Abstract
Genomic sequences and expressed sequence tag data for a diverse group of fungi (Saccharomyces cerevisiae, Schizosaccharomyces pombe, Aspergillus nidulans, Neurospora crassa, and Cryptococcus neoformans) provided the opportunity to accurately characterize conserved intronic elements. An examination of large intron data sets revealed that fungal introns in general are short, that 98% or more of them belong to the canonical splice site (ss) class (5′GU…AG3′), and that they have polypyrimidine tracts predominantly in the region between the 5′ ss and the branch point. Information content is high in the 5′ ss, branch site, and 3′ ss regions of the introns but low in the exon regions adjacent to the introns in the fungi examined. The two yeasts have broader intron length ranges and correspondingly higher intron information content than the other fungi. Generally, as intron length increases in the fungi, so does intron information content. Homologs of U2AF spliceosomal proteins were found in all species except for S. cerevisiae, suggesting a nonconventional role for U2AF in the absence of canonical polypyrimidine tracts in the majority of introns. Our observations imply that splicing in fungi may be different from that in vertebrates and may require additional proteins that interact with polypyrimidine tracts upstream of the branch point. Theoretical protein homologs for Nam8p and TIA-1, two proteins that require U-rich regions upstream of the branch point to function, were found. There appear to be sufficient differences between S. cerevisiae and S. pombe introns and the introns of two filamentous members of the Ascomycota and one member of the Basidiomycota to warrant the development of new model organisms for studying the splicing mechanisms of fungi.
Based on studies with a limited number of organisms, fungal genes appear to differ from those of higher eukaryotes in that fungal genes have relatively long exons and short introns (14, 37, 53). From these data, we hypothesized that fungi as a group would have exon and intron features that are similar as well as different from those of higher eukaryotes. Our approach to testing this hypothesis was to compare the exon and intron characteristics of two well-studied members of the Ascomycota group of fungal organisms, the budding yeast Saccharomyces cerevisiae and the fission yeast Schizosaccharomyces pombe (7, 12, 20, 30, 34, 68), with the intron and exon characteristics of three additional fungi for which genomic sequences and expressed sequence tag (EST) data are available: two filamentous members of the Ascomycota, Aspergillus nidulans and Neurospora crassa (http://www.broad.mit.edu/annotation/fungi/aspergillus/, http://www.broad.mit.edu/annotation/fungi/neurospora/, and http://www.genome.ou.edu/fungal.html) (19, 36, 72), and a member of the Basidiomycota, Cryptococcus neoformans (http://www-sequence.stanford.edu/group/C.neoformans/ and http://www.genome.ou.edu/cneo.html). The availability of genomic sequences and EST data for the latter three fungi permitted the establishment of large intron and exon databases for these organisms by aligning ESTs to genomic sequences. The five fungi selected for the comparisons in this study represent a diverse set of fungal organisms. C. neoformans, a member of the phylum Basidiomycota, diverged from members of the Ascomycota 500 million years ago (26); S. cerevisiae and S. pombe diverged from one another 400 million years ago (61); and N. crassa and A. nidulans diverged from S. cerevisiae 350 million years ago (5). The large volume of sequence data now available for these selected fungi allowed us to examine more comprehensive data sets for comparing fungal intron and exon splicing elements than were previously available.
Fungal gene expression, like that in higher eukaryotes, depends on accurate splicing via the coordinated efforts of a spliceosome. In metazoan systems, spliceosomes are composed of five small nuclear RNAs and over 60 proteins that function as splicing factors (40). The spliceosome coordinates with conserved cis elements in the intron to identify correctly the 5′ and 3′ splice sites (ss) (40). These cis elements consist of the 5′ and 3′ ss, found universally to be predominantly GU and AG, respectively (10, 44); the branch point A and surrounding motif (9, 48, 65); and polypyrimidine tracts (13). In the vertebrate model, early in spliceosome assembly, U1 snRNP binds to the 5′ ss, and a heterodimeric protein in the U2 complex, U2AF, binds to the polypyrimidine tract near the 3′ ss via one subunit, U2AF65, and to the 3′ ss via the other subunit, U2AF35 (40). These splicing complexes facilitate correct excision of the intron sequences and joining of the exon sequences, events which are necessary to obtain an mRNA that can be translated into a functional protein (40). Intron and exon features differ between groups of eukaryotes, and these differences influence the mechanism of recognition of ss, excision of introns, and joining of adjacent exons (14, 39). Information content in the introns at the 5′ and 3′ ss and the branch site, information content in the exon regions adjacent to the introns, and the locations of the polypyrimidine tracts are parameters that vary between groups of eukaryotic organisms and have the potential to influence splicing mechanisms.
In this study, we analyzed intron size distributions, distances from the branch point to the 3′ or 5′ ss, the information content of the ss and the branch site, the information content of the exon regions adjacent to the introns, the distribution of polypyrimidine tracts within the introns, and homologs of selected proteins that have been associated with spliceosomes. We confirmed that fungal introns are typically short and exons are long relative to their mammalian counterparts. The information content needed for splicing is found in fungal introns. Yeast introns have a broader length distribution and a higher information content than introns of the two filamentous fungi and the C. neoformans strain that we analyzed. Since fungal introns are short and have polypyrimidine tracts primarily in the region between the 5′ ss and the branch point, we suspect that the splicing mechanisms of fungi differ from the generally accepted splicing mechanisms described for metazoans. Homologs of U2AF proteins that have been associated with spliceosomes were found in all of the studied fungi except for S. cerevisiae. In addition, homologs of spliceosomal proteins such as Nam8p, a yeast U1 snRNP, and TIA-1, a splicing regulator in metazoans that is associated with splicing of introns with polypyrimidine tracts upstream of the branch site, were found in the fungi. Together, our findings suggest that further studies of fungal splicing mechanisms focusing on novel or nonclassical mechanisms are needed, since the available evidence indicates significant differences between fungi and metazoans.
MATERIALS AND METHODS
Organisms surveyed and sources for genomic sequences and ESTs.
Sources for the genomic sequences and EST data used in this study were as follows: S. pombe, complete genomic database (68) and ESTs from GenBank and dbEST (4) (http://www.ncbi.nlm.nih.gov/); A. nidulans, genomic assembly 1 from Broad Institute (Cereon) (http://www.broad.mit.edu/annotation/fungi/aspergillus/) and ESTs (36) from the University of Oklahoma (http://www.genome.ou.edu/fungal.html); N. crassa, complete genomic sequence (19) assembly 3 from Broad Institute (http://www.broad.mit.edu/annotation/fungi/Neurospora/) and ESTs (72) from the University of Oklahoma (http://www.genome.ou.edu/fungal.html); and C. neoformans strain B3501, genomic sequence from Stanford University (http://www-sequence.stanford.edu/group/C.neoformans/) and ESTs from the University of Oklahoma (http://www.genome.ou.edu/cneo.html).
From the S. cerevisiae complete genomic sequence (20), an annotated intron sequence database was constructed at the Ares Laboratory (23); we downloaded the database from http://www.cse.ucsc.edu/research/compbio/yeast_introns.html. For the calculation of information in the intron and exon regions, the complete genomic sequence was downloaded from GenBank (http://www.ncbi.nlm.nih.gov/), and the predicted coding sequences were downloaded from the Saccharomyces genome database (http://www.yeastgenome.org/). Introns and exons for Histoplasma capsulatum and Coccidioides immitis were obtained by downloading the preformatted GenBank 132 exon-intron database (http://mcb.harvard.edu/gilbert/eid/) (55); purging was done with exon-intron database filter_exp_keywl.p1 and filter_exp_keyw2.pl to remove genes that were identified only in silico. The intron and exon selection and validation methods were described previously (15).
Definitions applied.
The International Union of Pure and Applied Chemistry standard abbreviations for nucleotides were used throughout this work (28). Briefly, they are as follows: A, adenine; C, cytosine; G, guanine; T, thymine; U, uridine; Y, thymine, uridine, or cytosine; R, adenine or guanine; W, adenine, thymine, or uridine; M, adenine or cytosine; S, guanine or cytosine; K, guanine, thymine, or uridine; and N, adenine, guanine, cytosine, thymine, or uridine. To be called a consensus nucleotide, the nucleotide frequency must exceed 40% at the given position. For a degenerate call, the second nucleotide must occur more than 30% of the time, and the frequencey must be equal to or greater than two times the frequency of the third most frequently found nucleotide at that position (57).
Preparation of the cDNA library.
