selected genes used in qRT-PCR were designed as shown in Additional file 3: Table S3. The cDNA was transcribed from 1 μg of total RNA using the TOYOBO ReverTra Ace qPCR RT Master Mix (TOYOBO, Japan) in 10 μL of reaction mixture. The qRT-PCR was performed with the Roche LightCycler480 Real-Time Detection System (Roche) with Roche SYBR Green I (Roche, USA). The thermal profile for SYBR Green I RT-PCR was 95 °C for 15 min, followed by 40 cycles of 95 °C for 10s and 55 °C for 30s and 72 °C for 30s. Each plate was repeated three times in independent runs for all reference and selected genes. The reference gene (Actin) was used for normalization. The comparative CT method (2-ΔΔCT method) was used to analyze the expression levels of the different genes [97].
SNPs of candidate gene analysis
Picard-tools v1.96 and samtools v0.1.18 were used to sort, mark duplicated reads and reorder the bam alignment results of each sample. GATK2 software was used to perform SNP calling.
Statistical analysis
All of the experiments analyzed using data comparisons were repeated three times. Statistical analyses were performed using variance (ANOVA) followed by Duncan’s new multiple range tests with SPSS version 17.0 (SPSS, Chicago, IL, USA). A significance level of p < 0.01 was applied.
Acknowledgements
We thank Novogene Bioinformatics Technology Co., Ltd (Beijing, China) for its service in RNA-Seq and transcriptome analysis. We are grateful to Hongmin Zhang from Temple University for his assistance with English editing of the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31372024), China Agriculture Research System (CARS-30) and Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2015-ZFRI).
Availability of data and materials
The datasets used and analysed during the current study available from the corresponding author on reasonable request.
Authors’ contributions
LS, XF, YZ and CL conceived and designed the study. JJ and HS collected berry skin, prepared RNA samples. LS and XF extracted and detected anthocyanins from berry skin. LS and YZ performed the transcriptome analysis and qRT-PCR. LS and CL wrote and revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- 4CL
4-coumaroyl-coA synthase
- 5GT
UDP-glucose:anthocyanin-5-O-glucosyltransferase
- AAT
Anthocyanin acyltransferases
- ABC
ATP binding cassette
- AM
anthoMATEs
- ANR
Anthocyanidin reductase
- bHLH
Basic helix–loop-helix proteins
- bp
Base pair
- C4H
Cinnamate-4-hydroxylase
- cDNA
Complementary DNA
- CHI
Chalcone isomerase
- CHS
Chalcone synthase
- coum
Coumaryl
- cv
Cultivar
- Cy
Cyanidin
- DAF
Day after flowering
- DEG
Differential expression gene
- DFR
Dihydroflavonol 4-reductase
- diglu
Diglucoside
- Dp
Delphinidin
- ESI
Electrospray ionization
- ESI
Electrospray ionization
- F3′5′H
Flavonoid-3′5′-hydroxylase
- F3′H
Flavonoid-3′-hydroxylase
- F3H
Flavanone-3β-hydroxylase
- FLS
Flavonol synthase
- glu
Glucoside
- GO
Gene Ontology
- GST
Glutathione S-transferase
- LAR
Leucoanthocyanidin reductase
- LDOX
Leucoanthocyanidin dioxygenase
- MATE
Toxic compound extrusion
- MRM
Multiple reaction monitoring
- MRM
Multiple reaction monitoring
- MRP
Multidrug resistance-associated protein
- Mv
Malvidin
- OMT
O-methyltransferase
- PAL
Phenylalanine ammonia lyase
- Pg
Pelargonidin
- Pn
Peonidin
- Pt
Petunidin
- qRT-PCR
Quantitative real time poly-merase chain reaction
- QTL
Quantitative trait locus
- RPKM
Reads Per Kilobase of exon model per Million mapped reads
- SAM
S-adenosyl-L-methionine
- SCPL
Serine carboxypeptidase-like
- SNP
Single nucleotide polymorphism
- UFGT
UDP-glucose:flavonoid-3-O-glucosyltransferase
- UPLC
Ultra Performance Liquid Chromatography
Additional files
B2vsW2.DEG_GO_enrichment_result. (XLS 147 kb)
Differential expression UGTs between W3 and W1. (XLS 29 kb)
Primers for qRT-PCR. (XLS 25 kb)
Contributor Information
Lei Sun, Email: slcaas@163.com.
