Summary
Lipoxygenases are critical enzymes in the biosynthesis of families of bioactive lipids including compounds with important roles in the initiation and resolution of inflammation and in associated diseases such as diabetes, cardiovascular disease and cancer. Crystals diffracting to high resolution (1.9 Å) were obtained for a complex between the catalytic domain of leukocyte 12-lipoxygenase and the isoform specific inhibitor, 4-(2-oxapentadeca-4-yne)phenylpropanoic acid (OPP). In the three-dimensional structure of the complex, the inhibitor occupied a new U-shaped channel open at one end to the surface of the protein and extending past the redox active iron site that is essential for catalysis. In models, the channel accommodated arachidonic acid defining for the first time, the binding site for the substrate of the catalyzed reaction. There was a void adjacent to the OPP binding site connecting to the surface of the enzyme and providing a plausible access channel for the other substrate, oxygen.
Introduction
Polyunsaturated fatty acid metabolism culminates in the formation of numerous bioactive lipids, the eicosanoids, with specific roles in the inflammatory process (Funk, 2001). The prostaglandins and thromboxanes stem from the cyclooxygenase-initiated branch, whereas leukotrienes, lipoxins, HETEs, and other compounds are formed by lipoxygenase inaugurated events. The ultimate pattern of these metabolites has important consequences for human health and disease. Lipoxygenases catalyze the oxygenation of arachidonic acid in both a regiospecific and a stereospecific fashion producing hydroperoxide products primarily at the 5-, 8-, 12-, and 15- positions (Brash, 1999). The catalyzed reaction requires the redox participation of non-heme iron (Funk et al., 1990). The various isoenzymes are not only position specific, but also species, tissue and cell specific, making them difficult to target with inhibitors. It might be possible to design specific inhibitors, but structural studies of mammalian lipoxygenases that would reveal differences that could be exploited for this purpose have been few. There are currently only two examples of mammalian lipoxygenases for which three-dimensional structures were determined.
The three-dimensional structure for a complex between rabbit reticulocyte 15-lipoxygenase and an inhibitor was first published in 1997 (Gillmor et al., 1997). A subsequent analysis of the original X-ray diffraction data led to publication of a more detailed description of the structure in 2008 (Choi et al., 2008). Very recently, the structure of a stabilized human 5-lipoxygenase was reported in the absence of any ligand (Gilbert et al., 2011). Crystallographically determined structures for lipoxygenases from soybeans and coral were also published previously (Minor et al., 1996; Skrzypczak-Jankun et al., 1997; Neau et al., 2009). The lipoxygenases all consist of two domains, a smaller N-terminal β-barrel, known as a membrane-associating C2 domain, and a larger C-terminal domain composed for the most part of long α-helixes, and containing the redox-active iron necessary for the catalyzed reaction. A substantial body of knowledge concerning lipoxygenase catalysis has also been published, based on molecular manipulations, kinetics, and spectroscopic studies of specific isoenzymes (Ivanov et al., 2010; Nagel et al., 2006; Gafney, 1996). However, since 1997, there has been no new structural information regarding ligand-binding for this important class of enzymes. For example, there is no structural basis for something as fundamentally important as how the substrate binds.
Classifying the lipoxygenase sequences is a challenge because the isoenzymes can be both species and cell-type specific. Porcine leukocyte 12-lipoxygenase, the subject of the research in this report, is more like human leukocyte 15-lipoxygenase (Sigal et al., 1988) (86% identical) and rabbit reticulocyte 15-lipoxygenase (O’Prey et al., 1989) (79% identical) than human platelet 12-lipoxygenase (Funk et al., 1990) (66% identical) or human 5-lipoxygenase (Dixon et al., 1988) (41% identical). Because the reticulocyte and leukocyte 12- and 15-lipoxygenases have closely related sequences and catalytic behavior, producing similar ratios of 12- and 15-hydroperoxides under comparable conditions, these isoenzymes are sometimes referred to as 12/15-lipoxygenases (Dobrian et al., 2011). The porcine leukocyte enzyme was first isolated and later cloned for expression in yeast, insect cells, and ultimately E. coli (Yokoyama et al., 1986; Reddy et al., 1994a; Reddy et al., 1994b; Richards and Marnett, 1997; Rapp et al., 2009). A class of isoform specific inhibitors was discovered for this 12-lipoxygenase which led to a thorough kinetic analysis of the effect of 4-(2-oxapentadeca-4-yne)phenylpropanoic acid (OPP) on catalysis (Gorins et al., 1996; Richards, et al. 1999; Moody and Marnett, 2002). The results showed that the compound bound to the free (both Fe(II) and Fe(III)) form of the enzyme and the substrate bound form, all with high affinity.
Crystallization experiments on the mammalian lipoxygenases were previously hampered by an inherent flexibility in the protein. The working hypothesis for the basis for this effect is that the two domains can adopt numerous conformations in solution relative to one another, ranging from closely associated compact structures to minimally associated extended conformations. Evidence for the existence of this degree of flexibility was found in small angle X-ray scattering measurements on several mammalian lipoxygenases (Shang et al., 2011). The effect was also manifest for leukocyte 5-lipoxygenase, for which crystallization was only realized with a version of the protein modified to stabilize a compact configuration (Gilbert et al., 2011). The successful strategy adopted here was to truncate the N-terminal C2 β–barrel domain altogether, and investigate the crystallization behavior of the iron-containing C-terminal domain. There were several reports on truncated lipoxygenases previously, the mini-LOXs, but those constructs tended to have limited stability and were not crystallized (Maccarrone et al., 2004; Walther et al., 2002). A C-terminal domain for porcine leukocyte 12-lipoxygenase was discovered that retained catalytic activity. Crystals for this protein were obtained, but only in the presence of the isoform specific inhibitor, OPP. The crystals diffracted X-rays to high resolution affording a three-dimensional structure for a complex providing numerous insights into substrate-binding and catalysis. The position of the inhibitor in the complex adjacent to the redox-active iron cofactor identified the first substrate-binding site for any lipoxygenase. There was a void connected to the surface of the enzyme providing oxygen access to the site of catalysis. The structure of the complex provides a basis for understanding the regio- and stereo-selectivity of the catalyzed reaction, the structure-function behavior of inhibitors, and the results of numerous molecular manipulations previously conducted on the enzyme.
