Abstract
Plastids (photosynthetic organelles of plants and algae) are known to have spread between eukaryotic lineages by secondary endosymbiosis, that is, by the uptake of a eukaryotic alga by another eukaryote. But the number of times this has taken place is controversial. This is particularly so in the case of eukaryotes with plastids derived from red algae, which are numerous and diverse. Despite their diversity, it has been suggested that all these eukaryotes share a recent common ancestor and that their plastids originated in a single endosymbiosis, the so-called “chromalveolate hypothesis.” Here we describe a novel molecular character that supports the chromalveolate hypothesis. Fructose-1,6-bisphosphate aldolase (FBA) is a glycolytic and Calvin cycle enzyme that exists as two nonhomologous types, class I and class II. Red algal plastid-targeted FBA is a class I enzyme related to homologues from plants and green algae, and it would be predicted that the plastid-targeted FBA from algae with red algal secondary endosymbionts should be related to this class I enzyme. However, we show that plastid-targeted FBA of heterokonts, cryptomonads, haptophytes, and dinoflagellates (all photosynthetic chromalveolates) are class II plastid-targeted enzymes, completely unlike those of red algal plastids. The chromalveolate enzymes form a strongly supported group in FBA phylogeny, and their common possession of this unexpected plastid characteristic provides new evidence for their close relationship and a common origin for their plastids.
Plastids, the photosynthetic organelles of plants and algae and their nonphotosynthetic derivatives, are known to have originated through endosymbiosis involving a cyanobacterium (11). This so-called primary endosymbiosis gave rise to the primary plastids found in glaucophytes, red algae, green algae, and plants. All other algal groups acquired photosynthesis by secondary endosymbiosis, which involves the uptake of one of these eukaryotic primary algae by another eukaryote, and the conversion of this alga into an organelle (1, 18). Secondary endosymbiosis must have taken place more than once, because some secondary plastids are derived from green algae (those of euglenids and chlorarachniophytes), while others are derived from red algae (those of cryptomonads, haptophytes, heterokonts, and dinoflagellates). Apicomplexan parasites also have a plastid that is likely of red algal ancestry, although this has been controversial (for a review, see reference 30).
Exactly how many secondary endosymbiotic events have occurred has also proven contentious, with estimates ranging from two to seven (1, 4, 7). Among those algae with red algal secondary plastids, there is a great diversity of both host and plastid characteristics, which originally led to the belief that these plastids arose through many independent endosymbioses. This conclusion was apparently supported by phylogenetic analyses of both plastid and host gene sequences, which consistently failed to unite these groups but provided no strong alternative (6, 15, 22, 25). Based on the perceived complexity of plastid gain, it was nevertheless proposed that all these groups and their aplastidal relatives originated from a common ancestor and that their plastids are derived from a single, common endosymbiotic event involving a red alga (5). This collection of lineages, dubbed “chromalveolates,” comprised the cryptomonads, haptophytes, and heterokonts (collectively, chromists) and ciliates, dinoflagellates, and apicomplexa (collectively, alveolates). This hypothesis raises two obvious predictions: molecular sequences from both host lineages and from their plastids should prove to be closely related, and nonphotosynthetic chromalveolate lineages (e.g., ciliates) once had a plastid or perhaps still retain a relict organelle.
The first molecular evidence supporting the chromalveolate hypothesis was the phylogeny of the plastid-targeted enzyme, glyceralydehyde-3-phosphate dehydrogenase (GAPDH). Like most plastid proteins, GAPDH is encoded in the nuclear genome and posttranslationally targeted to the plastid by an N-terminal targeting peptide. Moreover, GAPDH activity is required in both the plastid and cytosol of plants and algae, so nuclear genomes would be predicted to encode two distinct genes: one for the eukaryotic cytosolic protein and one for the cyanobacterium-derived, plastid-targeted enzyme. In plants, euglenids, green algae, and red algae, this expectation is borne out: the plastid-targeted GAPDH is cyanobacterial while the cytosolic enzyme is closely related to other eukaryotic homologues. However, plastid-targeted GAPDH in apicomplexans, cryptomonads, dinoflagellates, heterokonts, and haptophytes are not cyanobacterial but, instead, are more closely related to cytosolic GAPDH homologues (8, 9, 14, 19) and probably evolved from a gene duplication of the cytosolic paralogue (9). Such events, called endosymbiotic gene replacements, are known from other genes but are relatively rare. Accordingly, the common origin of all chromalveolate plastid-targeted GAPDH homologues provided strong evidence that their plastids originated from a common endosymbiotic event (9).
