Abstract
The primary purpose of this study was to determine if methicillin-resistant Staphylococcus aureus (MRSA) strains could be identified in the milk of dairy cattle in a Paso del Norte region dairy of the United States. Using physiological and PCR-based identification schemes, a total of 40 Staph. aureus strains were isolated from 29 raw milk samples of 133 total samples analyzed. Pulsed-field gel electrophoresis after digestion with the SmaI enzyme revealed that the 40 confirmed strains were represented by 5 pulsed-field types, which each contained 3 or more strains. Of 7 hospital strains isolated from cows undergoing antibiotic therapy, 3 demonstrated resistance to 3 or more antimicrobial classes and displayed similar pulsed-field gel electrophoresis patterns. A secondary purpose of this study was to elucidate the evolutionary relationships of strains isolated in this study to genomically characterized Staph. aureus strains. Therefore, Roche 454 GS (Roche Diagnostics Corp., Dallas, TX) pyrosequencing was used to produce draft genome sequences of an MRSA raw milk isolate (H29) and a methicillin-susceptible Staph. aureus (PB32). Analysis using the BLASTn database (http://blast.ncbi.nlm.nih.gov/) demonstrated that the H29 draft genome was highly homologous to the human MRSA strain JH1, yet the β-lactamase plasmid carried by H29 was different from that carried by JH1. Genomic analysis of H29 also clearly explained the multidrug resistance phenotype of this raw milk isolate. Analysis of the PB32 draft genome (using BLASTn) demonstrated that this raw milk isolate was most related to human MRSA strain 04-02981. Although PB32 is not a MRSA, the PB32 draft genome did reveal the presence of a unique staphylococcal cassette mec (SCCmec) remnant. In addition, the PB32 draft genome revealed the presence of a novel bovine staphylococcal pathogenicity island, SaPIbovPB32. This study demonstrates the presence of clones closely related to human and (or) bovine Staph. aureus strains circulating in a dairy herd.
Keywords: Staphylococcus aureus, methicillin-resistant, mastitis, genomics
INTRODUCTION
Staphylococcus aureus is responsible for life-threatening human infections acquired in hospitals and within the community (Richards et al., 2001; Moran et al., 2006). The first methicillin-resistant Staph. aureus (MRSA) infection was described in 1961 (Jevons, 1961) and since then, human infections caused by multi-drug-resistant MRSA have become common (Waness, 2010). The MRSA phenotype results from the acquisition of the staphylococcal cassette chromosome mec (SCCmec), which harbors the mecA gene that encodes the low-affinity penicillin-binding protein 2a (PBP2a) (Katayama et al., 2000). The structure of numerous SCCmec elements reveals that this element is following multiple evolutionary trajectories [for review, see Ito et al. (2001), Li et al. (2011), and Shore et al. (2011)].
Staphylococcus aureus is also a major cause of bovine mastitis worldwide, which results in substantial economic losses for the dairy industry. Methicillin-resistant Staph. aureus-causing bovine mastitis was first reported in 1972 (Devriese et al., 1972) and MRSA transmission appears to occur between animals, and from animals to humans (Lee, 2003; Voss et al., 2005; Lozano et al., 2011a). Carriage of MRSA by livestock has been shown to correlate with the MRSA colonization of farmers and farm families, veterinarians, and health-care workers (Voss et al., 2005; Juhász-Kaszanyitzky et al., 2007; Wulf et al., 2008). Sequencing of 7 housekeeping gene amplicons has been used to group MRSA isolates into clonal complexes (CC) using “based upon related sequence types,” or BURST, analysis (Enright et al., 2000). These analyses have revealed that certain strains, referred to as livestock-associated MRSA, belong to a small number of CC, with the most prevalent being CC398, and it is known that CC398 strains also cause infections in humans (Voss et al., 2005; Salmenlinna et al., 2010; Lozano et al., 2011b). A recent report suggests that CC398 strains emerged from a human evolved methicillin-susceptible Staph. aureus (MSSA) strain and that livestock-associated MRSA sublineages arose during antimicrobial selection in livestock (Price et al., 2012).
