Abstract
Caspases are the key mediators of apoptotic cell death via their proteolytic activity. When caspases are activated in cells to levels detectable by available technologies, apoptosis is generally assumed to occur shortly thereafter. Caspases can cleave many functional and structural components to cause rapid and complete cell destruction within a few minutes. However, accumulating evidence indicates that in normal healthy cells the same caspases have other functions, presumably at lower enzymatic levels. Studies of non-apoptotic caspase activity have been hampered by difficulties with detecting low levels of caspase activity and with tracking ultimate cell fate in vivo. Here, we illustrate the use of an ultrasensitive caspase reporter, CaspaseTracker, which permanently labels cells that have experienced caspase activity in whole animals. This in vivo dual color CaspaseTracker biosensor for Drosophila melanogaster transiently expresses red fluorescent protein (RFP) to indicate recent or on-going caspase activity, and permanently expresses green fluorescent protein (GFP) in cells that have experienced caspase activity at any time in the past yet did not die. Importantly, this caspase-dependent in vivo biosensor readily reveals the presence of non-apoptotic caspase activity in the tissues of organ systems throughout the adult fly. This is demonstrated using whole mount dissections of individual flies to detect biosensor activity in healthy cells throughout the brain, gut, malpighian tubules, cardia, ovary ducts and other tissues. CaspaseTracker detects non-apoptotic caspase activity in long-lived cells, as biosensor activity is detected in adult neurons and in other tissues at least 10 days after caspase activation. This biosensor serves as an important tool to uncover the roles and molecular mechanisms of non-apoptotic caspase activity in live animals.
Keywords: Molecular Biology, Issue 117, Caspase, biosensor, Drosophila, apoptosis, non-apoptotic, brain, neurons, CaspaseTracker
Introduction
Caspases are cysteine proteases that mediate apoptotic cell death by cleaving many intracellular proteins after key aspartate residues. For example, initiator caspases activate effector caspases, derepress DNA nucleases, cleave cytoskeletal components and alter the lipid composition of cell membranes to rapidly dismantle cells and stimulate their recognition and engulfment by neighboring cells that dispose of the cell corpses.1-4 It is estimated that billions of cells die per day in the human body, and apoptosis is an important mechanism of chemotherapy-induced tumor cell death.5 A different set of caspases can cause cell death by distinct non-apoptotic processes to stimulate innate immunity.6 Therefore, most research on caspases has focused on their pro-death functions.
Interestingly, early evidence in the field revealed that the same caspases responsible for promoting cell death also have non-death functions. Pioneering studies have demonstrated that caspases are involved in diverse cellular functions in healthy cells, including the regulation of cell proliferation and migration during embryogenesis.7-9 Caspases are required for spermatid individualization in Drosophila10,11, for blocking an alternative necroptotic cell death pathway in mice12,13, and for microRNA processing in C. elegans.14,15 In perhaps the longest-lived cells, neurons, caspases and other apoptotic machinery are implicated in the regulation of neuronal activity by pruning synaptic endings, a process believed to be essential to strengthen other synapses for learning and memory.16-18 It is possible that caspases facilitate synaptic pruning by a type of mini-apoptosis of tiny neuronal projections without whole cell death.19 However, caspases may have alternative functions unrelated to apoptosis-like events.20,21 Dual roles in life and death are not unique to caspases; BCL-2 family proteins and cytochrome c have roles in cellular energetics in healthy cells but are also part of the core apoptotic pathway that is activated by many types of cell stress.22-25 Although not proven, it seems logical that evolution has linked day-jobs to death-jobs within the same molecules to ensure timely elimination of unfit or undesirable cells.
At present, the molecular mechanisms of non-apoptotic caspase activity are not understood, and the extent of non-apoptotic caspase activity during embryonic development and in adult tissues is also not known. A major challenge is the difficulty in distinguishing day-jobs from death-jobs of caspases. In contrast to apoptosis and pyroptosis, when caspase activity is amplified by a proteolytic cascade, the day-jobs of caspases are expected to occur at much lower levels of enzymatic activity, likely below detection by many available technologies.
Prior to the work presented here, others developed a variety of caspase biosensors for different purposes. The SCAT biosensors (e.g., ECFP-DEVD-Venus) rapidly detect real-time caspase activity in cultured cells and animal tissues using FRET.26,27 Upon caspase cleavage, the nuclear-targeted GFP moiety of Apoliner (mCD8-RFP-DQVD-nucGFP) undergoes subcellular relocalization within minutes when its plasma membrane-tether is cleaved by caspases.28 Similarly, ApoAlert-pCaspase3-Sensor (NES-DEVD-YFP-NLS) relocalizes from the cytosol to the nucleus upon caspase cleavage.29,30 More recently, the chromophore in iCasper was cleverly engineered to fluoresce when cleaved by caspases, permitting detection of biosensor activity in real time in neurons of Drosophila embryos, but primarily in association with developmental cell death.31 Caspase-dependent death of olfactory neurons during aging was demonstrated by immuno-detection of the caspase-cleaved form of CPV biosensors (e.g., mCD8-PARP-Venus).32,33 Importantly, the activated form of caspase-3 was detected in the absence of cell death by sensitive immunostain in spines of cultured neurons, and in the soma using the caspase-dependent fluorescence of the nuclear CellEvent reporter dye, but difficulties were encountered due to photo-toxicity, although cell death was delayed until after spine elimination.19 Thus, new caspase biosensors are needed to detect and track cells with basal caspase activity in vivo.
To overcome these difficulties, we generated a novel dual color caspase biosensor, designated CaspaseTracker. This strategy combines a modified version of the Drosophila caspase-sensitive Apoliner biosensor28 with the Drosophila G-TRACE FRT recombinase system34 to permanently label and track cells in vivo.35 The Gal4-activated G-TRACE system allows very low levels of caspases to activate CaspaseTracker, resulting in RFP expression in the cytoplasm and permanent nuclear-targeted GFP expression in any cell that has ever experienced caspase activity.35 This system can label cells throughout life in whole animals using Drosophila melanogaster, a tractable and widely used model system for the study of caspases and cell death.36-38
Protocol
1. Preparation of CaspaseTracker Flies
To prepare CaspaseTracker (DQVD) flies for experiments, perform this cross: UBI-CaspaseTracker x G-TRACE (UAS-RFP; UAS-FLP; Ubi>Stop>GFP-nls), by transferring 7-10 virgin female (or male) flies carrying the caspase biosensor substrate mCD8-DIAP1-Gal4 driven by the ubiquitin promoter35 together with the same number of male (or female) G-TRACE flies, which have the second chromosome CyO balancer to avoid lethality of the homozygous combination of G-TRACE (UAS-RFP, UAS-FLP, and Ubi>Stop>GFP-nls)34. Place flies in a fresh food vial with fresh yeast paste as a protein source.