Results and Discussion
The catalytic domain of 12-lipoxygenase formed crystals with OPP that diffracted X-rays to high resolution
Sparse matrix screening of crystallization conditions did not produce diffraction quality crystals for full-length 12-lipoxygenase or the protein containing any of numerous molecular modifications to amino acids (potentially exposed Cys residues) or clusters of amino acids (surface entropy reduction) carried out to enhance crystallization. Altogether, at least 10,000 trials were performed. The C-terminal domain (residues 112–663) cloned and expressed in E. coli was catalytically active and reasonably stable. A version of the catalytic domain containing an N-terminal six His-tag and serines substituting for cysteines at positions 210 and 292 was screened for crystallization in the presence and absence of the isoform specific inhibitor, OPP. Incubation of the modified 12-lipoxygenase catalytic domain with solid OPP followed by exposure to 10% PEG 20,000 in 0.1 M MES buffer at pH 6.5 containing 20% glycerol resulted in crystals of a complex. While the crystallization occurred consistently under carefully controlled conditions, the omission of any aspect of the protocol resulted in no crystallization. For example, there was a thrombin cleavage site engineered to allow for removal of the N-terminal six His-tag. The protein devoid of the tag did not crystallize under any conditions. Most importantly, no crystallization was ever observed in the absence of the inhibitor.
Large crystals were obtained from the optimized crystallization conditions, but these were invariably multiples that produced overlapping diffraction patterns. Small crystals irradiated through a 20 μm beam collimator on ID-D of the LS-CAT at the Advanced Photon Source provided single-crystal X-ray diffraction patterns extending to 1.9 Å resolution. The structure was determined by the molecular replacement method using the revised structure of 15-lipoxygenase (PDB ID 2P0M) for the starting model. Data collection and refinement values are collected in Table 1.
Table 1.
Data collection and refinement statistics
| 12-Lipoxygenase catalytic domain-inhibitor complex | |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 83.45, 181.54, 91.61 |
| a, b, g (°) | 90.00, 92.86, 90.00 |
| Resolution (Å) | 48.97-1.89 (1.99-1.89)* |
| Rmerge | 0.098 (0.284) |
| I/sI | 9.0 (3.8) |
| Completeness (%) | 99.5 (98.3) |
| Redundancy | 3.6 (3.4) |
| Refinement | |
| Resolution (Å) | 48.97-1.89 (1.94-1.89) |
| No. unique reflections | 214717 (15209) |
| Rwork/Rfree | 0.17/0.21 (0.21/0.25) |
| No. atoms | 18964 |
| Protein | 17580 |
| Ligand | 104 |
| Solvent | 1276 |
| Fe | 4 |
| B-factors | 22.42 |
| Protein | 22.15 |
| Ligand | 26.40 |
| Solvent | 11.86 |
| Fe | 20.41 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.026 |
| Bond angles (°) | 1.974 |
Number of crystals = 1.
Values in parentheses are for highest-resolution shell.
See also Supplementary Figure S1.
The inhibitor was located in a U-shaped channel open on one end to the protein surface
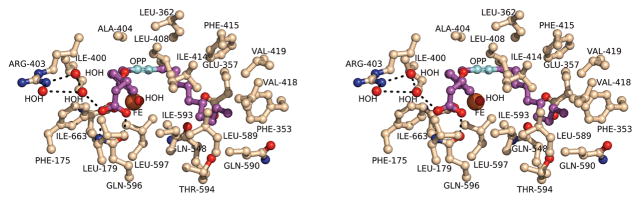
The isoenzyme specific inhibitor, OPP, was found in all four monomers of the protein in the asymmetric unit. In the structure of the complex, OPP was associated with the catalytic domain of 12-lipoxygenase in a U-shaped channel adjacent to the non-heme iron site that was open at one end to the surface of the protein (Figure 1). The carboxylate of the inhibitor formed hydrogen-bonding interactions with Gln-596 at the opening of the channel (Figure 2). Arg-403 was close by, but three water molecules mediate the interaction with the inhibitor. In addition, the occupancies of the carboxylate groups for the OPPs were refined to be 0.5 suggesting that the orientation could be flexible except for the presence of this one dominant conformation. Otherwise, the contacts between OPP and the protein were all hydrophobic interactions. For example, Leu-179 was located in the channel opening in a position to facilitate the penetration of the hydrophobic inhibitor. Gly-407, and Leu-597 extended this hydrophobic surface into the channel. Phe-175 was perpendicular to the aromatic ring of the inhibitor, and at a distance of 3.2 Å was in a position to form a CH-π interaction (Brandl et al., 2001). Ile-400, Leu-408, Ala-404, Ile-593, and Ile-663 were interacting with C10-C15 of OPP. The inhibitor-binding site was terminated in a pocket of hydrophobic residues including Phe-353, Val-418, Val-419, Phe-415, and Ile-414 (Figure 2).
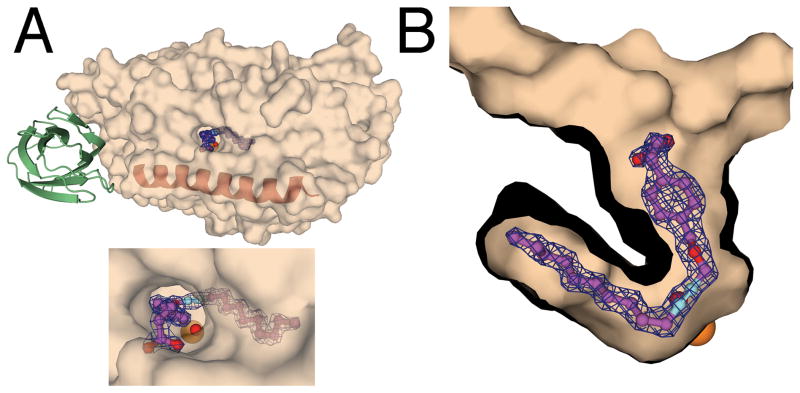
Figure 1.
