Abstract
Chromosome segregation in human oocytes is error prone, resulting in aneuploidy, which is the leading genetic cause of miscarriage and birth defects. The study of chromosome behavior in oocytes from model organisms holds much promise to uncover the molecular basis of the susceptibility of human oocytes to aneuploidy. Drosophila melanogaster is amenable to genetic manipulation, with over 100 years of research, community, and technique development. Visualizing chromosome behavior and spindle assembly in Drosophila oocytes has particular challenges, however, due primarily to the presence of membranes surrounding the oocyte that are impenetrable to antibodies. We describe here protocols for the collection, preparation, and imaging of meiosis I spindle assembly and chromosome behavior in Drosophila oocytes, which allow the molecular dissection of chromosome segregation in this important model organism.
Keywords: Genetics, Issue 116, Drosophila, oocyte, immunofluorescence, fluorescence in situ hybridization (FISH), meiosis, spindle, microtubule, chromosome segregation
Introduction
The study of meiosis is sometimes described as the "genetics of genetics". This is because the fundamental properties of chromosome inheritance and independent assortment are carried out through the segregation of chromosomes during gamete production. An important demonstration of the chromosome theory of inheritance came in 1916 from the work of Calvin Bridges in Drosophila melanogaster1. This and other classical genetics studies in Drosophila contributed greatly to our understanding of genetics. Cytological examination of meiotic chromosomes in Drosophila oocytes, however, has been challenging. This is primarily because immunofluorescence of late-stage Drosophila oocytes, when the spindle assembles and chromosomes are oriented for segregation, is hampered by the presence of membranes that render the oocyte impenetrable to antibodies.
Despite this challenge, Drosophila oocytes remain an attractive model for the study of chromosome behavior and spindle assembly. This is because of the powerful genetic tools available in Drosophila, but also because the oocytes arrest at metaphase I, when the chromosomes are oriented and the spindle is fully formed. This facilitates the collection and examination of large numbers of oocytes at this important stage of cell division. In addition, a simple model organism that is amenable to genetic manipulation for the study of oocyte chromosome segregation can provide an important contribution to our understanding of human reproductive health. Errors in chromosome number are the leading genetic cause of miscarriage and birth defects in humans2. A majority of these errors can be traced to the oocyte and are correlated with increasing maternal age. The average age of mothers in the U.S. has been increasing, making this a major public health concern.
We describe here methods for the cytological examination of Drosophila oocytes, including a demonstration of how to remove the oocyte membranes. These methods are modifications of protocols first described by Theurkauf and Hawley3, Zou et al.4, and Dernburg et al.5. We also include methods for the enrichment of different stages of oocytes, based on a protocol first described by Gilliland et al.6. Finally, we add instructions for the drug treatment of Drosophila oocytes. Together, these methods allow the cytological investigation of oocyte chromosome segregation and spindle assembly in Drosophila.
Protocol
Note: Procedures are performed at room temperature unless otherwise noted. Temperature-controlled incubators are used to maintain temperatures for fly rearing and crosses unless otherwise noted.
1. Preparations
- Prepare Flies.
- Prometaphase-enriched oocyte collections.
- Clear adult flies from healthy, young stock or cross cultures. Age bottles for two days at 25 °C. NOTE: Generally two healthy bottles will suffice, although more may be needed for some cross cultures.
- After two days, collect ~100 to 300 females (who are 0 to 2 days old at this point) from the bottles. The females do not need to be virgins. Add a dab of yeast paste to the side of a vial and place 30 females and 10 to 15 males each into the yeasted vials. Age vials for two days at 25 °C.
- Metaphase-enriched Oocyte Collections.
- Collect ~100 to 300 females from stock or cross cultures. Add a dab of yeast paste to the side of a vial and place 30 females each (with no males added) into the yeasted vials. Age vials for three to five days at 25 °C.
- Prepare Solutions. Note: Solutions may be stored indefinitely at room temperature, unless otherwise noted.
