Abstract
We demonstrate a method for size based capture of viable circulating tumor cell (CTC) from whole blood, along with the release of these cells from chip for downstream analysis and/or culture. The strategy employs the use of a novel Parylene C membrane slot pore microfilter to capture CTC and a coating of poly (N-iso-propylacrylamide) (PIPAAm) for thermoresponsive viable release of the captured CTC. The capture of live cells is enabled by leveraging the design of a slot pore geometry with specific dimensions to reduce the shear stress typically associated with the filtration process. While the microfilter exhibits a high capture efficiency, the release of these cells is non-trivial. Typically, only a small percentage of cells are released when techniques such as reverse flow or cell scraping are used. The strong adhesion of these epithelial cancer cells to the Parylene C membrane is attributable to non-specific electrostatic interaction. To counteract this effect, we employed the use of PIPAAm coating and exploited its thermal responsive interfacial properties to release the cells from the filter. Blood is first filtered at room temperature. Below 32 °C, PIPAAm is hydrophilic. Thereafter, the filter is placed in either culture media or a buffer maintained at 37 °C, which results in the PIPAAm turning hydrophobic, and subsequently releasing the electrostatically bound cells.
Keywords: Cancer Research, Issue 116, Circulating Tumor Cell, CTC culture, PIPAAm, Microfilter, Live Cell Capture, Thermoresponsive Release, Precision Medicine, Personalized Medicine, Translational Medicine, Liquid Biopsy
Introduction
Metastatic disease is responsible for most cancer deaths. Developing prognostic and companion diagnostic biomarker for metastasis is crucial in cancer management and treatment. Circulating tumor cells (CTC) play a central role in tumor dissemination and metastasis. Furthermore, being easily accessible as a 'liquid biopsy' biomarker, CTC in cancer patients' peripheral blood has been rising as a 'hotbed' for cancer biomarker research. CTC have been well validated as a prognostic biomarker in various cancer settings, including breast, prostate and colorectal cancer1-3. However, recent advances in CTC field has indicated that the mere enumeration of these rare cells has limited clinical utility, as shown in interventional clinical trials4. Thus, there is an emerging need for technologies that allow for molecular and functional characterization of CTC. Currently, only few technologies exist that allow for non-antigen biased, viable capture and release of CTC, enabling robust downstream molecular and functional analysis5,6. Majority of these microfabricated devices are coupled to microfluidic platforms and thus have a limiting factor in the amount of blood that that can be processed, which ranges from 2-4 ml7-10 . CTC are rare events in a single tube of blood draw (7.5 ml), therefore further reducing the amount of blood that can be processed, greatly hinders the chances of capturing and isolating these cells of interest.
We have developed two types of Parylene C membrane microfilter devices for the capture of CTC which exploit the size differences between larger tumor cells and the smaller normal blood cells11,12. We have previously reported on the round pore enumeration filter and compared it with a FDA-approved platform, where the microfilter was shown to be superior in CTC capture efficiency for cancer patient blood samples13,14. However, a limitation of the round filter is the necessity to use a formaldehyde-based fixative prior to filtration. This process preserves the cells morphology while allowing them to withstand the shear stress and pressure during the filtration process. While enumeration and molecular studies can be performed on-chip13, the fixative impairs the ability to perform functional characterization. To address this limitation, we developed a slot pore filter that negates the necessity to fix cells prior to filtration (Figure 1). The slot pore geometry (6 µm width x 40 µm length slot pores) allows the tumor cells to be captured while only partially occluding a pore and thus still permitting free passage for other blood cells and alleviating the rise in pressure that would lead to cell damage and eventual bursting15,16 The slot pore cartridge is composed of 2 acrylic pieces which sandwich the slot pore filter between the top and bottom piece with Polydimethylsiloxane (PDMS) acting as a gasket to provide a leak proof seal14,15 (Figure 1).