While GAPDH phylogenies may be difficult to explain by any other means, GAPDH is only a single gene, and such an ancient and major event in eukaryotic evolution would ideally be supported by many sources of evidence. Phylogenetic analysis of multiple plastid-encoded genes provided support for the descent of cryptomonads and haptophyte and heterokont plastids from a common red algal ancestor (32). Unfortunately, data from apicomplexans and dinoflagellates could not be included in this study due to the unusual nature of their plastid genomes (31, 33). As a result, any additional evidence for this major event in the history of eukaryotes will likely have to come from cytosolic and nuclear-encoded, plastid-targeted proteins.
Here we describe a novel gene replacement event in chromalveolates that involves the plastid-targeted fructose-1,6-bisphosphate aldolase (FBA). FBA is a key metabolic enzyme catalyzing the cleavage of B-fructose-1,6-phosphate to d-glyceraldehyde-3-phosphate and dihydroxyacetone phosphate in glycolysis and the reverse reaction in gluconeogenesis. In the plastid, FBA catalyzes the condensation of fructose-1,6-bisphosphate from dihydroxyacetone phosphate and GAP as well as the condensation of sedoheptulose-1,7-bisphosphate from erythrose-4-phosphate and GAP in the Calvin cycle. There are two distinct and unrelated classes of FBA: class I is found primarily in eukaryotes, and class II is found primarily in bacteria, including cyanobacteria (27). As with GAPDH, the Calvin cycle FBA in the plastid would be expected to be derived from the cyanobacterial endosymbiont and would therefore be a class II enzyme. However, plants, green algae, red algae, and chlorarachniophytes have two class I isoforms: a cytosolic protein active in glycolysis and gluconeogenesis and a plastid-targeted protein active in the Calvin cycle, which is thought to have arisen from a duplication of the cytosolic homologue (12).
Since the red algal plastid-targeted FBA is a class I enzyme related to plastid homologues of green algae and plants, it would be predicted that the plastid-targeted proteins of all algae with secondary plastids of red algal ancestry would be related to the class I red algal genes. However, the plastid-targeted FBA from the heterokonts (diatoms) is a class II enzyme, completely unrelated to the expected red algal homologue (17, 26). This enzyme is evidently derived from a gene replacement, so it marks an excellent potential test for the history of chromalveolate plastids. If the plastids of various chromalveolates arose independently, then they should contain typical red algal plastid-targeted class I FBA. Alternatively, if the chromalveolates and their plastids share a common origin, then other chromalveolate plastid-targeted FBAs should be related to the class II type A FBA found in diatoms. We have characterized genes for putative plastid-targeted FBA from cryptomonads, haptophytes, and dinoflagellates and found that they are all class II genes related to those of diatoms, supporting a common origin for their plastids. This represents the second such rare event to support the single origin of these plastids, and it supports the notion that the ancestor of chromalveolates was photosynthetic.
MATERIALS AND METHODS
Characterization of new FBA genes.