- Prepare Modified Robb's Buffer (5x): 500 mM HEPES, 500 mM sucrose, 275 mM sodium acetate, 200 mM potassium acetate, 50 mM glucose, 6 mM magnesium chloride, and 5 mM calcium chloride. Use 10 N 11:8 sodium hydroxide:potassium hydroxide to bring pH to 7.4. Sterilize by filtration; do not autoclave. Store at -20 °C. Thaw as needed to prepare ~200 ml of 1x Robb's per oocyte prep.
- Fixation Solutions.
- Option #1: Prepare formaldehyde/heptane fixation. Prepare Fixation Buffer: 1x phosphate-buffered saline (PBS) plus 150 mM sucrose. To use, make fresh with 687.5 µl Fixation Buffer and 312.5 µl 16% formaldehyde per oocyte prep. CAUTION: Wear gloves while using formaldehyde solutions in a fume hood. Dispose of waste according to institutional guidelines.
- Option #2: Prepare formaldehyde/cacodylate fixation. Prepare Fix Mix: 250 mM sucrose, 100 mM potassium acetate (pH 7.5), 25 mM sodium acetate (pH 7.0), and 25 mM EGTA (pH 8.0). To use, make fresh with 400 µl Fix Mix, 100 µl potassium cacodylate (1 M, pH 7.2), and 500 µl 16% formaldehyde per oocyte prep. CAUTION: Potassium cacodylate contains Arsenic.
- Prepare PBS/Triton X-100: 1x PBS plus 1% or 0.05% Triton X-100. Store at 4 °C.
- Prepare PBS-Tween 20-Bovine Serum Albumin (BSA) (PTB): 1x PBS, 0.5% BSA (w/v), and 0.1% Tween-20. May be stored at 4 °C for one week.
- Fluorescence In Situ Hybridization (FISH) Solutions.
- Prepare 20X Sodium chloride-Sodium Citrate (SSC): 3 M sodium chloride and 0.3 M sodium citrate.
- Prepare 2x SSC-Tween-20 (SSCT): 2x SSC plus 0.1% Tween-20. Make fresh, ~20 ml per oocyte prep.
- Prepare formamide solutions: 2x SSC, 0.1% Tween-20, plus formamide. Make fresh, 1 ml 20% formamide, 0.5 ml 40% formamide, and 2 ml 50% formamide per oocyte prep. CAUTION: Wear gloves while using formamide solutions in a fume hood. Dispose of waste according to institutional guidelines.
- Prepare hybridization solution: 2x SSC, 50% formamide, and 10% dextran sulfate (w/v). Store at 4 °C.
- FISH Probes.
- Order oligonucleotides (see Table 1 for sequences) with HPLC purification and desired 5' fluorescent modification (e.g., Cy3 or Cy5). Resuspend in Tris-EDTA (TE) at 50 ng/µl. NOTE: Protect oligos from light exposure at all times.
2. Collection of Late-stage Drosophila Oocytes
As Drosophila oocytes with membranes intact will stick to plastic and glass, pre-coat the inside of one 5 ml tube and one Pasteur pipet per oocyte prep with PTB.
Anesthetize all ~100 to 300 yeasted flies with carbon dioxide and add to a blender containing ~100 ml 1x Robb's Buffer. Pulse three times (~1 sec each). Keep oocytes in Robb's for <20 min to avoid activation. NOTE: Alternatively, oocytes may be hand-dissected from females. The advantage of this method is that it requires less females. However, care must be taken to limit exposure to carbon dioxide to only a few minutes to avoid artifacts associated with hypoxia7.
Filter through large mesh (~1,500 µm) into 250 ml beaker to remove large body parts. If many intact abdomens remain on mesh, re-grind material using additional Robb's Buffer, and filter again. Let settle ~2 min, then aspirate off top layer, removing as many of the large body parts as possible.
Filter through small mesh (~300 µm) into a 250 ml beaker. Rinse remaining oocytes out of first beaker using additional Robb's and coated Pasteur pipet. Let settle ~3 min; oocytes will settle out. Aspirate off all but ~10 ml.
Pour as much of the 10 ml as will fit into coated 5 ml tube. Let settle, remove liquid, and repeat with remainder. Rinse remaining oocytes out of beaker using additional Robb's and coated Pasteur pipet. Let settle in 5 ml tube for ~3-5 min.