While the capture efficiency of the slot pore filter is high, (Table 1), the captured CTC are bound to the Parylene C membrane by strong non-specific electrostatic interactions instead of extracellular matrix (ECM) mediated adhesion15. Methods such as reverse flow or the use of cell scrapers fail to effectively release the cells from the filter, or result in cell damage and cell death. We explored an unconventional use of PIPAAm to formulate a release strategy15. PIPAAm is a polymer that undergoes a reversible lower critical solution temperature (LCST) phase transition at a solution temperature of 32 °C17. Traditionally, this property of PIPAAm has been widely explored for tissue engineering applications. Typically, cells are cultured on PIPAAm coated surfaces at 37 °C when PIPAAm is hydrophobic. The cells can then be detached as a sheet when the culture temperature is shifted to below 32 °C, where the PIPAAm coated surface becomes hydrated17,18. We exploited this thermal property by performing the filtration process at room temperature (below 32 °C) and then enabling cell release by placing the filter in culture media maintained at 37 °C. At this temperature, the PIPAAm polymer layer becomes hydrophobic, thereby releasing the electrostatically bound cells15 (Figure 1).
Although the temperature responsive method as well as other methods have been successfully implemented to achieve viable CTC capture and release19-21, one key potential drawback shared by these technologies reported is that they all employ an antigen-dependent principle for CTC capture. Antigen based CTC capture, as shown previously, may lead to biased CTC analysis11,14. For example, many affinity-based technologies employ antibody that binds EpCAM for CTC capture. However, CTC has been shown to express various levels of EpCAM, leading to omission of EpCAM low and EpCAM negative CTC by these technologies. Also, limitations may occur when CTC from non-epithelial origin is of interest, such as CTC in melanoma and sarcoma settings. Thus, a technology that allows for viable CTC capture and release without potential bias introduced by antigen based capture is highly desirable.
Importantly, the microfilter capture device is purely sized based and the release strategy is agnostic to the presence of certain surface markers. We believe that the employment of the PIPAAm coated microfilter will help to expand our understanding of the metastatic process, through providing the ability to effectively and efficiently capture and release CTC for downstream analyses. This may potentially expose new molecules for which novel targeted systemic therapies might be directed as well as provide a biomarker that can be monitored easily and aid in cancer patient management.
Protocol
Ethics Statement: To protect the rights of human subjects, blood samples were obtained following an informed consent under protocols approved by the University of Miami institutional review boards under IRB 20150020. NOTE: Blood to be filtered for CTC capture should be collected in an EDTA tube to prevent coagulation.
1. Coating the Microfilter with Poly (N-iso-propylacrylamide) (PIPAAm)
Weigh out PIPAAm to prepare a 10% w/v solution in butanol. Mix using a vortex until the solution is clear.
Cut plastic microscope slides into roughly 12 mm x 12 mm squares either with a pair of scissors or sharp blade. Alternatively use a guillotine. NOTE: These squares will serve as a holder for the filters during the spin coating process.
Using a sharp pair of straight edge scissors, cut the slot pore microfilter wafer into 8 mm x 8 mm squares.
Using a polyimide film tape secure the cut filters onto the plastic squares previously cut in step 1.3. Apply the film only to the edge and the corner of the filter so that at least 7 mm X 7 mm of filter area is un-covered by the tape.
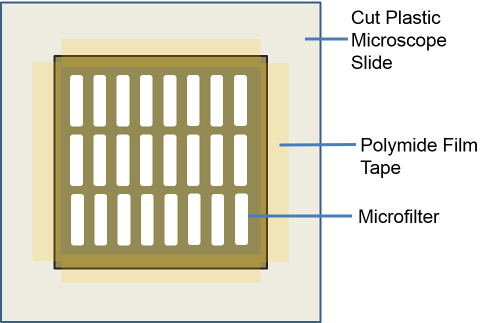 Figure 2: Representative Illustration of the Microfilter Set-up for PIPAAm Coating. 8 mm x 8 mm microfilter is positioned onto the 12 mm x 12 mm plastic square cut from a plastic microscope slide. Polyimide tape is used to secure the microfilter in place to spin coat the PIPAAm.Reproduced with permission from reference15. Please click here to view a larger version of this figure.