C. neoformans strain B3501 was kindly provided by J. Kwon-Chung (National Institutes of Health, Bethesda, Md.). C. neoformans yeast cells were cultured in yeast extract-peptone-dextrose broth at 30°C with shaking for 16 h. Following incubation and washing of the yeast cells, RNA was isolated by using a Mini-BeadBeater 8 apparatus (Biospec Products, Bartlesville, Okla.) in combination with 0.5-mm Zr/Si beads and an RNeasy kit (Qiagen, Santa Clarita, Calif.) according to the manufacturer's directions. The integrity of the total RNA was confirmed by formaldehyde-agarose gel electrophoresis. Poly(A)+ RNA was purified from total RNA by using a PolyATtract kit (Promega, Madison, Wis.). The cDNA library was constructed by synthesizing cDNA from poly(A)+ RNA by using a cDNA synthesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions, except that priming for first-strand synthesis was done with a cocktail of three 1-base-anchored poly(dT)-containing primers. The three primers consisted of a protected XhoI restriction site followed by a 5-nucleotide (nt) tag sequence (GACAC), an 18-base poly(dT) sequence, and an A, a C, or a G. Thus, the three primers differed only in the final base (A, C, or G). Following second-strand synthesis, the cDNA ends were made blunt, and EcoRI linkers were ligated. The cDNAs were digested with XhoI, size selected (400 bp and greater), directionally ligated into predigested Uni-ZAP XR (Stratagene), and packaged with Gigapack III Gold packaging extracts. The titer of the resulting cDNA primary library was determined. Samples of the primary library were subjected to mass excision by using ExAssist helper phage (Stratagene), and individual clones were picked from the resulting primary library for sequencing.
Procedures for sequencing of the cryptococcal cDNA library.
A C. neoformans double-stranded DNA template was isolated from the selected clones in a 96-well sample format by using a cleared-lysis method (72). For sequencing, approximately 0.2 μg of DNA was used with 20 pM universal M13 forward primer (5′-TGTAAAACGACGGCCAGT-3′) or T3 primer (5′-CGAAATTAACCCTCACTAAAG-3′) and 2 μl of ABI BigDye terminator mixture (PE-ABI 4303150) diluted 1:3 with 5× TM buffer (400 mM Tris-HCl [pH 9.0], 10 mM MgCl2). Thermocycling was done for 1 cycle of 95°C for 30 s, 60 cycles of 95°C for 10 s, 50°C for 5 s, and 60°C for 4 min, and holding at 4°C. DNA in the reactions was ethanol precipitated. Electrophoresis was performed by using ABI 3700 sequencers with a POP5 polymer for 2 h 50 min at an EP voltage of 6.5 kV and an EP current of 550 mA. Electrophoresis sequencing data were transferred to networked Sun workstations.
C. neoformans EST database preparation and assembly.
The EST database was constructed as follows. The sequences obtained with the universal forward primer were designated 3′ ESTs and given a .f1 suffix, while the sequences generated with the T3 primer were designated 5′ ESTs and given a .r1 suffix. A piped set of scripts was used in a semiautomatic process to screen each sequence for overall base quality by using Phred (P. Green, http://www.phrap.org/) and to remove vector, mitochondrial, ribosomal, and Escherichia coli contaminating sequences. The sequences passing the screen were termed high-quality ESTs and were subjected to a BLASTX search of the nonredundant protein (nr) database in GenBank. A FASTA sequence file of the ESTs and corresponding cloning and sequence data has been placed at http://www.genome.ou.edu/.
The 3′ ESTs were assembled separately by using Phrap (P. Green, http://www.phrap.org/) with a minmatch of 14 and a minscore of 80 in a cumulative fashion in order to monitor the level of clone redundancy. Both the 3′ and the 5′ ESTs were assembled by using Phrap as described above. The EST contigs were examined for chimeric sequences in a cursory fashion by examining any ESTs that did not align in the expected pattern of 5′ EST and reverse complement 3′ EST. Misaligned sequences were removed, and the entire database was reassembled. All members of the assembled EST database were examined for homology to the GenBank nr database by batch analysis. The assembled C. neoformans EST database and BLAST results were placed at http://www.genome.ou.edu/ in separate directories. The C. neoformans B3501 EST database contained 1,965 contigs and 1,168 singlets.
Intron and exon database construction.
The intron and exon databases for all organisms except S. cerevisiae were created by using FELINES (15). Briefly, the genomic sequence databases were formatted by using formatdb (2). Each FASTA-formatted genomic sequence and each FASTA-formatted EST were placed into separate files by using all2many.pl (J. D. White and B. A. Roe, unpublished data; http://www.genome.ou.edu/informatics.html). Next, two list files were created for each organism, one containing the names of all of the EST files created above and one containing all of the genomic sequence files created above. A FELINES option file was customized for each organism and contained the following parameters, which were constant between organisms: BLAST e-value, 0.001; minimum HSP value, 50; splice site scoring matrix, v; minimum intronless exon length, 300; intron length range, 20 to 2,000; acceptable splicing classes, GUAG, GCAG, AUAC, AUAG, and AUAA; minimum exon number, 1; mRNA and genomic identity minimum percentage, 90; minimum mRNA coverage percentage, 80; maximum number of mRNA gaps, 10; and maximum number of mRNA mismatches, 200. The intron and exon sequence databases then were constructed by running wiscrs.pl to create the Spidey alignment files and gumbie.pl in the default-filtered mode to extract the intron and exon sequences into their respective databases (15).
Branch sites.
Branch sites were identified by using the icat.pl program (15). The sequence CURAY was chosen as the primary motif, UURAY was chosen as the secondary motif, and a modified YURAY motif in which either the first, third, or fifth position was allowed to be any nucleotide was chosen as the alternate motif. The icat.pl program searched for each regular expression of the motifs and then chose the 3′-most instance of the motif in each intron sequence based on branch site motifs previously described for metazoans and S. cerevisiae (6, 9, 23).
Polypyrimidine tracts.
Polypyrimidine tracts were defined as at least six consecutive nonadenine nucleotides containing no fewer than three uridines (13, 54, 59, 60) and were identified by using FELINES perl scripts, cattracts.pl, and icat.pl (15).
Information content determinations.
The information content of the intron and exon regions was determined by using the CONSENSUS utility (27). Briefly, FASTA sequences containing 20 nt of the 5′ end of each intron, 13 nt flanking the branch point A (9 nt to the 5′ side of the branch point A and 3 nt to the 3′ side of the branch point A) of each intron, 20 nt of the 3′ end of each intron, and 5 nt in the exon region adjacent to each intron were created. These sequences were formatted for use in the CONSENSUS utility by using FASTA consensus version 2c. A frequency matrix for the sequences was created by using make-matrix version 2.4. Information content then was calculated by using gmat-inf-gc version 2c. Finally, the P values for comparisons of individual alignment regions with random sequences were calculated by using P value version 3a. The background nucleotide distribution was assumed to be completely random for all of the organisms studied.
Homologs of spliceosomal proteins.
To identify fungal homologs of the human branch point binding protein (BBP), S. pombe U2AF65 and U2AF35 subunits, S. cerevisiae Nam8p, and Homo sapiens TIA-1 and TIAR, the individual protein homolog sequences were compared to the genomic sequence and EST databases of S. pombe, A. nidulans, N. crassa, and C. neoformans B3501 by using TBLASTN. All of the sequences used were from GenBank, and the accession numbers were as follows: U2AF65 and U2AF35, homolog accession numbers CAB46760 and Q09176, respectively; BBP, accession number AF073779 1; Nam8p, accession number NP_0011954; S. pombe CSX1, accession number NP_594243; H. sapiens TIA-1, accession number NP_071505; and H. sapiens TIAR, accession number NP_003243. Translation of the DNA sequences was done by using FGENSH at the Softberry website (http://www.softberry.com/) with either the S. pombe or the N. crassa matrix provided or by using the TBLASTN output when the FGENSH matrix was inadequate. Domain searches were performed by using the National Center for Biotechnology Information con-served domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (41) and the protein family website (http://www.sanger.ac.uk/Software/Pfam/index.shtml) (3), which uses a hidden Markov model-based similarity search.
Multiple alignments of spliceosomal proteins.
Multiple alignments were created by using xced (version 3.93), an X-windows-based multiple-alignment program (http://www.biophys.kyoto-u.ac.jp/∼katoh/programs/align/xced/), to align the sequences and ClustalX (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/, version 1.83), a local stand-alone windowed version of ClustalW, to produce the multiple alignments (29). The scoring matrix used was JTT200PAM, with an AO of +0.06, an OGC of 2.4, and an EGC of 0.0. Percent identity was calculated by using MacVector with ClustalW pairwise alignment in the slow alignment mode with the default parameters.
Phylograms.