The aim of this study was primarily to characterize Staph. aureus strains isolated from raw milk samples of mastitic and healthy dairy cattle from a Paso del Norte region dairy, in an effort to identify the presence of MRSA. Using the draft genomes of 1 raw milk MRSA isolate and 1 MSSA isolate, we also report the evolutionary relatedness of these strains to previously sequenced Staph. aureus strains.
MATERIALS AND METHODS
Isolation and Characterization of Staph. aureus Strains
Samples of raw milk were collected from 33 hospital cows, some receiving antibiotic treatment, and 100 healthy cows in 2 milking parlors (A and B) at a local dairy. The isolation of Staph. aureus strains was carried out as follows: briefly, 100 μL of PBS-diluted milk samples were spread onto Baird-Parker agar (Becton Dickinson and Co., Sparks, MD) plates and incubated at 37°C for 48 h. Resulting colonies were then subjected to Gram staining, a catalase test with 3% hydrogen peroxide, and a coagulase test using rabbit plasma (Becton Dickinson and Co.). Presumptive Staph. aureus isolates were further scrutinized on mannitol salt agar (Acumedia Manufacturers Inc., Lansing, MI) plates. Luria broth (LB; Becton Dickinson and Co.) cultures of all presumptive isolates were then grown overnight and glycerol (20% vol/vol) was added to aliquots, which were then stored at −80°C. Working cultures of these isolates were maintained on LB agar plates at 4°C. Total DNA from 20-mL LB overnight cultures of 40 suspected Staph. aureus milk isolates, 2 positive-control Staph. aureus laboratory strains (COL and ATCC 25923), and a negative control culture (Staphylococcus epidermidis ATCC 12228) were isolated as previously described (Riordan et al., 2006). These DNA samples were then scrutinized by a PCR protocol designed to detect the Staph. aureus-specific nucA nuclease gene or for mecA as previously described (Murakami et al., 1991; Brakstad et al., 1992).
Antimicrobial Susceptibility
Antimicrobial susceptibility was determined using the disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI, 2008). Inducible clindamycin resistance was determined as previously described (McDougal et al., 2003). Vancomycin MIC were determined using E-test strips (AB Biodisk North America Inc., Piscataway, NJ) according to the manufacturer’s recommendations.
Pulsed-Field Gel Electrophoresis
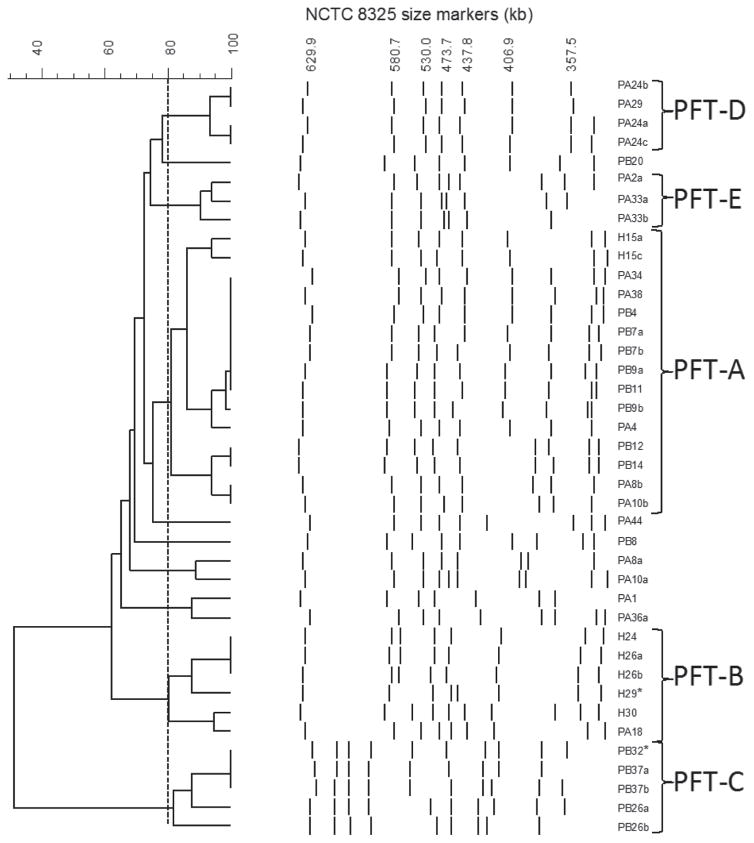
Overnight cell suspensions were embedded in 1.5% (wt/vol) low-melting point agarose (Sigma-Aldrich, St. Louis, MO) plugs, treated with lysostaphin and proteinase K (Sigma-Aldrich), and were then digested with SmaI (New England BioLabs Inc., Ipswich, MA). The plugs were electrophoresed in a 1% (wt/vol) agarose gel for 23 h at 6 V/cm, with an initial switch time of 1 s and final switch time of 30 s at 11.3°C using the CHEF DR III system (Bio-Rad Laboratories Inc., Hercules, CA). Band patterns were compared using the Fingerprinting II Informatix software (Bio-Rad Laboratories Inc.) to determine the clonal relatedness (O’Brien et al., 2005). Strains with 80% or greater pattern similarity were considered clonal (Tenover et al., 1995). Strain NCTC 8325 (NCTC, National Collection of Type Cultures) chromosomal DNA digested with SmaI was used as the size marker (Figure 1).
