Abstract
Macronuclei and micronuclei of ciliates have related genomes, with macronuclei developing from zygotic micronuclei through programmed DNA rearrangements. While Paramecium tetraurelia wild-type strain 51 and mutant strain d48 have the same micronuclear genome, qualitative differences between their macronuclear genomes have been described, demonstrating that programmed DNA rearrangements could be epigenetically controlled in ciliates. Macronuclear chromosomes end downstream of gene A (A51 Mac ends) and at the 5′ end of gene A (Ad48 Mac ends) in strains 51 and d48, respectively. To gain further insight into the process of chromosome end formation, we performed an extensive analysis of locus A rearrangement in strains d48 and 51, in strain d12, which harbors a gene A deletion, and in interstrain cross progeny. We show that (i) allele Ad12 harbors a deletion of >16 kb, (ii) A51 Mac ends distribute over four rather than three DNA regions, (iii) strains d48 and 51 display only quantitative differences (rare Ad48 and A51 Mac ends do form in strains 51 and d48, respectively), (iv) the level of A51 Mac ends is severalfold enhanced in d12- and d48-derived progeny, and (v) this level inversely correlates with the level of Ad48 Mac ends in the d48 parent. Together, these data lead to a model in which the formation of Ad48 Mac ends is epigenetically controlled by a d48 factor(s). We propose that the d48 factor(s) may be derived from RNA molecules transcribed from the Ad48 Mac ends and encompassing the truncated A gene and telomeric repeats.
Ciliate cells harbor two types of nuclei that ensure separate functions. Macronuclei that are transcriptionally active carry out the vegetative functions of these unicellular eukaryotes. Micronuclei that are transcriptionally silent during vegetative growth provide genetic continuity between sexual generations. In the course of conjugation, old maternal macronuclei do not replicate their DNA (they will be ultimately lost), while micronuclei enter meiosis. New macronuclei and micronuclei then develop from mitotic copies of zygotic nuclei. Macronuclear genomes result mainly from micronuclear genome fragmentation into short DNA molecules, at the ends of which telomeric repeats are added, excision of internal eliminated sequences (IESs) that are eventually eliminated, and massive DNA amplification that accounts for the large size (and the name) of macronuclei (20, 21). Once developed (the whole process takes just a few hours), macronuclear genomes are clonally stable throughout the cellular life span. Although chromosome breakage occurs in a wide range of genomes, the frequency with which this takes place in ciliates is unique and provides an opportunity to study the underlying regulation and mechanisms.
In the two species of Paramecium, P. primaurelia and P. tetraurelia, the micronuclear genome is organized into 60 to 90 chromosomes with an average size of 2 Mb, while macronuclear chromosomes range in size from 50 to 800 kb (note that although macronuclear DNA molecules are called chromosomes, they are acentromeric) (10, 19). Each micronuclear genome, therefore, has to undergo 500 to 1,000 events of chromosome end formation in order to successfully develop into a macronuclear genome.
Chromosome breakage in Paramecium has been studied during the formation of macronuclear chromosome ends (Mac ends) at five loci. At all loci, Mac ends display heterogeneity between and within macronuclei (a ploidy level of ∼1,000 n characterizes the P. primaurelia and P. tetraurelia macronuclear genomes as a result of the massive amplification of the micronuclear genome). At P. tetraurelia loci A (6) and B (23, 24) and at P. primaurelia loci ψG (2) and Tennessee 1 (14), telomeric repeats are not added at a single nucleotide position but rather at multiple positions that are dispersed over a limited number—from two to four—of ∼1.0-kb DNA regions. These DNA regions are separated by a few kilobases. At P. primaurelia locus G, the vast majority (∼99%) of Mac ends map within a single region (1, 15).
No cis-acting determinant of the chromosome breakage process has been identified in Paramecium. No conserved sequence could be characterized at, or close to, the telomeric repeat addition sites. In one case, chromosome breakage and IES excision are alternative DNA rearrangements, suggesting that some IESs act as cis-acting determinants for DNA fragmentation (1). This could be done either by providing accurate sites to cut or by inducing cut sites in their flanking sequences. In another case, chromosome breakage is linked to the elimination of repeated sequences via a mechanism distinct from IES excision; this suggests that chromosome breakage and repeat sequence elimination may also be related phenomena (14).
P. tetraurelia locus A has been a paradigm for studying Paramecium Mac end formation since 1988. In wild-type strain 51, Mac ends were mapped within three regions located downstream of the A gene that codes for surface antigen A (A51 Mac ends) (6). In mutant strain d48, an immunologically A− strain recovered following strain 51 mutagenesis, Mac ends were mapped at the 5′ end of gene A (Ad48 Mac ends) (6). Nevertheless, the micronuclear genome of strain d48 had been shown to be wild type, thereby providing the first indication that DNA fragmentation in Paramecium could be determined by an epigenetic mechanism (3).
In order to analyze Mac end formation at locus A, macronucleoplasm transplantations and crosses were performed between strains d48 and 51.
Macronucleoplasm from strain 51 cells restored gene A expression to injected d48 cells and their autogamous progeny (autogamy is a self-fertilization process; it is the only sexual reproduction process in P. tetraurelia clonal cell lines) (9, 13). If cells of strain 51 were in their first stages of macronuclear development, cytoplasm transfer could similarly restore gene A expression to injected d48 cells and their autogamous progeny (13). Neither macronucleoplasm nor cytoplasm from d48 cells affected gene A expression of the injected strain 51 autogamous cells (13). This led to a model in which A51 Mac end formation is epigenetically determined and relies on the capacity for gene A core and/or downstream sequences from the maternal macronuclei to export a diffusing factor(s) (here called 51 factor[s]) to the developing macronucleus (9). Note that Paramecium maternal and developing macronuclei inhabit the same cytoplasm until full development of the latter.
Cross d48 × 51 produced d48-derived F1 progeny that were mostly immunologically A− and 51-derived F1 progeny that were immunologically A+ (5, 6, 22). In the presently accepted model, a defect in antigen A detection in the d48-derived F1 progeny reflects the formation of Mac ends at the 5′ end of gene A as a consequence of failures in the generation of 51 factors in the d48 parent and in its import from the strain 51 parent (9, 17). The model predicts identical macronuclear genomes in the d48 parental cell lines arising from autogamy events and their F1 progeny arising from cross events.
The above-described model, which postulates the requirement for 51 factors for A51 Mac end formation, cannot account for the results reported from crosses involving strain d12. Most, if not all, Mac A ends in strain d12 have been mapped upstream of the 5′ end of gene A, and d12 macronuclear genomes have been shown to lack gene A. Since strain d12 lacked the 51 factors, the d12-derived F1 progeny were expected to be A−. Nevertheless, most of the F1 progeny from cross d12 × 51 were A+ while cross d12 × d48 produced mostly A+ and A− F1 progeny (22). The model also does not account for the fact that half of the A− F1 progeny obtained from crosses d12 × d48 and d48 × 51 yielded A+ F2 progeny (6, 22). In contrast to the prediction of the model, these data suggest differences between the macronuclear genomes of the d48 parents and the F1 progeny. Although the presence of a fragment defined by two primers located 512 and 1,361 bp upstream of the 5′ end of gene A was examined in some progeny, a systematic molecular characterization of F1 progeny for gene A content and Mac A ends has not been performed.
