Abstract
Foodborne infections in the US caused by Vibrio species have shown an upward trend. In the genus Vibrio, V. parahaemolyticus is responsible for the majority of Vibrio-associated infections. Thus, accurate differentiation among Vibrio spp. and detection of V. parahaemolyticus is critically important to ensure the safety of our food supply. Although molecular techniques are increasingly common, culture-depending methods are still routinely done and they are considered standard methods in certain circumstances. Hence, a novel chromogenic agar medium was tested with the goal of providing a better method for isolation and differentiation of clinically relevant Vibrio spp. The protocol compared the sensitivity, specificity and detection limit for the detection of V. parahaemolyticus between the new chromogenic medium and a conventional medium. Various V. parahaemolyticus strains (n=22) representing diverse serotypes and source of origins were used. They were previously identified by Food and Drug Administration (FDA) and Centers for Disease Control and Prevention (CDC), and further verified in our laboratory by tlh-PCR. In at least four separate trials, these strains were inoculated on the chromogenic agar and thiosulfate-citrate-bile salts-sucrose (TCBS) agar, which is the recommended medium for culturing this species, followed by incubation at 35-37 °C for 24-96 hr. Three V. parahaemolyticus strains (13.6%) did not grow optimally on TCBS, nonetheless exhibited green colonies if there was growth. Two strains (9.1%) did not yield the expected cyan colonies on the chromogenic agar. Non-V. parahaemolyticus strains (n=32) were also tested to determine the specificity of the chromogenic agar. Among these strains, 31 did not grow or exhibited other colony morphologies. The mean recovery of V. parahaemolyticus on the chromogenic agar was ~96.4% relative to tryptic soy agar supplemented with 2% NaCl. In conclusion, the new chromogenic agar is an effective medium to detect V. parahaemolyticus and to differentiate it from other vibrios.
Keywords: Infection, Issue 117, Vibrio parahaemolyticus, chromogenic media, assay development, TCBS, selective and differential media, detection, isolation, sensitivity, specificity, Standard Plate Count
Introduction
As a member of the Vibrio genus, V. parahaemolyticus is a Gram-negative, non-spore-forming, curved, rod-shaped bacterium. It exhibits high motility in both liquid and semi-solid environments. Most V. parahaemolyticus strains are non-pathogenic to humans, yet the pathogenic subtypes have caused epidemics and pandemics, hence this species is considered to be an important foodborne pathogen in many countries1,2. The incidence of Vibrio infection in the US has shown an upward trend since 20003. Among Vibrio spp., V. parahaemolyticus is the most frequently reported species causing illnesses in the US4,5. Other clinically relevant species include V. alginolyticus, V. vulnificus, V. cholerae, etc. A small percentage of the illnesses is caused by multiple species simultaneously.
V. parahaemolyticus is a natural inhabitant of marine water and therefore widely distributed in marine waters throughout the world including the estuaries. The species was discovered in 1950 following an outbreak of food poisoning in Japan. In the US, the species was first isolated in seawater, sediments, and shellfish in the Puget Sound region6,7. Filter feeders in marine habitats, such as bivalve shellfish, can harbor V. parahaemolyticus as part of their natural flora8. As such, V. parahaemolyticus infections in human are often linked to the consumption of contaminated seafood, especially raw or undercooked shellfish. A less common route of entry occurs when open wound is exposed to seawater, leading to skin infection. Most V. parahaemolyticus strains do not cause human disease, yet certain subtypes harboring virulence factors such as thermostable direct hemolysin (TDH) are pathogenic. The most prevalent symptoms of foodborne V. parahaemolyticus infection are diarrhea and abdominal pain, followed by nausea, vomiting, and fever. Headache and chills are also reported. The median incubation period is 15 hr, but can be up to 96 hr after consumption of sufficient amount of pathogenic strains9. The illness lasts from two to three days. The gastroenteritis symptoms caused by V. parahaemolyticus are largely self-limiting and therefore special treatment is not necessary. Mild cases of gastroenteritis can be effectively treated by oral rehydration. More severe illnesses can be treated by antibiotics such as tetracycline or ciprofloxacin10. Mortality rate is about 2% for gastroenteritis cases, but may be as high as 29% for those who develop bloodstream infection or septicemia. Any person who consumes seafood or has open wound exposed to seawater is at risk of V. parahaemolyticus infection. The more severe form of illnesses, life-threatening septicemia, is more common in a subpopulation with underlying medical conditions11, which include alcoholism, liver disease, diabetes, renal disease, malignancy, and other conditions leading to a weakened immune response. Notably, this group of individuals is also at a higher risk for contracting severe illnesses caused by V. vulnificus, which can be found in natural habitats similar to V. parahaemolyticus.
V. parahaemolyticus is routinely isolated using thiosulfate-citrate-bile salts-sucrose (TCBS) agar as a selective and differential medium. Enrichment in alkaline peptone water may precede isolation on TCBS agar. Presumptive colonies on TCBS are then further tested in an array of biochemical tests and/or molecular assays targeting the presence of species-specific genes. PCR-based methods are often used to confirm the identities of V. parahaemolyticus by amplifying the thermolabile hemolysin gene, tlh12.