Xiucai Fan, Email: fanxiucai@caas.cn.
Ying Zhang, Email: zhangying05@caas.cn.
Jianfu Jiang, Email: jiangjianfu@caas.cn.
Haisheng Sun, Email: sunhaisheng@caas.cn.
Chonghuai Liu, Email: liuchonghuai@caas.cn.
References
- 1.Sparvoli F, Martin C, Scienza A, Gavazzi G, Tonelli C. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.) Plant Mol Biol. 1994;24:743–755. doi: 10.1007/BF00029856. [DOI] [PubMed] [Google Scholar]
- 2.Castellarin S, Di Gaspero G, Marconi R, Nonis A, Peterlunger E, Paillard S, Adam-Blondon A, Testolin R. Colour variation in red grapevines (Vitis vinifera L.): genomic organisation, expression of flavonoid 3′-hydroxylase, flavonoid 3′,5′-hydroxylase genes and related metabolite profiling of red cyanidin-/blue delphinidin-based anthocyanins in berry skin. BMC Genomics. 2006;7:12. doi: 10.1186/1471-2164-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazza G, Francis FJ. Anthocyanins in grapes and grape products. Crit Rev Food Sci Nutr. 1995;35:341–371. doi: 10.1080/10408399509527704. [DOI] [PubMed] [Google Scholar]
- 4.Holton TA, Cornish EC. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell. 1995;7:1071–1083. doi: 10.1105/tpc.7.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang Z, Wu B, Fan P, Yang C, Duan W, Zheng X, Liu C, Li S. Anthocyanin composition and content in grape berry skin in Vitis germplasm. Food Chem. 2008;111:837–844. doi: 10.1016/j.foodchem.2008.04.069. [DOI] [Google Scholar]
- 6.Arozarena I, Ayestarán B, Cantalejo M, Vera M, Abril I, Casp A. Anthocyanin composition of Tempranillo, Garnacha and Cabernet Sauvignon grapes from high- and low-quality vineyards over two years. Eur Food Res Technol. 2002;214:303–309. doi: 10.1007/s00217-001-0471-0. [DOI] [Google Scholar]
- 7.He J, Liu Y, Pan Q, Cui X, Duan C. Different Anthocyanin Profiles of the Skin and the Pulp of Yan73(Muscat Hamburg × Alicante Bouschet) Grape Berries. Molecules. 2010;15:1141–1153. doi: 10.3390/molecules15031141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Race EJ, Shrikhande AJ. Characterization of Anthocyanins in Grape Juices by Ion Trap Liquid Chromatography − Mass Spectrometry. J Agr Food Chem. 2003;51:1839–1844. doi: 10.1021/jf0260747. [DOI] [PubMed] [Google Scholar]
- 9.Zhu L, Zhang Y, Lu J. Phenolic contents and compositions in skins of red wine grape cultivars among various genetic backgrounds and originations. Inte J Mol Sci. 2012;13:3492–3510. doi: 10.3390/ijms13033492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czemmel S, Heppel SC, Bogs J. R2R3 MYB transcription factors: key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma. 2012;249:109–118. doi: 10.1007/s00709-012-0380-z. [DOI] [PubMed] [Google Scholar]
- 11.Jez JM, Noel JP. Reaction mechanism of chalcone isomerase: pH dependence, diffusion control, and product binding differences. J Biol Chem. 2002;277:1361–1369. doi: 10.1074/jbc.M109224200. [DOI] [PubMed] [Google Scholar]
- 12.Waters DLE, Holton TA, Ablett EM, Lee LS, Henry RJ. cDNA microarray analysis of developing grape (Vitis vinifera cv. Shiraz) berry skin. Funct Integr Genomics. 2005;5:40–58. doi: 10.1007/s10142-004-0124-z. [DOI] [PubMed] [Google Scholar]
- 13.Bogs J, Ebadi A, McDavid D, Robinson SP. Identification of the Flavonoid Hydroxylases from Grapevine and Their Regulation during Fruit Development. Plant Physiol. 2005;140:279–291. doi: 10.1104/pp.105.073262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gollop R, Farhi S, Perl A. Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Sci. 2001;161:579–588. doi: 10.1016/S0168-9452(01)00445-9. [DOI] [Google Scholar]