Here, we report an extensive molecular analysis of the gene A core and its surrounding sequences within the genomes of strains d12, d48, and 51, as well as within those from the F1 progeny of interstrain crosses. Taken together, our data lead to a model contrasting with the present belief that Mac end formation at the 5′ end of gene A results from a lack of cross talk between the maternal and zygotic macronuclei. In this new model, Mac end formation at the 5′ end of gene A results from cross talk between the two nuclei via the production of a d48 factor(s).
Experiments performed in the ciliate Tetrahymena thermophila have led to a model for the epigenetic regulation of genome rearrangements in which widespread transcription from the zygotic nuclei and matching of the micronuclear transcripts against the maternal macronuclear genome participate in macronuclear genome development (18). In this study, we show that Paramecium maternal macronuclei do not behave as passive filters but actively produce rearrangement markers. Our data suggest that the d48 factors are derived from RNA molecules transcribed from the promoter of the truncated A gene and encompassing the adjacent telomeric repeats. Model mechanisms are discussed.
MATERIALS AND METHODS
Cell lines and genetic analysis.
P. tetraurelia strains d12, d48, and 51 have been extensively used in genetic studies of the surface antigen system (6, 22). Karyonidal cell cultures (a karyonidal cell macronuclear genome results from a single developmental event) were used for Mac end analysis and for crosses. Conjugant pairs were picked and transferred into 200-μl drops of bacterized medium. Exconjugants were isolated following separation. Cultures were derived from the two sister progeny that regularly arose from each exconjugant. The progeny parental origin was ascertained by determining the mating type, which is maternally inherited in P. tetraurelia. Cross d48 × 51 displayed pa2/pa+ allele segregation. The recessive allele pa2 dictates backward swimming. All F1 progeny had a wild-type phenotype, demonstrating successful karyogamy; allele pa2 and pa+ segregation was verified for four of the F2 progeny. Cross d48 × d12 displayed nd7-1/nd7+ allele segregation. Allele nd7-1 harbors an Ssp1 restriction site (25). This site was used to demonstrate successful karyogamy in F1 progeny. Crosses and cell cultures were performed at 27°C in grass infusion (wheat grass powder; Ines International, Lawrence, Kans.) inoculated with Klebsellia pneumoniae and supplemented with 0.4 μg of β-sistosterol/ml.
DNA extraction.
Cells were centrifuged, and the pellets were resuspended in 1 volume of their own culture medium before being added to 2 volumes of lysis solution (0.44 M EDTA [pH 9.0], 0.5% sodium dodecyl sulfate [SDS], 0.5% N-laurylsarcosine [Sigma], and 1 mg of proteinase K [Merck]/ml) at 55°C. The lysates were incubated for at least 5 h, gently extracted with phenol, and dialyzed against TE (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]).
DNA analysis, amplification, and sequencing.
DNA restriction and electrophoresis were carried out according to standard methods. Twenty micrograms of DNA was loaded on agarose gels, separated by electrophoresis, and blotted onto Hybond N+ membranes (Amersham) in 0.4 N NaOH after depurination in 0.25 N HCl. Hybridization was carried out overnight in 7% SDS-0.5 M sodium phosphate-1% bovine serum albumin at 60°C. The membranes were washed at 60°C in 0.1% SDS and 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M Na citrate, pH 7.0). Hybridization signals were revealed and quantified using Fuji and Molecular Dynamics phosphorimagers. Fragments IB and 11D were subcloned from lambda inserts. Other fragments were generated in PCRs performed on lambda inserts. The lambda inserts had been recovered from the screening of a micronuclear phage library (22). Oligonucleotide sequences and positions are shown in Table 1. Polymerase reactions and PCRs were performed either with enzyme Tfl (Promega) or the PCR Mix Extensor (ABgene). PCR products were purified using the Geneclean (Bio 101, Inc.) or Qiaquick (Qiagen) procedure. Fragments were labeled by using a random-priming kit (Boehringer Mannheim). In the analysis of cross d12 × d48, the 5′ end of gene A and downstream sequences were revealed by two probes, 11D and TA95-TB92, respectively. Since the two probes do not necessarily hybridize with the same efficiency, ratios between the 5′ end of gene A and downstream sequences were established in two steps. First, signals corresponding to the 5′ end of gene A and downstream sequences were quantified on strain 51 DNA, and their ratio was established and used to define a 100% A51 Mac end content. Second, signals corresponding to the 5′ end of gene A and downstream sequences were quantified on parent d48 and progeny DNAs, and their ratio was established and compared to that from strain 51 DNA.
TABLE 1.
Oligonucleotide sequences and positions
| Oligonucleotide | Sequence | Positiona |
|---|---|---|
| Locus A | ||
| TA3 | AATGAATTTTTCATCAATTCTATGTGAGAG | 1859-1888 |
| TB7 | AAACAATTATAATGTTTCATTCTATTTATC | 1570-1598 |
| TB8 | GTTCTTCCATTAACAGCTTCCCTTTTCTTT | 1536-1567 |
| TA17 | GTCGATTTGATATACTATATAGCCAATAAG | 16577-16606 |
| TB18 | GCACTAATCATTGCTTCTTGATTTAAGTAT | 17023-17050 |
| TB92 | TTCAATTTTTTGCTATAACTGTACTATT | 10781-10808 |
| TA95 | ATAATGTTGCATTATTTTTCTCCTGAC | 9985-10012 |
| TA96 | CTATCCATTTTTACTAAATATATCTACAAC | 11499-11529 |
| TB97 | ATCTTCAGATAATCGTTATAAAGACATC | 12438-12468 |
| Other loci | ||
| β-Tubulin A | CGTTCATATTCAAGGAGGACAATGTGG | 11-37 |
| β-Tubulin B | TCATGTTGGATTCAGCTTCAGTGAATT | 1299-1723 |
| ND7 A2 | GAGGATTATTATAATTAACCAGATTTGG | 1021-1048 |
| ND7 B2 | TTCGTTTCTTTTTTCTAGTTCCTACTTTT | 1393-1422 |
| Telomeric | CAACCCCAACCCCAACCCAAACCCCAA |
Coordinates of oligonucleotides mapping at locus A are given relative to the 5′ end of gene A except for oligonucleotides TA17 and TB18, located downstream of gene A. Coordinates of oligonucleotides TA17 and TB18 are given relative to the 3′ end of gene A. Coordinates of oligonucleotides mapping within the β-tubulin and ND7 genes are given relative to the gene 5′ ends.
RESULTS
Mac ends at locus A differ quantitatively but not qualitatively between strains d48 and 51.
Programmed rearrangements of the micronuclear genome regularly bring about Mac ends within DNA regions that are located 8, 13, or 26 kb downstream of gene A in the P. tetraurelia wild-type strain 51 (6). In strain d48 (a mutant strain recovered following X-ray mutagenesis of strain 51 and immunological screening for the nonexpression of surface antigen A), programmed breakage of the micronuclear genome regularly occurs within a region located at the 5′ end of gene A (3, 6). Nevertheless, the d48 micronuclear genome is identical to the strain 51 micronuclear genome, indicating that Mac end formation in strain d48 and/or strain 51 is epigenetically controlled (6).
As a first attempt to investigate chromosome breakage at P. tetraurelia locus A, we performed a systematic characterization of Mac ends over a 52-kb region that encompassed the 10-kb A gene and the upstream 30-kb and downstream 12-kb sequences in strains d48 and 51 (Fig. 1A).