The location of the OPP binding site in complex with the 12-lipoxygenase catalytic domain. A. A semitransparent surface representation of the catalytic domain indicates where OPP was found in the 2Fo-Fc map of the catalytic domain. The position of the N-terminal domain modeled from the structure of 15-lipoxygenase is presented in green. The maroon helix represents the position of helix α2. B. A cutaway surface representation shows the 2Fo-Fc map and the structure of OPP. The orange sphere indicates the position of the iron atom. The red sphere indicates the position of the iron-associated water/hydroxide. The 2Fo-Fc maps were contoured at 1.5 σ.
Figure 2.
The specific interactions between OPP and the 12-lipoxygenase catalytic domain. A stereodiagram representing the amino acid side chains interacting with OPP.
Arachidonic acid was modeled into the electron density for OPP in the 2Fo-Fc map, illustrating that the U-shaped channel could serve as the substrate-binding site. The position of arachidonic acid generated in this way is illustrated in Figure 3. In this model, C10 was the closest substrate carbon to Fe-OH with a C to O distance of 2.9 Å. The C10-H to FeOH-O distance would be on the order of only 1.8 Å. Kinetic isotope studies predicted a very short distance for the H-atom abstraction reaction for lipoxygenases (Nagel et al., 2006). The orientation of the arachidonic acid molecule in the model was also consistent with the stereochemical outcome for the catalyzed reaction. If hydrogen atom abstraction at C10 took place from the back to be consistent with the location of the FeOH, and the oxygen molecule reacted at C12 from the front as arachidonic acid is oriented in the figure, the result would be the formation of the observed 12(S)-hydroperoxide.
Figure 3.
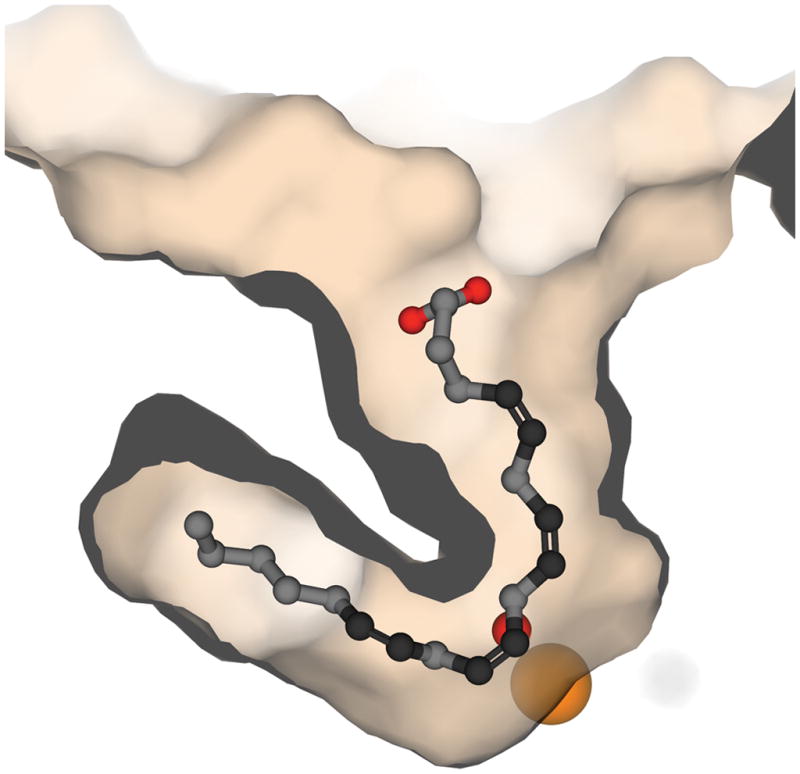
Arachidonic Acid modeled in place of the inhibitor, OPP, indicates the likely orientation for the substrate leading to catalysis.
An internal void in the structure of 12-lipoxygenase provides a path for oxygen from the surface to the active site
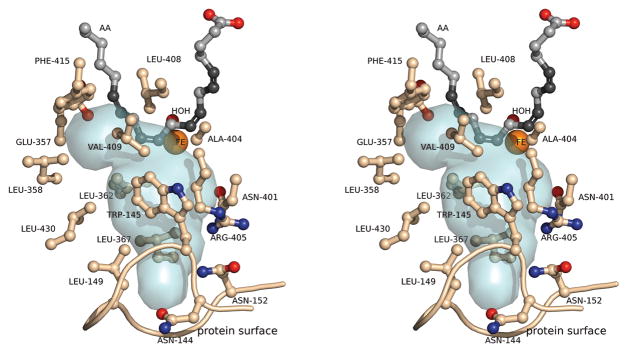
The orientation for the addition of oxygen described in the previous section is consistent with the presence of an extended void in the structure from the non-heme iron site to the surface of the protein that intersects the substrate channel providing a plausible dioxygen pathway for lipoxygenase catalysis (Figure 4). This channel has an amphiphilic character with hydrophilic amino acid side chains close to the surface, and hydrophobic side chains close to the active site. This distribution may facilitate the movement of oxygen along the channel to the substrate-binding site. Two amino acids have been implicated in the delivery of oxygen to the non-heme iron site, Leu-367 and Ala-404. Modification of the leucine at position-367 to phenylalanine led to an increase in KM for oxygen in rate measurements on the rabbit reticulocyte 15-lipoxygenase-catalyzed reaction (Saam et al., 2007). The structure confirms the hypothesis that this amino acid side chain lies in a catalytically relevant path for the oxygen molecule to the active site. The stereochemistry and regiochemistry of the lipoxygenase-catalyzed reaction was sensitive to the presence of alanine (vs. glycine) at position-404 (Cofa et al., 2004). The hypothesis was that the side chain methyl substituent impeded approach of oxygen toward one end of the pentadienyl radical intermediate, e.g. the 8-position, enforcing the observed stereochemical outcome, S, at the 12-position. The model of the three-dimensional structure including arachidonic acid shows that Ala-404 is immediately adjacent to the 8-position of the substrate blocking the access of the proposed oxygen channel to C8 as proposed.