Figure 1.
Pulsed-field gel electrophoresis (PFGE) patterns of SmaI-digested chromosomal DNA of Staphylococcus aureus strains investigated and dendrogram of percent relatedness derived from the patterns. Asterisks represent the 2 strains that were sequenced. NCTC = National Collection of Type Cultures; PFT = pulsed-field type.
Genome Sequencing
Initially, total DNA was isolated from overnight LB cultures of MRSA strain H29 and MSSA strain PB32. The genomes of both isolates were sequenced (>20× overall coverage) using Roche 454 GS (FLX titanium; Roche Diagnostics Corp., Dallas, TX) pyrosequencing according to the manufacturer’s instructions. All reads were assembled into contigs using the Newbler assembler 2.3 (454 Life Sciences Corp., Branford, CT). The draft genomes were then uploaded into the Rapid Annotations using Subsystems Technology (RAST) server for annotation (Aziz et al., 2008). Contigs for both genomes were then compared with other genomes in the databases using BLASTn (http://blast.ncbi.nlm.nih.gov/) and EvoPrinter (http://evoprinter.ninds.nih.gov/). Sequence types (ST) and allele numbers were determined with the Staph. aureus Multi Locus Sequence Typing (MLST) website (http://saureus.mlst.net), using sequences obtained from 454 sequencing.
RESULTS AND DISCUSSION
Growth on Baird Parker and mannitol salt agar, Gram stain, and catalase reaction presumptively identified 40 Staph. aureus isolates from 29 milk samples. More than one isolated colony was chosen from samples that showed variable reactions on the selective media above (designated by strain number followed by a, b, or c). All 40 of these isolates were coagulase and nucA positive. These isolates included 7 from hospital cows (H strains), 18 from parlor A cows (PA strains), and 15 from parlor B cows (PB strains). None of the strains investigated demonstrated resistance to vancomycin by disk diffusion. All H strains except H15c were oxacillin resistant, yet all 7 H strains were mecA positive. One H strain (H30) also demonstrated erythromycin resistance, whereas 3 H strains (H24, H26a, H26b) demonstrated resistance to ciprofloxacin and erythromycin. Strain H29 demonstrated resistance to both of these antimicrobials, as well as tetracycline and imipenem, and inducible clindamycin resistance. None of the PA and PB strains were methicillin resistant or mecA positive. Strain PA18, however, demonstrated ciprofloxacin and erythromycin resistance, whereas PB26b demonstrated resistance to fusidic acid.