Regardless of the choice of confirmation methods, it is important to have an effective medium to isolate and differentiate V. parahaemolyticus from other marine vibrios in the first place. TCBS has routinely been used to differentiate species within the Vibrio genus according to their abilities to ferment sucrose12. Positive fermentation reaction is accompanied by a color change of the pH indicator Bromothymol blue. V. parahaemolyticus colonies are fairly distinctive on TCBS, exhibiting blue to green color. However, this medium cannot easily differentiate V. alginolyticus and V. cholerae. Sucrose-fermenting Proteus species may produce yellow colonies resembling V. cholerae or V. alginolyticus13. On initial isolation on TCBS, V. parahaemolyticus may also be misidentified as Aeromonas hydrophila, Plesiomonas shigelloides, and Pseudomonas spp14. Strains with delayed sucrose fermentation may be confused with other sucrose nonfermenting Vibrio13, which include V. parahaemolyticus. TCBS was found to be not sensitive against Escherichia coli, Pseudomonas putrefaciens, among others. Several other species yield green to gray colonies which are potentially confused with V. parahaemolyticus or V. vulnificus15. As a result, it is desirable to develop alternative culture media with better sensitivity and specificity toward detecting and isolating V. parahaemolyticus and other closely related species.
Several media alternatives have been recently developed. In addition to the inclusion of selective agents, most incorporate chromogenic substrates to differentiate species based on their differential enzymatic activities. For example, indoxyl-β-glucoside and indoxyl-β-galactoside have been used as the chromogenic substrates to differentiate V. parahaemolyticus colonies (which appear bluish-green) from those of V. cholerae (purple) due to their differential abilities to produce β-glucosidase and β-galactosidase16. Different formulations of chromogenic agar developed by several groups have been evaluated and were reported to perform comparably to or better than TCBS17,18,19. An advantage of using a chromogenic medium is that the coloring of the surrounding medium is minimal thereby facilitating the isolation of particular colonies. In this study, we evaluated the ability of a newly formulated chromogenic medium to detect and isolate V. cholerae, V. parahaemolyticus, and V. vulnificus; with a special focus on its ability to differentiate V. parahaemolyticus from other species.
Protocol
1. Media and Culturing of Microbial Strains
NOTE: Use aseptic techniques in all experiments. Use sterile materials. Sterilize all containers, tools and reagent prior to use. Autoclave all waste materials prior to disposal because they are considered biohazardous. Autoclave temperature and time combination is ≥121 °C x ≥15 min for all of the following procedures.
- To make ~1-L tryptic soy agar (TSA), first add 1 L deionized water in a 2-L Erlenmeyer flask containing a magnetic stir bar. Use a flask that is at least two times larger than the final volume. Add 30 g of tryptic soy broth powder and 20 g agar granules into the flask. NOTE: Use 2% agar instead of 1.5% to limit swarming of some Vibrio spp.
- Mix thoroughly by turning on the stirrer. While stirring, turn on the heat to boil the mixture. Remove the flask from the heater as soon as the mixture begins to boil. Loosely cover the flask with a tin foil. Tape the foil to secure it to the flask before autoclave.
- To make tryptic soy broth (TSB), omit the agar from the recipe in step 1.1. NOTE: May use bottles instead of Erlenmeyer flask.
- To make tryptic soy agar supplemented with 2% sodium chloride or NaCl (TSAS), add 20 g of NaCl in the mixture prior to stirring and heating. To make tryptic soy broth supplemented with 2% NaCl (TSBS), omit the agar, add 20 g of NaCl in the mixture prior to stirring and heating.
To make brain heart infusion (BHI) agar, suspend 37 g of BHI powder and 15 g agar granules in 1 L of purified water. Heat with frequent agitation to dissolve the powder. Autoclave. Omit the agar to make BHI broth.
To make TCBS agar, suspend 89 g of the TCBS powder in 1 L of purified water. Heat with frequent agitation and boil for 1 min to completely dissolve the powder. Do not autoclave.
- For all agar media, cool the hot agar to 45-50 °C in a water bath. Arrange empty Petri plates in stacks of five to six plates. Starting from the bottom of the stack, pour the molten agar into each Petri plate to reach about half full. Close the Petri plate lid after pouring. Allow the agar to solidify by letting the plates sit at room temperature.
- Use the agar plates the next day or after 12 hr. Store unused plates in a refrigerator for up to two weeks. Before use, remove plates from the refrigerator and equilibrate them at room temperature for at least 15 min. NOTE: One-liter agar makes ~45 agar plates. Allow the agar plates to dry sufficiently on the day of preparation, and equilibrate them to room temperature after cold storage to effectively reduce the spreading of the colonies.
Obtain chocolate and chromogenic agar plates and equilibrate them at room temperature before each experiment.
- Subculture all 54 microbial strains shown in Table 1 every few days.
- Use a sterile inoculating loop to transfer cultures from a frozen stock or a previous batch to nonselective media such as BHI, TSB/TSA or chocolate agar. Grow halophilic Vibrio spp. on TSBS/TSAS.
- To check the purity of the culture, streak all strains in a pattern that would allow for observation of isolated colonies. For example, use a three-phase streaking pattern to dilute a large amount of bacteria to smaller amount, eventually yielding isolated colonies.
Incubate the plates upside-down at 35-37 °C for up to 48 hr. For Campylobacter spp., incubate tubes or platesin a closed-lid jar containing a gas pouch to produce a microaerophilic environment. Observe colony morphology after incubation. Pure cultures should yield colonies that exhibit similar colony morphology. NOTE: Incubate all plates upside-down to prevent condensed water droplets formed on the underside of the lid from falling on the colonies.
2. Species Determination by PCR
- Conduct tlh-PCR to confirm the identity of V. parahaemolyticus strains. Use primers tlh-F (5' AAA GCG GAT TAT GCA GAA GCA CTG 3') and tlh-R (5' GCT ACT TTC TAG CAT TTT CTC TGC 3') to amplify a 450-bp fragment of the thermolabile hemolysin gene20.