Phylograms were prepared by using a website-based implementation of ClustalW (http://clustalw.genome.jp/). The pairwise parameters used were as follows: fast/approximate, K-tuple size, 1; window size, 5; gap penalty, 3; number of top diagonals, 5; and scoring method, percentage. The multiple-alignment parameters used were as follows: gap open penalty, +10.0; gap extension penalty, 0.05; weight transition, no; hydrophobic residues for proteins, GPSNDQERK; hydrophilic gaps, yes; and weight matrix, BLOSUM.
Statistics.
Intron lengths were compared by using the Kruskal-Wallis test with Dunn's multiple-comparison posttest. Correlation coefficients were calculated by using the Correl function in the OpenOffice.org spreadsheet (http://www.openoffice.org/).
Nucleotide sequence accession numbers.
The ESTs determined here were submitted to dbEST under accession numbers CF182795 through CF194965.
RESULTS
Genome characteristics and data sets for the five organisms in the survey.
For this study, S. cerevisiae and S. pombe were selected as model fungal organisms because complete genomic sequence data were available (20, 68). Genomic sequence and EST data for the two additional members of the Ascomycota and the member of the Basidiomycota recently became available, making a comparative study of conserved intron and exon cis elements among these five organisms possible.
Characteristics of the genomes for the five organisms and the derived intron and exon data sets used in this study are shown in Table 1. The genome size for this group of fungi ranged from 12 to 43 Mb (Table 1). The estimated mean number of introns per gene varied considerably among the five fungi—the lowest being found in S. cerevisiae (0.04) and the highest being found in C. neoformans (2.42) (Table 1). The total number of introns in the data set for each organism ranged from 253 (approximately 100% of the S. cerevisiae introns) to 5,725 (approximately 36.5% of the C. neoformans introns). The intron data sets for S. pombe (1,280 introns), A. nidulans (2,115 introns), and N. crassa (1,897 introns) were sizable but represented only 27.1, 17.3, and 10.8%, respectively, of the estimated total introns (Table 1). With the exception of the S. cerevisiae data set, intron and exon data sets were based on alignments of ESTs with genomic sequences for each respective organism, allowing accurate intron-exon boundary predictions.
TABLE 1.
Characteristics of genomes, introns, and exons
| Organism | Estimated no. of genes in genome (genome size, in kb) | Estimated gene density (no. of genes/kb)a | Estimated no. of introns in genome | Estimated mean no. of introns/gene | No. of introns surveyed | Estimated % of total introns in the organismb | Meanc (ranged) intron length, in nt | No. of exons surveyed | Meanc (ranged) exon length, in nt |
|---|---|---|---|---|---|---|---|---|---|
| S. cerevisiae | 5,773 (12,070)e | 0.48 | 253f | 0.04g | 253 | 100.0 | 256 (52-1,002) | NDm | ND |
| S. pombe | 4,940 (13,800)g | 0.36 | 4,742g | 0.96g | 1,286 | 27.1 | 107 (35-817) | 3,944 | 330 (13-1,109) |
| A. nidulans | 12,000 (28,500)h | 0.42 | 11,520i | 0.96j | 1,997 | 17.3 | 73 (27-1,903) | 3,612 | 361 (6-2,309) |
| N. crassa | 10,082 (43,000)k | 0.23 | 17,139i | 1.7k | 1,850 | 10.8 | 119 (46-1,740) | 3,694 | 356 (8-4,370) |
| C. neoformans | 6,482 (18,500)l | 0.35 | 15,686i | 2.42j | 5,697 | 36.3 | 69 (35-1,978) | 7,548 | 207 (7-1,642) |
Calculated by dividing the estimated number of genes in the genome by the genome size in kilobases for each organism.
Number of introns in the survey divided by the estimated number of introns in the genome for each organism × 100.
Mean number of nucleotides in introns or exons in the survey, calculated by using get_inex.pl (15).
Range in number of nucleotides in introns or exons.
Ares Laboratory YIDB (23).
See reference 68.
Data are from reference 36 (http://www.ncbi.nlm.nih.gov/mapview/).
Estimated mean number of introns per gene times estimated number of genes in the genome of the organism.
Each contig in the EST database was defined as a gene, and then the numbers of introns in each gene (determined by alignment) were counted and averaged.
See reference 19 (http://www.ncbi.nlm.nih.gov/mapview/).
Fungal Genome Initiative Whitepaper (http://www.ncbi.nlm.nih.gov/mapview/).
ND, not done.
Intron length (size).
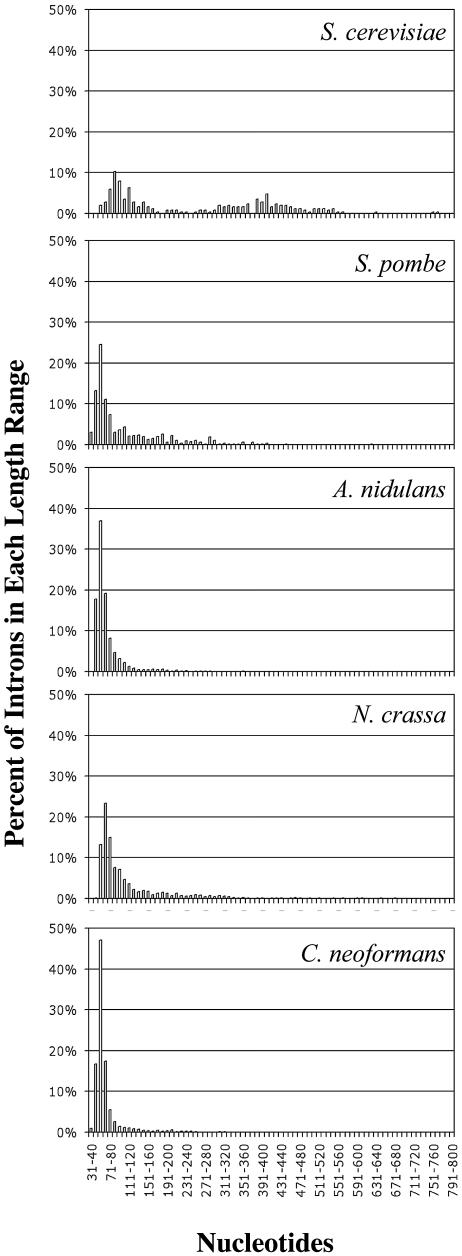
Fig. 1 shows the ranges of intron sizes for each of the five fungi. The mean intron length for S. cerevisiae is significantly larger than those for the other fungi in the study (P < 0.001). As reported by others, the size distribution of S. cerevisiae introns has a distinct bimodal pattern, with approximately 25% of the S. cerevisiae introns falling in the size range of 401 to 2,000 nt (38, 52, 62). The size distribution of S. pombe introns, with a mean length of 107 nt (Table 1), was more similar to those of the other three fungi (Fig. 1). A. nidulans, N. crassa, and C. neoformans had narrow ranges of intron lengths, with a dominant peak distribution between 50 and 70 nt (Fig. 1).
FIG. 1.
Comparison of intron length distributions among five fungi. The length ranges (in nucleotides) are shown on the x axes, and the percentages of introns in each length range are shown on the y axes.
Intron consensus elements.
Aligning ESTs with genomic sequences allows the accurate identification of exon-intron boundaries and the subsequent determination of conserved intron cis elements. The conserved sequence elements of splicing that have garnered the most interest are the 5′ ss, the 3′ ss, the branch point, and the polypyrimidine tracts. We surveyed the introns in our databases for the presence of these elements.
The utility that we used for generating our intron data sets, FELINES (15), filtered out all identified sequences that did not conform to the 5′GU…AG3′, 5′GC…AG3′, or 5′AU…AC3′ dinucleotide ss pairs. The percentages of sequences not meeting these criteria were 17% for S. pombe, 11% for A. nidulans, 19% for N. crassa, and 7% for C. neoformans. The majority (98 to 99.9%) of the remaining introns from all five fungi in this study had the canonical 5′GU…AG3′ donor-acceptor ss pairs. The percentages of the 5′GC…AG3′ class for S. cerevisiae, A. nidulans, N. crassa, and C. neoformans were 1.19, 1.15, 0.86, and 1.98%, respectively. These percentages are higher than that found for the mammalian 5′GC…AG3′ class (0.56%) based on a GenBank representation (11). S. pombe 5′GC…AG3′ introns, at 0.08%, were the exception. In the human genome, 5′AU…AC3′ introns represent 0.04% of the total (11). We found that 0.09% or 2 of the A. nidulans introns were in the 5′AU…AC3′ ss class. In both cases, two or more ESTs showing excellent alignment with the genomic sequence defined the A. nidulans 5′AU…AC3′ introns. The remaining intron data sets had no 5′AU…AC3′ introns.