Figure 2: Representative Illustration of the Microfilter Set-up for PIPAAm Coating. 8 mm x 8 mm microfilter is positioned onto the 12 mm x 12 mm plastic square cut from a plastic microscope slide. Polyimide tape is used to secure the microfilter in place to spin coat the PIPAAm.Reproduced with permission from reference15. Please click here to view a larger version of this figure.
Place the microfilter that is secured on the plastic square, onto the vacuum chuck of the spin-coater.
Program the spin-coater to the following recipe: Start, 500 rpm for 10 sec, 6,000 rpm for 60 sec, 500 rpm for 10 sec, Stop.
Dispense enough 10% w/v PIPAAm solution (prepared in 1.1) to completely cover the microfilter surface using a standard plastic transfer pipette and start the spin coater.
Remove the PIPAAm coated microfilter from the spin-coater after the coating process is completed. Leave the filter attached to the plastic square during storage. NOTE: The coated-filter can be stored at room temperature for up to 3 months.
2. Assembly of Filtration Cassette with PIPAAm Coated Microfilter
Rehydrate PIPAAm coated microfilter at room temperature by placing the microfilter still secured on the plastic square into a petri dish. Add 1x PBS until the microfilter is fully submerged and let stand for 5 min.
After the hydration step, release the filter from the plastic square by cutting the polyimide film tape using a pair of scissor. NOTE: Take note of which side of the filter has the PIPAAm coating.
Assemble the microfilter into the filtration cassette, by sandwiching the microfilter between the top and bottom acrylic piece along with the 2 Polydimethylsiloxane (PDMS) pieces acting as a gasket/seal. Clamp the acrylic cassette with clips to secure the filter and provide a tight seal. NOTE: The PIPAAm coating should be facing upwards, so as the sample is filtered through, the cells will be captured on PIPAAm coated surface of the filter (Figure 1).
3. Filtration of Blood Sample with Capture and Release of CTC from the Microfilter
As each syringe pump brand has its own user interface, refer to the user manual of the pump and program the syringe pump to flow at a rate of 75ml/hr and set the volume to 20 ml.
Warm 3 ml of McCoy's (commercially available) cell culture media (prepared by adding in 10% Fetal Bovine Serum and 1% Penicillin and Streptomycin) to 37 °C. NOTE: The media choice will be based on the patient's cancer type or else if this is a model system experiment where cultured cells are to be spiked into healthy donor blood, use the media the cells are normally cultured in. If unsure, please refer to culturing guide/method provided by the company the cells were purchased from.
Add 7.5 ml of commercially available Hank's Balanced Salt Solution (HBSS) to the 7.5 ml of blood sample and aspirate this diluted blood into a 25 ml syringe. NOTE: Adjust the volume of HBSS so that the blood sample is diluted with equal volume of 1:1 with HBSS. E.g. if 10 ml of blood is used, then mix with equal volume of 10 ml HBSS to achieve the 1:1 dilution.
Engage the filtration cassette with the syringe and place it onto the syringe pump. Place a 50 ml tube at bottom to collect the flow through and start the pump. NOTE: The filtration should take roughly 10- 12 min. All filtration steps are to be carried out at Normal Room Temperature (20-25 °C) including step 3.8
After filtration is complete, disengage the filtration cassette taking note which side is the top that has the cells caught on the PIPAAm-coated surface. Aspirate 1 ml of the warm culture media into a new syringe.
Re-engage the filtration cassette with the syringe containing the media, but at this time engage the bottom end of the cassette so that the PIPAAm coated-surface of the filter is facing away from the syringe. NOTE: This will be to release any cells that may be embedded into the slot pores by applying gentle reverse flow.
Place the syringe back onto the syringe pump and hold a small petri dish or 6-well plate right under the filtration cassette. Set the flow rate to 100 ml/hr and start the pump.