Messenger RNAs encoding class II FBA were identified from ongoing expressed sequence tag (EST) projects from the dinoflagellate Heterocapsa triquetra (strain CCMP 449), the cryptomonad Guillardia theta (strain CCMP 327), and the haptophyte Isochrysis galbana (strain CCMP 1323). ESTs were searched for both class I and II FBA genes, and transcripts for all distinct genes were isolated and completely sequenced. Rhodomonas sp. (strain CCMP 632) and Phaeodactylum tricornutum (strain CCMP 1327) were grown in f/2-Si medium at 16°C (12-h light and 12-h dark cycle). Cells were harvested at 4 weeks, and Rhodomonas RNA was prepared by using Trizol (Invitrogen). cDNA was synthesized from a poly(T) primer and used as a template for reverse transcription-PCR with degenerate primers designed to class II FBAs (GARGARGAYGGNGTNCAYAA and GGRTCRTARTAYTTYTTRTT). PCR products were cloned and sequenced; the sequence was used to design a Rhodomonas-specific primer for 3′ rapid amplification of cDNA ends, and these products were cloned and sequenced to retrieve a complete 3′ end sequence. DNA was prepared as described (16), and the class I FBA was amplified by using primers CGCAGTTGCAAACTAGATACGATG and GCTGGAGGCCGATGAGGCTTTCTTC. Sequences encoding class I and class II FBAs were also identified and assembled from the Thalassiosira pseudonana (diatom) genome sequence and EST data (genome.jgi-psf.org/thaps1/thaps1.home.html). Class I and II FBAs were also identified in EST sequences from Porphyra yezoensis and Gracilaria gracilis, and class I FBAs were identified and assembled from the Tetrahymena thermophila (www.tigr.org/tdb/e2k1/ttg/) and Paramecium tetraurelia (paramecium.cgm.cnrs-gif.fr) genomic and EST sequencing projects and from the Amphidinum carterae, Alexandrium tamarense, and Lingulodinium polyedrum EST projects.
Sequence analysis.
New amino acid sequences were added to existing class I and class II FBA alignments and examined for N-terminal extensions. Extensions were analyzed for characteristics consistent with signal peptides by both neural network and hidden Markov model methods by using SignalP version 3.0 software (2). Phylogenies were inferred by using distance and maximum likelihood. For distance analyses, the WAG substitution matrix was used with site-to-site rate variation modeled on a discrete gamma distribution with eight rate categories and invariable sites; the alpha parameter and the proportion of invariable sites were estimated from the data by TREE-PUZZLE version 5.0 (29). Trees were inferred from the corrected distances by a weighted neighbor-joining method (WEIGHBOR version 1.0.1) (3) and Fitch-Margoliash (FITCH version 3.6a) (10). Bootstrapped distances were calculated by using PUZZLEBOOT version 1.0.3 (M. Holder and A. Roger, www.tree-puzzle.de) with the alpha parameter and the proportion of invariable sites estimated by TREE-PUZZLE. Protein maximum likelihood trees were inferred by using PhyML version 2.1b1 (13) with site-to-site rate variation modeled on a discrete gamma distribution, and the shape parameter alpha and proportion of invariable sites were estimated from the data. Protein maximum likelihood trees were also inferred for class II type A FBA by using ProML version 3.6a (10) with global rearrangements and the rates and frequencies estimated by TREE-PUZZLE. Maximum likelihood bootstraps were performed as described above. Class I analyses used 318 characters, global class II analyses used 237 characters, and class II type A alone used 297 characters. Partial sequences from the cryptomonad Rhodomonas sp. and the dinoflagellate Karenia brevis were also analyzed by using 266 characters. All alignments are available upon request.
RESULTS AND DISCUSSION
Distribution and phylogeny of chromalveolate class I FBA.
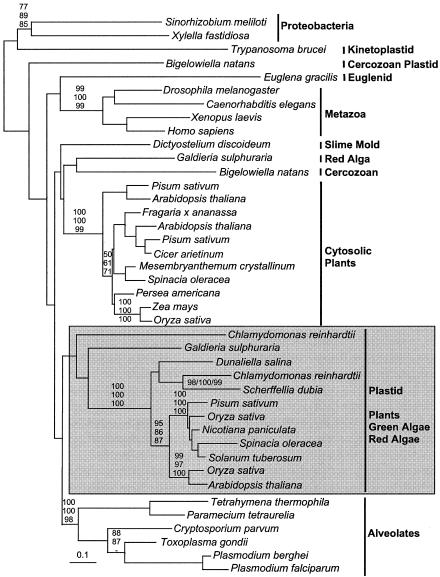
Class I FBA genes were found in the genomes of the ciliates Paramecium and Tetrahymena and in EST data from the dinoflagellates Amphidinum, Alexandrium, and Lingulodinium. However, class I FBA was not found in the genome of the diatom Thalassiosira (estimated to be 95% complete). Partial class I FBAs were also found in EST projects from the red algae Gracilaria and Porphyra. In the class I FBA phylogeny (Fig. 1), the apicomplexa and ciliates form a strongly supported group. The dinoflagellate EST sequences were significantly truncated, but in phylogenies where these fragments were included, they branched with ciliates and apicomplexa, as expected (data not shown). The Lingulodinium sequence included the 5′ end, which did not encode a leader. This demonstrates that all three major alveolate groups contain a cytosolic class I FBA, as is found in most other eukaryotes. The remainder of the class I phylogeny is much as has been previously described (12, 26). In particular, the plastid FBA of red algae, green algae, and plants group together with moderate support, and EST sequences from both Porphyra and Gracilaria branched with the plastid-targeted gene from Galdieria with good support, considering their short length (70 to 75%) (data not shown). Interestingly, one Porphyra EST was related to the divergent class of plastid-targeted FBA from Chlamydomonas and Bigelowiella (data not shown).