- Use a sterile inoculating loop to transfer a few isolated colonies of each V. parahaemolyticus strain from TSAS to 5 ml of TSBS. Incubate at 35-37 °C for 16-24 hr.
- Centrifuge cultures at 14,000 x g for 1 min. Remove supernatant and wash the pellet twice with phosphate buffered saline (PBS). Boil the suspension for 3 min to yield cell lysate. NOTE: V. parahaemolyticus is easy to be lysed. Therefore lysis reagent is not required. It is also possible to use a bit of colony directly as the template.
- Perform PCR in a 25-µl reaction volume. Prepare a reaction mixture containing a final concentration of 1x PCR buffer, 1.5 mM MgCl2, 100 µM of each dNTP, 1 µM of each primer, 1 U Taq Polymerase and 1 µl of cell lysate. After preincubation at 94 °C for 5 min, run 35 amplification cycles of 94 °C for 30 sec, 58 °C for 30 sec, and 72 °C for 60 sec21.
- Load aliquots of 5-µl amplicon in 1.5% agarose gels. Turn on power supply to start electrophoresis. Visualize the presence or absence of amplicons under UV illumination after ethidium bromide staining.
3. Growth on Selective and Differential Media
Two to four days before the experiment, streak all microbial strains shown in Table 1 on nonselective medium (TSAS, BHI or chocolate agar) for colony isolation. Incubate plates at 35-37 °C for 48 hr. Check the purity of the cultures by observing colony morphology after incubation. Pure cultures should yield colonies that exhibit similar colony morphology.
Transfer a few isolated colonies from Step 3.1 into 5 ml of broth. Incubate tubes at 35-37 °C for 16-24 hr. NOTE: Use young colonies, which are less than four days old, to prepare overnight cultures in all experiments.
Streak a loopful of overnight cultures on selective and differential media (TCBS and chromogenic agar) for colony isolation. Incubate plates at 35-37 °C for up to 96 hr.
Record the overall growth of all strains by examining both the culture density on the plate and the size of isolated colonies. Record the color of colonies under ambient and/or UV light. Note other characteristics of isolated colony such as the elevation, margin and form.
4. Recovery Assay
Select a representative subset of V. parahaemolyticus strains (n=14) that encompass different serotypes and origins of isolation22. Inoculate young cultures from the plates into 5 ml of TSBS. Incubate the tubes at 35-37 °C for 16-24 hr.
- Conduct a Standard Plate Count method of the overnight cultures as described below.
- Vortex to mix the overnight cultures well. Make a 10-fold or 10-1 dilution by transferring 100 µl of the overnight culture into a tube containing 900 µl PBS. Vortex to mix well.
- Use a new pipet tip to transfer 100 µl from the 10-1 dilution tube to another tube containing 900 µl PBS. Vortex to mix well. This constitutes 10-2 dilution. Repeat the process sequentially to obtain 10-7 dilution.
- Using the 10-4 to 10-7 dilution tubes, plate 100 µl each on the chromogenic, TCBS and TSAS agar plates. NOTE: The dilution factor (df) on the plate becomes 10-5 to 10-8, respectively.
- Spread the aliquots evenly on the agar surface. NOTE: It is fine to use the same spreader per strain on the same medium, as long as the most diluted aliquot is spread first (i.e., from 10-8 to 10-5). Do not use the same spreader for different media.
- Incubate plates at 35-37 °C for up to 96 hr. Count colonies on the plates. Ignore plates bearing colonies that are too numerous to count (tntc) or less than 25. Calculate CFU/ml according to the following:
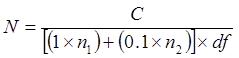 Where N = the number of cells in the undiluted tube, expressed as CFU/ml or CFU/g C = the total number of colonies counted on plates that bear 25-300 colonies n1 = the number of plate(s) where counted colonies are from the lower df
n2 = the number of plate(s) where counted colonies are from the subsequent 10-fold dilution df = the lower dilution factor (i.e., more concentrated dilution)
Where N = the number of cells in the undiluted tube, expressed as CFU/ml or CFU/g C = the total number of colonies counted on plates that bear 25-300 colonies n1 = the number of plate(s) where counted colonies are from the lower df
n2 = the number of plate(s) where counted colonies are from the subsequent 10-fold dilution df = the lower dilution factor (i.e., more concentrated dilution)
Compare CFU/ml among different media. Use CFU/ml on TSAS as 100%, calculate the % recovery of V. parahaemolyticus grown on the chromogenic and TCBS agar.
5. Competition Assay
- Choose a subset of strains that exhibit different colony morphologies on the chromogenic and TCBS agar. NOTE: This way, it will be possible to count colonies originated from V. parahaemolyticus only, despite the presence of other species in the inoculum.
- Choose a V. parahaemolyticus strain that yields the expected turquoise and cyan colonies on TCBS and the chromogenic agar, respectively.
- Choose a non-V. parahaemolyticus and a non-Vibrio species that do not grow on either of these media, or exhibit different colony color. NOTE: For example, V. metschnikovii grows very weakly on TCBS and did not grow on the chromogenic agar. Shigella sonnei does not grow on TCBS but yields magenta colonies on the chromogenic agar.
- After selection of the strains above, prepare overnight broth cultures using isolated colonies grown on nonselective media.
- Make overnight cultures of V. parahaemolyticus and V. metschnikovii by transferring a few isolated colonies from TSAS to 5 ml of TSBS. Incubate at 35-37 °C for 16-24 hr.
- Make overnight cultures of Shigella sonnei by transferring a few isolated colonies from BHI agar to 5 ml of BHI broth. Incubate at 35-37 °C for 16-24 hr.