Pulsed-field gel electrophoresis of SmaI-digested DNA separated the 40 strains into multiple pulsed-field types (PFT). Pulsed-field type A was the most frequently isolated PFT, representing 37.5% (n = 15) of all isolates (Figure 1). Pulsed-field type B contained the largest number of H strains (n = 5) and PA18, all of which demonstrated resistance to 2 or more of the antimicrobials investigated. Pulsed-field type C, D, and E all contained 2 or more strains isolated from a single milk sample, which indicates the clonality of the colonies isolated from these milk samples. In 2 instances where more than a single Staph. aureus colony was chosen from 1 milk sample (PA8a and PA8b; PA10a and PA10b), pulsed-field gel electrophoresis analysis revealed that the SmaI restriction fragment length polymorphisms were different (Figure 1). Both PA8b and PA10b clustered within the PFT-A strains, whereas PA8a and PA10a clustered close to PFT-A strains (Figure 1). This suggests that more than 1 Staph. aureus clone was present in each of these milk samples.
We next chose 2 strains for 454 sequencing. One was H29, which was a representative of the most widely distributed MRSA clone (PFT-B) and the other was PB32, which was an MSSA clone from PFT-C that was most related to the PFT-B MRSA clones identified in this study.
The draft genome of MRSA strain H29, derived from 119 contigs (all >200 bp in length), consists of 2,844,315 bp, which encodes for 2,664 protein-coding genes. The MLST website revealed that strain H29 is ST5 (1,4,1,4,12,1,10), an ST that contains human MRSA isolates and is found within CC5. The presence of ST5 MRSA strains circulating in Paso del Norte area hospitals located near the dairy investigated has previously been reported (O’Brien et al., 2005; Delgado et al., 2007).
Overall, the H29 draft genome (accession no. PRJ-NA179361) proved to be 100% identical over 97% of the human MRSA strain JH1 genome (accession no. NC009632) by BLASTn comparisons. Staphylococcus aureus JH1 is also an ST5 strain that was isolated from a patient with endocarditis and the JH lineage is capable of developing intermediate resistance to vancomycin (Sieradzki et al., 2003; Mwangi et al., 2007). A vancomycin E-test demonstrated that H29 does not demonstrate vancomycin-intermediate resistance.
Contig 1 (27,139 bp) of the H29 draft genome represents a β-lactamase plasmid that had only 42% nucleotide similarity to plasmid pSJH101 (accession no. NC009619) found in JH1, yet was 99.5% identical to plasmid SAP048A (27,268 bp; accession no. GQ900406) found in Staph. aureus strain NE 3809, a human clinical blood isolate from Nebraska. Plasmid SAP048A is a representative of pIB485-like plasmids that carry ΔTn552 encoding β-lactamase and a cluster of staphylococcal enterotoxin genes (sed, sej, and ser) that have contributed to outbreaks of Staph. aureus foodborne illness [for review, see Gustafson and Wilkinson (2005) and Shearer et al. (2011)]. In addition, contig 34 of H29 demonstrated 97% nucleotide identity to a previously described enterotoxin gene cluster (egc) operon that encodes 5 enterotoxin genes (seo, sem, sei, sen, and seg) and 2 pseudogenes (ϕent1 and ϕent2) located on the νSaβ genomic island (Jarraud et al., 2001; Kuroda et al., 2001). The H29 draft genome also contains the SC-Cmec ST II, which harbors a erythromycin resistance gene (erm; Ito et al., 2001), a Tn916-like transposon containing the tetracycline resistance gene tetM (de Vries et al., 2009), and a mutation in the DNA gyrase gene (gyrA) that leads to a 84S →84L alteration in GyrA that has previously been tied to ciprofloxacin resistance (Schmitz et al., 1998).
The draft genome of MSSA strain PB32 (accession no. PRJNA179544) derived from 80 contigs consists of 2,808,519 bp, which encodes 2,632 protein-coding genes. The MLST sequences derived from the draft genome confirmed PB32 as an ST124 (3,1,1,37,1,5,3) strain of CC97, which is represented by bovine as well as shared bovine/human strains (Smith et al., 2005). There do not appear to be any readily identifiable plasmid sequences in the PB32 draft genome.