5′ ss (donor sequence).
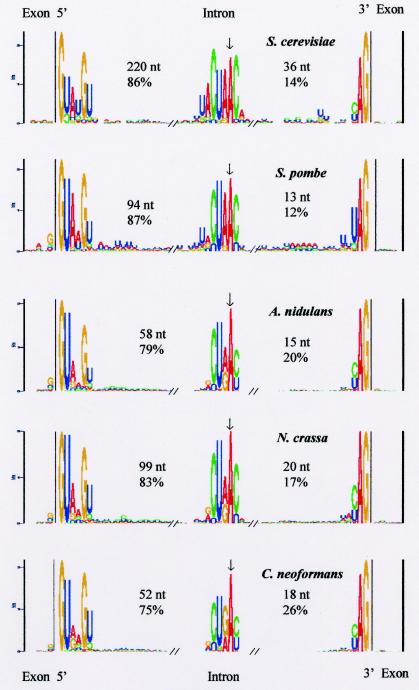
The consensus sequence for the region of the 5′ ss for each of the fungal organisms in this study is shown as a component of the structure logos in Fig. 2. The fungal consensus sequence derived from the five organisms, based on 11,083 introns and including the 5′GU…AG3′, 5′GC…AG3′, and 5′AU…AC3′ classes, was found to be NG|GURWGU (where the vertical bar indicates the splice junction and underlining indicates the first two bases of the intron), which was more degenerate than the metazoan 5′ ss consensus sequence, CAG|GUAAGU (9, 35). The fungal +4 base was an A or a U, a variation from the metazoan consensus sequence. When the two yeasts were excluded, the two filamentous members of the Ascomycota and the member of the Basidiomycota had a 5′ ss consensus sequence (NG|GURAGU) that was closer to the metazoan consensus sequence. For the yeast 5′ ss, a separate and longer consensus sequence (NG|GUAWGUW) could be constructed. U12-type introns have a 5′ ss motif (RUAUCCUUU) that differs from that of U2-type introns (9, 58), but this motif was not present in our data sets.
FIG. 2.
Sequence structure logos derived from the introns and exons in the database for each of the five fungi. The consensus sequence structure logos (21) for the 3′ end of the exon adjacent to the intron, the 5′ donor ss, the branch site, the 3′ acceptor ss, and the 5′ end of the exon adjacent to the intron are shown from left to right. The x axes show the nucleotide positions in the multiple alignments. The y axes reflect the frequency of occurrence of a base (bits), and the relative heights of the letters within the columns are proportional to the frequencies of the letters in the columns in the multiple alignments. Only columns in which the frequencies are significantly above random variation are shown.
3′ ss (acceptor sequence).
The metazoan 3′ ss consensus sequence, YAG, was also characteristic of all five organisms in this study (Fig. 2). We identified as well an extended fungal 3′ ss consensus sequence, WNYAG; in addition, when S. cerevisiae introns were excluded, a less degenerate sequence, UNYAG, could be defined. In addition, the 3′ ss sequences of S. cerevisiae and S. pombe introns had an extended A/U tract 5′ of the YAG acceptor site (Fig. 2). This finding was confirmed by determining the A/U contents in the region between the branch point and the 3′ ss for S. cerevisiae and S. pombe, which were 66 and 72%, respectively; the other three organisms had between 55 and 58% A/U contents in the same region.
Branch site consensus sequence.
The branch site is a key intron consensus sequence element required for lariat formation during the splicing process (50). The metazoan and S. cerevisiae branch site consensus sequences have been determined to be YNCURAY and UACUAAC, respectively, where the underlined A is the branch point (6). The reported branch site consensus sequence for S. pombe, CURAY, is more degenerate than that for S. cerevisiae (71).
Greater than 98% of the introns in our data sets could be shown to have potential branch sites by using the FELINES icat.pl script (15). As a training and validation step, the icat.pl script identified branch sites in 100% of the 253 S. cerevisiae introns (15) from the Ares Laboratory yeast intron database (YIDB) (23). The consensus sequences for the putative branch sites for each organism are shown in Fig. 2. From these data, we could derive a general fungal branch site consensus sequence of RCURAY, where the underlined A is the branch point. An earlier survey of a small number of annotated introns of H. capsulatum and C. immitis, two dimorphic pathogenic fungi, also showed that their branch site consensus sequences were in agreement with the general fungal branch site sequence defined above (data not shown).
U12-type introns have a unique branch site consensus sequence, UCCUURAC (9, 67), which was not found in the yeast data sets but which was present in 0.08% of A. nidulans introns, 0.22% of N. crassa introns, and 0.11% of C. neoformans introns.
The branch point A could be localized to a position between 13 and 36 nt from the 3′ end of the intron and between 52 and 220 nt from the 5′ ss, depending on the organism (Fig. 2). There was a very high correlation between the total number of nucleotides in an intron and the number of nucleotides in the region from the 5′ ss to the branch point A. Thus, variability in intron length seems to be a function of the distance between the 5′ ss and the branch point (Table 2).
TABLE 2.
Correlation of intron length in nucleotides with the number of nucleotides between the branch point and the 5′ or 3′ splice junction
| Organism | Correlation coefficient for total no. of nt in intron with no. of nt in the following region:
|
|
|---|---|---|
| 5′ to branch point | 3′ to branch point | |
| S. cerevisiae | 0.99 | 0.15 |
| S. pombe | 1.00 | 0.09 |
| A. nidulans | 0.99 | 0.16 |
| N. crassa | 0.98 | 0.23 |
| C. neoformans | 0.97 | 0.37 |
Intron and exon information content.
Information content is a measure of the sequence conservation in a given region. Lim and Burge (39) have shown that short introns of Drosphila melanogaster and Caenorhabditis elegans contain basically all of the information needed for recognition of the splicing machinery. Considering that most fungal introns are short, it might be anticipated that the information for splicing resides mainly in the introns at the highly conserved regions defining the 5′ ss, branch site, and 3′ ss and thus fits the intron definition mechanism (39). Consequently, one of our objectives was to determine whether there is indeed a substantially higher information content in the introns than in the flanking exon regions of the representative fungi. The sequence structure logos in Fig. 2 show that the sequences in the three intron regions of all of the fungi represented are more highly conserved than those in the exon regions flanking the introns.
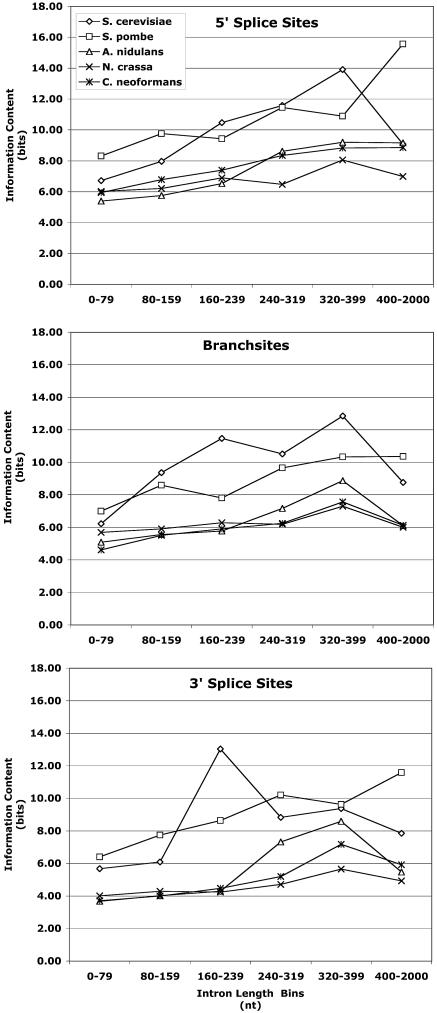
Having found that the introns of S. cerevisiae have a broader size distribution than those of the other fungi (Fig. 1), we also wondered whether the information content in the fungal introns and exons changes as the length of the introns increases. To address this question, we analyzed the sequence conservation (information content in bits) in the introns in 20 nt at the 5′ or 3′ ss and in 13 nt at the branch site and in the exons in 5 nt adjacent to the 5′ and 3′ ss of the five fungi in this study. The introns were divided into six bins with increasing length ranges, and P values were calculated as described by Hertz and Stormo (27). From comparisons of bits of information in a random sequence to bits of information in conserved regions, we found that the information content for each intron length range for each organism was significantly higher than that in a random sequence for the number of nucleotides represented (P < 0.001) (Fig. 3). S. cerevisiae and S. pombe had the highest information content in each conserved region of the introns, irrespective of the intron length range, relative to the other three fungi in this study (Fig. 3). The information content in the exon regions was higher than that in a random sequence for exons adjacent to introns that were less than 240 nt long for each organism (P < 0.01). The numbers and percentages of introns in each length range are shown in Table 3.