For each strain, perform a dilution series similar to Steps 4.2.1 and 4.2.2. Plate appropriate dilutions on TSAS or BHI to determine CFU/ml of the overnight culture, using the equation shown in Step 4.2.5. NOTE: Typically, 100 µl from the 10-5 to 10-7 dilution tubes works for most cultures. Use the CFU/ml values to back calculate the exact amount of cells used in next step. The calculated CFU/ml values are obtained after incubation, although the following steps are performed on the same date as Step 5.3.
Using the overnight cultures and dilution tubes in Steps 5.2 and 5.3, mix different amounts of a V. parahaemolyticus strain and a non-V. parahaemolyticus species. For example, mix 500 µl of the 10-5 dilution tube of V. parahaemolyticus with 500 µl of the overnight cultures of V. metschnikovii. NOTE: This mixture simulates high microflora background. To simulate a low microflora background, mix 500 µl each of the 10-5 dilution tube from both species.
Spread 100 µl of the mixture each on the chromogenic, TCBS and TSAS agar plates.
- After incubation at 35-37 °C for up to 96 hr, count colonies of V. parahaemolyticus and the non-V. parahaemolyticus species based on their difference in growth and colony morphology on the chromogenic and TCBS agar. NOTE: For example, if the non-V. parahaemolyticus species does not grow on the chromogenic and TCBS agar, all colonies will be of V. parahaemolyticus. If the non-V. parahaemolyticus species grows on both media, only the turquoise colonies on TCBS and cyan colonies on chromogenic agar will be of V. parahaemolyticus. The non-V. parahaemolyticus species may or may not exhibit similar colony morphology to V. parahaemolyticus on TSAS.
- If the non-V. parahaemolyticus species grows similarly to V. parahaemolyticus on this nonselective medium, divide the colony count by two to obtain numbers for V. parahaemolyticus only. Compare the actual colony count with the expected count derived from Step 5.3.
6. Effects of Oyster Homogenates
- Weigh ≥50 g oyster meat from ≥12 molluscan shellfish including meat and liquor.
- Add equal amount of PBS to the oyster meat and liquor. Blend the mixture at high speed for 90 sec. This constitutes 2-1 diluted oyster homogenate.
- Add 100 g of 2-1 diluted oyster homogenate to 400 g of PBS. Use a scale to measure the weight, not volume. Blend the mixture at high speed for 1 min. Autoclave the oyster homogenate. NOTE: This will be the oyster homogenate used for spiking.
- Repeat the Recovery assay (Step 4) in the presence of oyster homogenate.
- After the 500 g oyster homogenate cools down, add 100 µl of V. parahaemolyticus overnight cultures grown in TSBS to it. Determine the actual amount of V. parahaemolyticus cells in the inoculum by conducting the Standard Plate Count procedures described in Step 4.2.
- Repeat the Competition assay (Step 5) in the presence of oyster homogenate.
- After the 500 g oyster homogenate cools down, add 100 µl each of overnight cultures of V. parahaemolyticus and non-V. parahaemolyticus to it. Determine the actual amount of bacterialcells in the inoculum by conducting the Standard Plate Count procedures described in Step 4.2
Mix the bacterial cells with oyster homogenate well by using a homogenizer. NOTE: After mixing, the oyster homogenate containing the intentionally added cells is called spiked oyster homogenate.
Make dilutions of the spiked oyster homogenate to obtain 10-1 to 10-3 dilution tubes according to the procedures described in Step 4.2.2. Spread 100 µl of each dilution onto the chromogenic, TCBS and TSAS agar. Incubate plates at 35-37 °C for up to 96 hr.
Compare the actual colony count on chromogenic and TCBS agar with the expected colony count deduced from Steps 6.2.1 and 6.3.1. NOTE: For example, if a tube of V. parahaemolyticus overnight culture contains 108 CFU/ml, an inoculum of 100 μl means that 107 cells are added to the 500-g of oyster homogenate, yielding 5 x 104 cells/g. After dilution and plating, the plate having df = 10-2 should yield 500 colonies; while that having df = 10-3 should yield 50 colonies. These are the expected colony counts.
Representative Results
In this study, 54 microbial strains were assembled, which included 22 strains within the V. parahaemolyticus species, 19 other Vibrio species, and 13 non-Vibrio species (Table 1). Most V. parahaemolyticus strains were either received from FDA, CDC or other state health departments. They represent diverse serotypes and isolation sources. These strains were previously identified by the regulatory agencies. We further confirmed the identities of these V. parahaemolyticus by conducting a tlh-PCR21,22.
| Species | No. of strains tested | Growth and color of colonies | |
| TCBS | Chromogenic medium | ||
| Aeromonas hydrophila | 2 | No growth | 1 strain no growth; |
| 1 strain magenta color | |||
| Candida albicans | 1 | Weak growth, yellow | No growth |
| Campylobacter jejuni | 1 | No growth | No growth |
| Escherichia coli | 1 | No growth | Weak growth, magenta |
| Proteus mirabilis | 1 | No growth | No growth |
| Pseudomonas aeruginosa | 2 | Turquoise | Yellow |
| Staphylococcus aureus | 1 | Weak growth, yellow | No growth |
| Salmonella choleraesuis | 1 | No growth | No growth |
| Shigella boydii | 1 | No growth | No growth |
| Shigella flexneri | 1 | No growth | No growth |
| Shigella sonnei | 1 | No growth | Weak growth, magenta |
| Vibrio alginolyticus | 3 | Yellow or turquoise | Yellow |
| V. cholerae | 4 | Yellow or turquoise | Magenta |
| V. damsela | 1 | Turquoise | Cobalt |
| V. fisheri | 1 | Weak growth, yellow | No growth |
| V. fluvialis | 1 | Turquoise | Cobalt |
| V. furnissii | 1 | Yellow | Yellow |
| V. hollisae | 1 | Turquoise | Yellow |
| V. metschnikovii | 1 | No growth | No growth |
| V. mimicus | 1 | Turquoise | Magenta |
| V. parahaemolyticus | 22 | Most strains turquoise | Most strains cyan; few clear |
| V. proteolyticus | 1 | Turquoise | Cobalt |
| V. vulnificus | 4 | Turquoise or clear | Magenta |
Table 1. Microbial species used in this study and their growth characteristics on the selective and differential media. Color calling was based on a color wheel. Cyan is similar to teal; magenta is similar to pinkish lavender, turquoise is similar to green, yellow encompasses olive and brown.