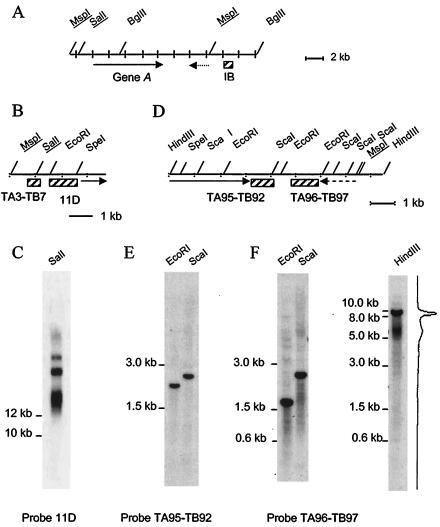
FIG. 1.
Mac end characterization in P. tetraurelia strain 51. (A) Restriction map of locus A in micronuclear genome 51. (B) Enlargement of restriction map of gene A 5′ end. (C) DNA 51 was restricted with SalI and hybridized with probe 11D. (D) Enlargement of restriction map of gene A 3′ end. (E) DNA 51 was restricted with EcoRI or ScaI and hybridized with probe TA95-TB92. (F) DNA 51 was restricted with EcoRI, HindIII, or ScaI and hybridized with probe TA96-TB97. A graph from a phosphorimager screen exposure is shown on the right. (A, B, and D) Probes are drawn as hatched rectangles. The solid arrow represents the A gene coding region. The dashed arrow represents a long minisatellite open reading frame (ORF) that identified poly(A) transcripts (4). However, since this ORF is part of a highly repetitive family, it is not possible to know whether the minisatellite sequence from locus A is specifically transcribed. MspI and SalI sites common to panels A, B, and D are underlined.
Previous characterization of Mac ends in strain 51 had been performed across a short DNA interval by using a fragment lying >6 kb downstream of gene A as a probe.
Here, we first used fragment 11D, located 0.2 to 1.2 kb upstream of the 5′ end of gene A, to probe strain 51 DNA digested with SalI (Fig. 1B). Fragment 11D revealed four signals of ≥12 kb (Fig. 1C). The three upper signals correspond to the telomeric fragments ending within DNA regions located 8, 13, and 26 kb downstream of gene A (20, 22, and 38 kb downstream of the SalI site, respectively). The lower-molecular-weight smear identified a new class of fragments that may correspond to restriction fragments with internal heterogeneity. Alternatively, this signal may represent a new family of telomeric fragments ending at the 3′ end of the A gene.
Next, we used fragments TA95-TB92 and TA96-TB97, which map 0.2 to 1.0 and 2.0 to 3.0 kb downstream of gene A, respectively, to hybridize strain 51 DNA digested with EcoRI or ScaI (Fig. 1D). Probe TA95-TB92 revealed only restriction fragments (Fig. 1E). In contrast, probe TA96-TB97 revealed a restriction fragment and a superimposed smear on each restricted DNA (Fig. 1F). Since analysis of EcoRI, SalI, and ScaI restrictions all identified a universal restriction region 1.7 to 3.7 kb downstream of gene A, this region must define telomeres. A51 Mac ends therefore form across four, rather than three, DNA regions downstream of gene A.
Superimposition of discrete and smeared signals indicated that the DNA region across which the newly characterized Mac ends had formed overlapped the tested EcoRI and ScaI sites. In order to characterize the whole length of the region across which macronuclear ends form, probe TA96-TB97 was hybridized to HindIII-restricted strain 51 DNA (Fig. 1F). This revealed a 5- to 7-kb smear identifying telomeric fragments produced from Mac ends mapping 2 to 4 kb downstream of gene A and a 10-kb signal representing a restriction fragment produced from Mac ends formed 8, 13, and 26 kb downstream of gene A.
A previous analysis of macronuclear d48 genomes mapped Mac ends at the 5′ end of gene A (6), but further PCR experiments suggested that macronuclear d48 genomes harbor a few full-length copies of gene A (22). To quantify their copy numbers and determine whether they are associated with wild-type A51 Mac ends, d48 DNA was restricted with enzyme MspI and probed with fragment TA3-TB7, located 1.3 to 1.6 kb upstream of gene A (Fig. 1A and B and 2A). A smear of 2.1 to 4.5 kb was revealed, thus identifying telomeric fragments ending −0.4 to +2.0 kb from the 5′ end of gene A (Fig. 2B). Upon overexposure, fragment TA3-TB7 also faintly revealed a >12-kb signal in d48 DNA that could identify A51 Mac ends (data not shown). This supported the notion that macronuclear d48 genomes may harbor a few full-length gene A copies.
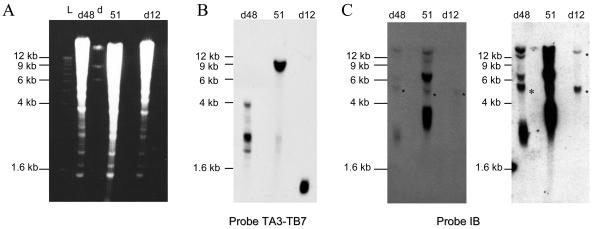
FIG. 2.
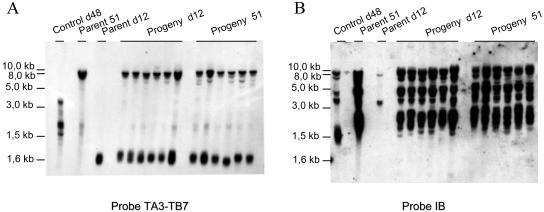
Mac end characterization in strains d12 and d48. (A) DNAs were restricted with enzyme MspI and visualized with ethidium bromide after electrophoresis. (B) Fragment TA3-TB7 hybridization. (C) Fragment IB hybridization. The right part of the figure shows an overexposure of the hybridization. PCR performed on d12 DNA using primers TA17 and TB18 that map within fragment IB failed to amplify any product, even though PCR products could be recovered from a 1,000-fold dilution of strain 51 DNA (Fig. 4C). Therefore, the two faint bands that are marked by asterisks identify cross-hybridizing sequences. In d48 and 51 DNAs, the upper cross-hybridizing sequence is masked by fragments derived from gene A downstream sequences. Size markers are a 1-kb ladder (Boehringer) (L) and lambda-HindIII DNA (d).
The same Southern blot was hybridized with fragment IB located 7.0 to 8.5 kb downstream of gene A. Fragment IB revealed four signals in the d48 and 51 tracks, although the relative intensities of these signals were very different (Fig. 2C). Signals of 2 to 4, 6 to 7, and >12 kb identified Mac ends located 8, 13, and 26 kb downstream of gene A, respectively. A signal of ≫12 kb probably identified additional Mac ends located ≫26 kb downstream of gene A. Therefore, full-length gene A copies in macronuclear d48 genomes are associated with A51 Mac ends.
Fragment TA3-TB7 also revealed a faint 3.0- to 4.5-kb smear on strain 51 DNA (Fig. 2B). PCR products were generated using a primer that maps 0.7 kb upstream of the 5′ end of gene A and a telomeric primer (data not shown). This demonstrated that strain 51 harbors Ad48 Mac ends, although in small quantities. We therefore concluded that strains d48 and 51 have no qualitative differences in their Mac ends at locus A, only quantitative differences.
Rearranging locus A in P. tetraurelia promotes the formation of four, and perhaps five, classes of Mac ends (Fig. 3). Ad48 Mac ends, located at the 5′ end of gene A, account for ∼99 and 1% of Mac A ends in the above-mentioned d48 and 51 cell lines, respectively. The Mac ends called here A51.1, A51.2, A51.3, and A51.4 are located 2 to 4, ∼8, >13, and >26 kb downstream of gene A, respectively (see Discussion). No Mac end could be identified across the 30 kb upstream of gene A (L. Amar, unpublished data). The four signals identified by probe 11D in Fig. 1C show a 4:2:1:1 ratio from the lower to the upper band. Similar ratios characterized all the DNAs we tested. The A51.1 Mac ends characterized in this study thus account for ∼50% of the Mac ends that form downstream of gene A.