3. Drug Treatments (Optional)
Coat a second 5 ml tube with PTB for each oocyte prep. Add appropriate solvent (control) or drug to 1 ml Robb's each for each oocyte prep (Table 2).
Split oocytes into second coated 5 ml tube. Let settle, remove liquid, and add 1 ml Robb's plus solvent into one tube and 1 ml Robb's plus drug into second tube. Nutate for appropriate amount of time for drug treatment (Table 2). Let settle.
4. Fixation
Aspirate off all liquid and immediately add 1 ml Fix.
- Option #1: Formaldehyde/heptane fixation (Fixation Buffer plus 5% formaldehyde).
- Fix for 2.5 min on a nutator. Add 1 ml heptane and vortex 1 min. Let settle ~1 min.
- Remove all liquid, and then add 1 ml 1x PBS. Vortex 30 sec. Let settle ~1 min.
- Remove all liquid, and then fill tube with 1x PBS. NOTE: Oocytes may be used immediately or kept on the nutator for several hours at room temperature.
- Option #2: Formaldehyde/cacodylate fixation (Fix Mix plus 8% formaldehyde and 100 mM cacodylate).
- Fix for 6 min on a nutator. Let settle 2 min. Remove liquid, and then fill tube with 1x PBS. NOTE: Oocytes may be used immediately or kept on the nutator for several hours at room temperature.
5. Removing Membranes ("Rolling")
Using coated Pasteur pipet, add ~500 to 1,000 oocytes to the frosted (sand-blasted) part of a glass slide. Remove all body parts and extraneous material using forceps. Do not let oocytes dry out; add 1x PBS as necessary.
Place a coverslip on top of the oocytes and gently "roll" oocytes until all membranes are removed (dragging the edge of the coverslip across the oocytes works best.) Periodically check progress under the microscope, adding more 1x PBS as necessary. Take care as too much pressure will destroy the oocytes. NOTE: For immunofluorescence only, continue with Step 6. For FISH (with or without immunofluorescence), continue with Step 7.
6. Antibody Staining of Drosophila Oocytes
- Extraction and Blocking
- Rinse rolled oocytes into a 15 ml conical tube containing ~15 ml PBS/1% Triton X-100. Nutate oocytes in this solution for no less than 1.5 hr and no more than 2 hr. This step allows antibody penetration.
- Let oocytes settle ~2 min. Remove liquid along with as many membranes as possible, then add PBS/0.05% Triton X-100. NOTE: Membranes will settle slower than rolled oocytes.
- Let oocytes settle ~2 min, then remove all but ~1 ml liquid. Transfer oocytes to graduated 1.5 ml tube and remove remaining liquid. Add 1 ml PTB for blocking and nutate for 1 hr.
- Antibody Staining
- Pre-absorption of secondary antibodies against embryos. NOTE: This step eliminates background staining from non-specific antibody interactions with Drosophila proteins.
- Collect and fix Drosophila embryos (~25 µl) per typical procedure8. Store in methanol at -20 °C.
- Remove methanol from embryos, add 800 µl methanol plus 200 µl 1x PBS, and nutate for 15 min. Remove 500 µl of supernatant and replace with 500 µl 1x PBS. Then nutate for 15 min. Repeat twice (for a total of ~1 hr of washes), and then finish in PTB. NOTE: Embryos may be used immediately or kept on the nutator for several hr at room temperature.
- Remove liquid, fill with PTB to 200 µl per oocyte prep, and add fluorescently-labeled secondary antibodies at appropriate dilutions (keeping in mind that the final volume will be 300 µl; see step 7.4.) Nutate at 4 °C overnight (preferable) or at room temperature for 3 to 4 hr. NOTE: The pre-absorbed antibodies will be used in Step 6.2.4. NOTE: Keep samples in the dark as much as possible once fluorescent antibodies have been added.
- Remove liquid from oocytes, fill with PTB to 300 µl, and add primary antibodies at appropriate dilutions. Nutate at 4 °C overnight (preferable) or at room temperature for 3 to 4 hr.
- Wash oocytes four times for 15 min each with 1 ml PTB.