Incubate files at 18 °C for 5 to 7 days (maximum 2 weeks) and then remove the parent flies from the vial to avoid overcrowding with new progeny, and to set up new breeding vials.
Continue incubation at 18 °C until progeny flies eclose. Select the non-CyO [non-curly wing] progeny of the correct genotype of CaspaseTracker, which are transgenic for CaspaseTracker and G-TRACE elements35.
Simultaneously, generate control parental non-transgenic w118 flies and caspase-insensitive (DQVA) flies in parallel to verify specific CaspaseTracker RFP and GFP fluorescence.
Perform PCR genotyping to confirm files with CaspaseTracker using the primers: 5'-TCCCCCGGGCTGCAGGAATTC, 3'-TGGAATTGGGGTACGTCTAGA, producing a 3,897 bp product. For genotyping the G-Trace loci, use the following primers for GFP (474 bp), 5'-CAC GAC TTC TTC AAG TCC GCC ATG CCC G, 3'-CTT GTA CAG CTC GTC CAT GCC GAG AGT GAT C; for RFP (258 bp), 5'-GGC TGC TTC ATC TAC AAG GTG AAG TTC ATC GG, 3'-GAT GTC CAG CTT GGA GTC CAC GTA GTA GTA GC; and for Flpase (655 bp), 5'- CCACCTAAGGTGCTTGTTCGTCAGTTTGTGG, 3'-GCC TAC TAA CGC TTG TCT TTG TCT CTG TCA C. For genotyping Gal4 in the pUWR vector (554 bp), 5'-GAA GCA CAC CTT CGC ATC GCT CAG TCA CGC, 3'-TGG AAT TGG GGT ACG TCT AGA.
2. Tissue Preparation, Staining and Mounting
For fly dissections, prevent dissected tissues from sticking to plastic pipette tips and centrifuge tubes, coat tips and tubes with 1% bovine serum albumin (BSA) dissolved in water.
- Dissect flies on a silicone cushion using forceps.
- To avoid damaging the tips of forceps on a hard surface when performing the tissue dissection, perform dissection on a silicon surface. For making silicone dissection plates, melt the silicon in the commercial kit according to manufacturer instructions (Materials List). After melting the silicone, add 3 to 4 mL to a 60 mm diameter tissue culture dish. Silicon dishes can be reused.
- Anesthetize a fly with CO2, and transfer it to a silicone plate with 1 mL of cold PBS. Introduce the CO2 to a vial containing the fly using a blowgun, and then transfer the anesthetized fly to a Pad that has a CO2 supply.
- Use a pair of forceps to hold the fly head, and a second pair of forceps to pull the thorax in the opposite direction to disconnect the fly head from the body covering without damaging the connection between the head and foregut.
- Use 2 pairs of forceps to remove the wings and legs from the thorax.
- Use a pair of forceps to hold the thorax, and another pair of forceps to pull the abdomen to separate it from the thorax again without damaging the connection between the foregut and midgut.
- Dissect internal organs including brain, gut, malpighian tubules and ovaries as previously demonstrated.39-41
- Remove all pieces of the cuticle and the fat bodies with forceps as these tissues produce strong autofluorescence. Patience and practice are required to prepare a complete organ system as shown.
Transfer dissected tissues to 1.5 mL centrifuge tubes, and fix tissues with 0.5 mL of 4% paraformaldehyde in PBS (phosphate-buffered saline) at RT for 20 to 30 min with rotation in the dark to avoid bleaching RFP and GFP in the tissues. Avoid prolonged fixation that can compromise subsequent staining results.
Remove the paraformaldehyde by gentle pipetting and wash 3 times with 0.5 mL PBST (PBS + 0.1% Triton X-100) at RT.
- Permeabilize the tissues with 0.5 mL PBST at 4 °C overnight with gentle shaking. Shorter incubations may cause incomplete permeabilization and compromise staining results.
- If it is necessary to reduce background staining, consider incubating tissues in 0.5 mL PBST with 1% BSA, instead of PBS.
Wash the tissues 3 times with 0.5 mL PBST.
- To stain nuclei and filamentous actin (F-actin), apply 0.5 mL PBST with 10 µg/mL of Hoechst 33342 blue nuclear dye and 0.3 µM Alexa Fluor 633 Phalloidin F-actin stain and incubate simultaneously with tissues for 1 h at RT with gentle rotation in the dark. Avoid prolonged staining as this will increase the background.
- For immunostaining, incubate fixed and permeabilized tissues with 0.1% Triton X-100 in 0.5 mL PBS overnight at 4 °C, stain with 1:100 dilution of anti-ELAV antibody or with 1:200 dilution of anti-caspase-3 overnight at 4 °C (300 µL). Follow by the corresponding secondary antibody diluted 1:100 and incubate for 1 to 3 h at RT. See Materials List for specific antibody information.
Wash the tissues 3 times, each for 5 min in 0.5 mL PBST with gentle rotation.
With a pipet, remove all PBST, and then add 200 µL anti-bleach mounting agent (see materials) to completely cover tissues for 1-3 h at RT, or 4 °C overnight. Optimal tissues that have fully absorbed the mounting agent will sink to the bottom of the tube.
Pre-clean the glass slide with water or 70% ethanol and transfer the tissues with mounting agent to the glass slide.
Carefully place a glass cover slip over the tissues. Apply petroleum jelly to the glass slide and the cover slip to avoid destroying the tissue by overcompression. Remove extra mounting agent with tissue paper.
Seal the cover slip by applying nail polish all along the edges of the cover slip to avoid leakage of mounting agents from the tissues.
3. Confocal Microscopy
Use an inverted confocal epi-fluorescence microscope. However, up-right microscopes can be also used. To image tissues using tiling, maintain stable room temperature to avoid drift of focus and shift of x-y plane due to the thermal expansion and contraction of the components. Environmental control systems and focus drift compensation systems help to avoid this problem due to loss of thermo-equilibrium of the microscope system.