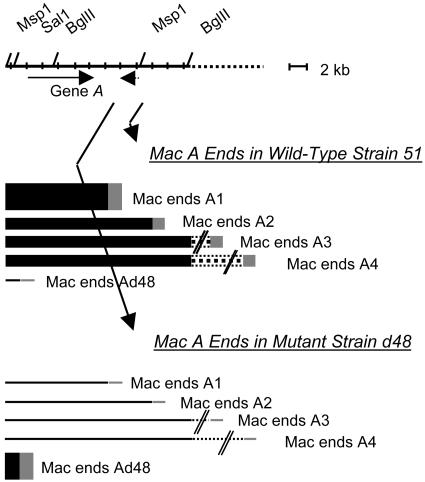
FIG. 3.
Schematic representation of Mac end formation in P. tetraurelia strains d48 and 51. Both strains harbor the wild-type micronuclear 51 genome but display quantitative differences in Mac end formation at locus A. Differences in Mac end levels are represented by differences in line thickness. Ad48, A51.1, and A51.2 Mac ends were mapped from comparisons of the micronuclear and macronuclear genomes. A51.3 and A51.4 Mac ends were mapped from macronuclear-genome analysis (6) and this work. Putative A51.5 Mac ends mapping ≫26 kb downstream of gene A are not shown. Uncharacterized DNA is shown as hatched lines; the micronuclear sequence downstream of the BglII site on the right-hand side has not been characterized and could harbor long eliminated sequences. The solid arrow represents the A gene coding region; the dotted arrow represents a long minisatellite open reading frame that identified poly(A) transcripts.
Allele Ad12 harbors a >16-kb deletion.
Strain d12 had been crossed with strains d48 and 51 in previous studies aimed at investigating the control of Mac end formation at locus A. Strain d12 was recovered from the same mutagenesis screen as strain d48 (3). In contrast to strain d48, the d12 micronuclear genome was reported to harbor a mutant Ad12 allele. However, there has been some disagreement over Mac end formation in this strain. Some reports suggested that all Mac ends form 1.3 kb upstream of gene A (8, 23) and that allele Ad12 harbors a deletion (5). One report tentatively identified a few copies of gene A in the d12 macronuclear genome and characterized a variant in which Mac ends formed at the 5′ end of gene A (22).
Hybridization of MspI-restricted d12 DNA with fragment TA3-TB7 as a probe revealed a single 1.4- to 1.6-kb smear (Fig. 2B). This suggested that all Mac ends map 1.6 kb upstream of gene A. To further characterize Ad12 Mac ends, we performed two independent single-strand PCRs on d12 DNA using the telomeric oligonucleotide as the only primer. The polymerization products generated from Mac A ends were then selectively sequenced by using oligonucleotide TA3 as a primer. The two sequences showed colinearity with the micronuclear sequence across 318 nucleotides (Fig. 4A). Beyond this position, only telomeric repeats could be found. These data revealed that, rather than being dispersed over a short DNA region, most, if not all, Ad12 Mac ends form at one position 1,569 nucleotides upstream of the 5′ end of gene A (Fig. 4B).
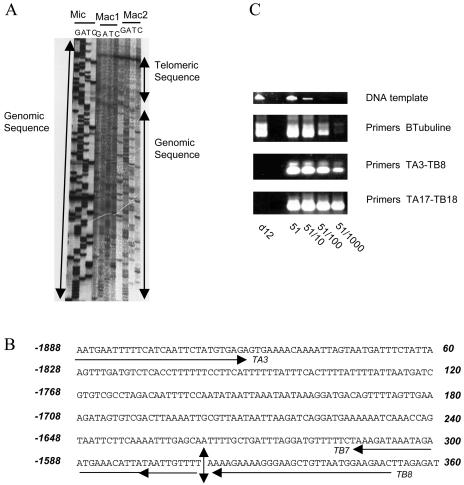
FIG. 4.
Mac end characterization in strain d12. (A) Sequencing of two single-strand PCR products. Two independent PCR products (lanes Mac1 and Mac2) were electrophoresed along with micronuclear A51 sequence (lane Mic). Sequences were colinear over 318 bases. Only the last 50 bp are shown here. (B) Deletion Ad12 characterization. Micronuclear A51 sequence is shown with the oligonucleotides used to characterize micronuclear and macronuclear d12 genomes. The vertical arrow indicates the d12 deletion point. Coordinates are given relative to the sequencing oligonucleotide TA3 on the right side and relative to the 5′ end of gene A on the left side. (C) PCR analysis of genome d12. Oligonucleotides TA3 and TB8, TA17 and TB18, or β-tubulin A and β-tubulin B were used to check for the presence of the corresponding sequences in d12 genomes. Oligonucleotide TB8 maps immediately downstream of the putative deletion point (B). Oligonucleotides TA17 and TB18 map in fragment IB (Fig. 1A) 8 kb downstream of the gene A translation stop site. One microliter of DNAs d12 and 51 (lanes d12 and 51) and 1 μl of 10-, 100-, and 1,000-fold-diluted 51 DNA (lanes 51/10, 51/100, and 51/1000) are shown colored with ethidium bromide at the top of the figure. Oligonucleotides β-tubulin A and β-tubulin B were able to generate 1.3-kb products in d12 and 51 DNAs, as well as in 10-, 100-, and 1,000-fold-diluted 51 DNA. Although PCR products of 0.35 and 0.50 kb could be efficiently recovered from undiluted, 10-, 100-, and 1,000-fold-diluted 51 DNAs when oligonucleotides TA3-TB8 and TA17-TB18, respectively, were used, no PCR product could be generated with d12 DNA.
As the upper sequence from the PCR products was faint (Fig. 4A) and could therefore have masked rare ends mapping downstream of position −1569, we checked for such rare ends in the d12 macronuclear genome and for the presence of the corresponding sequence in the d12 micronuclear genome. PCR was performed on d12 DNA using primers TA3 and TB8 (oligonucleotide TB8 maps immediately downstream of position 1569 [Fig. 4B]) or primers TA17 and TB18, which map downstream of gene A (Table 1). In both cases, we failed to amplify any product from d12 DNA, even though PCR products could be recovered from a 1,000-fold dilution of strain 51 DNA (Fig. 4C).
Compared to the strain 51 micronuclear genome, the d12 micronuclear genome lacks >16 kb, including the 10-kb A gene, 1,569 bp of upstream DNA, and at least 7 kb of downstream DNA. This deletion is likely to be terminal, as this would account for the very unusual feature of Ad12 Mac ends: Ad12 Mac ends do not show variability but apparently map at a single nucleotide position, −1569 relative to the 5′ end of gene A.
Chromosome breakage in progeny from cross d12 × d48 escapes parental patterns.
In light of the newly identified macronuclear and micronuclear differences among strains d12, d48, and 51, we examined Mac end formation at locus A in cross progeny. Conjugation in Paramecium is a reciprocal process that results in the formation of genetically identical zygotic nuclei in the two exconjugants. This reciprocity allows the analysis of the two types of maternal and developing macronucleus relationships in the same cross.
We first looked at Mac end formation in progeny from cross d12 × d48. We isolated the six progeny of three successful conjugation events with micronuclear genomes harboring the Ad12 and A51 alleles.