Four separate trials were conducted to determine the growth and colony morphology of these strains on the selective and differential media — TCBS and the chromogenic agar. TCBS is the conventional medium used for the isolation of some Vibrio species, including V. cholerae and V. parahaemolyticus12. Color variation of colonies grown on TCBS was yellow, turquoise (green) or clear (Figure 1).
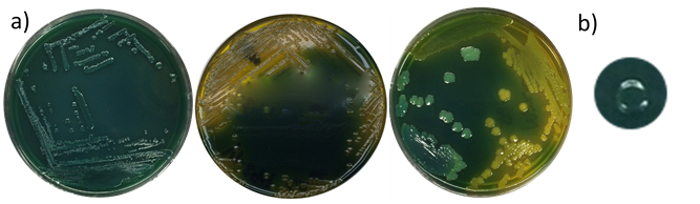 Figure 1. Colony morphology of Vibrio spp. on TCBS agar. V. parahaemolyticus (turquoise), V. cholerae (yellow), and a mixed inoculation of the two species (a). Colonies of V. parahaemolyticus appear turquoise, with a circular, entire, and convex morphology (b). Please click here to view a larger version of this figure.
Figure 1. Colony morphology of Vibrio spp. on TCBS agar. V. parahaemolyticus (turquoise), V. cholerae (yellow), and a mixed inoculation of the two species (a). Colonies of V. parahaemolyticus appear turquoise, with a circular, entire, and convex morphology (b). Please click here to view a larger version of this figure.
The ability of a new chromogenic medium to select for clinically relevant Vibrio species from food and environmental samples was tested. Additionally, the ability of this medium to simultaneously distinguish these species from each other was evaluated. Colony color observed on the chromogenic agar were cobalt, cyan, magenta, yellow or clear (Figure 2). As shown in Table 1, both TCBS and the chromogenic agar exhibited certain degrees of selectivity against non-Vibrio microorganisms.
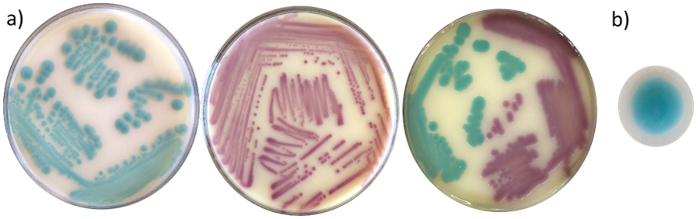 Figure 2. Colony morphology of Vibrio spp. on the newly developed chromogenic agar.V. parahaemolyticus (cyan), V. cholerae (magenta), and a mixed inoculation of the two species (a). Colonies of V. parahaemolyticus appear cyan, with a circular, entire, and convex morphology (b). Please click here to view a larger version of this figure.
Figure 2. Colony morphology of Vibrio spp. on the newly developed chromogenic agar.V. parahaemolyticus (cyan), V. cholerae (magenta), and a mixed inoculation of the two species (a). Colonies of V. parahaemolyticus appear cyan, with a circular, entire, and convex morphology (b). Please click here to view a larger version of this figure.
Following the Standard Plate Count method on overnight cultures, colonies of V. parahaemolyticus were observed on the chromogenic agar plates receiving the highest dilution factor (i.e., df = 10-8). These results were comparable to those on a nonselective medium (TSAS). This suggests that the detection limit of the chromogenic medium is similar to nonselective media, which is approximately 10 cells or lower in the absence of food matrix. The detection capacity for other Vibrio species such as V. alginolyticus, V. fluvialis and V. damsel was also decent compared to the nonselective medium. However, some strains of V. cholerae, V. vulnificus and V. mimicus yielded 10- to 100-fold less CFU on the chromogenic agar. The selective agents used in the chromogenic or other selective media inevitably inhibit some cells. Injured cells, for instance, could not recover in selective media. Nevertheless, the slightly poorer detection ability is a non-issue in most routine procedures that employ an enrichment step. Small amount of microorganisms in the food or environmental sample would multiply to a high level in the enrichment broth, surpassing the detection limit. Enrichment in alkaline peptone water is often done to determine the prevalence of Vibrio species in environmental samples12.
In the presence of oyster homogenate, the chromogenic agar continued to display a good degree of recovery. In other words, a large proportion (>70%) of V. parahaemolyticus cells were able to grow on the chromogenic agar, unaffected by the presence of oyster matrix (Figure 3a). The growth and recovery of V. parahaemolyticus cells was also not affected by the presence of another Vibrio species (Figure 3b). This is an important attribute because environmental samples are bound to contain different Vibrio species, whichare at high numbers especially after an enrichment procedure.