Figure 4.
The proposed oxygen access channel for 12-lipoxygenase. A stereodiagram from the structural model with arachidonic acid in the active site. Arachidonic acid is depicted in gray/black. The void adjacent to the substrate binding channel is a pale blue surface.
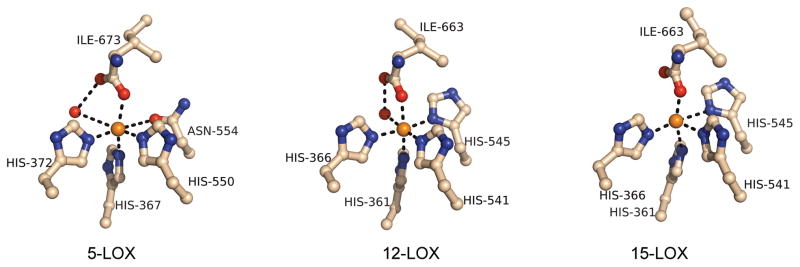
The iron-site in 12-lipoxygenase was a well-organized octahedral ligand field
The redox active, non-heme iron atom in 12-lipoxygenase was coordinated by four histidines (His-361, His-366, His-541, and His-545), the main chain carboxylate from the C-terminus (Ile-663), and one water/hydroxide in a pseudo-octahedral configuration (Figure 5). The coordinated water molecule was also stabilized by a hydrogen bond with the non-ligated oxygen atom (O2) of the terminal carboxylate. This additional bonding resulted in small B-factors for the water molecules (9, 12, 18, and 13 Å2 for the four chains A–D in the asymmetric unit) that were comparable in magnitude to the B-factors of their bonded iron atoms (9, 9, 17, and 12 Å2, respectively). The iron-ligand distances in the crystal structure, which averaged 2.23 ± 0.06 Å, are collected in Table 2 along with values for comparison from 5-lipoxygenase and 15-lipoxygenase. The triple bond of the inhibitor was adjacent to the non-heme site at a distance of roughly 4 Å from the iron. The metal-ligated water molecule was on the channel surface at a distance of 3.05 Å from the triple bond.
Figure 5.
A comparison of the iron coordination environments for the mammalian lipoxygenases.
Table 2.
Iron coordination sphere ligand distances in Å
| 12-LOX | 5-LOX (A) | 5-LOX(B) | 15-LOX (A) | 15-LOX (B) | |
|---|---|---|---|---|---|
| Fe···His-361 | 2.1 | 2.2 | 2.2 | 2.2 | 2.6 |
| Fe···His-366 | 2.2 | 2.1 | 2.3 | 2.4 | 2.1 |
| Fe···His-541 | 2.1 | 2.2 | 2.1 | 2.4 | 2.3 |
| Fe···His-545 | 2.3 | 3.2 | 3.2 | 2.3 | 2.6 |
| Fe···Ile-663 | 2.3 | 2.2 | 2.2 | 2.2 | 2.0 |
| Fe···H2O | 2.2 | 3.6 | none | none | none |
| H2O···Ile-663 | 2.5 | 3.0 | none | none | none |
The residue numbering corresponds to the sequences of 12 and 15-lipoxygenase. The corresponding residues in 5-LOX are His-367, His-372, His-550, Asn-554 and Ile-673, respectively. In both 5- and 15-LOX there are two molecules (A and B) in the asymmetric unit with significant differences in the distances.
The iron site in the 12-lipoxygenase catalytic domain was more ordered than the iron sites in 5- or 15-lipoxygenase. For example, in 5-lipoxygenase a water molecule was not as tightly coordinated to the iron and was not in a position to be as effectively stabilized by hydrogen bonding to the C-terminal Ile, resulting in a B-factor of 36 in one of the monomers in the structural model. No water molecule at all was observed in the non-heme iron site of the second monomer in the 5-lipoxygenase structure, or in the structure of 15-lipoxygenase (Choi et al., 2008; Gilbert et al., 2011). Further, the iron-binding site in 12-lipoxygenase had a distorted octahedral geometry, but significantly less distorted than in the previously reported structures. The average deviation from ideal octahedral geometry for the ligand-iron-ligand angles was 4.5° roughly half the value observed in other structures. What is most striking about the structures of the iron sites in the various lipoxygenases is how different they can be. This trend reinforces the notion that flexibility or mobility of the iron and its coordination sphere represents an aspect of the structure that is relevant to catalysis (Vahedi-Faridi et al., 2004). The 12-lipoxygenase-OPP structure also clearly illustrates that the substrate to iron distance can be very short. This is consistent with the working hypothesis for the mechanism of the catalyzed reaction, which includes H-atom tunneling that can only take place over very short distances (Nagel et al., 2006).
The induced fit mechanism for binding of inhibitor does not apply to porcine leukocyte 12-lipoxygenase
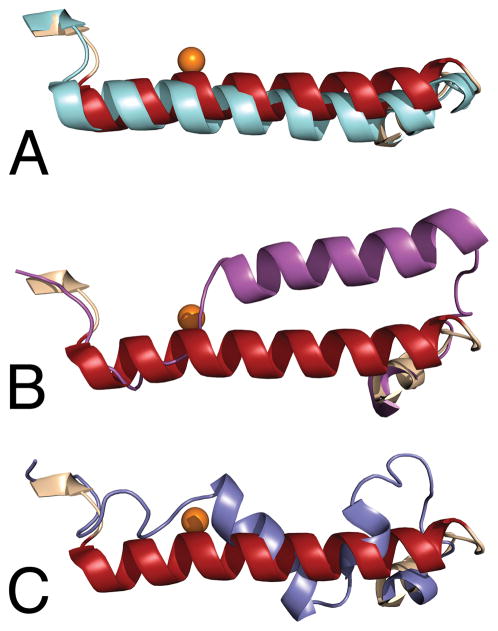
The three dimensional structure of the 12-lipoxygenase catalytic domain was different from the structures of 5- and 15- lipoxygenase (Choi et al., 2008; Gilbert et al., 2011), in an important aspect, the position and conformation of the amino acids making up helix α2. A comparison of the mammalian lipoxygenases in this region of their structures is presented in Figure 6. The reevaluation of the original X-ray diffraction data for rabbit 15-lipoxygenase concluded that the crystals of the enzyme-inhibitor complex contained an open, apo form and a closed form with the inhibitor present near the non-heme iron site (Choi et al., 2008). The differences in the structures were primarily related to the position and conformation of the amino acids that make up helix α2. The position of this helix in the 12-lipoxygenase catalytic domain was much more similar to the location in the open, apo structure of 15-lipoxygenase (Figure 6A) than to the inhibitor bound closed form (Figure 6B). The positions of these amino acids in the 5-lipoxygenase structure were quite distinctive (Gilbert et al., 2011). They were found in a short, three turn helix in a unique orientation flanked on both sides by extended loops (Figure 6C). The orange spheres in the figures represent the positions of the iron atoms, illustrating that these differences in structure were in the portion of the protein covering the active site. The interpretation for the presence of two conformations in 15-lipoxygenase was an induced fit mechanism for inhibitor binding (Choi et al., 2008). The new structure of the 12-lipoxygenase catalytic domain indicated that a conformational change of this kind was not a requirement for inhibitor binding, because OPP was bound directly to the “open” conformation of the protein.