Place the slide on the stage of the microscope. Take a few test images using low resolution (125 x 125 pixels) and determine the appropriate number of tiles needed to cover the entire region of interest (ROI).
Set the microscope scanning system to capture images with scanning resolution such as 500 x 500 pixels. Select the objective such as a 20X NA 0.8 or a 63X NA 1.4 Plan-Apochromat objective for analyzing tissues through glass coverslips, so that each of the images (tiles) overlaps by 20% on all sides.
Set up the image sequence from longest to shortest excitation wavelengths, as the long wavelength light causes lower photo-bleaching. The sequence from longest to shortest excitation wavelength is: (i) 633 nm for Phalloidin F-actin and for antibody to activated caspases, (ii) 561 nm for RFP, (iii) 488 nm for GFP, and (iv) 405 nm for Hoechst nuclear stain.
During imaging, minimize exposure of cells to fluorescent laser excitation during imaging process to avoid photo-bleaching by reducing the laser intensity to obtain high quality images of tissues.
After images are collected, merge the images using microscope software.
Representative Results
There are two key components that allow CaspaseTracker to detect caspase activity in normal healthy cells (Figure 1a). The first of these is a 146 amino acid caspase-cleavable polypeptide modeled after the caspase biosensor Apoliner (Figure 1b).28 This polypeptide is derived from DIAP1 (Drosophila inhibitor of apoptosis) containing a single naturally occurring caspase site that is cleaved during apoptosis typically by the caspase DrICE.42,43 DrICE is equivalent to caspase-3 in mammals and cleaves most known cellular substrates.4,32 Characteristic of caspases, DIAP1 and its derived polypeptide are cleaved after a specific aspartic acid, Asp20 located within the caspase recognition sequence 17-DQVD-20, and mutation of the obligatory Asp20 residue to Ala (DQVA) abolishes cleavage.42,43 Similar to Apoliner,28 this DIAP1 fragment is anchored in the cytoplasm at the plasma membrane via mouse CD8 (alpha chain amino acids 1-220), a commonly used tool in Drosophila. The CD8 membrane anchor prevents any tethered cargo bearing a nuclear translocation sequence (nucGFP in the case of Apoliner) from translocating to the nucleus in the absence of caspase activity.28 The second key component of CaspaseTracker is the Drosophila G-TRACE system in which the yeast transcription factor Gal4 induces the expression of flippase (FLP) recombinase (and simultaneously induces RFP).34 Flippase excises a stop codon leading to permanent expression of nucGFP. To make G-TRACE responsive to caspase activity, it was combined with the Apoliner system by tethering Gal4, which is required to activate G-TRACE, to the plasma membrane-anchored caspase-cleavable DIAP1 fragment of Apoliner (Figure 1a).35
We made additional modifications to the Apoliner component of CaspaseTracker to improve utility. Most important, it is critical that the caspase biosensor itself does not inhibit caspases, potentially preserving cells that would otherwise die. Although the Apoliner transgene did not cause obvious phenotypes in Drosophila28, as would be expected of a potent caspase inhibitor, even the occasional preservation of cells would defeat the purpose of identifying normal cells with caspase activity. However, the DIAP1 fragment in Apoliner contains a motif (135-DICG-138) that was subsequently shown to potently inhibit DrICE, the same caspase that cleaves DIAP1 at Asp20.43 The mechanism of caspase inhibition was revealed by a crystal structure in which DIAP1 Asp135 is bound into the DrICE active site as a caspase substrate mimic (but is not cleaved).43 Biochemical analysis demonstrated that an Arg substitution at Asp135 abolishes DrICE-inhibitory function.43 Therefore, CaspaseTracker was engineered with the same D135R mutation to avoid caspase inhibition (Figure 1b).35 Similar to Apoliner, the naturally occurring DIAP1 Asn-Asn sequence at the de novo N-terminus following cleavage at Asp20 was changed to Gly-Val in CaspaseTracker to prevent degradation of Gal4 by the N-end rule upon caspase cleavage (Figure 1b).28 In addition, we fused a myc-tag to the C-terminus of Gal4 for ready detection of CaspaseTracker expression.35 Of necessity, Apoliner's own RFP and nucGFP cassettes were deleted to make it compatible with G-TRACE, which also contains RFP and nucGFP.28,35
Because Gal4 bears its own nuclear localization signal and potently activates transcription via its target DNA response element (UAS) when fused to other transgenes in Drosophila, presumably only a few molecules of Gal4 released by cytoplasmic caspases are sufficient to turn on RFP expression within a few hours (estimated ≤12 h). Because biosensor activity requires de novo transcription and translation of RFP, it does not report real-time enzymatic activity. Although CaspaseTracker RFP will be significantly degraded likely within a day after caspase activity ceases, nucGFP will be expressed by the ubiquitin promoter for the life of the cell and its progeny.
To first demonstrate that CaspaseTracker is responsive to caspase activation in vivo, adult CaspaseTracker biosensor flies were exposed to cell death-inducing stimuli. The Drosophila ovary is a well-studied cell death model as egg chambers undergo programmed cell death when animals are stressed, a presumed mechanism of matching environmental conditions to progeny production.37,44,45 To induce cell death, biosensor flies were cold-shocked 1 h at -7 °C (freezing the animals), and ovaries were dissected from live animals the next day (Figure 2a). In contrast to untreated biosensor flies, the egg chambers after cold shock were noticeably smaller and exhibited robust red (recent) and green (permanent) biosensor activity, verifying biosensor activation after a cell death stimulus (Figure 2b, merge). Biosensor activity was detected in the nuclei of both germ cells (nurse cells and oocytes) and somatic cells (follicle cells) in egg chambers, but only in the treated flies.35 Morphologies characteristic of apoptosis were also readily observed in biosensor-positive egg chambers including nuclear fragmentation, DNA condensation and degradation (loss of blue stain). Biosensor activity can be detected in the same cells with both nuclear condensation (Hoechst) and active caspases (immunostain), but the most intense staining for active caspases occurred in areas where both nuclear DNA and caspase biosensor signals were diminished or degraded (Figure 2b).46,47 The cell death mechanisms occurring after cold-shock are not well characterized, and other death mechanisms may be at play. However, similar results were obtained following amino acid starvation, which is known to cause apoptosis.35
To determine if CaspaseTracker could detect non-apoptotic caspase activity, the internal organs of healthy 1-day-old flies were meticulously dissected by removing the autofluorescent cuticle and fat. Tissues from a single representative fly were fixed, mounted, stained with Hoechst to mark nuclei and with Phalloidin stain for F-actin to detect muscle cells. In contrast to healthy egg chambers, which lack biosensor activation, the F-actin-stained muscle cells between egg chambers had both red and green biosensor-activity (Figure 3a, inset). However, it is possible that the biosensor also labels other cells. Many other healthy tissues and organs of optimally reared 1-day-old flies exhibited prominent caspase biosensor activity presumably reflecting basal non-apoptotic caspase function (Figure 3a). Biosensor-positive cells appeared morphologically normal and occurred in organized patterns in tissues suggesting that specific subsets of cells activate caspases, for example stripes of biosensor-positive cells wrapping the hindgut (Figure 3a, inset).