FIG. 3.
Information content at the 5′ ss, the branch site, and the 3′ ss for introns in various length ranges. The information content (bits) in 20 nt at the beginning of the intron (top panel), 13 nt around the branch point (9 nt on the 5′ side of the branch point A and 3 nt on the 3′ side) (middle panel), and 20 nt at the end of the intron (bottom panel) was determined for introns in each length range.
TABLE 3.
Number of introns in each nucleotide length range for five fungi
| Organism | No. of introns in the following nt length range (% of total introns):
|
||||||
|---|---|---|---|---|---|---|---|
| 0-2,000 | 0-79 | 80-159 | 160-239 | 240-319 | 320-399 | 400-2,000 | |
| S. cerevisiae | 301 | 44 (14.6) | 114 (37.9) | 7 (2.3) | 16 (5.3) | 57 (18.9) | 63 (20.9) |
| S. pombe | 1,279 | 752 (58.8) | 275 (21.5) | 136 (10.6) | 83 (6.5) | 20 (1.6) | 13 (1.0) |
| A. nidulans | 2,092 | 1,715 (82.0) | 287 (13.7) | 60 (2.9) | 11 (0.5) | 7 (0.3) | 12 (0.6) |
| N. crassa | 1,891 | 952 (50.3) | 607 (32.1) | 154 (8.1) | 97 (5.1) | 32 (1.7) | 49 (2.6) |
| C. neoformans | 5,598 | 4,890 (87.4) | 500 (8.9) | 132 (2.4) | 36 (0.6) | 14 (0.3) | 26 (0.5) |
With the exception of introns that were longer than 400 nt for S. cerevisiae, the information content in the 5′ ss, branch site, and 3′ ss clearly increased as the intron length increased for S. cerevisiae and S. pombe (Fig. 3). There was some increase in information content in the 5′ ss region and the 3′ ss region as the intron length increased for A. nidulans and C. neoformans, but the increase in information content as the intron length increased was not as great as that for S. cerevisiae and S. pombe, and the increase did not continue into the 400- to 2,000-nt range for A. nidulans and C. neoformans (Fig. 3, top and bottom panels). A. nidulans introns showed a moderate increase in information content at the branch site as the intron size increased up to 399 nt (Fig. 3, middle panel). For N. crassa and C. neoformans, the information content at the branch site increased very slightly as the intron length increased, especially in the intron length ranges where most of the introns are found (<240 nt) (Fig. 3, middle panel).
The information content in the exon regions adjacent to each intron was determined for each bin of introns for each organism and was found to be minimal or absent in some cases (Fig. 2; see also Fig. S1 in the supplemental material). The 5 nt in the exon bordering the 5′ end of the intron had a significant amount of information compared to that in a random sequence (P < 0.01), except for exons adjacent to S. cerevisiae 400- to 2,000-nt introns and exons adjacent to A. nidulans and C. neoformans introns longer than 240 nt (Fig. 2; see also Fig. S1A in the supplemental material). In the 5 nt in the exon adjacent to the 3′ ss, the highest information content was observed in exons adjacent to S. cerevisiae introns shorter than 320 nt; however, for introns 320 nt long or longer, the highest information content was observed in exons adjacent to S. pombe introns (see Fig. S1B in the supplemental material). The information content for A. nidulans, N. crassa, and C. neoformans exons adjacent to the 3′ ss was lower than that for S. cerevisiae and S. pombi exons but above that in a random sequence for exons adjacent to introns shorter than 240 nt. For this latter group of fungi, only N. crassa exons adjacent to the 3′ ss of introns longer than 240 nt had information content significantly higher than that in a random sequence (P < 0.02) (see Fig. S1B in the supplemental material).
Polypyrimidine tracts.
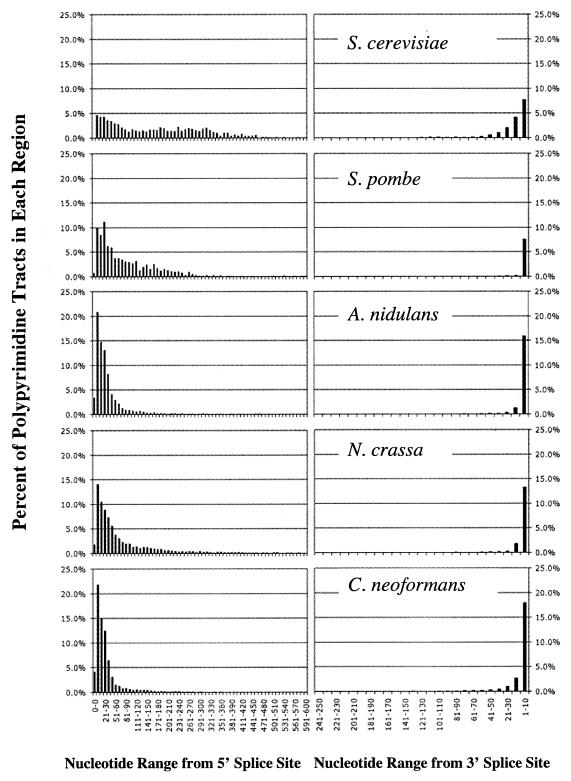
Polypyrimidine tracts in mammalian introns are conserved elements typically found near the 3′ ss, and they function as a binding site for spliceosomal protein U2AF65 (49). We screened for polypyrimidine tracts in the introns in our data sets by using a minimal definition of six consecutive nucleotides with at least 3 U's and no A's (13, 15, 59). We defined two intron regions, the region from the 5′ ss to the branch point and the region from the branch point to the 3′ ss, screening both for the presence of polypyrimidine tracts. From our results, introns could be placed into four classes based on the locations of the polypyrimidine tracts with reference to the branch point. Table 4 shows the percentages of polypyrimidine tracts in each of the four classes. Figure 4 shows the distribution of the distances (in nucleotides) from the 5′ ss or the 3′ ss to the branch point for polypyrimidine tracts for each organism. The majority of introns in all five organisms (83.2 to 93.7%) had polypyrimidine tracts in the region from the 5′ ss to the branch point (Table 4). Surprisingly, 48 to 62% of the introns in S. pombe, A. nidulans, N. crassa, and C. neoformans had polypyrimidine tracts only in the region from the 5′ ss to the branch point. Additionally, most of the polypyrimidine tracts in this region were located close to the 5′ ss (Fig. 4, left panels). In the region from the 3′ ss to the branch point, where one might expect to find polypyrimidine tracts, based on metazoan introns, we found polypyrimidine tracts in only 27.6 to 68.8% of the introns (Table 4 and Fig. 4, right panels).
TABLE 4.
Percentages of introns with polypyrimidine tracts in various regions
| Organism | % of introns with polypyrimidine tracts in the following region:
|
|||
|---|---|---|---|---|
| 5′ to branch point only | Branch point to 3′ only | Both | Neither | |
| S. cerevisiae | 28.5 | 3.6 | 65.2 | 2.8 |
| S. pombe | 62.1 | 1.6 | 26.0 | 10.4 |
| A. nidulans | 52.8 | 5.4 | 31.2 | 10.6 |
| N. crassa | 47.5 | 4.8 | 41.9 | 5.7 |
| C. neoformans | 47.9 | 7.4 | 35.3 | 9.4 |
FIG. 4.
Distributions of polypyrimidine tracts between the 5′ ss and the branch point A and between the branch point A and the 3′ ss for S. cerevisiae, S. pombe, A. nidulans, N. crassa, and C. neoformans. The data are presented as percentages of polypyrimidine tracts at the distance range (in nucleotides) from the 5′ ss to the branch point A (left panels) or the 3′ ss to the branch point A (right panels).
Spliceosomal proteins.
Having found fungal intron characteristics that differed from those of mammalian introns, we were interested in determining whether these fungal genomes contained genes similar to the mammalian genes encoding binding elements involved in spliceosome formation. We focused on protein splicing factors involved in early steps of spliceosome formation, particularly those involved in commitment complex formation at the branch point.
(i) BBP.