Figure 6.
A comparison of the positions of helix α2 in the structures of the mammalian lipoxygenases. In each panel, the position of the 12-lipoxygenase-OPP complex is in maroon, and the position of the iron atom is indicated by an orange sphere. A. Open conformation: apo 15-lipoxygenase structure in cyan. B. Closed conformation: 15-lipoxygenase-RS75091 complex in magenta. C. 5-Lipoxygenase structure in slate.
The inhibitors in the structures of 12- and 15- lipoxygenases adopted quite different orientations in their respective binding sites. This was consistent with the fact that a significant conformational change was necessary to account for inhibitor binding to 15-lipoxygenase (Choi et al., 2008). A comparison of the orientations of the two inhibitor molecules is provided in Figure 7. Alignment of the two protein structures was conducted using secondary structural elements. The OPP and U-shaped channel for 12-lipoxygenase as a transparent surface representation are shown in approximately the same orientation as in Figure 1B. The relative position of RS75091 in the 15-lipoxygenase structure is shown for comparison. In the structure of 15-lipoxygenase, the inhibitor occupied a large “boot-shaped” cavity that was not connected to the surface of the protein. While OPP and RS75091 were both found close to the iron site, further similarities were not evident. The common structural features of the inhibitors, the carboxylate groups, aromatic rings, and hydrophobic tails were all found in completely different places in the two structures. The geometry enforced by the ortho substitution pattern in RS75091 in all likelihood accounts for the different modes of binding. For example, this feature would prevent entry to the U-shaped channel by direct insertion. The structure of inhibitor-bound 15-lipoxygenase has been used as a starting point in molecular modeling and docking studies for the purpose of inhibitor discovery (Toledo et al., 2010). The structure of the 12-lipoxygenase complex will provide a template for such studies based on a much more mechanistically relevant channel. Further, the results with 12-lipoxygenase and OPP compared to 15-lipoxygenase and RS75091 indicated that there may be a structural basis for distinguishing between isoenzymes with inhibitors.
Figure 7.
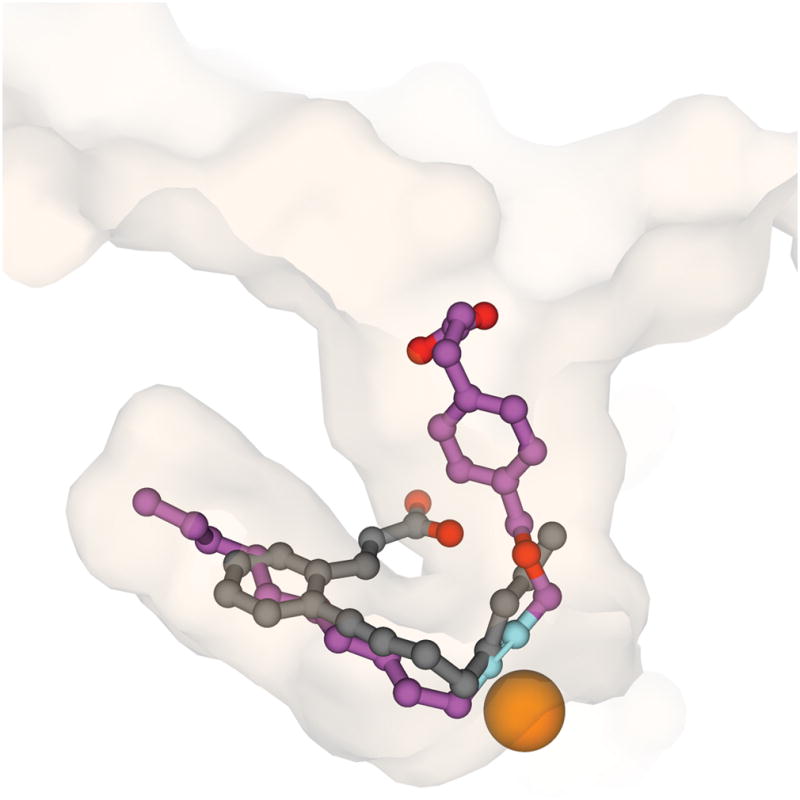
A comparison of the positions of OPP (magenta) in the complex with 12-lipoxygenase in approximately the same orientation as Figure 1B and RS75091 (gray) in the complex with 15-lipoxygenase. The cutaway surface of 12-lipoxygenase is in semi-transparent representation. The surface of 15-lipoxygenase is not shown for clarity.
The structure of the complex is informative regarding the mode of inhibition and the mechanism of the catalyzed reaction
OPP binds to both the Fe(III) and Fe(II) forms of leukocyte 12-lipoxygenase but much more tightly to the Fe(II) form (KIFe(II) = 70 nM; KIFe(III) = 2000 nM) (Moody and Marnett, 2002). The tight-binding to the Fe(II) form accounts for OPP’s mechanism of inhibition, which is preventing activation of the enzyme by hydroperoxide-dependent oxidation of Fe(II) (inactive) to Fe(III) (active) enzyme. This implies that the U-shaped channel is also the site of fatty acid hydroperoxide binding because iron oxidation requires direct contact between the metal center and the hydroperoxide group.