FIG. 1.
Protein maximum likelihood phylogeny of class I FBA. Plastid-targeted FBA from plants, green algae, and red algae are boxed and shaded; all other groups are indicated to the right. Note that fragments of dinoflagellate class I genes are also known, and these branch with other alveolates. Numbers at nodes indicate bootstrap support from (top to bottom or left to right) maximum likelihood (PhyML), Fitch-Margoliash, and weighted neighbor-joining.
In addition to the commonly studied class I FBAs shown in Fig. 1, two other distinct and distantly related subgroups of class I genes are known, one primarily archaeal and the other primarily in gram-positive eubacteria (28). We identified an EST from the diatom Phaeodactylum that shares a high degree of similarity with these gram-positive FBAs. The identity of this sequence was confirmed by amplification from Phaeodactylum genomic DNA, which revealed the presence of a typical eukaryotic spliceosomal intron (data not shown). The significance of such a gene in a eukaryote is unclear since the role of this enzyme in bacteria is unknown, but it does appear to be a case of bacteria-to-eukaryote lateral gene transfer.
Distribution and phylogeny of chromalveolate class II FBAs.
Between one and three class II FBA genes were characterized from each organism where they were sought, and their characteristics are summarized in Table 1. Plastid-targeted proteins in these organisms are preceded by bipartite leaders which initiate with a signal peptide (18, 21). The characteristics of the signal peptides are generally predictable, and these predictions and their statistical significance are also given in Table 1. The Heterocapsa C1 (Table 1) sequence also included two distinctive hydrophobic domains sandwiching a short region rich in hydroxylated amino acids (in this case, threonine), which is another distinguishing feature of plastid-targeting peptides in dinoflagellates (23). In addition, class II FBA ESTs were identified from the dinoflagellate Karenia and the red algae Porphyra and Gracilaria. The Porphyra EST includes the amino terminus, which does not encode a leader. Interestingly, the complete genome of Cyanidioschyzon merolae (20) lacks any class II FBA, which implies that the red algae likely acquired this class II enzyme after the divergence of cyanidiales (Cyanidioschyzon) but before the divergence of bangiophytes (Porphyra) and florideophytes (Gracilaria) from one another.
TABLE 1.
Summary of chromalveolate class II FBA sequences
| Sequence | Full length | Signal prediction (probability)a | Inferred location |
|---|---|---|---|
| Heterocapsa CY | Yes | No (0.027) | Cytosol |
| Isochrysis CY | Yes | No (0.084) | Cytosol |
| Thalassiosira CY | Yes | No (0.000) | Cytosol |
| Phytophthora CY | Yes | No (0.000) | Cytosol |
| Heterocapsa C1 | Yes | Yes (0.969) | Plastid |
| Guillardia C1 | Yes | Yes (0.998) | Plastid |
| Rhodomonas C1 | No | Plastidb | |
| Isochrysis C1 | Yes | Yes (0.996) | Plastid |
| Karenia C1 | No | Plastidb | |
| Thalassiosira C1 | No | Plastidb | |
| Phaeodactylum C1 | Yes | Yes (0.998) | Plastid |
| Odontella C1 | Yes | Yes (1.000) | Plastid |
| Thalassiosira C2 | Yes | Yes (0.998) | Plastid |
| Phaeodactylum C2 | Yes | Yes (0.995) | Plastid |
Consistent prediction based on neural network and hidden Markov model (HMM) methods. Number corresponds to the signal peptide probability based on HMM prediction.