BBP would be expected to be found in all organisms that form spliceosomes, and candidate BBP homologs have been identified for S. pombe and N. crassa as well as for humans and S. cerevisiae. Analysis of the multiple-sequence alignment of BBPs allowed identification of the expected homologs of BBP in A. nidulans and C. neoformans (see Fig. S2A in the supplemental material). All of the BBP homologs had the MSL5 domain characteristic of BBPs (63) and an adjacent AIR domain, a ring Zn finger motif associated with posttranslational modification (http://www.ncbi.nlm.nih.gov/COG/). A phylogram of the fungal BBP homologs (see Fig. S2B in the supplemental material) shows clustering of S. pombe, A. nidulans, and N. crassa BBPs, which have between 23 and 33% identity with C. neoformans BBP, between 15 and 30% identity with S. cerevisiae BBP, and between 20 and 31% identity with human BBP, whereas C. neoformans BBP has 25% identity with human BBP and 27% identity with S. cerevisiae BBP.
(ii) U2AF.
U2AF is a heterodimeric protein consisting of a U2AF65 subunit and a U2AF35 subunit. It binds to the polypyrimidine tract at the 3′ end of the intron, associates with the 3′ acceptor site early in the establishment of the metazoan spliceosome, and is essential for correct splicing (69). Homologs for human U2AF65 and U2AF35 subunits have been identified in S. pombe (66). S. cerevisiae has Mud2p, a functional equivalent of U2AF65, but no U2AF35 homolog (1).
To search for fungal U2AF homologs, sequences of both the human and the S. pombe U2AF large and small subunits were used to screen A. nidulans, N. crassa, and C. neoformans genomes and their EST databases. Homologs were found in all of the fungal data sets examined.
The human U2AF65 subunit contains three RNA recognition motifs (RRM) and a serine- and arginine-rich region (43). The C. neoformans, N. crassa, and A. nidulans U2AF65 subunit homologs also contain three RRM; however, the S. pombe U2AF65 subunit homolog has only two RRM, and Mud2p, the U2AF65 functional homolog in S. cerevisiae, has only one RRM (56) (see Fig. S3 in the supplemental material).
U2AF35 subunit homologs also were observed in all fungi except for S. cerevisiae. The U2AF35 subunit homologs in all of the fungi that had them were similar; all of them contained one RRM, a KOG2202 domain, and two Zn finger domains (see Fig. S4 in the supplemental material).
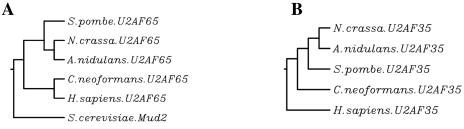
A phylogram for the U2AF65 splicing factor homologs and Mud2p of S. cerevisiae shows, as would be expected, that Mud2p falls outside the cluster containing the other fungal proteins, having only 14 to 17% identity with the other fungal U2AF65 homologs (Fig. 5A). The filamentous Ascomycota (A. nidulans and N. crassa) U2AF65 homologs have between 27 and 28% identity with the C. neoformans U2AF65 homolog and between 29 and 30% identity with the S. pombe U2AF65 homolog (Fig. 5A). The S. pombe, A. nidulans, and N. crassa U2AF65 homologs as well as the C. neoformans U2AF65 homolog have between 25 and 29% identity with the human U2AF65 protein, indicating the U2AF65 homologs of these fungi are more similar to the human U2AF65 protein than to Mud2p, the functional equivalent in S. cerevisiae.
FIG. 5.
(A) Phylogram for U2AF65 protein homologs and Mud2p. (B) Phylogram for U2AF35 protein homologs.
S. pombe U2AF35 clusters with the filamentous members of the Ascomycota (A. nidulans and N. crassa) (Fig. 5B). The U2AF35 subunit homolog of the member of the Basidiomycota (C. neoformans) clusters away from those of S. pombe, A. nidulans, and N. crassa (Fig. 5B). However, all of the fungal U2AF35 homologs represented in Fig. 5B have 48 to 50% identity with the human U2AF35 protein.
(iii) Nam8p, TIA-1, and TIAR homologs.
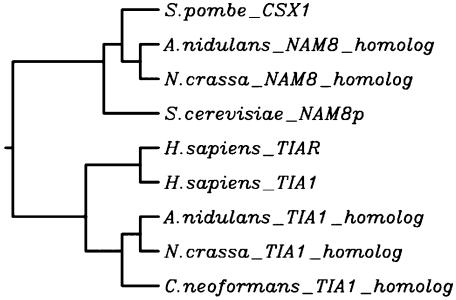
Having found high percentages of fungal introns with polypyrimidine tracts only in the region from the 5′ ss to the branch point, we searched for homologs of another family of RNA binding proteins that have been implicated in the splicing of introns that have U-rich regions upstream of the branch point. Nam8p, TIA-1, and TIAR are proteins that share homologies in RRM (22, 25, 31, 47, 64), are involved in stabilizing the functional association of U1 snRNP with the 5′ ss, and have activities dependent on polypyrimidine regions downstream of the 5′ ss (17, 22, 25, 47). By examining the fungal genomes for homologs to the three proteins, we found Nam8p homologs in the S. pombe, A. nidulans, and N. crassa genome data sets and TIA-1 homologs in the A. nidulans, N. crassa, and C. neoformans genome data sets (see Fig. S5 in the supplemental material). All of the homologs have the characteristic KOGO148 domain found in TIA-1 and TIAR, which includes three RRM (see Fig. S5 in the supplemental material). Consequently, these homologs are candidates for factors that could bind to 5′ polypyrimidine tracts or that could associate with other proteins in the commitment complex. The phylogram in Fig. 6 shows that the filamentous fungi have both a Nam8p homolog with 25 to 27% identity and a TIA-1 homolog with 14 to 25% identity, whereas S. pombe has only a Nam8p homolog (19% identity) and C. neoformans has only a TIA-1 homolog (29% identity). Except for the S. pombe CSX1 protein (51), there are limited biological data on the functions of this group of fungal RNA binding protein homologs.
FIG. 6.
Phylogram for Nam8p, TIA-1, and TIAR homologs.
DISCUSSION
Our analyses of the large data sets for introns and exons described here for a diverse group of fungi provide basic information about the intron and exon lengths (sizes) and a detailed representation of conserved intron and exon sequence motifs. Fungal introns have many of the characteristics associated with metazoan introns, such as the canonical donor-acceptor ss pair 5′GU…AG3′. Although we were able to define for the 5′ ss, 3′ ss, and branch sites of fungal introns a general consensus sequence that differed subtly from the metazoan consensus sequences in the respective regions, the differences in these sequence alone did not suggest that fungi use a novel splicing mechanism. The number of nucleotides between the branch point A and the 3′ ss in fungal introns is within the range found for mammalian introns—11 to 40 nt (54). Thus, the branch sites in fungal introns are positioned appropriately to function in splicing in a manner similar to that of branch sites in metazoan introns.
On the other hand, we detected features of fungal introns that set them apart from metazoan introns. Our large data set confirmed what had been surmised from the limited available data that fungal introns are characteristically short, with mean intron lengths ranging from 69 nt for C. neoformans to 256 nt for the model yeast S. cerevisiae (7). This length difference is accounted for by the distance from the 5′ ss to the branch point. We also found that the information content in fungal introns at the 5′ and 3′ ss and the branch site is substantial compared to the information content of the exon regions adjacent to the introns. These findings, in conjunction with the fact that fungal introns are short, suggest that splicing in fungi fits the intron definition model (39).
Of the five fungi surveyed, S. cerevisiae introns have the broadest length distribution pattern, followed by S. pombe introns. Furthermore, introns of these two yeasts have the highest intron information content of the five fungi studied. N. crassa, A. nidulans, and C. neoformans introns fall into a narrow length range, with peak numbers of introns within the size range of 50 to 70 nt. The latter three fungi have less information in their introns than is found in the introns of the two yeasts. Lim and Burge reported that for the short introns of S. cerevisiae, the nematode C. elegans, the fruit fly D. melanogaster, the dicot plant Arabidopsis thaliana, and the primate H. sapiens, the intron length distribution peaks occurring at higher numbers of nucleotides indicate that there must be increasing bits of information in the introns for accurate identification of the introns (39). These observations suggested to us that as the mean lengths of the short introns surveyed by Lim and Burge (39) increased, so did the information content in the introns. In keeping with this idea, Fields has demonstrated that the short C. elegans introns (<75 nt) have less information content at the 5′ ss than the somewhat rare long C. elegans introns (>75 nt) (16).