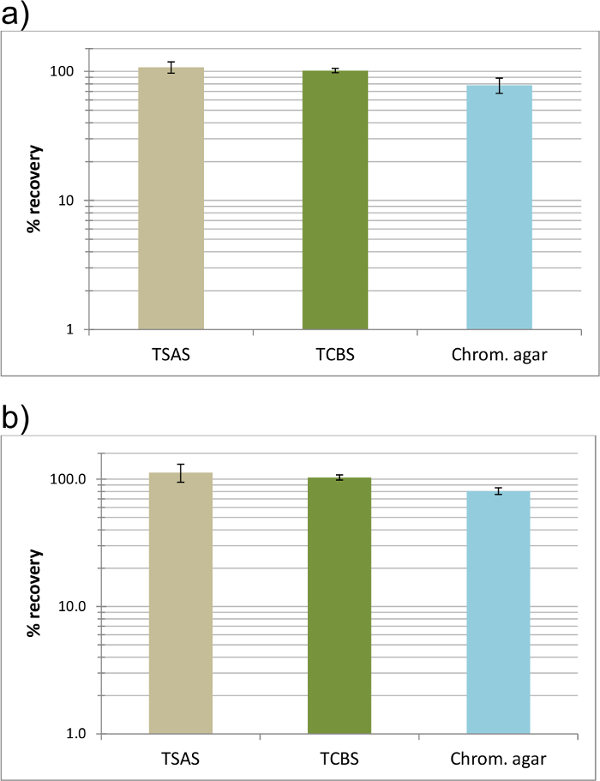 Figure 3. Percent recovery of a V. parahaemolyticus strain in oyster homogenate with and without a bacterial competitor. Recovery (mean ± SD) was calculated according to the observed vs the expected CFU count on the agar plates. Expected CFU count was calculated based on the bacterial load in the inoculum. High recovery of a V. parahaemolyticus strain was observed on all media without competition (a). Similar level of recovery was observed when high levels of V. metschnikovii cells was present (b). Please click here to view a larger version of this figure.
Figure 3. Percent recovery of a V. parahaemolyticus strain in oyster homogenate with and without a bacterial competitor. Recovery (mean ± SD) was calculated according to the observed vs the expected CFU count on the agar plates. Expected CFU count was calculated based on the bacterial load in the inoculum. High recovery of a V. parahaemolyticus strain was observed on all media without competition (a). Similar level of recovery was observed when high levels of V. metschnikovii cells was present (b). Please click here to view a larger version of this figure.
To further compare the chromogenic and TCBS agar, the sensitivity and specificity were calculated. Table 2 shows the ability of these media to accurately identify V. parahaemolyticus. Sensitivity is related to the percentage of true positive, which means the V. parahaemolyticus strains yielded the expected colony morphology on the media. Specificity is related to the percentage of true negative, which means non-V. parahaemolyticus strains should exhibit poor to no growth, or a different colony morphology than V. parahaemolyticus. The sensitivity and specificity for TCBS to identify V. parahaemolyticus were 86.4% and 71.8%, respectively. In comparison, the sensitivity and specificity of the chromogenic medium were 90.9% and 96.9%, respectively.
| a) | ||
| TCBS agar | V. parahaemolyticus | Non-V. parahaemolyticus |
| Good growth and turquoise colonies | 19 | 9 |
| Poor growth or non-turquoise colonies | 3 | 23 |
| b) | ||
| Chromogenic agar | V. parahaemolyticus | Non-V. parahaemolyticus |
| Good growth and cyan colonies | 20 | 1 |
| Poor growth or non-cyan colonies | 2 | 31 |
Table 2. Sensitivity and specificity of TCBS (a) and chromogenic (b) agar in the detection of V. parahaemolyticus. The identities of V. parahaemolyticus were determined previously by biochemical test and tlh-PCR.
Discussion
This study focuses on culture media development and evaluation. Conventionally, TCBS is the selective and differential medium used for isolating and detecting V. parahaemolyticus, V. cholerae and V. vulnificus12. However, limitations have been reported for this medium, such as the inability to differentiate V. cholerae from other Vibrio species. Sucrose and pH indicator are the differentiation agents of TCBS. Thus, acid production by sucrose fermenter causes color change of the medium. The coloring of the medium is a drawback of TCBS because it may obscure observation of colony morphology. The newly developed chromogenic medium utilizes high pH and salinity to suppress the growth of non-Vibrio species found in many marine samples. The new chromogenic medium has a final pH of 8.6±0.2. Per liter of deionized water, it consists of 10 g of peptone, 10 g of sea salts mixture, 10 g of ox bile, 10 g of sodium thiosulfate, 5 g of yeast extract, 5 g of sodium citrate, 2.2 g of sodium carbonate, 2 g of lactose, 0.5 g of sodium pyruvate, 0.3 g of a chromogenic mix and 15 g of agar. The chromogenic mix allows the differentiation among Vibrio spp. according to their differential abilities to produce certain enzymes. Instead of altering the color of the TCBS agar medium, different Vibrio spp. would exhibit different colony color on the new chromogenic medium. In comparison, a commercially available chromogenic medium contains 15 g of agar, 8 g of peptone and yeast extract, 51.4 g of salts, 0.3 g of a chromogenic mix and a final pH of 9.0±0.2.
The robustness of an assay evaluation depends on the sample size. Since this study emphasizes on the effectiveness of the new chromogenic medium to isolate and detect V. parahaemolyticus, it is important to amass many diverse V. parahaemolyticus strains. Previous studies comparing TCBS and new culture media often involved environmental or food samples containing unknown types and quantity of microorganisms23,24,25,26. Multiple colonies per sample were isolated yet the clonal nature of the isolates was often undetermined. Ideally, different strains should be tested in assay development otherwise the accuracy of the assay would be inflated. Strain identity can be established by conventional subtyping methods such as pulsed-field gel electrophoresis (PFGE) or multi-locus sequencing. V. parahaemolyticus strains in this study were previously characterized by ribotyping and PFGE19.