In order to look for the level of Ad12 Mac ends relative to those of Ad48 and A51 Mac ends in their macronuclear polyploidy genomes, fragment TA3-TB7 was hybridized to MspI-SpeI-restricted DNAs (data not shown). Single signals were revealed in parental DNAs, a 1.4- to 1.6-kb smear identifying Ad12 Mac ends in parent d12 and a 2.5-kb restriction fragment identifying Ad48 and/or A51 Mac ends in parent d48. Two signals were revealed in progeny DNAs. In the first two pairs of progeny (progeny 1 and 2), the 1.4- to 1.6-kb smear and 2.5-kb restriction fragment showed a ratio of ∼1:1. This strongly suggested that alleles Ad12 and A51 had been equally amplified in the developing macronuclei from these progeny and that only Ad48 and A51 Mac ends had formed from allele A51. In the third pair of progeny (progeny 3), the two signals showed a ratio close to 2:1, and Ad12 Mac ends were predominant. This could be a consequence of the preferential amplification of allele Ad12. Alternatively, it could be a consequence of the breakage of some chromosomes amplified from allele A51 1.6 kb upstream of gene A.
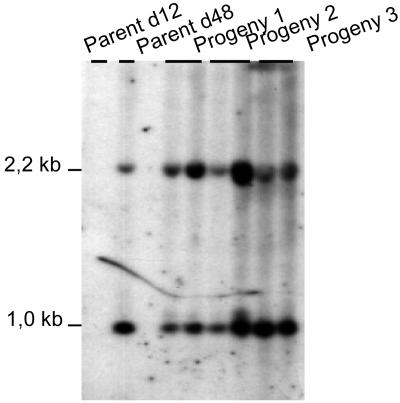
In order to quantify the relative levels of Ad48 and A51 Mac ends in the d48 parent and progeny macronuclei, DNAs were restricted with EcoRI and SpeI and probed with fragments 11D and TA95-TB92 (Fig. 1B and D). Restriction fragments of 1.2 and 2.2 kb were revealed, identifying Mac ends formed at the 5′ and 3′ ends of gene A, respectively, on all DNAs but d12 DNA (Fig. 5). Although the two probes did not necessarily hybridize with the same efficiency, we developed a quantification protocol to estimate the relative amounts of Mac A ends in parents and progeny (see Materials and Methods). In the d48 parental cell line and in progeny 3, ∼20% of the A chromosomes exhibited A51 Mac ends, while in progeny 1 and 2, 45% ± 4.5% of the chromosomes amplified from allele A51 ended downstream from the A gene 3′ end. Two out of the three progeny therefore showed a Mac end pattern different from that of their parents.
FIG. 5.
Mac end characterization in progeny from d12 × d48 cross. DNAs were restricted with EcoRI and SpeI and probed with fragments 11D and TA92-TB95 in order to identify restriction fragments of 1.0 and 2.2 kb from the A gene 5′ and 3′ ends, respectively. No signal was observed on parent d12, attesting to the lack of any corresponding sequence in d12 genomes.
Chromosome breakage in progeny from cross d12 × 51 sums up parental patterns.
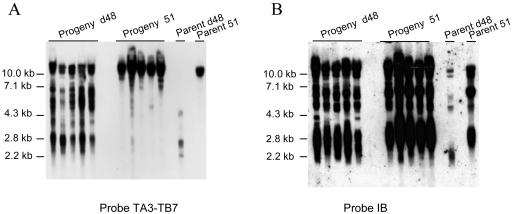
We then examined Mac end formation at locus A in progeny from cross d12 × 51. We isolated the 12 progeny of six successful conjugation events. DNAs from parents and progeny were restricted with MspI and hybridized with fragment TA3-TB7 (Fig. 6A). Signals of 1.4 to 1.6 and 10 to 12 kb were revealed in all progeny. A signal of 1.4 to 1. 6 kb identified telomeric fragments derived from Ad12 Mac ends; a signal of 10 to 12 kb identified telomeric fragments derived from A51.1 Mac ends and a restriction fragment derived from A51.2 to A51.4 Mac ends (fragments derived from A51.1 and A51.2 to A51.4 Mac ends could not be separated in this type of migration). The two signals showed a ratio close to 1:1, showing that alleles Ad12 and A51 had been equally amplified in their macronuclei. Hybridization of the same Southern blot with probe IB revealed signals identifying A51.2 to A51.4 Mac ends in all progeny (Fig. 6B; A51.1 Mac ends map upstream of probe IB). Signal ratios were similar in progeny and parent 51 (note that the two faint signals revealed by fragment IB in d12 DNA could identify only cross-hybridizing loci; no PCR product was generated from d12 DNA when primers TA17 and TB18, mapping within fragment IB, were used [Fig. 2C and 4C]). Chromosome breakage at locus A in progeny from cross d12 × 51, therefore, sums up the parental breakage patterns d12 and 51.
FIG. 6.
Mac end characterization in progeny from d12 × 51 cross. MspI-restricted DNAs were hybridized with (A) fragment TA3-TB7 or (B) fragment IB. Fragment IB identified two cross-hybridizing sequences of 3.5 and 8 kb in DNA from parent d12; in d48 and 51 and progeny DNAs, the upper cross-hybridizing sequence is masked by fragments derived from gene A downstream sequences (Fig. 2).
Chromosome breakage differs between parent d48 and its progeny in cross d48 × 51.
We also examined Mac A ends in progeny from cross d48 × 51. We isolated the 10 progeny from five successful conjugation events. Parent and progeny DNAs were restricted with MspI and hybridized with fragment TA3-TB7 (Fig. 7A). Parent 51 and its progeny similarly displayed a strong >10-kb signal identifying telomeric fragments derived from A51.1 Mac ends and a restriction fragment derived from A51.2 to A51.4 Mac ends. The 51-derived progeny also displayed signals of <10 kb that identified minor rearrangements at locus A. In contrast, parent d48 and its progeny displayed sharp differences. While probe TA3-TB7 revealed a 2- to 4-kb signal on parent d48, it revealed two signals of 2 to 7 and 10 to 12 kb in its progeny. Presumably, the 2- to 7-kb signal identified telomeric fragments derived from Ad48 Mac ends mapping from −0.5 to +4.5 kb relative to the 5′ end of gene A while the signal of 10 to 12 kb corresponded to fragments derived from A51 Mac ends. Hybridization of the same Southern blot with probe IB indeed revealed strong signals identifying abundant A51.2 to A51.4 Mac ends in all the d48-derived progeny (Fig. 7B). The 2- to 7- and 10- to 12-kb signals detected by probe TA3-TB7 showed a mean ratio of 3:1 in the progeny, indicating that A51 Mac ends accounted for ∼25% of the Mac A ends in the d48-derived progeny.
FIG. 7.
Mac end characterization in progeny from d48 × 51 cross. MspI-digested DNA was hybridized with (A) fragment TA3-TB7 or (B) fragment IB. Fragment IB identified two faintly cross-hybridizing sequences of 3.5 and 8 kb (Fig. 2).
DISCUSSION
Mapping of new Mac ends at P. tetraurelia locus A.