Pass all the media through the filter and collect the flow-through in the petri dish. Wait for all the media to pass through and then stop the pump and remove the syringe and filtration cassette from the pump.
Disengage the filtration cassette from the syringe and open it to retrieve the filter from the cassette. Place the filter with the PIPAAm surface facing down into the same petri dish used to collect the reverse flow when releasing the cells from the pores. Add the rest of the warm media and place the petri dish into a culture incubator set at 37 °C.
After 30 min all the cells will be released from the filter. However, leave filter in the petri dish for up to 24 hours to ensure all cells detach. NOTE: Alternatively, the released cells can be collected into a microcentrifuge tube to perform further downstream analysis such as PCR, ELISA, Western Blot to name a few examples.
After 24 hr in culture, carefully remove the microfilter using forceps. NOTE: The filter can be discarded at this time as the cells have been released from it.
Gently pipette the culture medium up and down using a 1,000 µl pipette, to re-suspend the erythrocytes and peripheral blood mononuclear cell (PBMC) that are caught on the filter and released into the plate with the CTC. Perform this action carefully and gently to avoid detaching the loosely-attached cancer cells. Carefully remove the entire medium containing the re-suspended erythrocytes and PBMC while leaving behind the attached-cancer cells and substitute with fresh medium pre-warmed to 37 °C. NOTE: This step can be repeated once if excessive erythrocytes are present after one wash.
4. CTC Viability Evaluation
Evaluate cultured CTC viability using a Live/Dead Assay8. NOTE: Numerous Live/Dead assays are available commercially. Following the manufacturer's protocol is highly advised. Please refer to the materials/reagents list for the commercial assay kit used for this experiment.
Representative Results
Using healthy donors' blood (obtained under a protocol approved by the University of Miami IRB 20150020 following an informed consent) spiked with cultured cancer cells, the thermoresponsive technique for release of viable circulating tumor cells (CTC),achieved capture, release and retrieval efficiency of 94% ± 9%, 82% ± 5% and 77% ± 5% respectively (Table 1)15. By comparison, the release and retrieval efficiency of uncoated filters were significantly lower (7% ± 1% release efficiency and 6% ± 1% retrieval efficiency) (Tables 1)15. In order to evaluate the viability of CTCs released from filter, ~1,000 SK-Br-3 cells were spiked into 7.5 ml of healthy donor's blood, filtered through the PIPAAm coated slot filter and released using the method described above. A Live/Dead assay was performed to evaluate the cell viability before spike into blood and after release. The pre-spike viability was 98% (592 out of 602 cells counted) and the viability of cells captured and released from blood was 95% (540 out 567 cells counted)15 (Figure 3A). In parallel, the cells captured and released post-filtration from the blood sample were cultured in McCoy's 5A culture medium. Images were taken at Day 3 and Day 10. As shown in Figure 3B, cells released from the filter not only remained viable, but expanded rapidly in culture, thus establishing their viability post filtration.
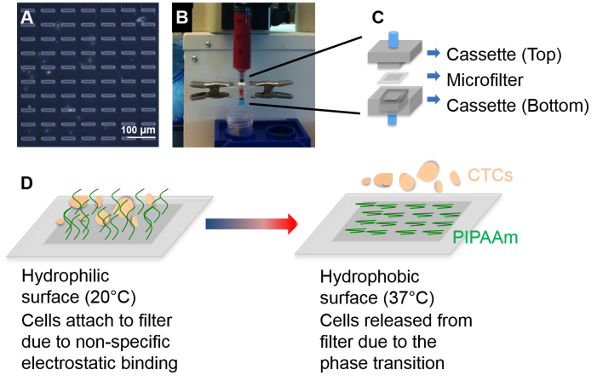 Figure 1: PIPAAm Coated Slot Filters to Capture and Release Circulating Tumor Cells from Blood. (Top panel) (A) Brightfield image of PIPAAm coated slot filter; (B) Filtration of whole blood for CTC capture. (C) Schematic of the microfilter cassette where the, PIPAAm coated slot filter is sandwiched between the top and bottom cassette. (Bottom panel) (D) Schematic of the process to release captured CTC from PIPAAm coated slot filters. Reproduced with permission from reference15. Please click here to view a larger version of this figure.