- Remove liquid from oocytes, add 200 µl of supernatant from embryos (pre-absorbed secondary antibodies). Then fill with PTB to 300 µl. Nutate at room temperature for 3 to 4 hr (preferable) or at 4 °C overnight.
- Wash oocytes once for 15 min with 1 ml PTB, remove liquid. Then add 0.5 µl Hoechst 33342 and 500 µl PTB and nutate for 7 min.
- Wash oocytes twice for 15 min each with 1 ml PTB. NOTE: Oocytes may be mounted on a slide immediately or may be stored in PTB at 4 °C until ready for imaging.
7. FISH (Continue from Step 5 Above)
Rinse rolled oocytes into a 15 ml conical tube containing ~15 ml 2x SSCT. Let oocytes settle ~2 min. Remove all but ~0.5 ml liquid along with as many membranes as possible. NOTE: Membranes will settle slower than rolled oocytes.
Transfer oocytes to 0.5 ml tube and remove remaining liquid. Successively add and remove 500 µl 20%, 40%, and 50% formamide solutions, nutating for 10 min in each solution. NOTE: Oocytes will settle slower with higher percentages of formamide.
Remove liquid, then add 500 µl 50% formamide solution, and nutate at 37 °C for 1 to 5 hr. NOTE: Longer incubations result in better probe penetration.
Remove liquid, leaving behind no more than ~100 µl oocytes, and add 36 µl hybridization solution plus 2 µl of probe (50 ng/µl) and 2 µl of water or 2 µl of a 2nd probe (Table 1).
Incubate at 91 °C for 3 min (FISH only) or 80 °C for 20 min (FISH plus immunofluorescence), followed by incubation in a 37 °C water bath overnight. NOTE: These steps may be performed in a thermocycler.
Do not remove liquid. Add 500 µl 50% formamide solution and nutate at 37 °C for 1 hr.
Let settle, remove liquid, add 500 µl 50% formamide solution, and nutate at 37 °C for 1 hr.
Let settle, remove liquid, add 500 µl 20% formamide solution, and nutate at room temperature for 10 min.
Perform three quick washes in 500 µl 2x SSCT (let settle, remove then add liquid, invert several times, repeat.) Let settle, remove liquid, then add 500 µl PTB and nutate for 4 hr.
8. Antibody Staining after FISH
Remove liquid from oocytes, fill with PTB to 300 µl, and add 10 µl anti-α-tubulin antibody conjugated to FITC. Alternatively, other antibodies can be used, following above protocol. Nutate at room temperature overnight.
Wash oocytes once for 15 min with 500 µl PTB, remove liquid, then add 0.5 µl Hoechst 33342 and 500 µl PTB and nutate for 7 min.
Wash oocytes twice for 15 min each with 500 µl PTB. NOTE: Oocytes may be mounted on a slide immediately or may be stored in PTB at 4 °C until ready for imaging. Keep oocytes in the dark as much as possible once fluorescent antibodies have been added.
| Repeat Name | Chromosome | Oligo Sequence* |
| 359 | X | GGGATCGTTAGCACTGGTAATTAGCTGC |
| AACAC | 2 | AACACAACACAACACAACACAACACAACACAACACAACAC |
| dodeca | 3 | CCCGTACTGGTCCCGTACTCGGTCCCGTACTCGGT |
| 1.686 | 2+3 | AATAACATAGAATAACATAGAATAACATAG |
| AATAT | 4(+Y) | AATATAATATAATATAATATAATATAATAT |
| *359 sequence from Eric Joyce, personal communication, other sequences from Sullivan et al.8 |
Table 1: FISH probes for Drosophila centromeric repeats.
| Drug | Solvent | Stock concentration | Final concentration | Time of treatment | Effect |
| colchicine | ethanol | 125 mM | 150 µM | 10 min or 30 min | destabilize non-kinetochore (10 min)9 or all (30 min) microtubules |
| paclitaxel | DMSO | 10 mM | 10 µM | 10 min | stabilize microtubules |
| Binucleine 2 | DMSO | 25 mM | 25 µM | 20 min | inhibit Aurora B kinase10 |
Table 2: Drug treatment.