- 15.Springob K, Nakajima J, Yamazaki M, Saito K. Recent advances in the biosynthesis and accumulation of anthocyanins. Nat Prod Rep. 2003;20:288–303. doi: 10.1039/b109542k. [DOI] [PubMed] [Google Scholar]
- 16.Dixon RA, Steele CL. Flavonoids and isoflavonoids-a gold mine for metabolic engineering. Trends Plant Sci. 1999;4:394–400. doi: 10.1016/S1360-1385(99)01471-5. [DOI] [PubMed] [Google Scholar]
- 17.Dixon RA, Xie D, Sharma SB. Proanthocyanidins-a final frontier in flavonoid research? New Phytol. 2005;165:9–28. doi: 10.1111/j.1469-8137.2004.01217.x. [DOI] [PubMed] [Google Scholar]
- 18.Xie D, Dixon RA. Proanthocyanidin biosynthesis–still more questions than answers? Phytochemistry. 2005;66:2127–2144. doi: 10.1016/j.phytochem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Tian L, Pang Y, Dixon RA. Biosynthesis and genetic engineering of proanthocyanidins and (iso)flavonoids. Phytochem Rev. 2008;7:445–465. doi: 10.1007/s11101-007-9076-y. [DOI] [Google Scholar]
- 20.Fujita A, Soma N, Goto-Yamamoto N, Shindo H, Kakuta T, Koizumi T, Hashizume K. Anthocyanidin reductase gene expression and accumulation of flavan-3-ols in grape berry. Amer J Enol Viticult. 2005;56:336–342. [Google Scholar]
- 21.Gargouri M, Manigand C, Maugé C, Granier T, Langlois d’Estaintot B, Cala O, Pianet I, Bathany K, Chaudière J, Gallois B. Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera. Acta Crystallogr D Biol Crystallogr. 2009;65:989–1000. doi: 10.1107/S0907444909025013. [DOI] [PubMed] [Google Scholar]
- 22.Boss PK, Davies C, Robinson SP. Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol Biol. 1996;32:565–569. doi: 10.1007/BF00019111. [DOI] [PubMed] [Google Scholar]
- 23.Boss PK, Davies C, Robinson SP. Analysis of the Expression of Anthocyanin Pathway Genes in Developing Vitis vinifera L. cv Shiraz Grape Berries and the Implications for Pathway Regulation. Plant Physiol. 1996;111:1059–1066. doi: 10.1104/pp.111.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ford CM, Boss PK, Hoj PB. Cloning and characterization of Vitis vinifera UDP-glucose: flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanidins in vivo. J Biol Chem. 1998;273:9224–9233. doi: 10.1074/jbc.273.15.9224. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi S, Ishimaru M, Ding CK, Yakushiji H, Goto N. Comparison of UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) gene sequences between white grapes (Vitis vinifera) and their sports with red skin. Plant Sci. 2001;160:543–550. doi: 10.1016/S0168-9452(00)00425-8. [DOI] [PubMed] [Google Scholar]
- 26.Bailly C, Cormier FCCO, Do CB. Characterization and activities of S-adenosyl-l-methionine: cyaniding 3-glucoside 3′-O-methyltransferase in relation to anthocyanin accumulation in Vitis vinifera cell suspension cultures. Plant Sci. 1997;122:81–89. doi: 10.1016/S0168-9452(96)04545-1. [DOI] [Google Scholar]
- 27.Ibrahim RK, Bruneau A, Bantignies B. Plant O-methyltransferases: molecular analysis, common signature and classification. Plant Mol Biol. 1998;36:1–10. doi: 10.1023/A:1005939803300. [DOI] [PubMed] [Google Scholar]
- 28.Yonekura-Sakakibara K, Nakayama T, Yamazaki M, Saito K. Modification and stabilization of anthocyanins. In: Gould K, Davies K, Winefield C, editors. Anthocyanins. New York: Springer; 2009. pp. 169–190. [Google Scholar]
- 29.Nakayama T, Suzuki H, Nishino T. Anthocyanin acyltransferases: specificities, mechanism, phylogenetics, and applications. J of Mol Catal B-Enzym. 2003;23:117–132. doi: 10.1016/S1381-1177(03)00078-X. [DOI] [Google Scholar]