Figure 1: PIPAAm Coated Slot Filters to Capture and Release Circulating Tumor Cells from Blood. (Top panel) (A) Brightfield image of PIPAAm coated slot filter; (B) Filtration of whole blood for CTC capture. (C) Schematic of the microfilter cassette where the, PIPAAm coated slot filter is sandwiched between the top and bottom cassette. (Bottom panel) (D) Schematic of the process to release captured CTC from PIPAAm coated slot filters. Reproduced with permission from reference15. Please click here to view a larger version of this figure.
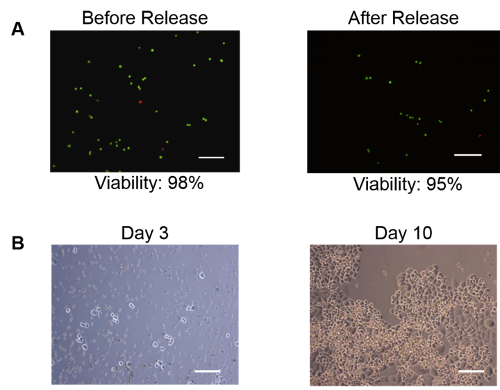 Figure 3: Capture, Release and Culture of Viable Tumor Cells. Breast cancer cells (SKBr-3) spiked in normal donor blood were captured and released using a PIPAAm coated microfilter. The cells remained viable post release and expanded in culture rapidly. (A) Live-dead assay performed on the cells released from the filter. 95% (540 out of 567 cells counted) of the cells showed to be viable (green) following the release. Dead cells are labelled in red. (B) Day 3 through day 10 culture of the cells released from PIPAAm coated slot filter. The cells remained viable and expanded in culture. Scale bar = 100 µm.Reproduced with permission from reference15. Please click here to view a larger version of this figure.
Figure 3: Capture, Release and Culture of Viable Tumor Cells. Breast cancer cells (SKBr-3) spiked in normal donor blood were captured and released using a PIPAAm coated microfilter. The cells remained viable post release and expanded in culture rapidly. (A) Live-dead assay performed on the cells released from the filter. 95% (540 out of 567 cells counted) of the cells showed to be viable (green) following the release. Dead cells are labelled in red. (B) Day 3 through day 10 culture of the cells released from PIPAAm coated slot filter. The cells remained viable and expanded in culture. Scale bar = 100 µm.Reproduced with permission from reference15. Please click here to view a larger version of this figure.
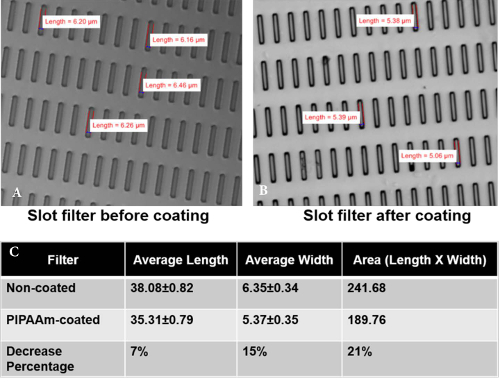 Figure 4: Measurement of Pore Parameters Pre- and Post- PIPAAm Coating. Brightfield images of slot filter (A) pre- and (B) post- PIPAAm coating. (C) Pore length was decreased by 7.3% ± 2.1% after PIPAAm coating and pore width was decreased by 15.3% ± 5.6% after PIPAAm coating. Reproduced with permission from reference15. Please click here to view a larger version of this figure.