Representative Results
The methods we have described here will result in the collection of late-stage Drosophila oocytes representing three stages of meiosis (Figure 1). Oocytes in prophase are distinguished by the presence of the nuclear envelope, which is visible by the lack of tubulin signal in the region surrounding the karyosome (Figure 1A). Prometaphase is the period after nuclear envelope breakdown during which the spindle assembles. During prometaphase, the karyosome assumes a distinctive shape, becoming elongated often with the 4th chromosomes apart from the main karyosome mass (Figure 1B). Prometaphase concludes with metaphase, at which Drosophila oocytes naturally arrest until ovulation. In metaphase oocytes, the karyosome has retracted into a round shape (Figure 1C).
We describe here methods to manipulate the speed of egg-laying of Drosophila females to enrich for oocytes either in prometaphase or metaphase. Oocytes from prometaphase-enriched collections frequently show the elongated karyosome (Figure 2A) while metaphase-enriched collections or collections from virgin Drosophila females, which almost completely eliminate egg-laying, primarily show the round karyosome (Figure 2B). Interestingly, prometaphase-enriched collections also typically show much more robust spindles, suggesting that both the karyosome and spindle change between prometaphase and metaphase in Drosophila oocytes9.
Figure 3 shows representative images from the two Protocols described here. Antibodies against α-tubulin (to show the spindle), CENP-C (to show centromeres), and INCENP (to show the central spindle) were used on oocytes fixed with formaldehyde/heptane (Figure 3A). Probes for the repetitive centromeric sequences on the 2nd chromosome (AACAC) and 3rd chromosome (dodeca) were used on oocytes fixed with formaldehyde/cacodylate and co-stained with an α-tubulin antibody according to the FISH protocol (Figure 3B). The AACAC probe appears as several clustered foci while the dodeca probe appears as a single focus per chromosome. Oocytes treated with 150 µM colchicine for 10 min were fixed with formaldehyde/heptane (Figure 3C). This treatment eliminates most non-kinetochore-microtubules, which results in all microtubules contacting the DNA at centromere-labeled foci. Oocytes treated with 10 µM paclitaxel for 10 min show excessive microtubules in the cytoplasm, but little effect on the spindle microtubules (Figure 3D). Oocytes treated with 25 µM Binucleine 2 for 20 min to inhibit Aurora B kinase show complete loss of spindle microtubules (Figure 3E).
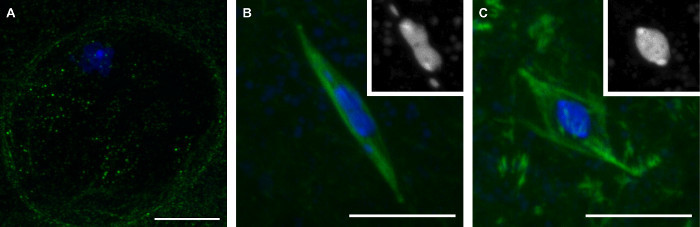 Figure 1:Three stages of mature Drosophila oocytes: Prophase, Prometaphase, and Metaphase. Confocal images of Drosophila oocytes in prophase (A), prometaphase (B), and metaphase (C) of meiosis I. DNA is shown in blue and tubulin is shown in green in merged images. Insets in (B) and (C) show DNA in white. Scale bars = 10 µm. Please click here to view a larger version of this figure.
Figure 1:Three stages of mature Drosophila oocytes: Prophase, Prometaphase, and Metaphase. Confocal images of Drosophila oocytes in prophase (A), prometaphase (B), and metaphase (C) of meiosis I. DNA is shown in blue and tubulin is shown in green in merged images. Insets in (B) and (C) show DNA in white. Scale bars = 10 µm. Please click here to view a larger version of this figure.