During assay development, sensitivity, specificity and detection limit are among the key factors to be investigated. Sensitivity, also known as accuracy or diagnostic sensitivity, is the positive percent agreement between the reference and test methods. Specificity, also known as precision or diagnostic specificity, is the negative percent agreement between the reference and test methods. In this study, we used PCR-based method as the reference method because the results were verified among different groups. TCBS and the chromogenic media were the test methods. To determine sensitivity, results from the V. parahaemolyticus strains were used; i.e., Sensitivity = 100% x True Positive / (True Positive + False Negative). On the other hand, results from non-V. parahaemolyticus strains were used to determine specificity; and hence, Specificity = 100% x True Negative / (True Negative + False Positive). To provide more reliable specificity results, non-V. parahaemolyticus strains should be isolated from an environment where sampling will be conducted. Our sensitivity and specificity calculations are based on documents written by government27, academia28 and scientific organization29 on a topic related to assay development and validation.
Based on our results, the chromogenic medium showed better performance than TCBS in the identification of V. parahaemolyticus. The overall percent agreement between the reference method and chromogenic medium is 94.4%, compared to 77.8% between the reference method and TCBS. The sensitivity and specificity of the chromogenic medium are 90.9% and 96.9% respectively, which are greater than those of TCBS (86.4% and 71.9%, respectively). Previous studies evaluating other chromogenic media for the detection of V. parahaemolyticus found that sensitivities ranged from 85 to 88% whereas specificities ranged from 94 to 95%23,24,25. However, these studies did not use the same calculation method as our study. Further, they sometimes used a different reference method or they combined results from both biochemical analyses and chromogenic medium as one test method. As a result, it is difficult to directly compare our results with these previous studies.
In addition to exhibiting a better performance in detecting V. parahaemolyticus, the chromogenic agar used in this study could also differentiate more Vibrio species than TCBS due to the inclusion of a chromogenic mix in the medium formula, yielding multiple colors. Although it was not the focus of this study to detect V. cholerae and V. vulnificus, it is worth to note that the magenta colonies exhibited by these species can be further differentiated by fluorescence under UV. A limitation to use the chromogenic agar for the detection of V. cholerae and V. vulnificus is its apparent low recovery for these species. To circumvent this issue, an enrichment step must be included. Another possible limitation of the chromogenic agar is its shorter shelf life than TCBS. Ongoing optimization of this new medium is required to maintain pH during storage.
Despite the popularity of molecular techniques, culture-depending methods are still valuable as they are often less costly and have a better detection limit. These culturing methods can be used as a screening tool. Conversely, they can be used to confirm the identity and viability of microorganisms following molecular analyses. To enumerate bacteria via a culture-depending approach, Standard Plate Count is recognized as the standard method. The equation shown in Step 4.2.5 is a more accurate way to determine CFU/g or CFU/ml than averaging CFUs from different dilution factors. It is important to note that all diluents and media throughout the procedures must contain sufficient salt to maintain the viability of halophilic bacteria such as V. parahaemolyticus. Incubation temperature and environment must be optimal for the bacterial growth.
The chromogenic medium is designed to detect Vibrio species that are important human pathogens. Therefore, further studies must be conducted to determine its effectiveness to detect other environmental Vibrio species that are not medically relevant. In future applications, the new chromogenic agar can be incorporated into routine testing of environmental samples for V. parahaemolyticus. This can be done by direct streaking of samples on the chromogenic agar to detect for the presence of presumptive V. parahaemolyticus. The chromogenic agar can also be used to enumerate V. parahaemolyticus following dilutions. Quantification can also be estimated by carrying out a Most Probable Number method using enrichment broth, followed by streaking on the chromogenic agar for confirmation.
Disclosures
Some media were generously provided by Hardy Diagnostics, Santa Maria, CA. Thorsen conducted this study while a student at California Polytechnic State University. He is currently an employee of Hardy Diagnostics.
Acknowledgments
We thank M. Channey, E. Chau, and K. Tomas for their assistance on the project. Project supplies were partially funded by California Polytechnic State University.
References
- Yeung PS, Boor KJ. Epidemiology, pathogenesis, and prevention of foodborne Vibrio parahaemolyticus infections. Foodborne Pathog. Dis. 2004;1(2):74–88. doi: 10.1089/153531404323143594. [DOI] [PubMed] [Google Scholar]
- Yeung PSM, Boor KJ. Epidemiology, molecular biology, and detection of foodborne Vibrio parahaemolyticus infections. In: Faruque SM, editor. Foodborne and Waterborne Bacterial pathogens: Epidemiology, Evolution and Molecular Biology. Caister Academic Press; 2012. pp. 153–184. [Google Scholar]
- Centers for Disease Control and Prevention. Vital Signs: Incidence and Trends of Infection with Pathogens Transmitted Commonly Through Food - Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 1996-2010. Morbidity and Mortality Weekly Report. 2011;10:1996–2010. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Summary of human Vibrio cases reported to CDC, 2008. 2008. Available from: https://stacks.cdc.gov/view/cdc/21591.