Approximately 500 to 1,000 breakage events occur during macronuclear-genome development in P. aurelia. In P. tetraurelia strain 51, breakage events had been shown to produce Mac ends 8, 13, and 26 kb downstream of gene A (6). We have named these A51.2, A51.3, and A51.4 Mac ends. By analyzing a 52-kb region that encompasses the 10-kb A gene, as well as 12 kb downstream and 30 kb upstream of it, we have characterized two new types of Mac ends in strain 51, Ad48, and A51.1 Mac ends. Ad48 Mac ends map at the 5′ end of gene A and account for very few of the Mac ends formed at this locus (1 to 2%). A51.1 Mac ends map 2 to 4 kb downstream of gene A. They overlap the 3′ end of a minisatellite sequence located 2.5 to 4.4 kb downstream of gene A (4) and account for about half of the Mac ends that form at locus A. While Ad48, A51.1, and A51.2 Mac ends were characterized by comparison of micronuclear and macronuclear genomes, A51.3 and A51.4 Mac ends were mapped by macronuclear-genome analysis (reference 6 and this work). The micronuclear sequence downstream of 51.2 Mac ends has been only partially characterized and could harbor long eliminated sequences. The distances separating A51.3 and A51.4 Mac ends from gene A could therefore be underestimated.
The original characterization of the wild-type 51 strain and mutant d48 strain described qualitative differences in their Mac A ends, although both strains do harbor the same micronuclear genome (6). We show here that strain d48, at least the presently available strain, displays only quantitative Mac end differences from strain 51 in rearranging locus A. While <2% of the A chromosomes have Ad48 Mac ends in macronuclear strain 51 genomes, 80 to 99% of them, depending on the d48 cell line we analyzed, exhibit Ad48 Mac ends in macronuclear d48 genomes.
Mutant strain d12 had been shown to harbor a mutant allele that has remained uncharacterized (3). Our work demonstrates that allele Ad12 displays a deletion that starts 1,569 bp upstream of the 5′ end of gene A and extends over >16 kb. Recovery of a d12 variant in which Mac A ends were mapped at the A gene 5′ end rather than 1.6 kb upstream from it has been described (22). Since no Mac ends are expected to form within a region that lacks the Ad12 allele, the d12 variant should in fact have been a contaminant. Nuclear transfer and genetic crosses were performed with cell lines derived from the d12 stock from which the d12 variant had been recovered (12, 22). Since it is unclear whether these experiments used the original d12 mutant, their data could not be discussed below.
Macronuclear d12 genomes do not harbor any of the Mac A ends characterizing macronuclear d48 and 51 genomes. Furthermore, Ad12 Mac ends are not dispersed over any DNA region but apparently map at a unique nucleotide position 1,569 bp upstream of the 5′ end of gene A. Together, these data suggest that allele Ad12 displays a terminal deletion that is simply carried over into the macronucleus.
Characterizing molecular features from F1 progeny.
Differences in rearranging locus A between strains d48 and 51, which harbor identical micronuclear genomes, demonstrated that Mac end formation in strain d48 and/or strain 51 is epigenetically controlled (6).
Macronucleoplasm transplantation experiments and phenotypic analysis of F1 progeny obtained from cross d48 × 51 led to a model in which accurate chromosome breakage downstream of gene A in developing macronuclei relies on the production of 51 factors by maternal strain 51 macronuclei. In that model and further developments, breakage pattern d48 is the consequence of a failure in the production and export of 51 factors (9, 13, 17, 22).
The above-mentioned model predicted that the macronuclear genomes from progeny obtained from interstrain crosses were similar to those of their parents (22). Using the new genomic information we had provided, we have made three key observations. First, d48 parents and their F1 progeny differ in their macronuclear contents, whatever the cross. Second, the progeny derived from d12 cells crossed with either d48 or 51 cells display molecular differences, although both progeny are Ad12/A51 heterozygotes. Third, the progeny derived from d48 cells crossed with either d12 or 51 cells have similar macronuclear genomes, although the former are Ad12/A51 heterozygotes while the latter are A51/A51 homozygotes.
Controlling DNA breakage at the 5′ end of P. tetraurelia gene A.
The d48 parent crossed with the 51 parent displayed only 2% A51 Mac ends, while its progeny displayed a 12-fold enhancement of these Mac ends, on average.
Export of 51 factors from parent 51 to parent d48 could account for the enhanced formation of A51 Mac ends in the d48-derived progeny. In that case, no enhanced formation of A51 Mac ends would be expected in the d48-derived progeny from a d12 × d48 cross (remember that strain d12 is devoid of any corresponding information). Nevertheless, the d48-derived progeny from the d12 × d48 cross that had equal levels of Ad12 Mac ends and Ad48-A51 Mac ends displayed a twofold enhancement of A51 Mac ends compared to their parent (Ad48-A51 Mac ends can arise only from allele A51; see below for a discussion of the impact of the relative levels of amplification from alleles Ad12 and A51 on enhancement in A51 Mac end formation).
Our data instead lead to a model in which the formation of Mac ends at the 5′ end of gene A is controlled by a d48 factor(s) and the partial rescue of A51 Mac ends in the d48-derived progeny is due to a partial lack of the d48 factor(s) as a consequence of its partial export from conjugant d48 to conjugant d12 or 51. The import of d48 factors in parent d12 could fit the partial formation of A51 Mac ends in its progeny. On the other hand, the lack of Ad48 Mac ends in the 51-derived progeny is likely to result from competition between sequences of the maternal and developing macronuclear genomes for the d48 factors. The 51 maternal macronuclear genome harbors ∼1,000 chromosomes that end downstream of gene A and therefore may sequester the imported d48 factors.
Sequestration of the d48 factors could also account for the observations made of macronuclear transfers between strains d48 and 51. Transfer of d48 macronucleoplasm did not promote any extinction of gene A expression in 51-derived progeny, probably because of a sequestration of the d48 factors by the maternal genome that displayed ∼1,000 A51 Mac ends (13). Sequestration of the d48 factors by A51 Mac end-harboring chromosomes from the maternal genome should also account for the rescue of gene A expression in the progeny from d48 cells that had been injected with 51 macronucleoplasm (9, 13). More than half of the macronucleoplasm was transferred in those experiments, thus providing ∼500 chromosomes for d48 factor sequestration.
Gene A expression rescue has also been described in autogamous progeny from d48 cells in which 1/10 of the cytoplasm from strain 51 cells at different stages of macronuclear development had been transferred (13). Eighteen percent of rescues were observed when cytoplasm was transplanted from cells at the stage of micronucleus swelling, and 73% were observed when cytoplasm was transplanted from cells at the stage of skein macronucleus and at later stages. If cytoplasm were devoid of any DNA material, rescue would have to rely on a DNA-independent mechanism, and sequestration of d48 factors might be the wrong hypothesis. However, it is unclear whether the transplanted cytoplasm from autogamous strain 51 cells had some A51 Mac ends available for d48 factor sequestration. The parental macronuclei expand to the whole cell in the course of sexual reproduction, first as an unwinding ribbon-like structure and then in the form of ∼30 nuclear fragments in which the DNA is transcribed but does not replicate. Could part of the ribbon, or a few nuclear fragments, have been part of the transferred cytoplasm? If d48 factors are produced, the facts that conjugation reduced the level of Ad48 Mac ends and transfer of autogamous cytoplasm rescued antigen A expression in d48-derived progeny suggest that the d48 factors are produced in relatively small quantities.
An active rather than a passive role for Paramecium parental macronuclei?
A model for cross talk between the maternal and developing macronuclei of ciliates has been proposed, based on studies of the process of IES excision in T. thermophila (18). The scan RNA model postulates that widespread transcripts of the micronuclear zygotic genome are chopped into small RNA molecules, which are matched against the genomes of the maternal macronucleus that behave as filters. Anything left over, such as transcripts from IESs, would diffuse to the developing macronuclei and induce epigenetic modifications, ultimately leading to IES excision. In the same way, anything left over, such as transcripts from sequences located downstream of breakage regions, could diffuse to the developing macronuclei and induce epigenetic modifications, ultimately leading to chromosome breakage, Mac end formation, and downstream sequence elimination.