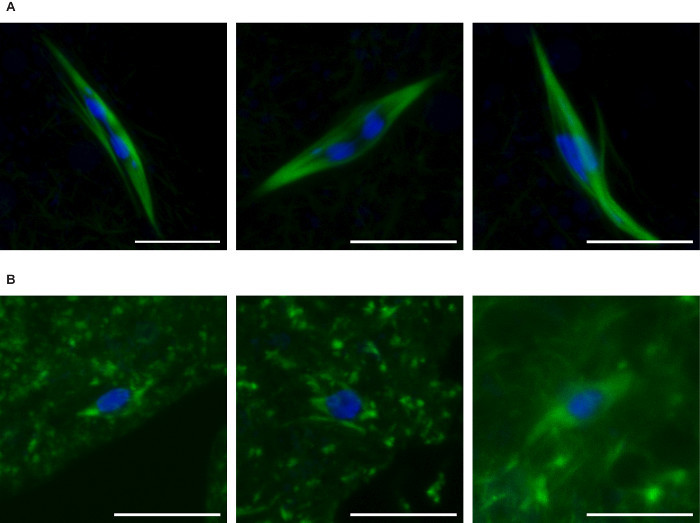 Figure 2:Prometaphase- vs. metaphase-enriched collections ofDrosophila oocytes. Confocal images of Drosophila oocytes from prometaphase-enriched (A) or metaphase-enriched (B) collections. DNA is shown in blue and tubulin is shown in green. Scale bars = 10 µm. Please click here to view a larger version of this figure.
Figure 2:Prometaphase- vs. metaphase-enriched collections ofDrosophila oocytes. Confocal images of Drosophila oocytes from prometaphase-enriched (A) or metaphase-enriched (B) collections. DNA is shown in blue and tubulin is shown in green. Scale bars = 10 µm. Please click here to view a larger version of this figure.
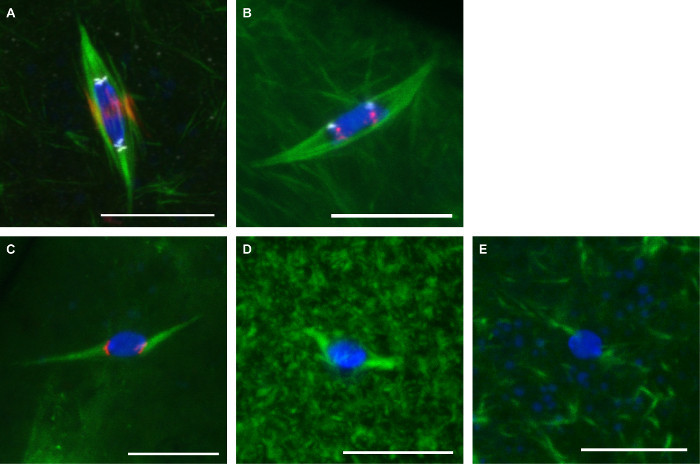 Figure 3:Antibody staining, FISH, and drug treatments of Drosophila oocytes. Confocal images of Drosophila oocytes with DNA in blue and tubulin in green. Scale bars = 10 µm. (A) Immunofluorescence in formaldehyde/heptane fixation with INCENP (central spindle) in red and CENP-C (centromeres) in white. (B) FISH in formaldehyde/cacodylate fixation with AACAC shown in red and dodeca shown in white. (C) Treatment with 150 µM colchicine for 10 min with CENP-C shown in red. Treatment with 10 µM paclitaxel for 10 min (D) or 25 µM Binucleine 2 for 20 min (E). Please click here to view a larger version of this figure.
Figure 3:Antibody staining, FISH, and drug treatments of Drosophila oocytes. Confocal images of Drosophila oocytes with DNA in blue and tubulin in green. Scale bars = 10 µm. (A) Immunofluorescence in formaldehyde/heptane fixation with INCENP (central spindle) in red and CENP-C (centromeres) in white. (B) FISH in formaldehyde/cacodylate fixation with AACAC shown in red and dodeca shown in white. (C) Treatment with 150 µM colchicine for 10 min with CENP-C shown in red. Treatment with 10 µM paclitaxel for 10 min (D) or 25 µM Binucleine 2 for 20 min (E). Please click here to view a larger version of this figure.