The structure of the bound OPP in the cavity fits beautifully with the structure-activity relationship for inhibition (Gorins et al., 1997). The number of carbons between the carboxylate of OPP and the phenyl ring could be 0–2 but when it was increased to 3, inhibition was lost. The number of carbons between the phenyl ring and the ether oxygen could only be one and the alkyl group could be C8–12 with C10 optimal. Inhibition was lost when the alkyl group was C14. Although the presence of the alkyne conferred optimal activity, trans-olefins and allenes were nearly as active whereas cis-olefins were much less active and saturated hydrocarbons were inactive. Examination of the structure of the OPP-12-lipoxygenase complex reveals that the alkyne is positioned immediately adjacent to the non-heme iron and the phenyl-propionate and long-chain alkyl groups provide maximal interactions between Gln-596 and the end of the U-shaped cavity. Interestingly, 5,8,11,14-eicosatetraynoic acid, the tetra-alkynyl analog of arachidonic acid, acts in a mechanistically similar fashion to OPP by preventing hydroperoxide-dependent activation of porcine leukocyte 12-lipoxygenase.
Experimental Procedures
Construction of the expression plasmid
pET-20-C210SC292S was obtained with the QuikChange multi site-directed mutagenesis kit using primers 5′-CAA CAG GAT TTT CTG GAG TGG CCA GAG CAA GCT-3′ and 5′-GCC AAT GTC ATC CTG AGT AGC CAG CAG TAC C-3′. The cDNA of the lipoxygenase catalytic domain (residues 112 to 663) was amplified by polymerase chain reaction using customized primers 5′-GAG CCT CCA TAT GGG CAC TGC CCG CAC AGT G-3′ and 5′-CTG GCT CGA GTC AGA TGG CCA CAC TGT TTT C-3′, was cleaved with NdeI and XhoI, and was then cloned into pET-28a via NdeI/XhoI restriction sites. The LCD gene was transferred from pET-28-HLCDS to expression vector pET-20b-HLCDS through XbaI/XhoI sites.
Expression and Purification of 12-lipoxygenase catalytic domain
The pET-20b-HLCDS plasmids were transformed into Rosetta 2 (DE3) cells. Freshly grown bacterial cultures (1 mL) from a single colony were first grown in 100 mL Lysogeny Broth (LB) containing ampicillin (100 μg mL−1) and chloramphenicol (34 μg mL−1) 4 h (OD600~ 1.2), and the entire culture was combined with 1000 mL Auto-induction Lysogeny Broth (Auto-LB) medium (Studier, 2005) [10 g tryptone, 5 g yeast extract, 3.3 g (NH4)2SO4, 6.8 g KH2PO4, Na2HPO4 7H2O 13.4 g, Glucose 0.5 g, α-lactose 2.0 g, MgSO4 0.15 g] with ampicillin (100 μg mL−1) and chloramphenicol (34 μg mL−1), and the cells were incubated 24 h at 30 °C with shaking (200 rpm). The cells were collected by centrifugation and re-suspended in phosphate buffered saline and collected again by centrifugation. The cells were opened by sonication in 200 mL lysis buffer containing, 10 mM Tris HCl, 1 mM TCEP, 60 μg mL−1 chicken egg white type II-O trypsin inhibitor, 100 μg mL−1 catalase, 20 μg mL−1 DNase I, pH 7.4 and 0.5 mg mL−1 chicken egg white lysozyme, and the supernatant of the lysate was obtain by ultracentrifugation at 33000 g. The supernatant was then loaded to a Ni-NTA column (bed volume ~5 mL) and washed with 100 mM TrisHCl pH 7.5, 10 mM imidazole, and eluted with 100 mM TrisHCl pH 7.5, 200 mM imidazole. The elution was dialyzed against 10 mM TrisHCl, 1mM TCEP buffer overnight and further purified on a Source 15Q (GE Life Science) anion exchange column using 0 to 40% gradient of 10 mM TrisHCl, 1 M NaCl. The chromatographic procedures were conducted on an AKTA FPLC (Amersham Biosciences) operated at 4 °C. An enzymatic assay was used to find the active fractions, and the purity of the isolated protein was evaluated by SDS PAGE in 10% Bis-Tris mini gels (NuPAGE, Invitrogen) using MES SDS running buffer.
Enzymatic assay
The maximum rate of the lipoxygenase-catalyzed reaction with arachidonic acid as substrate was determined spectrophotometrically at 234 nm in 3 mL of assay buffer (50 mM Tris-Cl, 0.03% Tween-20, pH 7.4) at 25 °C as previously described (22). Arachidonic acid was dissolved in absolute ethanol and was delivered by airtight 50 μL syringe. All final assay solutions contained 1% ethanol (v/v). The specific activity for the catalytic domain was 1.55 ± 0.38 μmol mg−1 min−1 compared with 8.14 ± 0.59 μmol mg−1 min−1 for full length 12-lipoxygenase prepared in the same fashion, or about 21% on a per molecule basis.
Crystallization and data collection
Purified protein was concentrated to 6 mg/mL using Millipore Amicon 50 kDa MCO filtration devices and then incubated with solid OPP for 16 hours. This solution was then further concentrated to 11~13 mg/mL. Crystals were grown by hanging-drop vapor diffusion method at 291 K by mixing 2 uL protein inhibitor and 2 μL reservoir solution containing 0.1 M MES pH 6.5, 5%~10% PEG-20,000, 20% glycerol. The crystals were flash frozen for shipment and data collection.