Figure 4: Measurement of Pore Parameters Pre- and Post- PIPAAm Coating. Brightfield images of slot filter (A) pre- and (B) post- PIPAAm coating. (C) Pore length was decreased by 7.3% ± 2.1% after PIPAAm coating and pore width was decreased by 15.3% ± 5.6% after PIPAAm coating. Reproduced with permission from reference15. Please click here to view a larger version of this figure.
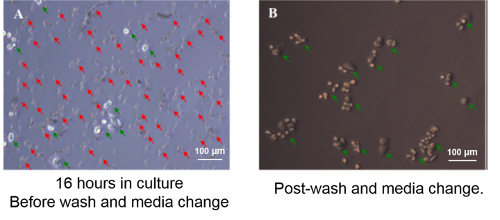 Figure 5: Contaminating Erythrocytes and Leukocytes are Removed by Gentle Washes. Approximately 1,000 SKBr-3 cells were retrieved from blood by PIPAAm coated slot filter and were plated on a 48-well plate. (A) At 16 hr in culture, SKBr-3 tumor cells adhered to culture plate (green arrows), whereas apoptotic erythrocytes and leukocytes were also settling at the bottom of the plate (red arrows). (B) Post-wash, non-adherent cells were removed, leaving adherent tumor cells on the plate (green arrows). Reproduced with permission from reference15. Please click here to view a larger version of this figure.
Figure 5: Contaminating Erythrocytes and Leukocytes are Removed by Gentle Washes. Approximately 1,000 SKBr-3 cells were retrieved from blood by PIPAAm coated slot filter and were plated on a 48-well plate. (A) At 16 hr in culture, SKBr-3 tumor cells adhered to culture plate (green arrows), whereas apoptotic erythrocytes and leukocytes were also settling at the bottom of the plate (red arrows). (B) Post-wash, non-adherent cells were removed, leaving adherent tumor cells on the plate (green arrows). Reproduced with permission from reference15. Please click here to view a larger version of this figure.
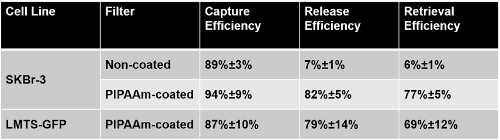 Table 1: Capture, Release and Retrieval Efficiency of PIPAAm Coated Slot Filters. Capture efficiency = cell numbers captured on filter before release / cell numbers spiked into blood. Release efficiency = cell numbers released from the filter / cell numbers captured on the filter before release. Retrieval efficiency = cell numbers released from filter / cell numbers spiked into blood.Reproduced with permission from reference15.
Table 1: Capture, Release and Retrieval Efficiency of PIPAAm Coated Slot Filters. Capture efficiency = cell numbers captured on filter before release / cell numbers spiked into blood. Release efficiency = cell numbers released from the filter / cell numbers captured on the filter before release. Retrieval efficiency = cell numbers released from filter / cell numbers spiked into blood.Reproduced with permission from reference15.
Discussion
The process of capturing viable CTC from whole blood and releasing them from the microfilter is relatively straightforward; however a few critical points are worth mentioning. It is imperative, as with all cell culture that a sterile condition is maintained through the whole process. The initial step of coating the filter with PIPAAm is critical, as the basis for the technique of releasing the cells from the filter is based on exploiting PIPAAm's temperature responsive interfacial properties. To ensure the filter has been coated effectively, measure the size of a pore (width and length of a slot pore) on the filter under a microscope prior to coating and then again measure a pore size post coating. The pore size should be decreased by 1µm on both the width and the length (Figure 4). As mentioned, the slot filter while capturing CTC efficiently, also captures some large PBMC cells, however these cells are non-adherent cells and thus during step 3.13 are removed from the culture leaving behind the adherent cancer cells (Figure 5).