Inferred based on phylogenetic relationships.
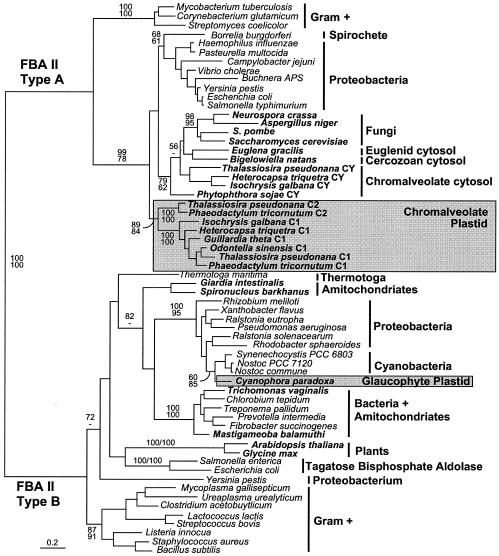
In a global phylogeny of class II FBA (Fig. 2), the previously recognized division between type A and type B genes is well supported (100%). The cyanobacteria and glaucophyte plastid genes (inferred to be the ancestral plastid FBA) are type B enzymes (24), while all of the chromalveolate sequences are type A enzymes. Within the type A group, the chromalveolate sequences are split into two groups: one branching with the cytosolic homologues from ascomycete fungi, a euglenid and a chlorarachniophyte, and the other forming a distinct lineage. Of the former group, full-length sequences from Thalassiosira and Isochrysis lack any evidence of a leader (see Fig. S1 to S5 in the supplemental material) and are therefore inferred to be cytosolic (CY). The Phytophthora gene may be related to this group, but its position in the phylogeny is not resolved. All of the full-length sequences falling in the other chromalveolate clade (C1 and C2 collectively) encode predicted plastid targeting leaders, and these are therefore inferred to be plastid targeted. In support of this, the Phaeodactylum proteins in this clade include leaders that have been experimentally demonstrated to direct the targeting of green fluorescent protein fusion proteins to the plastid (17). The diatoms have two distinct plastid-targeted FBAs, designated C1 and C2 (17). The C2 form has been found only in diatoms to date, whereas the C1 form is found in all four photosynthetic chromalveolate groups: cryptomonads, haptophytes, heterokonts, and dinoflagellates. Both clades are well supported as is their relationship to one another. In trees where the truncated red algal ESTs were included, they branched at the base of the eukaryotic cytosolic clade. They showed no specific affinity to the chromalveolates and grouped with one another with very strong support (maximum likelihood, 92%; distance, 87%) (data not shown).
FIG. 2.
Global protein maximum likelihood phylogeny of class II FBA. Plastid-targeted FBA from chromalveolates (type A) and glaucophytes (type B) are boxed and shaded, and all other groups are indicated to the right. Eukaryotes are named in bold. Numbers at nodes indicate bootstrap support from (top to bottom) maximum likelihood (PhyML) and weighted neighbor-joining.
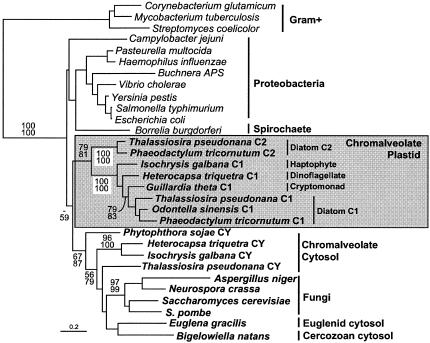
Type A and type B genes are homologous and readily aligned, but they are both characterized by type-specific insertions; so by focusing specifically on type A, the phylogenetic positions of the chromalveolate sequences can be examined more comprehensively by using more data. This phylogeny (Fig. 3) is similar in important characteristics to the type A topology in the global class II phylogeny. Most significantly, the chromalveolate plastid-targeted C1 clade is well supported in all analyses (100%), and the support for the relationship between the C1 and C2 clades is also moderate to strong (79 to 81%). When the truncated type A genes from the cryptomonad Rhodomonas and the haptophyte plastid-containing dinoflagellate Karenia are included, they also branch within the C1 clade (see Fig. S1 to S5 in the supplemental material). The position of the Karenia sequence is not sufficiently resolved to conclude whether it is derived from the haptophyte or dinoflagellate.