The best evidence that CaspaseTracker is a specific reporter of caspases is the lack of signal in flies expressing the control biosensor, which is identical to CaspaseTracker except for a single Asp to Ala amino acid mutation at the obligatory caspase cleavage site, changing DQVD to DQVA (Figure 1b and 3b). Except for an exceptionally rare cell (Figure 3b, green arrow), the only signal observed in the DQVA control biosensor flies are autofluorescent structures, such as mouth parts (Figure 3b) and ingested food in the crop and elsewhere (Figure 3b and 4), which are also observed in non-transgenic parental flies that lack the caspase biosensor.35 The CaspaseTracker-labeled cells occur at regular intervals along the malpighian (kidney) tubules and often exhibit both recent (red) and past (green) caspase activation simultaneously, while many cells in the brain, optic lobes, cardia, crop, midgut and oviduct are either red or green (Figures 4 and 5a).
For evidence that long-lived cells can survive caspase activity, the optic lobe was immunostained for the pan-neuronal marker ELAV (embryonic lethal abnormal vision).48 The nuclei of many neurons were double labeled with nuclear-targeted GFP from CaspaseTracker and ELAV, consistent with long-lived cells using caspases for non-death functions (Figure 5b). Furthermore, if the biosensor is turned off for 10 days (using Gal80ts), biosensor-positive cells persist for the duration in the gut (Figure 6a and b), brain and elsewhere (not shown). Using the caspase biosensor CaspaseTracker, we provide the first overview of basal caspase activity in whole animals.
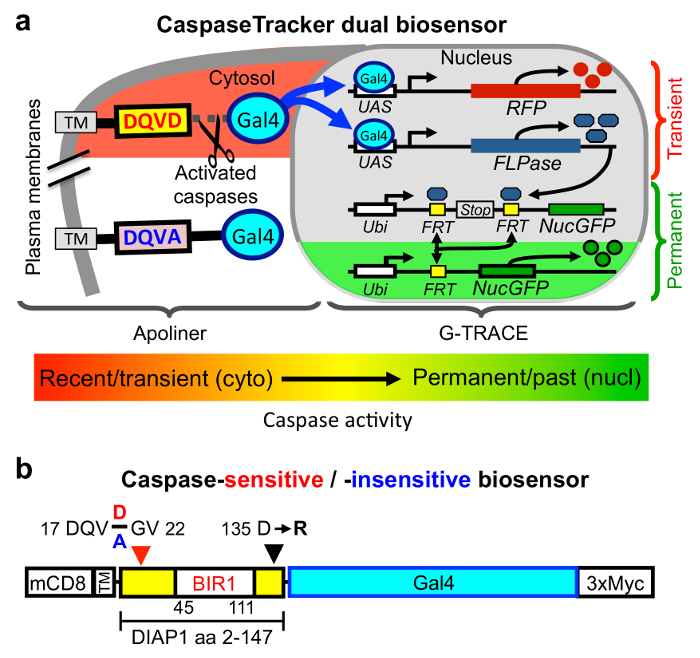 Figure 1: CaspaseTracker biosensor system for detecting non-apoptotic caspase activity. (a) Components of the Drosophila caspase biosensor. (b) The caspase-cleavable portion of CaspaseTracker biosensor system is composed of a plasma membrane anchor (mCD8), a fragment of DIAP1 containing a natural caspase-cleavage site Asp20 fused to the Gal4 transcription factor with a C-terminal 3x-myc tag and expressed via the ubiquitin promoter. Caspase cleavage results in translocation of Gal4 to the nucleus to induce the G-TRACE system. In a control biosensor, the requisite Asp20 residue in the 4-amino acid caspase cleavage site (DQVD) is changed to Ala (DQVA) to abolish caspase cleavage. Transgenic CaspaseTracker flies contain 4 transgenes. Please click here to view a larger version of this figure.
Figure 1: CaspaseTracker biosensor system for detecting non-apoptotic caspase activity. (a) Components of the Drosophila caspase biosensor. (b) The caspase-cleavable portion of CaspaseTracker biosensor system is composed of a plasma membrane anchor (mCD8), a fragment of DIAP1 containing a natural caspase-cleavage site Asp20 fused to the Gal4 transcription factor with a C-terminal 3x-myc tag and expressed via the ubiquitin promoter. Caspase cleavage results in translocation of Gal4 to the nucleus to induce the G-TRACE system. In a control biosensor, the requisite Asp20 residue in the 4-amino acid caspase cleavage site (DQVD) is changed to Ala (DQVA) to abolish caspase cleavage. Transgenic CaspaseTracker flies contain 4 transgenes. Please click here to view a larger version of this figure.