Overall, the PB32 draft genome (accession no. PRJ-NA179544) proved to be 100% identical over 94% of the human MRSA strain 04-02981 genome (accession no. CP001844.2) by BLASTn comparisons. Strain 04-02981 is a plasmidless ST225 (1,4,1,4,12,25,10) multidrug-resistant SCCmec ST II strain that was isolated in Köln (Germany) in 2004 (Nübel et al., 2010). Sequence type ST225, which was first described in the 1990s in the United States, represents a single locus variant of ST5 strains and the genome of 04-02981 is also collinear with the JH1 genome (Nübel et al., 2010).
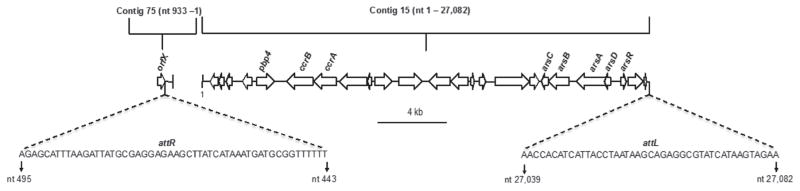
Staphylococcus aureus SCCmec insertion is carried out by site-specific recombination between the attB site on the chromosome and the attS site on SCCmec (Ito et al., 1999; Katayama et al., 2000). This recombination event results in copies of the att sites at each end of SC-Cmec, with attR within orfX and attL at the other end of the SCCmec (Wang and Archer, 2010). A 27,082-bp segment within contig 15 (215,480 bp) contains several genes previously described in SCCmec elements as well as the attL attachment site (Figure 2). These genes include the recombinase genes ccrA and ccrB, pbp4, and an arsenic resistance operon (arsA-D, arsR). It has been reported that all ST225 and JH strains carry the same ccrB gene (Nübel et al., 2010). The ccrB gene of PB32, however, only demonstrated 93% nucleotide identity to the ccrB gene of both strains 04-02981 and JH1. The mecA gene and the mecA controlling genes (mecI and mecRI) were not present in the draft genome of PB32. In the PB32 draft genome, orfX is found on one end of contig 75 (130,864 bp) with attR attachment site (Figure 2), suggesting that contig 15 and contig 75 belong together. Staphylococcus aureus strains that possess chromosomally located remnants of SCCmec determinants, including some with a pbp4 gene, have been reported (Luong et al., 2002; Corkill et al., 2004; Donnio et al., 2007; Shore et al., 2008; Wong et al., 2010; Lindqvist et al., 2012) and we suspect that sequences within contigs 15 and 75 described here represent a novel SCCmec remnant. It is of interest to note that similar to PB32, ST225 MSSA isolates can also carry SCCmec remnants (Nübel et al., 2010).
Figure 2.
Cartoon representing open reading frames (ORF) of the staphylococcal cassette chromosome mec (SCCmec) remnant found within contigs 15 and 75 of Staphylococcus aureus strain PB32. Direction of the arrows represents the orientation of the ORF; attachment sites and their respective nucleotide sequences and positions are indicated. nt = nucleotide.