FIG. 3.
Protein maximum likelihood phylogeny of class II type A FBA. Plastid-targeted FBA from chromalveolates are boxed and shaded, and all other groups are indicated by brackets to the right. Eukaryotes are named in bold. Numbers at nodes indicate bootstrap support from (top to bottom) maximum likelihood as determined by ProML and PhyML.
Implications of chromalveolate plastid-targeted FBA.
If plastids derived from secondary endosymbiosis with a red alga acquired FBA by direct descent, then their genes would be related to the plastid genes of red algae, which is to say a class I FBA (12). However, all known putative plastid-targeted FBAs from chromalveolates are class II genes. If they acquired their FBA from the original cyanobacterial endosymbiont, it would be a class II type B gene, but all chromalveolate plastid FBAs are type A. These conclusions are simple and very strongly supported by the phylogeny. Together they reveal that photosynthetic chromalveolates all acquired their plastid-targeted FBA in common from some source other than that of all other plastids, which is most easily explained by the common and unique origin of all chromalveolate plastids.
The evolution of FBA is marked by a number of unusual evolutionary events, including lateral gene transfers, gene duplications (paralogy), and recompartmentalization of nonorganellar proteins to organelles (12, 26). Given this complexity, the patchy distribution of eukaryotes in the type A tree, and the almost certainly underestimated distribution of eukaryotic class II FBA, it is difficult to know the original source of the chromalveolate plastid-targeted FBA with absolute confidence. Nevertheless, the clade of cytosolic enzymes (the CY clade) suggests that a gene duplication in the ancestor of chromalveolates is one potential source, although the CY clade is not demonstrably related to the C1 or C2 plastid-targeted enzymes. Similarly, chromalveolate genes may be derived from a cytosolic red algal enzyme (perhaps from the nucleus of their endosymbiont), hinted at by the red algal ESTs. Whatever the ultimate source, the chromalveolates acquired a class II FBA from somewhere, likely duplicated it, retargeted one copy to the plastid by the addition of signal and transit peptides, and then lost their original plastid-targeted class I enzyme. If the source was a red alga, then both the close relationship of the red algal ESTs to one another (87 to 92% support) and the close relationship between the chromalveolate C1 enzymes (100% support) argue against the likelihood of multiple independent origins from different red algae. For this to be the case, the whole stream of events outlined above would have to have taken place twice independently from two sources closer to chromalveolates than either Gracilaria or Porphyra. This is highly unlikely and also inconsistent with what we know of red algal diversity since Gracilaria and Porphyra (whose class II enzymes are closely related), together with Cyanidioschyzon and Galdieria (where only class I enzymes are known), collectively represent the three major divisions of red algae.
It is also interesting that all three major groups of alveolates possess cytosolic class I FBA. If the class II FBA was acquired in the common ancestor of chromalveolates, then alveolates have either reacquired a class I FBA or have retained both class I and II enzymes for some time (dinoflagellates still have both). If chromalveolate plastids were not derived from a single endosymbiosis, then one would have to conclude that the dinoflagellates acquired both C1 and CY class II FBAs (and perhaps their plastid) from a chromist. While such possibilities remain to be disproven, the fact that cryptomonads, heterokonts, haptophytes, and dinoflagellates all share this highly distinctive genetic characteristic and that their putative plastid-targeted enzymes form a very strongly supported clade in FBA phylogeny are most easily explained by the common ancestry of these organisms and their plastids.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Canadian Institutes for Health Research (MOP-42517) and the Protist EST Program from Genome Atlantic/Genome Canada, which supported EST sequencing from Guillardia, Heterocapsa, and Isochrysis. P.J.K. is a scholar of the CIAR and a new investigator of the CIHR and MSFHR.