- 30.D’Auria JC. Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol. 2006;9:331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Bontpart T, Cheynier V, Ageorges A, Terrier N. BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytol. 2015;208:695–707. doi: 10.1111/nph.13498. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J. Flavonoid transport mechanisms: how to go, and with whom. Trends Plant Sci. 2015;20:576–585. doi: 10.1016/j.tplants.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Koes RE, Quattrocchio F, Mol JNM. The flavonoid biosynthetic pathway in plants: function and evolution. Bioessays. 1994;16:123–132. doi: 10.1002/bies.950160209. [DOI] [Google Scholar]
- 34.Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 36.He F, Pan Q, Shi Y, Duan C. Biosynthesis and Genetic Regulation of Proanthocyanidins in Plants. Molecules. 2008;13:2674–2703. doi: 10.3390/molecules13102674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He F, Mu L, Yan G, Liang N, Pan Q, Wang J, Reeves MJ, Duan C. Biosynthesis of Anthocyanins and Their Regulation in Colored Grapes. Molecules. 2010;15:9057–9091. doi: 10.3390/molecules15129057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owens CL. Pigments in Grape. In: Chen C, editor. Pigments in Fruits and Vegetables. New York: Springer; 2015. pp. 189–204. [Google Scholar]
- 39.Hichri I, Heppel SC, Pillet J, Léon C, Czemmel S, Delrot S, Lauvergeat V, Bogs J. The Basic Helix-Loop-Helix Transcription Factor MYC1 Is Involved in the Regulation of the Flavonoid Biosynthesis Pathway in Grapevine. Mol Plant. 2010;3:509–523. doi: 10.1093/mp/ssp118. [DOI] [PubMed] [Google Scholar]
- 40.Deluc L, Bogs J, Walker AR, Ferrier T, Decendit A, Merillon JM, Robinson SP, Barrieu F. The Transcription Factor VvMYB5b Contributes to the Regulation of Anthocyanin and Proanthocyanidin Biosynthesis in Developing Grape Berries. Plant Physiol. 2008;147:2041–2053. doi: 10.1104/pp.108.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kobayashi S, Ishimaru M, Hiraoka K, Honda C. Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta. 2002;215:924–933. doi: 10.1007/s00425-002-0830-5. [DOI] [PubMed] [Google Scholar]
- 42.Koshita Y, Kobayashi S, Ishimaru M, Funamoto Y, Shiraishi M, Azuma A, Yakushiji H, Nakayama M. Ananthocyanin regulator from grapes, VlmybA1-2, produces reddish-purple plants. J Jpn Soc Hort Sci. 2008;77:33–37. doi: 10.2503/jjshs1.77.33. [DOI] [Google Scholar]
- 43.Cutanda-Perez M, Ageorges A, Gomez C, Vialet S, Terrier N, Romieu C, Torregrosa L. Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol Biol. 2009;69:633–648. doi: 10.1007/s11103-008-9446-x. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304:982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi S, Goto-Yamamoto N, Hirochika H. Association of VvmybA1 gene expression with anthocyanin production in grape (Vitis vinifera) skin-color mutants. J Jpn Soc Hort Sci. 2005;74:196–203. doi: 10.2503/jjshs.74.196. [DOI] [Google Scholar]
- 46.Walker AR, Lee E, Bogs J, McDavid DAJ, Thomas MR, Robinson SP. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007;49:772–785. doi: 10.1111/j.1365-313X.2006.02997.x. [DOI] [PubMed] [Google Scholar]
- 47.This P, Lacombe T, Cadle-Davidson M, Owens CL. Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1. Theor Appl Genet. 2007;114:723–730. doi: 10.1007/s00122-006-0472-2. [DOI] [PubMed] [Google Scholar]
- 48.Fournier-Level A, Le Cunff L, Gomez C, Doligez A, Ageorges A, Roux C, Bertrand Y, Souquet JM, Cheynier V, This P. Quantitative Genetic Bases of Anthocyanin Variation in Grape (Vitis vinifera L. ssp. sativa) Berry: A Quantitative Trait Locus to Quantitative Trait Nucleotide Integrated Study. Genetics. 2009;183:1127–1139. doi: 10.1534/genetics.109.103929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fournier-Level A, Lacombe T, Le Cunff L, Boursiquot J, This P. Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.) Heredity. 2009;104:351–362. doi: 10.1038/hdy.2009.148. [DOI] [PubMed] [Google Scholar]