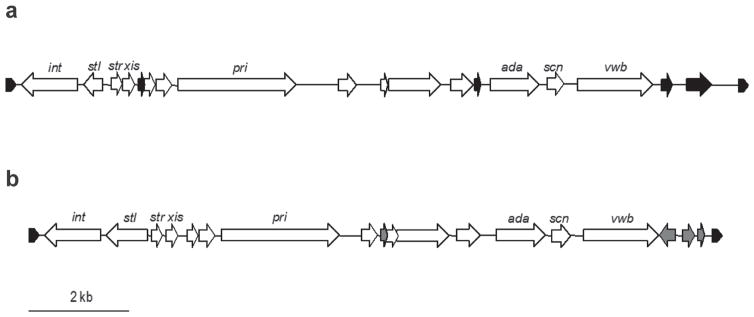
Strain PB32 contains another mobile genetic element, a novel bovine staphylococcal pathogenicity island (SaPIbov) located within contig 66 (127,041 bp), which will be referred to as SaPIbovPB32. Sequence comparison of SaPIbovPB32 (nucleotide 90,549–104,940) with other SaPIbov sequences demonstrated the greatest nucleotide identity to SaPIbov5 (accession no. HM228919) at 79% identity, followed by 66% identity to SaPIbov4 (accession no. HM211303; Viana et al., 2010). The SaPIbov5 (13,526 bp) was described in a Staph. aureus ST398 clone (Viana et al., 2010). Staphylococcus aureus strains that contain SaPIbov5 have been isolated from different animals including cows, sheep, and goats (Viana et al., 2010). The SaPIbovPB32 is 14,391 kb in length and is flanked by 21 bp direct repeats that are found in other SaPIbov sequences (Figure 3). Of the 18 open reading frames found in SaPIbov5, 11 were shared with SaPIbovPB32, which included the integrase-, excisionase-, and primase-like genes as well as a von Willebrand factor-binding gene (vwb). Strain PB32 also has a second vwb variant located on contig 12. Similar to PB32, it has been previously been reported that ruminant-associated Staph. aureus strains possess 2 variants of vwb, one located on SaPIbov and another located elsewhere on the chromosome (Guinane et al., 2010; Viana et al., 2010). The vwb gene contained within the SaPIbov is responsible for coagulation of ruminant-specific plasma and is suggested to play an important role in host adaptation (Guinane et al., 2010; Viana et al., 2010). The genome of strain 04-02981 does not demonstrate the presence of a SaPIBov sequence. Lastly, a gene encoding enterotoxin A (sea) that produces a product with 91% amino acid identity to the Staph. aureus strain COL SeaA (Gill et al., 2005) was located on contig 22 of the PB32 draft genome.
Figure 3.
Comparison of staphylococcal pathogenicity island PB32 (SaPIbovPB32; a) with SaPIbov5 (b). Direction of the arrows represents the orientation of the open reading frames (ORF). Open reading frames shared between SaPIbov5 and SaPIbovPB32 are indicated by white arrows. Black arrows in (a) indicate ORF found only in SaPIbovPB32 but not in the SaPIbov5, whereas the gray arrows in (b) indicate ORF found only in SaPIbov5. The hatched arrows at the ends of both sequences indicate the position of the 21-bp direct repeats (GAGTGGGAATAATTATATATA).
CONCLUSIONS
Pulsed-field gel electrophoresis analysis revealed clonally related Staph. aureus strains circulating within the dairy herd investigated. Strains represented by PFT-A, which included 13 MSSA strains and 2 mecA-positive strains (H15a and H15c), were the most widely distributed clones found within this herd. The presence of mecA-positive strains within PFT-A, including one that was also oxacillin resistant (H15a), suggests that a PFT-A clone has either gained or lost the mecA determinant. Collectively, genomic analysis corroborates the multidrug resistance phenotype displayed by H29 and revealed the presence of both a novel SaPIbov sequence and SCCmec remnant in PB32. Genome analysis also demonstrated that H29 carried enterotoxin genes on both plasmid and chromosome locations, whereas PB32 harbored a chromosomally located sea gene. This finding suggests that these strains are capable of causing staphylococcal food poisoning. In addition, although the draft genome of PB32 was most related to MRSA strain 04-02981, based on differences in SCCmec and SaPIbov content, ST, and ccrB sequences, PB32 and 04-02981 represent clearly diverged clones. Overall, our data demonstrates that H29 and PB32 represent novel clones of human and (or) bovine-related strains of Staph. aureus. We speculate that the unique genomic features of these strains possibly reflect the evolutionary trajectory they have taken in the dairy herd examined.