We thought it would be of interest to determine whether the intron length and the intron information content showed a similar relationship in the introns of the fungi. We found that with only a single intron size range exception, as the S. cerevisiae and S. pombe intron lengths increase, so does the information content. This pattern of an increase in information content with an increase in the size of the introns is more pronounced at the 5′ ss and 3′ ss of S. pombe introns than S. cerevisiae introns. However, the information content at the branch sites across most intron size ranges was higher for S. cerevisiae than for S. pombe. With N. crassa, A. nidulans, and C. neoformans introns, the direct correlation between information content and intron length was more subtle than that observed for the introns of the two yeasts. As mentioned earlier, the majority of the introns of N. crassa, A. nidulans, and C. neoformans fall into a more restricted length range than do the introns of S. cerevisiae and S. pombe; therefore, the information content necessary for effective splicing over this narrow range of intron lengths may not require a large increase in information content with an intron length increase. We did note a slight increase in information content for intron lengths between 240 and 2,000 nt for N. crassa, A. nidulans, and C. neoformans introns relative to what we saw at the lower nucleotide ranges. It is not completely clear why introns of the two yeasts have higher information content than do introns of N. crassa, A. nidulans, and C. neoformans. These differences may suggest subtle differences in splicing mechanisms between the yeasts and the three other fungi.
The most striking difference that we observed between the fungal introns and mammalian introns was the absence of polypyrimidine tracts between the 3′ ss and the branch point in a sizable population (31.3 to 72.5%) of introns from each organism in the study. Considering that we used a minimal definition for identifying polypyrimidine tracts, it is clear that polypyrimidine tracts are absent in many fungal introns downstream of the branch point. The polypyrimidine tracts that we did observe between the 3′ ss and the branch point are relatively weak and may function in a manner different from that of the classical polypyrimidine tracts defined for metazoans. These observations are consistent with the absence of polypyrimidine tracts between the branch point and the 3′ ss that also has been observed for certain Drosophila introns (46). Splicing mechanisms in small introns of Drosophila that lack a 3′-end polypyrimidine tract but instead have a pair of polypyrimidine tracts in the region from the 5′ ss to the branch point are different from those found in mammalians (17, 32, 33). Bon et al. also noted that poly(T) tracts were found in S. cerevisiae introns upstream of the branch site (7). Our observations in conjunction with those of Bon et al. (7) suggest that the classical splicing signals defined for metazoans may differ from those in fungi.
The classical splicing of metazoan pre-mRNA involves the recruitment of U1 snRNP to the 5′ ss and U2 snRNP with its two associated U2AF subunits. One of the two U2AF subunits, U2AF65, binds to the polypyrimidine tract in the 3′ ss region, and the other subunit, U2AF35, associates with the 3′ ss acceptor site to facilitate correct splicing in metazoan introns (8, 24, 42, 45, 69). In a small Drosophila intron that lacks the polypyrimidine tract in the 3′ end of the intron, the pair of polypyrimidine tracts in the region between the 5′ ss and the branch point are required for U2AF binding and efficient splicing (32, 33). Forch et al. (17) also have reported that U2AF65 promotes the recruitment of U1 snRNP to weak 5′ ss that have downstream U-rich sequences, and these authors have suggested that U2AF65 plays this role in splicing by binding to polypyrimidine tracts in the region from the 5′ ss to the branch site. We identified homologs of both U2AF subunits in A. nidulans, N. crassa, and C. neoformans, and U2AF65 and U2AF35 subunits have been found in S. pombe (66). S. cerevisiae has a functional equivalent of U2AF65 (Mud2p) but no U2AF35 homolog (1). These findings are consistent with the observations described above and suggest that the splicing mechanism in S. pombe, the filamentous ascomycetes, and C. neoformans may differ from that in metazoans but may be similar to that described for a small intron of Drosophila that lacks a 3′-end polypyrimidine tract (32, 33). Based on the observations that S. pombe and S. cerevisiae have higher information content in their introns than the other three fungi and that S. cerevisiae does not have a U2AF35 homolog, one may speculate that pre-mRNA splicing may be different in the yeasts and the other fungi. Our observations indicate a need for defining a new model fungal organism other than S. cerevisiae and possibly S. pombe that could be exploited for establishing how the U2AF subunit homologs function in protein-protein and protein-RNA interactions during pre-mRNA splicing.
Considering that splicing of the small introns of Drosophila have many characteristics that match those of fungal introns, the results of splicing studies with Drosophila could serve as an excellent guide for future studies of model fungal organisms. Another protein in Drosophila that is associated with splicing in pre-mRNA and that contains a 5′ ss with downstream polypyrimidine tracts is TIA-1 (18). A protein similar to TIA-1 is Nam8p of S. cerevisiae (47). Nam8p is associated with yeast U1 snRNP, and the activity of Nam8p is optimal when there are pyrimidine-rich sequences downstream of the 5′ ss (47, 70). Because TIA-1 and Nam8p function in splicing in introns that are similar to many of the introns characterized in our study, we screened for homologs of these proteins in genomic and EST data sets of the fungi used in this study. The finding of homologs of both TIA-1 and Nam8p in the A. nidulans and N. crassa data sets, a Nam8p homolog in S. pombe, and a TIA-1 homolog in C. neoformans suggests that splicing in these organisms is dependent on mechanisms similar to those described for S. cerevisiae or Drosophila, in which Nam8p or TIA-1 is involved. The mechanisms described for the splicing of small introns with polypyrimidine tracts upstream of the branch point may be unique to eukaryotic organisms other than vertebrates because Nam8p has no counterpart in mammalian U1 snRNP (22).
Taken together, our findings show that while there are significant similarities between introns of vertebrates and fungi, there are also some important differences that will have an impact on the mechanisms used for excising the introns. Fungi may be excellent model organisms for studying the splicing machinery needed for efficient splicing in groups of organisms that have short introns with polypyrimidine tracts only in the region downstream of the 5′ ss but upstream of the branch site. Within the fungal organisms studied here, the introns of S. cerevisiae and S. pombe, the two yeasts, were found to differ in many ways from the introns of the two filamentous ascomycetes and the one basidiomycete. Based on these differences among groups of fungi, it seems necessary to select for splicing studies a new model organism that more accurately reflects the characteristics of the filamentous ascomycetes.
Supplementary Material
Acknowledgments
Special thanks are due to James White (Advanced Center for Genome Technology, University of Oklahoma), who provided conceptual and computational assistance for many of the statistical measures.
This work was supported by grant AI-47079 from the National Institutes of Health and EPSCOR grant EPS9550478 from the National Science Foundation.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Abovich, N., X. C. Liao, and M. Rosbash. 1994. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 8:843-854. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. M., and E. L. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. M. Ostell, and D. L. Wheeler. 2003. GenBank. Nucleic Acids Res. 31:23-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berbee, M. L., and J. W. Taylor. 2000. Fungal molecular evolution: gene trees and geologic time, p. 229-243. In D. McLaughlin, E. G. McLaughlin, and P. A. Lemke (ed.), The mycota: systematics and evolution, vol. VII. Part B. Springer-Verlag KG, Berlin, Germany. [Google Scholar]
- 6.Berglund, J. A., K. Chua, N. Abovich, R. Reed, and M. Rosbash. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89:781-787. [DOI] [PubMed] [Google Scholar]
- 7.Bon, E., S. Casaregola, G. Blandin, B. Llorente, C. Neuveglise, M. Munsterkotter, U. Guldener, H. Mewes, J. V. Helden, B. Dujon, and C. Gaillardin. 2003. Molecular evolution of eukaryotic genomes: hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 31:1121-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burge, C. B., R. A. Padgett, and P. A. Sharp. 1998. Evolutionary fates and origins of U12-type introns. Mol. Cell 2:773-785. [DOI] [PubMed] [Google Scholar]
- 9.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosomes, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world—the nature of modern RNA suggests a prebiotic RNA. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Burset, M., I. A. Seledtsov, and V. V. Solovyev. 2000. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 28:4364-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burset, M., I. A. Seledtsov, and V. V. Solovyev. 2001. Splice DB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res. 29:255-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B. A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301:71-76. [DOI] [PubMed] [Google Scholar]