We thank N. M. Fast for critical reading of the manuscript and J. D. Palmer for helpful discussions and comments.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Archibald, J. M., and P. J. Keeling. 2002. Recycled plastids: a green movement in eukaryotic evolution. Trends Genet. 18:577-584. [DOI] [PubMed] [Google Scholar]
- 2.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 3.Bruno, W. J., N. D. Socci, and A. L. Halpern. 2000. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol. Biol. Evol. 17:189-197. [DOI] [PubMed] [Google Scholar]
- 4.Cavalier-Smith, T. 1999. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 46:347-366. [DOI] [PubMed] [Google Scholar]
- 5.Cavalier-Smith, T. 1998. A revised six-kingdom system of life. Biol. Rev. Camb. Philos. Soc. 73:203-266. [DOI] [PubMed] [Google Scholar]
- 6.Daugbjerg, N., and R. A. Andersen. 1997. Phylogenetic analyses of the rbcL sequences from haptophytes and heterokont algae suggest their chloroplasts are unrelated. Mol. Biol. Evol. 14:1242-1251. [DOI] [PubMed] [Google Scholar]
- 7.Delwiche, C. F. 1999. Tracing the thread of plastid diversity through the tapestry of life. Am. Nat. 154(Suppl.):S164-S177. [DOI] [PubMed] [Google Scholar]
- 8.Fagan, T., J. Woodland Hastings, and D. Morse. 1998. The phylogeny of glyceraldehyde-3-phosphate dehydrogenase indicates lateral gene transfer from cryptomonads to dinoflagellates. J. Mol. Evol. 47:633-639. [DOI] [PubMed] [Google Scholar]
- 9.Fast, N. M., J. C. Kissinger, D. S. Roos, and P. J. Keeling. 2001. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol. 18:418-426. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1993. PHYLIP (phylogeny inference package) version 3.5. University of Washington, Seattle, Wash.
- 11.Gray, M. W., and D. F. Spencer. 1996. Organellar evolution, p. 109-126. In D. M. Roberts, P. Sharp, G. Alderson, and M. A. Collins (ed.), Evolution of microbial life, vol. 54. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 12.Gross, W., D. Lenze, U. Nowitzki, J. Weiske, and C. Schnarrenberger. 1999. Characterization, cloning, and evolutionary history of the chloroplast and cytosolic class I aldolases of the red alga Galdieria sulphuraria. Gene 230:7-14. [DOI] [PubMed] [Google Scholar]
- 13.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 14.Harper, J. T., and P. J. Keeling. 2003. Nucleus-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase (GAPDH) indicates a single origin for chromalveolate plastids. Mol. Biol. Evol. 20:1730-1735. [DOI] [PubMed] [Google Scholar]
- 15.Helmchen, T. A., D. Bhattacharya, and M. Melkonian. 1995. Analyses of ribosomal RNA sequences from glaucocystophyte cyanelles provide new insights into the evolutionary relationships of plastids. J. Mol. Evol. 41:203-210. [DOI] [PubMed] [Google Scholar]
- 16.Keeling, P. J., and B. S. Leander. 2003. Characterisation of a non-canonical genetic code in the oxymonad Streblomastix strix. J. Mol. Biol. 326:1337-1349. [DOI] [PubMed] [Google Scholar]
- 17.Kilian, O., and P. G. Kroth. 2004. Presequence acquisition during secondary endocytobiosis and the possible role of introns. J. Mol. Evol. 58:712-721. [DOI] [PubMed] [Google Scholar]
- 18.Kroth, P. G. 2002. Protein transport into secondary plastids and the evolution of primary and secondary plastids. Int. Rev. Cytol. 221:191-255. [DOI] [PubMed] [Google Scholar]
- 19.Liaud, M. F., C. Lichtle, K. Apt, W. Martin, and R. Cerff. 2000. Compartment-specific isoforms of TPI and GAPDH are imported into diatom mitochondria as a fusion protein: evidence in favor of a mitochondrial origin of the eukaryotic glycolytic pathway. Mol. Biol. Evol. 17:213-223. [DOI] [PubMed] [Google Scholar]