Minor modifications to the protocol may be needed if different volumes of blood are to be filtered or in instances when the viscosity is extremely high. As mentioned in the protocol, it is critical to dilute blood 1:1 with HBSS. The viscosity of blood varies from patient to patient, however for the most part this will not affect the filtration. There will be certain instances when the viscosity is extremely high or large clots are present, in which case the syringe pump will not be able to force the blood through the filter. It is advised in these instances to stop the syringe pump, disengage the cassette from the syringe, open the top part of cassette, but leave the filter still attached to the bottom part. Large blood clots will be easily visible on the filter. Gently pipette 1 ml of HBSS onto the filter while holding the cassette at a slight angle to allow the clot to wash off. Once the clot has been removed, close the cassette, re-engage it to the syringe and continue filtration.
While the PIPAAm coated filters may be made in batches and stored for later use, we have found that the shelf life of these pre-coated filters is approximately 3 months. The effective capture and release of the cells may be decreased in part due to the PIPAAm degrading over time.
One of the limiting factors to establish a culture from patient CTC will lie in the number of CTC that are present in the blood sample. Low numbers will likely make the ability to expand the cells extremely difficult if not impossible. However, sequestering a sample with low cell counts into smaller culture areas may help, such as in a single well of 96 well plate instead of a 6 well plate. The identification of culture media, or modification of commercially available media, that will provide the optimal environment for these cells to grow is another challenge that will have to be overcome.
Molecular and cellular analyses of CTC provide valuable information for cancer prognosis and may help drive precision medicine15. The viability of the cells released from the capture filter is a key development toward the ability to culture patient CTC for drug sensitivity and screening tests. It has been long known that CTC tend to differ from the primary tumor and thus the necessity to treat both accordingly. As an example, Schneeweiss et al. reported that in metastatic breast cancer (MBC), antigen profiles of metastatic tissue and primary tumor differ in up to 20% of patients. Reassessment of predictive markers, including human epidermal growth factor receptor 2 (HER2) expression, might help to optimize MBC treatment. While tissue sampling is invasive and often difficult to repeat, CTC analysis requires only a blood sample and might provide an easy-to-repeat, real-time "liquid biopsy" approach22.
A major significance of the technique lies in the fact that while several platforms commonly used in CTC capture and analyses are limited in molecular and cellular analyses as a fixative is necessary for processing the sample, or the CTC are immobilized on platform. Further, affinity-based systems, which allow for viable CTC capture and release, can potentially be biased by the choice of target antigen15. Alternatively, the slot filter with the PIPAAm coating provides a label free platform that is effective and efficient in the capture and release of viable CTC from whole blood. This method provides the ability for further characterization of viable CTC, including single cell phenotypic and genomic analysis as well as ex vivo CTC culture.
Work is currently underway to employ this technology for clinical applications. The ability to effectively culture CTC from patient samples will lead to formulation of effective therapy regiments on a patient to patient basis, reveal new drug targets, and lead to development of effective chemotherapeutics.
Disclosures
Part of this work has been protected under U.S. Provisional Patent Application No. 62/219,808. Siddarth Rawal is a shareholder of Circulogix Inc. that commercially produces the filters and filter cassettes/cartridges used in this Article.
Acknowledgments
We thank all the patients who have donated blood samples to support this work. We thank Drs. Guiseppe Giaconne, Ritesh Parajuli, and Marc E. Lippman for their assistance in clinical sample acquirement, and Drs. Carmen Gomez, Ralf Landgraf, Stephan Züchner, Toumy Guettouche, Diana Lopez for their insightful discussions. Zheng Ao thanks partial support and assistance from the Sheila and David Fuente Graduate Program in Cancer Biology, Sylvester Comprehensive Cancer Center.