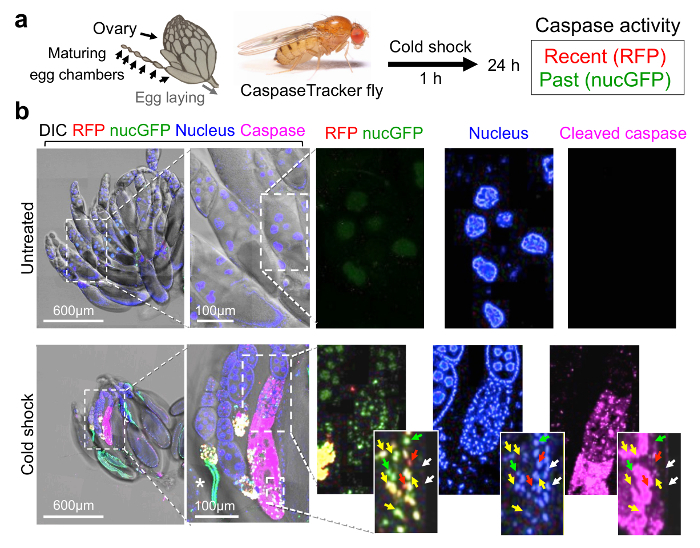 Figure 2: CaspaseTracker is activated by caspases during cell death. (a) Schematic of Drosophila ovary and flow diagram for cold shock-induced cell death. Drosophila ovary drawing (by Polan Santos) and Drosophila image (by Darren Obbard) are used with permission. (b) Egg chambers from the ovary of 6-day female CaspaseTracker flies fed with normal fly food for 6 days (untreated) or 1 day after cold shock (-7°C, 1 h, followed by 25°C for 24 h) to induce caspase activation in egg chambers. Enlargements of cold-treated ovary (bottom right frames) show three egg chambers in one developing ovariole chain (centered approximately vertically) with evidence of nuclear condensation (top egg chamber), nuclear fragmentation (middle egg chamber and insets) and degradation (lower egg chamber) based on Hoechst DNA stain (blue). Active caspases (anti-caspase-3, magenta) are detected in the middle and lower egg chambers. RFP and GFP biosensor activity (red, green and yellow arrows) occur in cells with nuclear DNA condensation that often stain diffusely with active caspases (insets). White arrows mark nuclei lacking both biosensor activity and active caspase immunostain. *Autofluorescence signals. These data are reproduced with permission from Tang et al., Sci Rep 2015. Please click here to view a larger version of this figure.
Figure 2: CaspaseTracker is activated by caspases during cell death. (a) Schematic of Drosophila ovary and flow diagram for cold shock-induced cell death. Drosophila ovary drawing (by Polan Santos) and Drosophila image (by Darren Obbard) are used with permission. (b) Egg chambers from the ovary of 6-day female CaspaseTracker flies fed with normal fly food for 6 days (untreated) or 1 day after cold shock (-7°C, 1 h, followed by 25°C for 24 h) to induce caspase activation in egg chambers. Enlargements of cold-treated ovary (bottom right frames) show three egg chambers in one developing ovariole chain (centered approximately vertically) with evidence of nuclear condensation (top egg chamber), nuclear fragmentation (middle egg chamber and insets) and degradation (lower egg chamber) based on Hoechst DNA stain (blue). Active caspases (anti-caspase-3, magenta) are detected in the middle and lower egg chambers. RFP and GFP biosensor activity (red, green and yellow arrows) occur in cells with nuclear DNA condensation that often stain diffusely with active caspases (insets). White arrows mark nuclei lacking both biosensor activity and active caspase immunostain. *Autofluorescence signals. These data are reproduced with permission from Tang et al., Sci Rep 2015. Please click here to view a larger version of this figure.
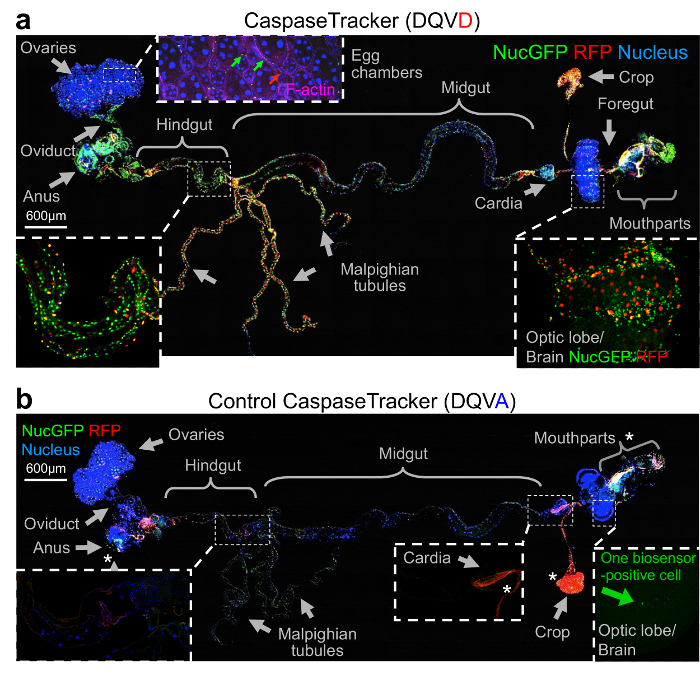 Figure 3: Evidence for widespread physiological caspase activity. (a) Merged confocal image of CaspaseTracker (DQVD) RFP and nucGFP from a single newly eclosed fly raised at 18 °C, dissected and stained with H33342 for nuclei and Phalloidin for F-actin (pink), and imaged with DIC. Phalloidin is shown only in the inset for egg chambers. (b) The same procedure and imaging conditions were followed for control CaspaseTracker (DQVA) flies. *Autofluorescent structures. These data are reproduced with permission from Tang et al., Sci Rep 2015. Please click here to view a larger version of this figure.
Figure 3: Evidence for widespread physiological caspase activity. (a) Merged confocal image of CaspaseTracker (DQVD) RFP and nucGFP from a single newly eclosed fly raised at 18 °C, dissected and stained with H33342 for nuclei and Phalloidin for F-actin (pink), and imaged with DIC. Phalloidin is shown only in the inset for egg chambers. (b) The same procedure and imaging conditions were followed for control CaspaseTracker (DQVA) flies. *Autofluorescent structures. These data are reproduced with permission from Tang et al., Sci Rep 2015. Please click here to view a larger version of this figure.