- 50.Huang Y, Bertrand Y, Guiraud J, Vialet S, Launay A, Cheynier V, Terrier N, This P. Expression QTL mapping in grapevine—Revisiting the genetic determinism of grape skin colour. Plant Sci. 2013;207:18–24. doi: 10.1016/j.plantsci.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 51.He PC. Viticulture. Beijing: China Agriculture Press; 1999. [Google Scholar]
- 52.Kong Q. Chinese Ampelography. Beijing: China Agricultural Scientific and Technical Press; 2004. [Google Scholar]
- 53.Wan Y, Schwaninger H, Li D, Simon CJ, Wang Y, Zhang C. A review of taxonomic research on Chinese wild grapes. VITIS-J Grapevine Res. 2008;47:81–88. [Google Scholar]
- 54.Duan C, He P, Kang J. The study of diglucoside anthocyanins in Chinese wild grape. Journal of Northwest Sci-Tech University of Agriculture and Forestry. 1997;25:23–28. [Google Scholar]
- 55.Zhao Q, Duan C, Wang J. Anthocyanins Profile of Grape Berries of Vitis amurensis, Its Hybrids and Their Wines. Int J Mol Sci. 2010;11:2212–2228. doi: 10.3390/ijms11052212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liang N, Pan Q, He F, Wang J, Reeves MJ, Duan C. Phenolic Profiles of Vitis davidii and Vitis quinquangularis Species Native to China. J Agr Food Chem. 2013;61:6016–6027. doi: 10.1021/jf3052658. [DOI] [PubMed] [Google Scholar]
- 57.Jiao J, Fu X, Liu C, Fan X, Zhang Y, Jiang J. Study of the relationship between the cultivars of Vitis vinifera and the white-fruited and hermaphrodite Chinese wild grapes. Mol Breeding. 2014;34:1401–1411. doi: 10.1007/s11032-014-0124-1. [DOI] [Google Scholar]
- 58.Jaillon O, Aury J, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 59.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Downey MO, Harvey JS, Robinson SP. Synthesis of flavonols and expression of flavonol synthase genes in the developing grape berries of Shiraz and Chardonnay (Vitis vinifera L.) Aust J Grape Wine R. 2003;9:110–121. doi: 10.1111/j.1755-0238.2003.tb00261.x. [DOI] [Google Scholar]
- 61.Fujita A, Goto-Yamamoto N, Aramaki I, Hashizume K. Organ-specific transcription of putative flavonol synthase genes of grapevine and effects of plant hormones and shading on flavonol biosynthesis in grape berry skins. Biosci Biotechnol Biochem. 2006;70:632–638. doi: 10.1271/bbb.70.632. [DOI] [PubMed] [Google Scholar]
- 62.Castellarin SD, Di Gaspero G. Transcriptional control of anthocyanin biosynthetic genes in extreme phenotypes for berry pigmentation of naturally occurring grapevines. BMC Plant Biol. 2007;7:46. doi: 10.1186/1471-2229-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arapitsas P, Oliveira J, Mattivi F. Do white grapes really exist? Food Res Int. 2015;69:21–25. doi: 10.1016/j.foodres.2014.12.002. [DOI] [Google Scholar]
- 64.Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012;69:1030–1042. doi: 10.1111/j.1365-313X.2011.04853.x. [DOI] [PubMed] [Google Scholar]
- 65.Jeong ST, Goto-Yamamoto N, Hashizume K, Esaka M. Expression of multi-copy flavonoid pathway genes coincides with anthocyanin, flavonol and flavan-3-ol accumulation of grapevine. VITIS-J Grapevine Res. 2008;47:135–140. [Google Scholar]
- 66.Goto-Yamamoto N, Wan GH, Masaki K, Kobayashi S. Structure and transcription of three chalcone synthase genes of grapevine (Vitis vinifera) Plant Sci. 2002;162:867–872. doi: 10.1016/S0168-9452(02)00042-0. [DOI] [Google Scholar]
- 67.Jeong ST, Goto-Yamamoto N, Kobayashi S, Esaka M. Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci. 2004;167:247–252. doi: 10.1016/j.plantsci.2004.03.021. [DOI] [Google Scholar]