References
- Cristofanilli M, Budd GT, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Scher HI, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- Cohen SJ, Punt CJa, et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol. 2009;20(7):1223–1229. doi: 10.1093/annonc/mdn786. [DOI] [PubMed] [Google Scholar]
- Smerage JB, Barlow WE, et al. Circulating Tumor Cells and Response to Chemotherapy in Metastatic Breast Cancer: SWOG S0500. J. Clin. Oncol. 2014;32(31):3483–3490. doi: 10.1200/JCO.2014.56.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach AJ, Kim JH, Arshi A, Hur SC, Di Carlo D. Automated cellular sample preparation using a Centrifuge-on-a-Chip. Lab chip. 2011;11(17):2827–2834. doi: 10.1039/c1lc20330d. [DOI] [PubMed] [Google Scholar]
- Ozkumur E, Shah AM, et al. Inertial Focusing for Tumor Antigen-Dependent and -Independent Sorting of Rare Circulating Tumor Cells. Sci. Transl. Med. 2013;5(179):179. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagrath S, Sequist LV, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa M, Kenmotsu H, et al. Size-Based Isolation of Circulating Tumor Cells in Lung Cancer Patients Using a Microcavity Array System. PLoS ONE. 2013;8(6) doi: 10.1371/journal.pone.0067466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat AAS, Hou HW, Li LD, Lim CT, Han J. Pinched flow coupled shear-modulated inertial microfluidics for high-throughput rare blood cell separation. Lab on a chip. 2011;11(11):1870–1878. doi: 10.1039/c0lc00633e. [DOI] [PubMed] [Google Scholar]
- Tan SJ, Lakshmi RL, Chen P, Lim W-T, Yobas L, Lim CT. Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosens. Bioelectron. 2010;26(4):1701–1705. doi: 10.1016/j.bios.2010.07.054. [DOI] [PubMed] [Google Scholar]
- Vona G, Sabile A, et al. Isolation by size of epithelial tumor cells a new method for the immunomorphological and molecular characterization of circulatingtumor cells. Am. J. Pathol. 2000;156(1):57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrinucci D, Bethel K, et al. Case study of the morphologic variation of circulating tumor cells. Human pathology. 2007;38(3):514–519. doi: 10.1016/j.humpath.2006.08.027. [DOI] [PubMed] [Google Scholar]
- Lin HK, Zheng S, et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin. Cancer Res. 2010;16(20):5011–5018. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Rawal S, et al. Clinical translation of a novel microfilter technology Capture, characterization and culture of circulating tumor cells. PHT. 2013. pp. 220–223.
- Ao Z, Parasido E, et al. Thermoresponsive release of viable microfiltrated Circulating Tumor Cells (CTCs) for precision medicine applications. Lab Chip. 2015;15:4277–4282. doi: 10.1039/c5lc01024a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Lu B, Tai YC, Goldkorn A. A cancer detection platform which measures telomerase activity from live circulating tumor cells captured on a microfilter. Cancer Res. 2010;70(16):6420–6426. doi: 10.1158/0008-5472.CAN-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano T, Bae YH, Jacobs H, Kim SW. Thermally on-off switching polymers for drug permeation and release. J. Control. Release. 1990;11(1-3):255–265. [Google Scholar]
- Yamada N, Okano T, Sakai H, Karikusa F, Sawasaki Y, Sakurai Y. Thermo-responsive polymeric surfaces; control of attachment and detachment of cultured cells. Die Makromol. Chemie, Rapid Commun. 1990;11(11):571–576. [Google Scholar]
- Deng Y, Zhang Y, et al. An integrated microfluidic chip system for single-cell secretion profiling of rare circulating tumor cells. Sci. Rep. 2014;4:7499. doi: 10.1038/srep07499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S, Zhao H, et al. Capture and stimulated release of circulating tumor cells on polymer-grafted silicon nanostructures. Adv. Mater. 2013;25(11):1547–1551. doi: 10.1002/adma.201203185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Zhou H, et al. Effective and selective cell retention and recovery from whole blood by electroactive thin films. ACS Appl. Mater. Interfaces. 2014;6(23):20804–20811. doi: 10.1021/am505072z. [DOI] [PubMed] [Google Scholar]
- Wallwiener M, Hartkopf AD, et al. The impact of HER2 phenotype of circulating tumor cells in metastatic breast cancer: a retrospective study in 107 patients. BMC cancer. 2015;15(1):403. doi: 10.1186/s12885-015-1423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