Discussion
Staging Drosophila Oocytes
Although an elongated karyosome is often seen in prometaphase oocytes, using karyosome shape to distinguish prometaphase from metaphase oocytes can be problematic. During prometaphase, the karyosome begins as a round shape, elongates, and then retracts to a round shape as the oocyte approaches the metaphase arrest. This means that many prometaphase oocytes do not have an elongated karyosome. In addition, if mutant or drug-treated oocytes are examined, karyosome shape may be affected, precluding using this method to stage oocytes. Because other markers to distinguish these stages are not currently available, we use the method described above to manipulate the speed of egg-laying of Drosophila females to enrich for oocytes either in prometaphase or metaphase. The rationale is that if oocytes spend a specific amount of time in prometaphase, and an indefinite amount of time arrested in metaphase, which lasts until egg-laying, then more rapid egg-laying will skew towards a higher percentage of prometaphase oocytes, while slower egg-laying will enrich for metaphase. The amount of effort required to collect virgin females or collect females over a shorter time frame than 2 days, as described by Gilliland et al.6, is not rewarded by an appreciable increase in prometaphase oocytes. It is also impossible to use the dorsal appendages to stage oocytes prepared with this protocol as dorsal appendages are removed along with the membranes. Although it is not possible to stage individual oocytes with 100% certainty using this method, the data from prometaphase-enriched or metaphase-enriched collections can be taken together to draw conclusions about these different stages9.
Drosophila oocytes are not fertilized until they pass through the oviduct; therefore, because the oocytes collected for the protocols described here examine oocytes taken before egg-laying, these oocytes have not been fertilized. Passage through the oviduct also causes the oocyte to activate and resume cell cycle progression. Methods to examine cell cycle stages after metaphase I exist11,12, but are outside the scope of this protocol.
Fixation Method Considerations
We describe two separate methods for the fixation of late-stage Drosophila oocytes. The formaldehyde/cacodylate fixation method was first described by Theurkauf and Hawley3 and the formaldehyde/heptane fixation method was first described by Zou et al.4. In addition, a third method (methanol fixation) is used by some13, but not described here. Removal of the oocyte membranes is somewhat more difficult after formaldehyde/heptane fixation compared to formaldehyde/cacodylate. This is because "rolling" depends on friction between the coverslip and oocyte, and formaldehyde/heptane fixation makes the oocytes more slippery. Despite this added difficulty, our first choice of fixation method when testing new antibodies is formaldehyde/heptane because it works best for the majority of antibodies that we have tried. On the other hand, we prefer formaldehyde/cacodylate fixation for FISH, primarily for the ease of membrane removal, although formaldehyde/heptane fixation has also been successful. Both of these methods preserve oocyte morphology better than methanol fixation, and therefore we recommend methanol fixation only in the cases of antibodies that are refractory to formaldehyde-based fixation methods.
The protocols described here focus on fixed imaging, in order to demonstrate effective methods for rendering mature Drosophila oocytes amenable to antibody penetration, namely through the removal of the oocyte membranes. Alternative methods for membrane removal (using forceps14 or sonication13) are possible; however, we find "rolling" to be the most efficient and reliable method. Techniques for live imaging have also been described elsewhere14-16 and are outside the scope of this protocol.
FISH Recommendations
FISH in Drosophila oocytes was originally described by Dernburg et al.5. This protocol was designed to provide optimal conditions for probe hybridization to the condensed oocyte chromosomes, but the original protocol was not optimal for immunofluorescence. The method described above includes adaptations we have made to enhance antibody staining of α-tubulin, which is primarily what we use when performing FISH. For other antibodies, lowering the temperature of the denaturation step (Step 7.5) from 80 °C may be necessary.
The probes listed here are for the repetitive sequences in the heterochromatin near the centromeres. Because these sequences are highly repetitive, the probes anneal to multiple places and the collective signals are quite strong. We have had little success with probes for sequences in the euchromatin, possibly because of the unique structure of the chromosomes while contained in the condensed karyosome.
Imaging
Mature Drosophila oocytes, like the oocytes of most organisms, are large, single cells. That means that there is a large volume of cytoplasm that contains a single nucleus. Fortunately, this nucleus, also known as a karyosome in Drosophila, can be readily identified as it is typically located just underneath the dorsal appendages close to the cortex. The dorsal appendages are removed along with the oocyte membranes, but the karyosome can still be located by finding the small divot in the oocyte cortex where the appendages once were. The best images are obtained from oocytes that have been mounted with this divot facing the coverslip since this places the karyosome closest to the objective. Although it may be possible to arrange the oocytes in this orientation prior to mounting, we prefer to mount many oocytes randomly and then search the slide for those in the preferred orientation.