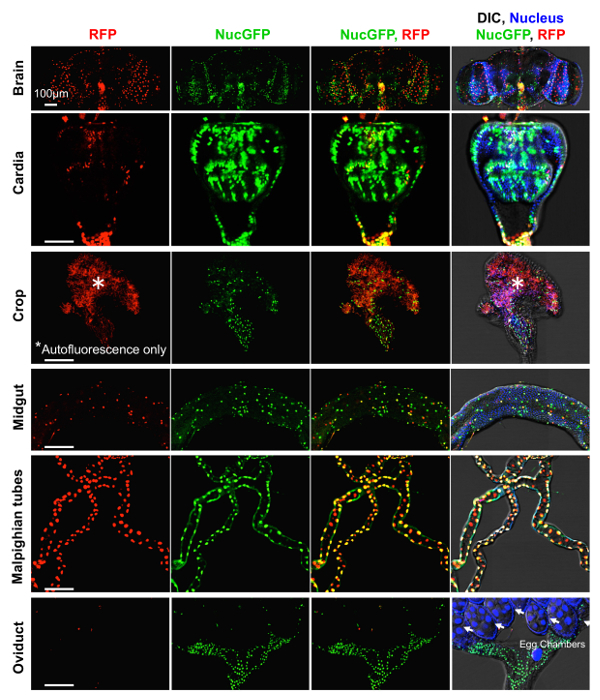 Figure 4: Basal caspase activity in many tissues. Higher magnifications of DIC and fluorescence confocal images of GFP (past) and RFP (recent) CaspaseTracker (DQVD) activity and Hoechst-stained nuclei in the brain, cardia, crop, midgut, Malpighian tubules and oviducts from Figure 3a. *Autofluorescent structures. These data are reproduced with permission from Tang et al., Sci Rep 2015. Please click here to view a larger version of this figure.
Figure 4: Basal caspase activity in many tissues. Higher magnifications of DIC and fluorescence confocal images of GFP (past) and RFP (recent) CaspaseTracker (DQVD) activity and Hoechst-stained nuclei in the brain, cardia, crop, midgut, Malpighian tubules and oviducts from Figure 3a. *Autofluorescent structures. These data are reproduced with permission from Tang et al., Sci Rep 2015. Please click here to view a larger version of this figure.
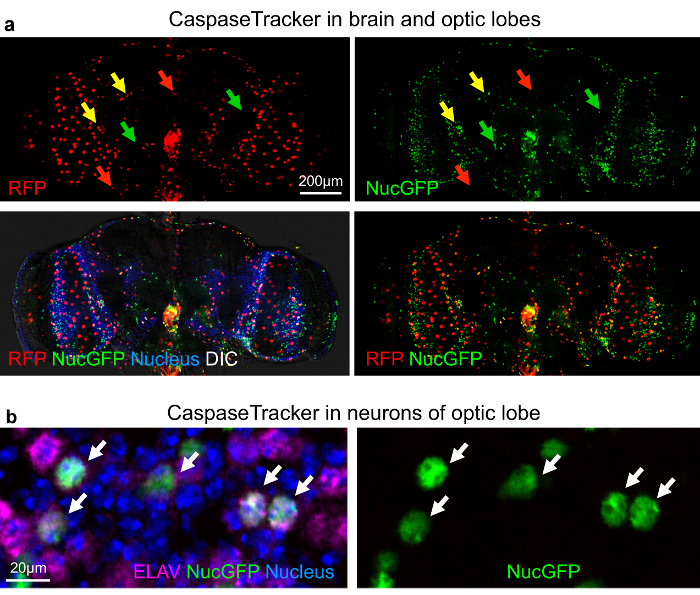 Figure 5: Non-apoptotic caspase activity in neurons of the Drosophila optic lobe. (a) DIC and confocal images with RFP (recent) and GFP (past) CaspaseTracker (DQVD) activity and Hoechst-stained nuclei in the brain. RFP+GFP dual-labeled cells (yellow arrows). (b) NucGFP colocalizes with immunostain for pan neuronal nuclear ELAV protein in the optic lobe of CaspaseTracker (DQVD) fly brain. Neurons with co-localized signals of NucGFP and ELAV are shown (white arrows). Note, RFP-labeled neurons are not present in this particular field. These data are reproduced with permission from Tang et al., Sci Rep 2015. Please click here to view a larger version of this figure.
Figure 5: Non-apoptotic caspase activity in neurons of the Drosophila optic lobe. (a) DIC and confocal images with RFP (recent) and GFP (past) CaspaseTracker (DQVD) activity and Hoechst-stained nuclei in the brain. RFP+GFP dual-labeled cells (yellow arrows). (b) NucGFP colocalizes with immunostain for pan neuronal nuclear ELAV protein in the optic lobe of CaspaseTracker (DQVD) fly brain. Neurons with co-localized signals of NucGFP and ELAV are shown (white arrows). Note, RFP-labeled neurons are not present in this particular field. These data are reproduced with permission from Tang et al., Sci Rep 2015. Please click here to view a larger version of this figure.
Discussion
Here we illustrate the construction and inner workings of CaspaseTracker that facilitate detection of widespread basal caspase activity in healthy tissues. The critical steps for detecting non-apoptotic caspase activity in vivo are: 1) generating flies with the biosensor transgene, 2) verifying caspase-specific reporter function with appropriate controls, 3) practicing dissection techniques to observe all internal organ systems of adult Drosophila, and 4) distinguishing biosensor activity from autofluorescent artifacts.
The natural caspase-cleavable polypeptide that serves as the caspase-specific sensor was modified to avoid any caspase-inhibitory activity by the biosensor. Further modifications were made to avoid rapid degradation of the Gal4 transcription factor, and the insertion of an epitope tag. We also describe whole body organ dissection of flies, immunostaining, mounting and confocal microscopy of fly tissues to detect non-apoptotic caspase activity.
While CaspaseTracker is ideal for detecting cells that survive with low levels of caspase activity, it is not a reporter of real-time enzyme activity, as several hours are required to turn on the biosensor. However, we have not determined the minimum time to detection, which is likely much shorter than needed for the applications described here. In contrast to CaspaseTracker, the major limitation of other caspase biosensors is their inability to detect low levels of caspase activity, as they are designed primarily to study cell death. Caspase activity is amplified following a cell death stimulus, causing massive cell demolition in less than 30 min.49 Therefore, detection of caspase activation is often assumed to be a marker of certain cell death.50,51 However, mounting evidence indicates that caspases have non-apoptotic, even non-degradative functions in healthy cells.7-21 The presumed low levels of spatially and temporarily restricted enzymatic activity of caspases that carry out their day-jobs are likely below detection by available technologies. This problem is overcome by CaspaseTracker, which likely requires few caspase cleavage events to generate robust signals, and does not require ongoing caspase enzymatic activity to permanently label these cells in vivo.