- 13.Coolidge, C. J., R. J. Seely, and J. G. Patton. 1997. Functional analysis of the polypyrimidine tract in pre-mRNA splicing. Nucleic Acids Res. 25:888-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deutsch, M., and M. Long. 1999. Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 27:3219-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drabenstot, S. D., D. M. Kupfer, J. D. White, D. W. Dyer, B. A. Roe, K. L. Buchanan, and J. W. Murphy. 2003. FELINES: a utility for extracting and examining EST-defined introns and exons. Nucleic Acids Res. 31:e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields, C. 1990. Information content of Caenorhabditis elegans splice site sequences varies with intron length. Nucleic Acids Res. 18:1509-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forch, P., L. Merendino, C. Martinez, and J. Valcarcel. 2003. U2 small nuclear ribonucleoprotein particle (snRNP) auxiliary factor of 65 kDa, U2AF65, can promote U1 snRNP recruitment to 5′ splice sites. Biochem. J. 372:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forch, P., O. Puig, N. Kedersha, C. Martinez, S. Granneman, B. Seraphin, P. Anderson, and J. Valcarcel. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol. Cell 6:1089-1098. [DOI] [PubMed] [Google Scholar]
- 19.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L.-J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Qui, P. Ianakiev, D. Bell-Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braum, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Strange-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. Li, R. J. Pratt, S. A. Osmani, C. P. C. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seiler, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 20.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546-567. [DOI] [PubMed] [Google Scholar]
- 21.Gorodkin, J., L. J. Heyer, and G. D. Stormo. 1997. Finding the most significant common sequence and structure motifs in a set of RNA sequences. Nucleic Acids Res. 25:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottschalk, A., J. Tang, O. Puig, J. Salgado, G. Neubauer, H. V. Colot, M. Mann, S. B., M. Rosbash, R. Luhrmann, and P. Fabrizio. 1998. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. RNA 4:374-393. [PMC free article] [PubMed] [Google Scholar]
- 23.Grate, L., and M. Ares, Jr. 2002. Searching yeast intron data at the Ares lab website. Methods Enzymol. 350:380-392. [DOI] [PubMed] [Google Scholar]
- 24.Graveley, B. R., K. J. Hertel, and T. Maniatis. 2001. The role of U2AF35 and U2AF65 in enhancer-dependent splicing. RNA 7:806-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guiner, C. L., F. Lejeune, D. Galiana, L. Kister, R. Breathnach, J. Stevenin, and F. D. Gatto-Konczak. 2001. TIA-1 and TIAR activate splicing of alternative exons with weak 5′ splice sites followed by a U-rich stretch on their own pre-mRNAs. J. Biol. Chem. 276:40638-40646. [DOI] [PubMed] [Google Scholar]
- 26.Heitman, J., A. Casadevall, J. K. Lodge, and J. R. Perfect. 1999. The Cryptococcus neoformans genome sequencing project. Mycopathologia 148:1-7. [DOI] [PubMed] [Google Scholar]
- 27.Hertz, G., and G. D. Stormo. 1999. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15:563-577. [DOI] [PubMed] [Google Scholar]
- 28.IUPAC-IUB Commission on Biochemical Nomenclature (CBN). 1971. Abbreviations and symbols for nucleic acids, polynucleotides and their constituents. Recommendations. Arch. Biochem. Biophys. 145:425-436. [Google Scholar]
- 29.Jeanmougin, F., J. D. Thompson, M. Goy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with ClustalX. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 30.Kaufer, N. F., and J. Potashkin. 2000. Survey and summary analysis of the splicing machinery in fission yeast: a comparison with budding yeast and mammals. Nucleic Acids Res. 28:3003-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawakami, A., Q. Tian, X. Duan, M. Streuli, S. F. Schlossman, and P. Anderson. 1992. Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. USA 89:8681-8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy, C. F., and S. M. Berget. 1997. Pyrimidine tracts between the 5′ splice site and branch point facilitate splicing and recognition of a small Drosophila intron. Mol. Cell. Biol. 17:2774-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy, C. F., A. Kramer, and S. M. Berget. 1998. A role for SRp54 during intron bridging of small introns with pyrimidine tracts upstream of the branch point. Mol. Cell. Biol. 18:5425-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kessler, M. M., Q. Zeng, S. Hogan, R. Cook, A. J. Morales, and G. Cottarel. 2003. Syst. discovery of new genes in the Saccharomyces cerevisiae genome. Genome Res. 13:264-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 36.Kupfer, D. 1999. Development, analysis and use of an expressed sequence tag database from the multicellular Ascomycete, Aspergillus nidulans. Ph.D. thesis. University of Oklahoma, Norman.
- 37.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. (Erratum, 412:566.) [DOI] [PubMed] [Google Scholar]
- 38.Lim, L. P., and P. A. Sharp. 1998. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol. Cell. Biol. 18:3900-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim, L. P., and C. B. Burge. 2001. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl. Acad. Sci. USA 98:11193-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maniatis, T., and B. Tasic. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236-243. [DOI] [PubMed] [Google Scholar]
- 41.Marchler-Bauer, A., J. B. Anderson, S. C. DeWeese, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcarcel. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402:838-841. [DOI] [PubMed] [Google Scholar]
- 43.Moore, M. J. 2000. Intron recognition comes of age. Nat. Struct. Biol. 7:14-16. [DOI] [PubMed] [Google Scholar]
- 44.Mount, S. M. 1982. A catalog of splice junction sequences. Nucleic Acids Res. 10:459-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mount, S. M. 2000. Genome sequence, splicing, and gene annotation. Am. J. Hum. Genet. 67:788-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mount, S. M., C. Burks, G. Hertz, G. D. Stormo, O. White, and C. Fields. 1992. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 20:4255-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puig, O., A. Gottschalk, P. Fabrizio, and B. Seraphin. 1999. Interaction of U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 13:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Query, C. C., M. J. Moore, and P. A. Sharp. 1994. Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev. 8:587-597. [DOI] [PubMed] [Google Scholar]
- 49.Reed, R., and T. Maniatis. 1985. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell 41:95-105. [DOI] [PubMed] [Google Scholar]
- 50.Reed, R., and T. Maniatis. 1988. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 2:1268-1276. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Gabriel, M. A., G. Burns, W. H. McDonald, V. Martin, J. R. Yates III, J. Bahler, and P. Russell. 2003. RNA-binding protein Csx1 mediates global control of gene expression in response to oxidative stress. EMBO J. 22:6256-6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodriguez-Medina, J. R., and B. C. Rymon. 1994. Prevalence and distribution of introns in non-ribosomal protein genes of yeast. Mol. Gen. Genet. 243:532-539. [DOI] [PubMed] [Google Scholar]
- 53.Romfo, C. M., C. J. Alvarez, W. J. Van Heeckeren, C. J. Webb, and J. A. Wise. 2000. Evidence for splice site pairing via intron definition in Schizosaccharomyces pombe. Mol. Cell. Biol. 20:7955-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roscigno, R. F., M. Weiner, and M. A. Garcia-Blanco. 1993. A mutational analysis of the polypyrimidine tract of introns. J. Biol. Chem. 268:11222-11229. [PubMed] [Google Scholar]
- 55.Saxonov, S., I. Daizadeh, A. Fedorov, and W. Gilbert. 2000. EID: the exon-intron database—an exhaustive database of protein-coding intron-containing genes. Nucleic Acids Res. 28:185-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schultz, J., R. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shapiro, M. B., and P. Senapathy. 1987. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 15:7155-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharp, P. A., and C. B. Burge. 1997. Classification of introns: U2-type or U12-type. Cell 91:875-879. [DOI] [PubMed] [Google Scholar]
- 59.Shelley, C. S., and F. E. Baralle. 1987. Deletion analysis of a unique 3′ splice site indicates that alternating guanine and thymine residues represent an efficient splicing signal. Nucleic Acids Res. 15:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh, R., J. Valcarcel, and M. R. Green. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268:1173-1176. [DOI] [PubMed] [Google Scholar]
- 61.Sipiczki, M. 2000. Where does fission yeast sit on the tree of life? Genome Biol. 1:1011-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spingola, M., L. Grate, D. Haussler, and M. Ares, Jr. 1999. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian, Q., M. Streuli, H. Saito, S. F. Schlossman, and P. Anderson. 1991. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell 67:629-639. [DOI] [PubMed] [Google Scholar]
- 65.Valcarcel, J., R. K. Gaur, R. Singh, and M. R. Green. 1996. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science 273:1706-1709. (Erratum, 274:21.) [DOI] [PubMed] [Google Scholar]
- 66.Wentz-Hunter, K., and J. Potashkin. 1996. The small subunit of the splicing factor U2AF is conserved in fission yeast. Nucleic Acids Res. 24:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Will, C. L., C. Schneider, A. M. MacMillan, N. F. Katopodis, G. Neubauer, M. Wilm, R. Luhrmann, and C. C. Query. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 20:4536-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wood, V., R. Gwilliam, M. A. Rajandream, M. Lyne, R. Lyne, A. Stewart, J. Sgouros, N. Peat, J. Hayles, S. Baker, et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 69.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832-835. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, D., and M. Rosbash. 1999. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 13:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, M. Q., and T. G. Marr. 1994. Fission yeast gene structure and recognition. Nucleic Acids Res. 22:1750-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, H., M. Nowrousian, D. Kupfer, H. V. Colot, G. Berrocal-Tito, H. Lai, D. Bell-Pedersen, B. Roe, A., J. J. Loros, and J. C. Dunlap. 2001. Analysis of expressed sequence tags from two starvation, time-of-day-specific libraries of Neurospora crassa reveals novel clock-controlled genes. Genetics 157:1057-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.