Structure Determination
Diffraction data were collected at 100 K at LS-CAT beamline 21-ID-D (λ=1.0781 Å) at the Advanced Photon Source, Argonne, Illinois using 20 μm beam collimator. Data were processed with iMosflm (Battye et al., 2011) and reduced by SCALA (Bailey, 1994). Crystals belonged to space group P21 (a = 83.450 Å, b=181.540 Å, c=91.610 Å, and β=92.86°). There were four monomers in one asymmetric unit (Table 1). The rms deviations for the alpha carbons in any two of the molecules ranged from 0.2188 to 0.3904 Å. There were no significant structural differences evident among the monomers. The C-terminal part of rabbit 15-lipoxygenase structure (PDB ID 2P0M, residues 112 to 663 of chain A) was used as the search model. Molecular replacement was performed with Phaser (McCoy et al., 2007). Mutations and manual model building were performed in COOT (Emsley et al., 2010). Refinement was continued in REFMAC with rigid body refinement and restrained refinement (Potterton et al., 2003; Skubak et al., 2004). Non-crystallographic symmetry was not applied during the refinement. The potential of phase bias was excluded by simulated annealing. Ligand was built using SMILE and waters were adding from COOT (Emsley et al., 2010). The simulated annealing OMIT maps were generated with Phenix (Adams et al., 2010). The void of the dioxygen channel was calculated with a sphere size of 1.2 Å using the program HOLLOW (Ho and Gruswitz, 2008). Ligand occupancy refinement was carried out in Phenix (Adams et al., 2010). An example of the Fo-Fc electron density for OPP is presented in Supplementary Figure S1. The final model has Rwork and Rfree 17.3 and 21.4, respectively (Table 1). The values of the Ramachandran plot for the final refinement of the structure were obtained with PROCHECK (92.3% most favored, 7.5% additional allowed, 0.2% generously allowed, and 0% outliers) (Laskowski et al., 1993). The illustrations were prepared with Pymol Molecular Graphics System, Version 1.3 (Schrödinger, LLC). All of the figures contain representations of monomer A.
Supplementary Material
Highlights.
The first crystal structure of 12-lipoxygenase was solved at high resolution
The position of an inhibitor identified the substrate binding/active site
A channel providing access for oxygen to the active site was also discovered
The structure accounts for virtually all features of catalysis and inhibition
Acknowledgments
This research was supported by funding from the National Institutes of Health (HL091482). The able assistance of the staff of LS-CAT at the Advanced Photon Source – Argonne National Laboratory, Keith Brister, Joseph Brunzelle, and David Smith, is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S. The CCP4 Suite – programs for protein crystallography. Acta Cryst D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Battye TGG, Kontogiannis L, Johnson O, Powella HR, Lesliea AGW. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Cryst D. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl M, Weiss MS, Jabs A, Sühnel J, Hilgenfeld R. C-H • • • π-interactions in proteins. J Mol Biol. 2001;307:357–377. doi: 10.1006/jmbi.2000.4473. [DOI] [PubMed] [Google Scholar]
- Brash AR. Lipoxygenases: Occurrence, functions, catalysis and acquisition of substrate. J Biol Chem. 1999;274:23679–23682. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- Choi J, Chon JK, Kim S, Shin W. Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data. Proteins Struct Funct Bioinf. 2008;70:1023–1032. doi: 10.1002/prot.21590. [DOI] [PubMed] [Google Scholar]
- Coffa G, Brash AR. A single active site residue directs oxygenation stereospecificity in lipoxygenases: stereocontrol is linked to the position of oxygenation. Proc Natl Acad Sci USA. 2004;101:15579–15584. doi: 10.1073/pnas.0406727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RAF, Jones RE, Diehl RE, Bennett CD, Kargman S, Rouzer CA. Cloning of the cDNA for human 5-lipoxygenase. Proc Natl Acad Sci USA. 1988;85:416–420. doi: 10.1073/pnas.85.2.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrian AD, Lieb DC, Cole BK, Taylor-Fishwick DA, Chakrabarti SK, Nadler JL. Functional and pathological roles of the 12- and 15-lipoxygenases. Prog Lipid Res. 2011;50:115–131. doi: 10.1016/j.plipres.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and Development of Coot. Acta Cryst D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk CD, Furci L, FitzGerald GA. Molecular cloning, primary sructure, and expression of the human platelet/erythroleukemia cell 12-lipoxygenase. Proc Natl Acad Sci USA. 1990;87:5638–5642. doi: 10.1073/pnas.87.15.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;249:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- Funk MO, Carroll RT, Thompson JF, Sands RH, Dunham WR. Role of iron in lipoxygenase catalysis. J Am Chem Soc. 1990;112:5375–5376. [Google Scholar]
- Gafney BJ. Lipoxygenases: structural principles and spectroscopy. Ann Rev Biophys, Biomol Struct. 1996;25:431–459. doi: 10.1146/annurev.bb.25.060196.002243. [DOI] [PubMed] [Google Scholar]
- Gilbert NC, Bartlett SG, Waight MT, Neau DB, Boeglin WE, Brash AR, Newcomer ME. The structure of human 5-lipoxygenase. Science. 2011;331:217–219. doi: 10.1126/science.1197203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor SA, Villasenor A, Fletterick R, Sigal E, Browner MF. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity. Nature Struct Biol. 1997;4:1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- Gorins G, Kuhnert L, Johnson CR, Marnett LJ. (Carboxyalkyl)benzyl propargyl ethers as selective inhibitors of leukocyte-type 12-lipoxygenases. J Med Chem. 1996;39:4871–4878. doi: 10.1021/jm9606047. [DOI] [PubMed] [Google Scholar]