The evidence presented indicates that CaspaseTracker is specific for caspases, though we cannot exclude the possibility that other unidentified Asp-specific proteases could activate CaspaseTracker. However, feeding flies with a cocktail of caspase-inhibitor peptides blocks CaspaseTracker activity (unpublished). We also emphasize the importance of avoiding biosensors that themselves inhibit caspases, and the importance of control biosensors and non-transgenic flies to avoid erroneous conclusions due to autofluorescence inherent to the insect cuticle, internal fat deposits, ingested food or other confounders.
Another challenge is distinguishing pro-death and non-death functions of caspases. Pro-death and non-death functions could be linked evolutionarily for the purpose of transitioning cells between a normal physiological state to an appropriately timed death, for example the expansion of immune cells to fight off infection and subsequent elimination of these cells to prevent cancer. Thus, we cannot eliminate the possibility that some of the observed basal caspase activity in internal organs is instead an attempt to promote cell death, seemingly consistent with the estimated tens of billions cell deaths occurring daily in the human body.52 However, the healthy morphology of cells expressing the biosensor strongly indicates that CaspaseTracker is capable of detecting caspase activity intended for normal day-jobs of caspases.
Caspases are suggested to have non-death roles such as cell proliferation. This is consistent with earlier attempts to generate mammalian cell lines stably expressing viral proteins that bind and directly inhibit cellular caspases (e.g., CrmA). While hundreds of cell colonies arose during drug selection with the empty control vector, unexpectedly zero colonies formed in parallel cultures transfected with the vector expressing caspase inhibitors (unpublished). Thus, caspases likely have widespread functions in normal cells that are currently underappreciated. This is further supported by the finding that basal biosensor activity in the CaspaseTracker fly is detected in many cell types across most organ systems, including neurons, in normal healthy flies. Healthy caspase activity in neurons may dismantle synaptic endings to promote normal brain function by cleaving the same substrates as during apoptosis, or their day-job functions may be entirely distinct from apoptosis-like activities. While low versus high enzymatic levels of active caspases may be a critical distinction between day-jobs and cell death, there are few knockin mutant animals with specific caspase-uncleavable substrates that support this possibility.17,53 However, recent progress indicates that caspase-8 inhibits necroptosis possibly by cleaving RIPK,54 synaptic depression and AMPA receptor endocytosis may be regulated by caspase cleavage of Gap43,55 and microRNA processing may be regulated by caspase cleavage of Dicer.14,15,56
There are many additional applications of this biosensor. To determine if caspases are activated at specific times or in specific tissues, expression of the biosensor can be readily regulated temporally (e.g., using Gal80ts) and in specific cell types (e.g., olfactory-specific Gal4 drivers). Using Gal80ts to suppress biosensor expression until emergence of adult flies still resulted in widespread biosensor activity, indicating that caspases can be activated at adult stages (Figure 6c and d).35 By replacing the DIAP1 fragment with other caspase substrates, other applications may include biosensors specific for different caspases and for detecting cleavage of specific substrates. The CaspaseTracker biosensor will enhance our understanding of the role of non-apoptotic caspase activities.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank Polan Santos and Darren Obbard for Drosophila illustrations in Fig. 2a, Marcelo Jacobs-Lorena for use of the JHMRI insectary. This work was supported by the Life Science Research Foundation fellowship (H.L.T.), University Grants Committee of the Hong Kong AoE/B-07/99 (M.C.F.), and NIH grants NS096677, NS037402 and NS083373 (J.M.H.). Ho Lam Tang is a Shurl and Kay Curci Foundation Fellow of the Life Sciences Research Foundation.
References
- Salvesen GS, Abrams JM. Caspase activation - stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene. 2004;23:2774–2784. doi: 10.1038/sj.onc.1207522. [DOI] [PubMed] [Google Scholar]
- Hay BA, Guo M. Caspase-dependent cell death in Drosophila. Annu Rev Cell Dev Biol. 2006;22:623–650. doi: 10.1146/annurev.cellbio.21.012804.093845. [DOI] [PubMed] [Google Scholar]
- Segawa K, et al. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 2014;344:1164–1168. doi: 10.1126/science.1252809. [DOI] [PubMed] [Google Scholar]
- Akagawa H, et al. The role of the effector caspases drICE and dcp-1 for cell death and corpse clearance in the developing optic lobe in Drosophila. Dev Biol. 2015. [DOI] [PubMed]
- Souers AJ, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med. 2013;19:202–208. doi: 10.1038/nm.3048. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Steller H. Apoptosis, stem cells, and tissue regeneration. Sci Signal. 2010;3:re8. doi: 10.1126/scisignal.3145re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman BT, Yuan J. Apoptotic and non-apoptotic roles of caspases in neuronal physiology and pathophysiology. Nat Rev Neurosci. 2012;13:395–406. doi: 10.1038/nrn3228. [DOI] [PubMed] [Google Scholar]
- Juraver-Geslin HA, Durand BC. Early development of the neural plate: new roles for apoptosis and for one of its main effectors caspase-3. Genesis. 2015;53:203–224. doi: 10.1002/dvg.22844. [DOI] [PubMed] [Google Scholar]
- Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- Kaplan Y, Gibbs-Bar L, Kalifa Y, Feinstein-Rotkopf Y, Arama E. Gradients of a ubiquitin E3 ligase inhibitor and a caspase inhibitor determine differentiation or death in spermatids. Dev Cell. 2010;19:160–173. doi: 10.1016/j.devcel.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, et al. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther C, et al. Caspase-8 regulates TNF-alpha-induced epithelial necroptosis and terminal ileitis. Nature. 2011;477:335–339. doi: 10.1038/nature10400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BP, et al. CED-3 caspase acts with miRNAs to regulate non-apoptotic gene expression dynamics for robust development in C. elegans. Elife. 2014;3:e04265. doi: 10.7554/eLife.04265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, et al. A novel mechanism underlies caspase-dependent conversion of the dicer ribonuclease into a deoxyribonuclease during apoptosis. Cell Res. 2014;24:218–232. doi: 10.1038/cr.