- 68.Gutha LR, Casassa LF, Harbertson JF, Naidu RA. Modulation of flavonoid biosynthetic pathway genes and anthocyanins due to virus infection in grapevine (Vitis vinifera L.) leaves. BMC Plant Biol. 2010;10:187. doi: 10.1186/1471-2229-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Castellarin SD, Pfeiffer A, Sivilotti P, Degan M, Pererlunger E, Gaspero GD. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant, Cell & Environ. 2007;30:1381–1399. doi: 10.1111/j.1365-3040.2007.01716.x. [DOI] [PubMed] [Google Scholar]
- 70.Holton TA, Brugliera F, Lester DR, Tanaka Y, Hyland CD, Menting JG, Lu C, Farcy E, Stevenson TW, Cornish EC. Cloning and expression of cytochrome P450 genes controlling flower colour. Nature. 1993;366:276–279. doi: 10.1038/366276a0. [DOI] [PubMed] [Google Scholar]
- 71.Brugliera F, Barri-Rewell G, Holton TA, Mason JG. Isolation and characterization of a flavonoid 3′-hydroxylase cDNA clone corresponding to the Ht1 locus of Petunia hybrida. Plant J. 1999;19:441–451. doi: 10.1046/j.1365-313X.1999.00539.x. [DOI] [PubMed] [Google Scholar]
- 72.Falginella L, Castellarin SD, Testolin R, Gambetta GA, Morgante M, Di Gaspero G. Expansion and subfunctionalisation of flavonoid 3′,5′-hydroxylases in the grapevine lineage. BMC Genomics. 2010;11:562. doi: 10.1186/1471-2164-11-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jeong ST, Goto-Yamamoto N, Hashizume K, Esaka M. Expression of the flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase genes and flavonoid composition in grape (Vitis vinifera) Plant Sci. 2006;170:61–69. doi: 10.1016/j.plantsci.2005.07.025. [DOI] [Google Scholar]
- 74.Pastore C, Zenoni S, Tornielli GB, Allegro G, Dal Santo S, Valentini G, Intrieri C, Pezzotti M, Filippetti I. Increasing the source/sink ratio in Vitis vinifera (cv Sangiovese) induces extensive transcriptome reprogramming and modifies berry ripening. BMC Genomics. 2011;12:631. doi: 10.1186/1471-2164-12-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soubeyrand E, Basteau C, Hilbert G, van Leeuwen C, Delrot S, Gomès E. Nitrogen supply affects anthocyanin biosynthetic and regulatory genes in grapevine cv. Cabernet-Sauvignon berries. Phytochemistry. 2014;103:38–49. doi: 10.1016/j.phytochem.2014.03.024. [DOI] [PubMed] [Google Scholar]
- 76.Matus JT, Loyola R, Vega A, Pena-Neira A, Bordeu E, Arce-Johnson P, Alcalde JA. Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J Exp Bot. 2009;60:853–867. doi: 10.1093/jxb/ern336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matus JT, Poupin MJ, Cañón P, Bordeu E, Alcalde JA, Arce-Johnson P. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.) Plant Mol Biol. 2010;72:607–620. doi: 10.1007/s11103-010-9597-4. [DOI] [PubMed] [Google Scholar]
- 78.Crifò T, Petrone G, Lo Cicero L, Lo Piero AR. Short Cold Storage Enhances the Anthocyanin Contents and Level of Transcripts Related to Their Biosynthesis in Blood Oranges. J Agr Food Chem. 2012;60:476–481. doi: 10.1021/jf203891e. [DOI] [PubMed] [Google Scholar]
- 79.Zhou H, Lin-Wang K, Wang H, Gu C, Dare AP, Espley RV, He H, Allan AC, Han Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015;82:105–121. doi: 10.1111/tpj.12792. [DOI] [PubMed] [Google Scholar]
- 80.Offen W, Martinez-Fleites C, Yang M, Kiat-Lim E, Davis BG, Tarling CA, Ford CM, Bowles DJ, Davies GJ. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. Embo J. 2006;25:1396–1405. doi: 10.1038/sj.emboj.7600970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peppi MC, Walker MA, Fidelibus MW. Application of abscisic acid rapidly upregulated UFGT gene expression and improved color of grape berries. VITIS-J Grapevine Res. 2008;47:11. [Google Scholar]