Future Applications
In the future it will be important to identify markers that reliably distinguish prometaphase I and metaphase I spindles. Being able to more reliably stage the oocytes will reduce the reliance on more labor intensive alternatives, such as imaging large numbers of stage-enriched oocytes or live imaging, which also has its staging problems. This method is a basic method which can be used to take advantage of advances in Drosophila genetics as new tools become available to modify or knock out gene products. This includes methods to directly target protein activity with new drugs or targeted degradation strategies. In addition, these methods are not limited to imaging meiotic spindles. Any structure within the oocyte, such as the actin cytoskeleton, could be analyzed using these methods.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank Christian Lehner for providing the CENP-C antibody and Eric Joyce for recommendations on FISH. Work in the McKim lab was funded by a grant from NIH (GM101955).
References
- Bridges CB. Non-disjunction as proof of the chromosome theory of heredity. Genetics. 1916;1(1):1–52. doi: 10.1093/genetics/1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol. 1992;116(5):1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Hallen MA, Yankel CD, Endow SA. A microtubule-destabilizing kinesin motor regulates spindle length and anchoring in oocytes. J Cell Biol. 2008;180(3):459–466. doi: 10.1083/jcb.200711031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86(1):135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- Gilliland WD, Hughes SF, Vietti DR, Hawley RS. Congression of achiasmate chromosomes to the metaphase plate in Drosophila melanogaster oocytes. Dev Biol. 2009;325(1):122–128. doi: 10.1016/j.ydbio.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Gilliland WD, et al. Hypoxia transiently sequesters mps1 and polo to collagenase-sensitive filaments in Drosophila prometaphase oocytes. PLoS One. 2009;4(10):e7544. doi: 10.1371/journal.pone.0007544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W, Ashburner M, Hawley RS. Drosophila Protocols. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Radford SJ, Hoang TL, Głuszek AA, Ohkura H, McKim KS. Lateral and End-On Kinetochore Attachments Are Coordinated to Achieve Bi-orientation in Drosophila Oocytes. PLoS Genet. 2015;11(10):e1005605. doi: 10.1371/journal.pgen.1005605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smurnyy Y, Toms AV, Hickson GR, Eck MJ, Eggert US. Binucleine 2, an isoform-specific inhibitor of Drosophila Aurora B kinase, provides insights into the mechanism of cytokinesis. ACS Chem Biol. 2010;5(11):1015–1020. doi: 10.1021/cb1001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald AP, Goralski TJ, Caulton JH. In vitro activation of Drosophila eggs. Dev Biol. 1983;98(2):437–445. doi: 10.1016/0012-1606(83)90373-1. [DOI] [PubMed] [Google Scholar]
- Page AW, Orr-Weaver TL. Activation of the meiotic divisions in Drosophila oocytes. Dev Biol. 1997;183(2):195–207. doi: 10.1006/dbio.1997.8506. [DOI] [PubMed] [Google Scholar]
- Tavosanis G, Llamazares S, Goulielmos G, Gonzalez C. Essential role for gamma-tubulin in the acentriolar female meiotic spindle of Drosophila. EMBO J. 1997;16(8):1809–1819. doi: 10.1093/emboj/16.8.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA, Komma DJ. Spindle dynamics during meiosis in Drosophila oocytes. J Cell Biol. 1997;137(6):1321–1336. doi: 10.1083/jcb.137.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies HJ, Clarkson M, Saint RB, Namba R, Hawley RS. In: Drosophila Protocols. Sullivan W, Ashburner M, Hawley RS, editors. Cold Spring Harbor Laboratory Press; 2000. pp. 67–85. [Google Scholar]
- Colombié N, et al. Dual roles of Incenp crucial to the assembly of the acentrosomal metaphase spindle in female meiosis. Development. 2008;135(19):3239–3246. doi: 10.1242/dev.022624. [DOI] [PMC free article] [PubMed] [Google Scholar]