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fannjiang Y, et al. BAK alters neuronal excitability and can switch from anti- to pro-death function during postnatal development. Dev Cell. 2003;4:575–585. doi: 10.1016/s1534-5807(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Ofengeim D, et al. N-terminally cleaved Bcl-xL mediates ischemia-induced neuronal death. Nat Neurosci. 2012;15:574–580. doi: 10.1038/nn.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Sheng M. Caspases in synaptic plasticity. Mol Brain. 2012;5:15. doi: 10.1186/1756-6606-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erturk A, Wang Y, Sheng M. Local pruning of dendrites and spines by caspase-3-dependent and proteasome-limited mechanisms. J Neurosci. 2014;34:1672–1688. doi: 10.1523/JNEUROSCI.3121-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DS, Okamoto H. Local caspase activation interacts with Slit-Robo signaling to restrict axonal arborization. J Cell Biol. 2013;203:657–672. doi: 10.1083/jcb.201303072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein-Rotkopf Y, Arama E. Can't live without them, can live with them: roles of caspases during vital cellular processes. Apoptosis. 2009;14:980–995. doi: 10.1007/s10495-009-0346-6. [DOI] [PubMed] [Google Scholar]
- Li P, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Lewis J, et al. Inhibition of virus-induced neuronal apoptosis by Bax. Nat Med. 1999;5:832–835. doi: 10.1038/10556. [DOI] [PubMed] [Google Scholar]
- Chen YB, et al. Bcl-xL regulates mitochondrial energetics by stabilizing the inner membrane potential. J Cell Biol. 2011;195:263–276. doi: 10.1083/jcb.201108059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi CH, et al. Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell. 2011;146:607–620. doi: 10.1016/j.cell.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K, Nagai T, Miyawaki A, Miura M. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J Cell Biol. 2003;160:235–243. doi: 10.1083/jcb.200207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto K, et al. Local initiation of caspase activation in Drosophila salivary gland programmed cell death in vivo. Proc Natl Acad Sci U S A. 2007;104:13367–13372. doi: 10.1073/pnas.0702733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardet PL, et al. A fluorescent reporter of caspase activity for live imaging. Proc Natl Acad Sci U S A. 2008;105:13901–13905. doi: 10.1073/pnas.0806983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HL, et al. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol Biol Cell. 2012;23:2240–2252. doi: 10.1091/mbc.E11-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbs A, Nimmervoll B, Sun JJ, Sava IE, Luhmann HJ. Control of programmed cell death by distinct electrical activity patterns. Cereb Cortex. 2011;21:1192–1202. doi: 10.1093/cercor/bhq200. [DOI] [PubMed] [Google Scholar]
- To TL, et al. Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc Natl Acad Sci U S A. 2015;112:3338–3343. doi: 10.1073/pnas.1502857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentin A, Arama E. Caspase levels and execution efficiencies determine the apoptotic potential of the cell. J Cell Biol. 2012;196:513–527. doi: 10.1083/jcb.201107133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara T, et al. Caspase inhibition in select olfactory neurons restores innate attraction behavior in aged Drosophila. PLoS Genet. 2014;10:e1004437. doi: 10.1371/journal.pgen.1004437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, et al. G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat Methods. 2009;6:603–605. doi: 10.1038/nmeth.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HL, Tang HM, Fung MC, Hardwick JM. In vivo CaspaseTracker biosensor system for detecting anastasis and non-apoptotic caspase activity. Sci Rep. 2015;5:9015. doi: 10.1038/srep09015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzanne M, Steller H. Shaping organisms with apoptosis. Cell Death Differ. 2013;20:669–675. doi: 10.1038/cdd.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins VK, Timmons AK, McCall K. Diversity of cell death pathways: insight from the fly ovary. Trends Cell Biol. 2013;23:567–574. doi: 10.1016/j.tcb.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkissian T, Timmons A, Arya R, Abdelwahid E, White K. Detecting apoptosis in Drosophila tissues and cells. Methods. 2014;68:89–96. doi: 10.1016/j.ymeth.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson WR, Hiesinger PR. Preparation of developing and adult Drosophila brains and retinae for live imaging. J Vis Exp. 2010. [DOI] [PMC free article] [PubMed]
- Wong LC, Schedl P. Dissection of Drosophila ovaries. J Vis Exp. 2006. p. e52. [DOI] [PMC free article] [PubMed]
- Tauc HM, Tasdogan A, Pandur P. Isolating intestinal stem cells from adult Drosophila midguts by FACS to study stem cell behavior during aging. J Vis Exp. 2014. p. e52223. [DOI] [PMC free article] [PubMed]
- Ditzel M, et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- Li X, Wang J, Shi Y. Structural mechanisms of DIAP1 auto-inhibition and DIAP1-mediated inhibition of drICE. Nat Commun. 2011;2:408. doi: 10.1038/ncomms1418. [DOI] [PubMed] [Google Scholar]
- Yi SX, Moore CW, Lee RE. Rapid cold-hardening protects Drosophila melanogaster from cold-induced apoptosis. Apoptosis. 2007;12:1183–1193. doi: 10.1007/s10495-006-0048-2. [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231:265–278. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 2010;17:534–539. doi: 10.1038/cdd.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty CE, Bergmann A. Detecting caspase activity in Drosophila larval imaginal discs. Methods Mol Biol. 2014;1133:109–117. doi: 10.1007/978-1-4939-0357-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP, Lisbin MJ, White K. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr Biol. 1996;6:1634–1641. doi: 10.1016/s0960-9822(02)70787-2. [DOI] [PubMed] [Google Scholar]
- Albeck JG, et al. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 2009;16:1093–1107. doi: 10.1038/cdd.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med. 2012;18:1630–1638. doi: 10.1038/nm.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR. Means to an end : apoptosis and other cell death mechanisms. Cold Spring Harbor Laboratory Press; 2011. [Google Scholar]
- Chau BN, et al. Signal-dependent protection from apoptosis in mice expressing caspase-resistant Rb. Nat Cell Biol. 2002;4:757–765. doi: 10.1038/ncb853. [DOI] [PubMed] [Google Scholar]
- Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13:2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, et al. The novel caspase-3 substrate Gap43 is involved in AMPA receptor endocytosis and long-term depression. Mol Cell Proteomics. 2013;12:3719–3731. doi: 10.1074/mcp.M113.030676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A, Shi Y, Kage-Nakadai E, Mitani S, Xue D. Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science. 2010;328:327–334. doi: 10.1126/science.1182374. [DOI] [PMC free article] [PubMed] [Google Scholar]