- 82.Hall D, Yuan XX, Murata J, De Luca V. Molecular cloning and biochemical characterization of the UDP-glucose: Flavonoid 3-O-glucosyltransferase from Concord grape (Vitis labrusca) Phytochemistry. 2012;74:90–99. doi: 10.1016/j.phytochem.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 83.Jánváry L, Hoffmann T, Pfeiffer J, Hausmann L, Töpfer R, Fischer TC, Schwab W. A double mutation in the anthocyanin 5-O-glucosyltransferase gene disrupts enzymatic activity in Vitis vinifera L. J Agr Food Chem. 2009;57:3512–3518. doi: 10.1021/jf900146a. [DOI] [PubMed] [Google Scholar]
- 84.He F, Chen W, Yu K, Ji X, Duan C, Reeves MJ, Wang J. Molecular and biochemical characterization of the UDP-glucose: Anthocyanin 5-O-glucosyltransferase from Vitis amurensis. Phytochemistry. 2015;117:363–372. doi: 10.1016/j.phytochem.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 85.Hugueney P, Provenzano S, Verries C, Ferrandino A, Meudec E, Batelli G, Merdinoglu D, Cheynier V, Schubert A, Ageorges A. A novel aation-dependent O-methyltransferase involved in anthocyanin methylation in grapevine. Plant Physiol. 2009;150:2057–2070. doi: 10.1104/pp.109.140376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lücker J, Martens S, Lund ST. Characterization of a Vitis vinifera cv. Cabernet Sauvignon 3′,5′-O-methyltransferase showing strong preference for anthocyanins and glycosylated flavonols. Phytochemistry. 2010;71:1474–1484. doi: 10.1016/j.phytochem.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 87.Fournier-Level A, Hugueney P, Verries C, This P, Ageorges A. Genetic mechanisms underlying the methylation level of anthocyanins in grape (Vitis vinifera L.) BMC Plant Biol. 2011;11:179. doi: 10.1186/1471-2229-11-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Costantini L, Malacarne G, Lorenzi S, Troggio M, Mattivi F, Moser C, Grando MS. New candidate genes for the fine regulation of the colour of grapes. J Exp Bot. 2015;66:4427–4440. doi: 10.1093/jxb/erv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van Buren J, Bertino J, Robinson W. The stability of wine anthocyanins on exposure to heat and 1167 light. Amer J Enol Vit. 1968;19:147–154. [Google Scholar]
- 90.Rinaldo A, Cavallini E, Jia Y, Moss SMA, McDavid DAJ, Hooper LC, Robinson SP, Tornielli GB, Zenoni S, Ford CM, et al. A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce most acylated anthocyanins present in grape skins. Plant Physiol. 2015 doi: 10.1104/pp.15.01255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao J, Dixon RA. The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 2010;15:72–80. doi: 10.1016/j.tplants.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 92.Conn S, Curtin C, Bezier A, Franco C, Zhang W. Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J Exp Bot. 2008;59:3621–3634. doi: 10.1093/jxb/ern217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gomez C, Terrier N, Torregrosa L, Vialet S, Fournier-Level A, Verries C, Souquet JM, Mazauric JP, Klein M, Cheynier V, et al. Grapevine MATE-Type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 2009;150:402–415. doi: 10.1104/pp.109.135624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klein M, Burla B, Martinoia E. The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. Febs Lett. 2006;580:1112–1122. doi: 10.1016/j.febslet.2005.11.056. [DOI] [PubMed] [Google Scholar]
- 95.Francisco RM, Regalado A, Ageorges A, Burla BJ, Bassin B, Eisenach C, Zarrouk O, Vialet S, Marlin T, Chaves MM, et al. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell. 2013;25:1840–1854. doi: 10.1105/tpc.112.102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 97.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analysed during the current study available from the corresponding author on reasonable request.