- Scallan E, et al. Foodborne illness acquired in the United States - major pathogens. Emerg. Infect. Dis. 2011;17(1):7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baross J, Liston J. Occurrence of Vibrio parahaemolyticus and related hemolytic vibrios in marine environments of Washington State. Appl. Microbiol. 1970;20(2):179–186. doi: 10.1128/am.20.2.179-186.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baross J, Liston J. Isolation of Vibrio parahaemolyticus from the Northwest Pacific. Nature. 1968;217(5135):1263–1264. doi: 10.1038/2171263a0. [DOI] [PubMed] [Google Scholar]
- Kueh CS, Chan KY. Bacteria in bivalve shellfish with special reference to the oyster. J. Appl. Bacteriol. 1985;59(1):41–47. doi: 10.1111/j.1365-2672.1985.tb01773.x. [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration. Bad Bug Book, Foodborne Pathogenic Microorganisms and Natural Toxins. Second Edition 2012.
- Qadri F, et al. Adaptive and inflammatory immune responses in patients infected with strains of Vibrio parahaemolyticus. J. Infect. Dis. 2003;187(7):1085–1096. doi: 10.1086/368257. [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration. Quantitative risk assessment on the public health impact of Vibrio parahaemolyticus in raw oysters. 2005. Available from: http://www.fda.gov/Food/ScienceResearch/ResearchAreas/RiskAssessmentSafetyAssessment/ucm050421.htm.
- Food and Drug Administration. Bacteriological analytical manual online. 2004. Available from: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070830.htm.
- MacFaddin JF. Media for isolation-cultivation-identification-maintenance of medical bacteria. Vol. 1. Baltimore, Md: Williams & Wilkins; 1985. [Google Scholar]
- Bottone EJ, Robin T. Vibrio parahaemolyticus: suspicion of presence based on aberrant biochemical and morphological features. J. Clin. Microbiol. 1978;8(6):760–763. doi: 10.1128/jcm.8.6.760-763.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotz MJ, Tamplin ML, Rodrick GE. Thiosulfate-citrate-bile salts-sucrose agar and its selectivity for clinical and marine vibrio organisms. Ann. Clin. Lab. Sci. 1983;13(1):45–48. [PubMed] [Google Scholar]
- Hara-Kudo Y, Nishina T, Nakagawa H, Konuma H, Hasegawa J, Kumagai S. Improved method for detection of Vibrio parahaemolyticus in seafood. Appl. Environ. Microbiol. 2001;67(12):5819–5823. doi: 10.1128/AEM.67.12.5819-5823.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddabra R, Piemont Y, Scheftel JM. Evaluation of a new chromogenic medium, chromID Vibrio, for the isolation and presumptive identification of Vibrio choleare and Vibrio parahaemolyticus from human clinical specimens. Eur. J. Clin. Microbiol. Infect. Dis. 2011;30(6):733–737. doi: 10.1007/s10096-010-1145-2. [DOI] [PubMed] [Google Scholar]
- Kodaka H, Teramura H, Mizuochi S, Saito M, Matsuoka H. Evaluation of the Compact Dry VP method for screening raw seafood for total Vibrio parahaemolyticus. J. Food. Prot. 2009;72(1):169–173. doi: 10.4315/0362-028x-72.1.169. [DOI] [PubMed] [Google Scholar]
- Su YC, Duan J, Wu WH. Selectivity and specificity of a chromogenic medium for detecting Vibrio parahaemolyticus. J. Food Prot. 2005;68(7):1454–1456. doi: 10.4315/0362-028x-68.7.1454. [DOI] [PubMed] [Google Scholar]
- Bej AK, Patterson DP, Brasher CW, Vickery MC, Jones DD, Kaysner CA. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods. 1999;36(3):215–225. doi: 10.1016/s0167-7012(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Yeung PSM, DePaola A, Kaysner CA, Boor KJ. A PCR assay for specific detection of the pandemic Vibrio parahaemolyticus O3:K6 clone from shellfish. J. Food Sci. 2003;68(4):1459–1466. [Google Scholar]
- Yeung PSM, Hayes MC, DePaola A, Kaysner CA, Kornstein L, Boor KJ. Comparative phenotypic, molecular, and virulence characterization of Vibrio parahaemolyticus O3:K6 isolates. Appl. Environ. Microbiol. 2002;68(6):2901–2909. doi: 10.1128/AEM.68.6.2901-2909.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Su Y-C. Comparison of a chromogenic medium with thiosulfate-citrate-bile salts-sucrose agar for detecting Vibrio parahaemolyticus. J. Food Sci. 2005;70:M125–M128. [Google Scholar]
- Pinto AD, Terio V, Novello L, Tantillo G. Comparison between thiosulphate-citrate-bile salt sucrose (TCBS) agar and CHROMagar Vibrio for isolating Vibrio parahaemolyticus. Food Control. 2011;22(1):124–127. [Google Scholar]
- Canizalez-Roman A, Flores-Villaseñor H, Zazueta-Beltran J, Muro-Amador S, Leòn-Sicairos N. Comparative evaluation of a chromogenic agar medium-PCR protocol with a conventional method for isolation of Vibrio parahaemolyticus strains from environmental and clinical samples. Can J Microbiol. 2011;57(2):136–142. doi: 10.1139/w10-108. [DOI] [PubMed] [Google Scholar]
- Kriem MR, et al. Prevalence of Vibrio spp. in raw shrimps (Parapenaeus longirostris) and performance of a chromogenic medium for the isolation of Vibrio strains. Lett Appl Microbiol. 2015;61(3):224–230. doi: 10.1111/lam.12455. [DOI] [PubMed] [Google Scholar]
- Food Drug Administration. Statistical guidance on reporting results from studies evaluating diagnostic tests. http://www.fda.gov/RegulatoryInformation/Guidances/ucm071148.htm. 2007.
- Burd EM. Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev. 2010;23(3):550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Organisation for Animal Health. Principles and methods of validation of diagnostic assays for infectious diseases. 2013. Available from: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/1.01.05_VALIDATION.pdf.