The scan RNA model cannot readily account for the establishment of the Mac end pattern at the A locus of P. tetraurelia strain 51, although it can account for its maintenance through generations. The further from gene A the sequences map, the less the corresponding micronuclear transcripts would be sequestered by the maternal macronuclear genome, since A51.1 to A51.4 Mac ends display a 4:2:1:1 ratio. As a consequence, sequences located downstream from A51.4 Mac ends in the developing macronuclear genome would be more heavily marked for elimination than those located close to A51.1 Mac ends. Note that although A51.1 Mac ends account for ∼50% of the Mac ends that form downstream from gene A, DNA breakage in the corresponding region does not necessarily account for the majority of the breakage events occurring downstream from gene A. A51.1 Mac end formation may result from breakages within regions of A51.1, A51.2, A51.3, and A51.4 Mac end formation on each chromatid. In that case, the region of A51.1 Mac end formation would in fact be less frequently broken than the region of A51.4 Mac end formation. According to the scan RNA model, the A51 allele produces similar RNA molecules in strains d48 and 51 and the RNA molecules derived from the A gene are not retained by the maternal d48 macronuclear genome but mark the corresponding sequences from the developing genome for elimination.
The scan RNA model, however, cannot account for the pattern of Mac A ends that we observed in the heterozygous Ad12/A51 progeny. According to the model, these F1 progeny should produce RNA molecules encompassing the micronuclear A gene. In the d12-derived progeny, these RNA molecules could not be sequestered by the maternal genome devoid of the corresponding sequences and should therefore mark the developing genome for Ad12 Mac end formation 1.6 kb upstream of the A gene. Nevertheless allele A51 produces only A51 Mac ends in cross d12 × 51 progeny.
Previous data demonstrated that the epigenetic factors controlling chromosome breakage at P. tetraurelia locus A have to be nucleic acids encoded by the locus itself (5, 16, 17). In contrast with the scan RNA model, in which the maternal d48 macronuclear genome has a passive role as a filter, our data suggest that the maternal d48 macronuclear genome plays an active role in the control of chromosome breakage at the 5′ end of gene A by producing d48 factors.
Could the d48 factors be DNA molecules derived from Ad48 Mac ends? This does not appear likely, since it would imply different properties of Ad12 Mac ends and Ad48 Mac ends; Ad12 Mac ends do not promote any regular epigenetic control of Mac end formation.
Could the d48 factors be RNA derived? As noted above, and in contrast with parental d48 macronuclei, parental d12 macronuclei lacked any regular effect on Mac end formation in d12-derived progeny in our hands. Ad12 Mac ends do not map within coding DNA but are located several hundred basepairs downstream of the 3′ end of a gene encoding a cleavage and polyadenylation specificity factor (Amar, unpublished), while the great majority of Ad48 Mac ends map within the first 400 bp of the 5′ end of gene A. This strongly suggests that the d48 factors are RNA molecules encompassing the 5′ end of gene A and adjacent telomeric repeats or derived molecules. These molecules could have an effect by themselves or in combination with some antisense transcripts produced from a telomeric embedded promoter. The wild-type 51 strain has a few Ad48 Mac ends. It is unclear whether the d48 factors specifically associate with the mutation as a result of the original mutagen-induced truncation of some macronuclear A chromosomes or whether the d48 factors are also produced in strain 51 cells.
Injection of recombinant plasmids harboring gene A segments into macronuclei from d48 cells rescued gene A expression in their autogamous progeny (11, 26, 27). Recombinant plasmids introduced into Paramecium macronuclei are randomly linearized, telomerized, and propagated as autonomously replicating DNA units. When recombinant plasmids devoid of Paramecium promoters are used, both sense and antisense RNA molecules are transcribed from cryptic promoters (7). Therefore, recombinant plasmids harboring gene A fragments are expected to produce antisense transcripts encompassing the A gene and telomeric repeats. These antisense transcripts could pair and neutralize Ad48 Mac end transcripts through RNA double-strand formation, thereby promoting a d48 factor-free environment. However, plasmids harboring fragments starting downstream of the A gene position 3092 could not induce any rescue of A expression in the progeny of the injected cells (11, 26). Transcripts from such plasmids are not expected to display A sequences overlapping Ad48 Mac ends, most of which map upstream of the A gene position 3092, but should display telomeric sequences. Nevertheless, these plasmids and their corresponding transcripts do not appear to be able to neutralize the putative d48 factors. Could d48 factor neutralization require transcript association, and thus proximity, on chromosomes rather than within the cytoplasm or nucleoplasm?
Chromosome breakage and DNA amplification in Paramecium.
Chromosome breakage has recently been proposed to be a consequence of the systematic elimination of multicopy sequences through a process distinct from IES excision (14). Although this model may apply to some loci, it may not be the rule. P. tetraurelia macronuclear genomes do harbor large gene families with several tens of members. P. tetraurelia macronuclear genomes also harbor at least one highly repeated minisatellite sequence, one member of which lies 3 to 4.5 kb downstream of gene A (4) and partially overlaps A51.1 Mac ends. However, this sequence appeared to be retained on A51.2 to A51.4 Mac ends, which account for 50% of the Mac ends formed at locus A.
As previously noted, chromosome breakage and sequence elimination in P. aurelia could be linked in two ways (5, 17). Mac end formation could result from a differential amplification of micronuclear sequences. Alternatively, Mac end formation could result from breakage of the replicated micronuclear genome and the selective elimination of some DNA fragments in the developing macronuclei.
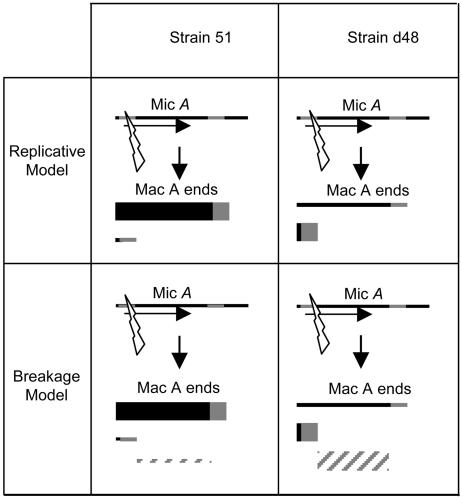
In the replicative model, Mac end formation at P. tetraurelia locus A would result from differences in the amplification of the micronuclear sequences lying on both sides of the 5′ end of gene A. The d48 factors would switch the 5′ end of gene A into a DNA replication terminator site or into a DNA replication initiation site used for unidirectional synthesis (Fig. 8). In the breakage model, DNA fragments of >10 kb should be produced by breakages within the DNA regions of Ad48 and A51 Mac end formation. These DNA fragments should be especially abundant in strain d48.
FIG. 8.
Models for Mac end formation at the 5′ end of P. tetraurelia gene A. In the breakage model, Mac end formation results from the breakage of the micronuclear genome at the 5′ end of gene A, as well as downstream from it, within DNA regions shown in grey. These events would produce DNA fragments of ∼10 kb (shown as hatched bars) that have to be selectively eliminated. Differences in levels of Mac ends and DNA fragments are represented by differences in line thickness. Breakage at the 5′ end of gene A required d48 factors, represented by jagged arrows. In the replicative model, Mac end formation results from differential amplification of micronuclear sequences on both sides of the 5′ end of gene A. The d48 factors could switch the 5′ end of the gene A region into a DNA replication terminator or into a DNA replication initiation site used for unidirectional synthesis. The solid arrow represents the A gene coding region.