- 20.Matsuzaki, M., O. Misumi, I. T. Shin, S. Maruyama, M. Takahara, S. Y. Miyagishima, T. Mori, K. Nishida, F. Yagisawa, Y. Yoshida, Y. Nishimura, S. Nakao, T. Kobayashi, Y. Momoyama, T. Higashiyama, A. Minoda, M. Sano, H. Nomoto, K. Oishi, H. Hayashi, F. Ohta, S. Nishizaka, S. Haga, S. Miura, T. Morishita, Y. Kabeya, K. Terasawa, Y. Suzuki, Y. Ishii, S. Asakawa, H. Takano, N. Ohta, H. Kuroiwa, K. Tanaka, N. Shimizu, S. Sugano, N. Sato, H. Nozaki, N. Ogasawara, Y. Kohara, and T. Kuroiwa. 2004. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428:653-657. [DOI] [PubMed] [Google Scholar]
- 21.McFadden, G. I. 1999. Plastids and protein targeting. J. Eukaryot. Microbiol. 46:339-346. [DOI] [PubMed] [Google Scholar]
- 22.Muller, K. M., M. C. Oliveira, R. G. Sheath, and D. Bhattacharya. 2001. Ribosomal DNA phylogeny of the Bangiophycidae (Rhodophyta) and the origin of secondary plastids. Am. J. Bot. 88:1390-1400. [PubMed] [Google Scholar]
- 23.Nassoury, N., M. Cappadocia, and D. Morse. 2003. Plastid ultrastructure defines the protein import pathway in dinoflagellates. J. Cell Sci. 116:2867-2874. [DOI] [PubMed] [Google Scholar]
- 24.Nickol, A. A., N. E. Muller, U. Bausenwein, M. G. Bayer, T. L. Maier, and H. E. Schenk. 2000. Cyanophora paradoxa: nucleotide sequence and phylogeny of the nucleus encoded muroplast fructose-1,6-bisphosphate aldolase. Z. Naturforsch. Sect. C 55:991-1003. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira, M. C., and D. Bhattacharya. 2000. Phylogeny of the Bangiophycidae (Rhodophyta) and the secondary endosymbiotic origin of algal plastids. Am. J. Bot. 87:482-492. [PubMed] [Google Scholar]
- 26.Rogers, M. B., and P. J. Keeling. 2003. Lateral gene transfer and re-compartmentalisation of Calvin cycle enzymes in plants and algae. J. Mol. Evol. 58:367-375. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez, L., D. Horner, D. Moore, K. Henze, T. Embley, and M. Müller. 2002. Fructose-1,6-bisphosphate aldolases in amitochondriate protists constitute a single protein subfamily with eubacterial relationships. Gene 295:51-59. [DOI] [PubMed] [Google Scholar]
- 28.Siebers, B., H. Brinkmann, C. Dorr, B. Tjaden, H. Lilie, J. van der Oost, and C. H. Verhees. 2001. Archaeal fructose-1,6-bisphosphate aldolases constitute a new family of archaeal type class I aldolase. J. Biol. Chem. 276:28710-28718. [DOI] [PubMed] [Google Scholar]
- 29.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 30.Williams, B. A. P., and P. J. Keeling. 2003. Cryptic organelles in parasitic protists and fungi. Adv. Parasitol. 54:9-67. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, R. J. M. I., P. W. Denny, D. J. Preiser, K. Rangachari, K. Roberts, A. Roy, A. Whyte, M. Strath, D. J. Moore, P. W. Moore, and D. H. Williamson. 1996. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol. 261:155-172. [DOI] [PubMed] [Google Scholar]
- 32.Yoon, H. S., J. D. Hackett, G. Pinto, and D. Bhattacharya. 2002. A single, ancient origin of the plastid in the Chromista. Proc. Natl. Acad. Sci. USA 99:15507-15512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, Z., B. R. Green, and T. Cavalier-Smith. 2000. Phylogeny of ultra-rapidly evolving dinoflagellate chloroplast genes: a possible common origin for sporozoan and dinoflagellate plastids. J. Mol. Evol. 51:26-40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.