- Ho BK, Gruswitz F. HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I, Heydeck D, Hofheinz K, Roffeis J, O’Donnell VB, Kuhn H, Walther M. Molecular enzymology of lipoxygenases. Arch Biochem Biophys. 2010;503:161–174. doi: 10.1016/j.abb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Maccarrone M, Di Venere A, van Zadelhoff G, Mei G, Veldink G, Rosato N, Finazzi-Agro A. Further structural and functional properties of mini-lipoxygenase, an active fragment of soybean lipoxygenase-1. Spectroscopy. 2004;18:331–338. [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Steczko J, Stec B, Otwinowski Z, Bolin JT, Walter R, Axelrod B. Crystal structure of soybean lipoxygenase L-1 at 1.4 angstrom resolution. Biochemistry. 1996;35:10687–10701. doi: 10.1021/bi960576u. [DOI] [PubMed] [Google Scholar]
- Moody JS, Marnett LJ. Kinetics of inhibition of leukocyte 12-lipoxygenase by the isoform-specific inhibitor 4-(2-oxapentadeca-4-yne)phenylpropanoic acid. Biochemistry. 2002;41:10297–10303. doi: 10.1021/bi020320q. [DOI] [PubMed] [Google Scholar]
- Nagel ZD, Klinman JP. Tunneling and dynamics in enzymatic hydride transfer. Chem Rev. 2006;106:3095–3118. doi: 10.1021/cr050301x. [DOI] [PubMed] [Google Scholar]
- Neau DB, Gilbert NC, Bartlett SG, Boeglin W, Brash AR, Newcomer ME. The 1.85 angstrom structure of an 8R-lipoxygenase suggests a general model for lipoxygenase product specificity. Biochemistry. 2009;33:7906–7915. doi: 10.1021/bi900084m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Prey J, Chester J, Thiele BJ, Janetzki S, Prehn S, Fleming J, Harrison PR. The promoter structure and complete sequence of the gene encoding the rabbit erythroid cell-specific 15-lipoxygenase. Gene. 1989;84:493–499. doi: 10.1016/0378-1119(89)90526-x. [DOI] [PubMed] [Google Scholar]
- Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Cryst D. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Rapp J, Xu S, Sharp AM, Griffith WP, Kim YW, Funk MO. EPR spectroscopy and electrospray ionization mass spectrometry reveal distinctive features of the iron site in leukocyte 12-lipoxygenase. Arch Biochem Biophys. 2009;490:50–56. doi: 10.1016/j.abb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy RG, Yoshimoto T, Yammamoto S, Funk CD, Marnett LJ. Expression of porcine leukocyte 12-lipoxygenase in a baculovirus/insect cell system and its characterization. Arch Biochem Biophys. 1994a;312:219–226. doi: 10.1006/abbi.1994.1302. [DOI] [PubMed] [Google Scholar]
- Reddy RG, Yoshimoto T, Yamamoto S, Marnett LJ. Expression, purification, and characterization of porcine leukocyte 12-lipoxygenase produced in the methylotrophic yeast Pichia pastoris. Biochem Biophys Res Commun. 1994b;205:381–388. doi: 10.1006/bbrc.1994.2676. [DOI] [PubMed] [Google Scholar]
- Richards KM, Marnett LJ. Leukocyte 12-lipoxygenase: Expression, purification, and investigation of the role of methionine residues in turnover-dependent inactivation and 5,8,11,14-eicosatetraynoic acid inhibition. Biochemistry. 1997;36:6692–6699. doi: 10.1021/bi963051a. [DOI] [PubMed] [Google Scholar]
- Richards KM, Moody JS, Marnett LJ. Mechanism of inhibition of porcine leukocyte 12-lipoxygenase by the isoform-specific inhibitor 4-(2-oxapentadeca-4-yne)phenylpropanoic acid. Biochemistry. 1999;38:16529–16538. doi: 10.1021/bi991336s. [DOI] [PubMed] [Google Scholar]
- Saam J, Ivanov I, Walther M, Hermann-Georg H, Kuhn H. Molecular dioxygen enters the active site of 12/15-lipoxygenase via dynamic oxygen access channels. Proc Natl Acad Sci USA. 2007;104:13319–13324. doi: 10.1073/pnas.0702401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang WF, Ivanov I, Svergun DI, Borbulevych OY, Aleem AM, Stehling S, Jankun J, Kuhn H, Skrzypczak-Jankun E. Probing dimerization and structural flexibility of mammalian lipoxygenases by small-angle X-ray scattering. J Mol Biol. 2011;409:654–668. doi: 10.1016/j.jmb.2011.04.035. [DOI] [PubMed] [Google Scholar]
- Sigal E, Craik CS, Highland E, Grunberger D, Costello LL, Dixon RAF, Nadel JA. Molecular cloning and primary structure of human 15-lipoxygenase. Biochem Biophys Res Commun. 1988;157:457–464. doi: 10.1016/s0006-291x(88)80271-7. [DOI] [PubMed] [Google Scholar]
- Skrzypczak-Jankun E, Amzel LM, Kroa BA, Funk MO. Structure of soybean lipoxygenase L3 and a comparison with its L1 isoenzyme. Proteins Struct Funct Genet. 1997;29:15–31. [PubMed] [Google Scholar]
- Skubak P, Murshudov GN, Pannu NS. Direct incorporation of experimental phase information in model refinement. Acta Cryst D. 2004;60:2196–2201. doi: 10.1107/S0907444904019079. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expression Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Toledo L, Masgrau L, Marechal JD, Lluch JM, Gonzalez-Lafont A. Insights into the mechanism of binding of arachidonic acid to mammalian 15-lipoxygenases. J Phys Chem B. 2010;114:7037–7046. doi: 10.1021/jp912120n. [DOI] [PubMed] [Google Scholar]
- Vahedi-Faridi A, Brault PA, Shah P, Kim YW, Dunham WR, Funk MO. Interaction between non-heme iron of lipoxygenases and cumene hydroperoxide: Basis for enzyme activation, inactivation, and inhibition. J Am Chem Soc. 2004;126:2006–2015. doi: 10.1021/ja0390855. [DOI] [PubMed] [Google Scholar]
- Walther M, Anton M, Wiedmann M, Fletterick R, Kuhn H. The N-terminal domain of the reticulocyte-type 15-lipoxygenase is not essential for enzymatic activity but contains determinants for membrane binding. J Biol Chem. 2002;277:27360–27366. doi: 10.1074/jbc.M203234200. [DOI] [PubMed] [Google Scholar]
- Yokoyama C, Shinjo F, Yoshimoto T, Yamamoto S, Oates JA, Brash AR. Arachidonate 12-lipoxygenase purified from porcine leukocytes by immunoaffinity chromatography and its reactivity with hydroperoxyeicosatetraenoic acids. J Biol Chem. 1986;261:6714–6721. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.