Neither of these models provides an easy explanation for the fact that Ad48 Mac ends map within an enlarged region in cross progeny when compared to those of parent d48 (Fig. 7B). However, the fragmentation model best accounts for the fact that, in the progeny from cross d12 × d48, the level of A51 Mac ends inversely correlates with the level of sequences amplified from allele A51. The progeny from this cross that displayed 70% of the Ad12 Mac ends did not have more A51 Mac ends than their d48 parent. It can be postulated that neither the parental macronuclear genome nor the developing macronuclear genome provided enough sequences to sequester the d48 factors and create the d48 factor-free environment required for some chromosomes to escape breakage at the 5′ end of gene A.
In most systems, epigenetic mechanisms modulate gene expression during development. In ciliates, epigenetic mechanisms modulate programmed genome rearrangement patterns that have to be viewed as the first level of genome expression in these organisms. In the case of P. tetraurelia locus A, Mac end formation at the 5′ end of gene A appears to be epigenetically controlled, perhaps by RNA molecules produced by the maternal macronuclear genome. Our data provide new horizons for investigating the molecules involved in the corresponding epigenetic mechanism.
Acknowledgments
We thank James Forney and John Preer for the gift of P. tetraurelia strains d12 and d48; Jean Cohen for that of strain nd7-1; Joy Steele, Louise Preer, and John Preer for providing the micronuclear genomic library; and Edith Heard for her critical reading of the manuscript.
This work was supported by the Centre National de la Recherche Scientifique, the Ministère de l'Education Nationale de la Recherche et de la Technologie (Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires), the Groupement de Recherches et d'Etudes sur les Génomes (grant no. 22/95), the Association pour la Recherche sur le Cancer (grant no. 1374), and grant no. 97N63/0016 from the Centre National de la Recherche Scientifique. K. Dubrana was the recipient of a doctoral fellowship from the Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Amar, L. 1994. Chromosome end formation and internal sequence elimination as alternative genomic rearrangements in the ciliate Paramecium. J. Mol. Biol. 236:421-426. (Erratum, 265:465, 1997.) [DOI] [PubMed] [Google Scholar]
- 2.Dubrana, K., and L. Amar. 2000. Programmed DNA under-amplification in Paramecium primaurelia. Chromosoma 109:460-466. [DOI] [PubMed] [Google Scholar]
- 3.Epstein, L. M., and J. D. Forney. 1984. Mendelian and non-Mendelian mutations affecting surface antigen expression in Paramecium tetraurelia. Mol. Cell. Biol. 4:1583-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forney, J., and K. Rodkey. 1992. A repetitive DNA sequence in Paramecium macronuclei is related to the beta subunit of G proteins. Nucleic Acids Res. 20:5397-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forney, J., F. Yantiri, and K. Mikami. 1996. Developmentally controlled rearrangement of surface protein genes in Paramecium tetraurelia. J. Eukaryot. Microbiol. 43:462-467. [Google Scholar]
- 6.Forney, J. D., and E. H. Blackburn. 1988. Developmentally controlled telomere addition in wild-type and mutant paramecia. Mol. Cell. Biol. 8:251-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galvani, A., and L. Sperling. 2001. Transgene-mediated post-transcriptional gene silencing is inhibited by 3′ non-coding sequences in Paramecium. Nucleic Acids Res. 29:4387-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Godiska, R., K. J. Aufderheide, D. Gilley, P. Hendrie, T. Fitzwater, L. B. Preer, B. Polisky, and J. R. Preer, Jr. 1987. Transformation of Paramecium by microinjection of a cloned serotype gene. Proc. Natl. Acad. Sci. USA 84:7590-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harumoto, T. 1986. Induced change in a non-Mendelian determinant by transplantation of macronucleoplasm in Paramecium tetraurelia. Mol. Cell. Biol. 6:3498-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones, W. 1956. Nuclear differentiation in Paramecium. Ph.D. thesis. Aberystwyth University, Aberystwyth, Wales.
- 11.Kim, C. S., J. R. Preer, Jr., and B. Polisky. 1994. Identification of DNA segments capable of rescuing a non-Mendelian mutant in paramecium. Genetics 136:1325-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi, S., and S. Koizumi. 1990. Characterization of Mendelian and non-Mendelian mutant strains by micronuclear transplantation in Paramecium tetraurelia. J. Protozool. 37:489-492. [Google Scholar]
- 13.Koizumi, S., and S. Kobayashi. 1989. Microinjection of plasmid DNA encoding the A surface antigen of Paramecium tetraurelia restores the ability to regenerate a wild-type macronucleus. Mol. Cell. Biol. 9:4398-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Mouel, A., A. Butler, F. Caron, and E. Meyer. 2003. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in paramecia. Eukaryot. Cell 2:1076-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer, E., F. Caron, and A. Baroin. 1985. Macronuclear structure of the G surface antigen gene of Paramecium primaurelia and direct expression of its repeated epitopes in Escherichia coli. Mol. Cell. Biol. 5:2414-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer, E. 1992. Induction of specific macronuclear developmental mutations by microinjection of a cloned telomeric gene in Paramecium primaurelia. Genes Dev. 6:211-222. [DOI] [PubMed] [Google Scholar]
- 17.Meyer, E., and S. Duharcourt. 1996. Epigenetic regulation of programmed genomic rearrangements in Paramecium aurelia. J. Eukaryot. Microbiol. 43:453-461. [DOI] [PubMed] [Google Scholar]
- 18.Mochizuki, K., N. Fine, T. Fujisawa, and M. Gorovsky. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110:689. [DOI] [PubMed] [Google Scholar]
- 19.Phan, H. L., J. Forney, and E. H. Blackburn. 1989. Analysis of Paramecium macronuclear DNA using pulsed field gel electrophoresis. J. Protozool. 36:402-408. [DOI] [PubMed] [Google Scholar]
- 20.Prescott, D. M. 1994. The DNA of ciliated protozoa. Microbiol. Rev. 58:233-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prescott, D. M. 2000. Genome gymnastics: unique modes of DNA evolution and processing in ciliates. Nat. Rev. Genet. 1:191-198. [DOI] [PubMed] [Google Scholar]
- 22.Rudman, B., L. B. Preer, B. Polisky, and J. R. Preer, Jr. 1991. Mutants affecting processing of DNA in macronuclear development in paramecium. Genetics 129:47-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott, J., C. Leeck, and J. Forney. 1994. Analysis of the micronuclear B type surface protein gene in Paramecium tetraurelia. Nucleic Acids Res. 22:5079-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott, J. M., K. Mikami, C. L. Leeck, and J. D. Forney. 1994. Non-Mendelian inheritance of macronuclear mutations is gene specific in Paramecium tetraurelia. Mol. Cell. Biol. 14:2479-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skouri, F., and J. Cohen. 1997. Genetic approach to regulated exocytosis using functional complementation in Paramecium: identification of the ND7 gene required for membrane fusion. Mol. Biol. Cell. 8:1063-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You, Y., K. Aufderheide, J. Morand, K. Rodkey, and J. Forney. 1991. Macronuclear transformation with specific DNA fragments controls the content of the new macronuclear genome in Paramecium tetraurelia. Mol. Cell. Biol. 11:1133-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.You, Y., J. Scott, and J. Forney. 1994. The role of macronuclear DNA sequences in the permanent rescue of a non-Mendelian mutation in Paramecium tetraurelia. Genetics 136:1319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]