Acknowledgments
All authors acknowledge the participation of the Roadrunner Genomics Facility at New Mexico State University (NMSU; Las Cruces) and NSF award no. DBI-0821806. All authors also acknowledge prior support from the National Institutes of Health: SC-1GM083882-01 (J. E. Gustafson), R25 GM07667-30 [NMSU Minority Access to Research Careers (MARC) program], S06-GM61222-05 [NMSU Minority Biomedical Research Support-Research Initiative for Scientific Enhancement (MBRS-RISE) program], the National Center for Research Resources (5P20RR016480-12), and the National Institute of General Medical Sciences [8P20GM103451; New Mexico Idea Network of Biomedical Research Excellence (INBRE) program].
Footnotes
This Whole Genome Shotgun project has been deposited at the DNA Data Bank of Japan/European Bioinformatics Institute/GenBank (DDBJ/EMBL/GenBank) under the accessions PRJNA179361 (strain H29) and PRJNA179544 (strain PB32). The versions described in this paper are the first versions, ANHW01000000.
References
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakstad OG, Aasbakk K, Maeland JA. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J Clin Microbiol. 1992;30:1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (Clinical and Laboratory Standards Institute) Documents M100–S18 in Performance standards for antimicrobial susceptibility testing; eighteenth informational supplement. CLSI; Wayne, PA: 2008. [Google Scholar]
- Corkill JE, Anson JJ, Griffiths P, Hart CA. Detection of elements of the staphylococcal cassette chromosome (SCC) in a methicillin-susceptible (mecA gene negative) homologue of a fucidin-resistant MRSA. J Antimicrob Chemother. 2004;54:229–231. doi: 10.1093/jac/dkh284. [DOI] [PubMed] [Google Scholar]
- de Vries LE, Christensen H, Skov RL, Aarestrup FM, Agersø Y. Diversity of the tetracycline resistance gene tet(M) and identification of Tn916- and Tn5801-like (Tn6014) transposons in Staphylococcus aureus from humans and animals. J Anti-microb Chemother. 2009;64:490–500. doi: 10.1093/jac/dkp214. [DOI] [PubMed] [Google Scholar]
- Delgado A, Riordan JT, Lamichhane-Khadka R, Winnett DC, Jimenez J, Robinson K, O’Brien FG, Cantore SA, Gustafson JE. Heterovancomycin-intermediate methicillin-resistant Staphylococcus aureus isolate from a medical center in Las Cruces, New Mexico. J Clin Microbiol. 2007;45:1325–1329. doi: 10.1128/JCM.02437-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriese LA, Van Damme LR, Fameree L. Methicillin (cloxacillin)-resistant Staphylococcus aureus strains isolated from bovine mastitis cases. Zentralbl Veterinärmed B. 1972;19:598–605. doi: 10.1111/j.1439-0450.1972.tb00439.x. [DOI] [PubMed] [Google Scholar]
- Donnio P-Y, Février F, Bifani P, Dehem M, Kervégant C, Wilhelm N, Gautier-Lerestif A-L, Lafforgue N, Cormier M, Le Coustumier A MR-MSSA Study Group of the Collège de Bactériologie-Virologie-Hygiène des Hôpitaux de France. Molecular and epidemiological evidence for spread of multiresistant methicillin-susceptible Staphylococcus aureus strains in hospitals. Antimicrob Agents Chemother. 2007;51:4342–4350. doi: 10.1128/AAC.01414-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Dur-kin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinane CM, Ben Zakour NL, Tormo-Mas MA, Weinert LA, Lowder BV, Cartwright RA, Smyth DS, Smyth CJ, Lindsay JA, Gould KA, Witney A, Hinds J, Bollback JP, Rambaut A, Penadés JR, Fitzgerald JR. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol Evol. 2010;2:454–466. doi: 10.1093/gbe/evq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson JE, Wilkinson B. Staphylococcus aureus as a food pathogen: The staphylococcal enterotoxins and stress response system. In: Griffiths M, editor. Understanding Pathogen Behavior: Virulence, Stress Response and Resistance. Woodhead Publishing Ltd; Cambridge, UK: 2005. pp. 331–357. [Google Scholar]
- Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2001;45:1323–1336. doi: 10.1128/AAC.45.5.1323-1336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999;43:1449–1458. doi: 10.1128/aac.43.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, Etienne J, Vandenesch F, Bonneville M, Lina G. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol. 2001;166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- Jevons MP. “Celbenin”-resistant staphylococci. BMJ. 1961;1:124–125. [Google Scholar]
- Juhász-Kaszanyitzky É, Jánosi S, Somogyi P, Dán A, van der Graaf-van Bloois L, van Duijkeren E, Wagenaar JA. MRSA transmission between cows and humans. Emerg Infect Dis. 2007;13:630–632. doi: 10.3201/eid1304.060833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- Lee JH. Methicillin (Oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environ Microbiol. 2003;69:6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Skov RL, Han X, Larsen AR, Larsen J, Sørum M, Wulf M, Voss A, Hiramatsu K, Ito T. Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother. 2011;55:3046–3050. doi: 10.1128/AAC.01475-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist M, Isaksson B, Grub C, Jonassen TØ, Hällgren A. Detection and characterisation of SCCmec remnants in multiresistant methicillin-susceptible Staphylococcus aureus causing a clonal outbreak in a Swedish county. Eur J Clin Microbiol Infect Dis. 2012;31:141–147. doi: 10.1007/s10096-011-1286-y. [DOI] [PubMed] [Google Scholar]
- Lozano C, Aspiroz C, Ara M, Gómez-Sanz E, Zarazaga M, Torres C. Methicillin-resistant Staphylococcus aureus (MRSA) ST398 in a farmer with skin lesions and in pigs of his farm: Clonal relationship and detection of lnu(A) gene. Clin Microbiol Infect. 2011a;17:923–927. doi: 10.1111/j.1469-0691.2010.03437.x. [DOI] [PubMed] [Google Scholar]
- Lozano C, Aspiroz C, Ezpeleta AI, Gómez-Sanz E, Zarazaga M, Torres C. Empyema caused by MRSA ST398 with atypical resistance profile, Spain. Emerg Infect Dis. 2011b;17:138–140. doi: 10.3201/eid1701.100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong TT, Ouyang S, Bush K, Lee CY. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J Bacteriol. 2002;184:3623–3629. doi: 10.1128/JB.184.13.3623-3629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: Establishing a national database. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi MM, Wu SW, Zhou Y, Sieradzki K, de Lencastre H, Richardson P, Bruce D, Rubin E, Myers E, Siggia ED, Tomasz A. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci USA. 2007;104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nübel U, Dordel J, Kurt K, Strommenger B, Westh H, Shukla SK, Žemličková H, Leblois R, Wirth T, Jombart T, Balloux F, Witte W. A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog. 2010;6:e1000855. doi: 10.1371/journal.ppat.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien FG, Lim TT, Winnett DC, Coombs GW, Pearson JC, Delgado A, Langevin MJ, Cantore SA, Gonzalez L, Gustafson JE. Survey of methicillin-resistant Staphylococcus aureus strains from two hospitals in El Paso, Texas. J Clin Microbiol. 2005;43:2969–2972. doi: 10.1128/JCM.43.6.2969-2972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Żmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. Staphylococcus aureus CC398: Host adaptation and emergence of methicillin resistance in livestock. mBio. 2012;4:e00520–12. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards C, Emori TG, Edwards J, Fridkin S, Tolson J, Gaynes R. Characteristics of hospitals and infection control professionals participating in the National Nosocomial Infections Surveillance System 1999. Am J Infect Control. 2001;29:400–403. doi: 10.1067/mic.2001.118408. [DOI] [PubMed] [Google Scholar]
- Riordan JT, O’Leary JO, Gustafson JE. Contributions of sigB and sarA to distinct multiple antimicrobial resistance mechanisms of Staphylococcus aureus. Int J Antimicrob Agents. 2006;28:54–61. doi: 10.1016/j.ijantimicag.2006.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmenlinna S, Lyytikäinen O, Vainio A, Myllyniemi AL, Raulo S, Kanerva M, Rantala M, Thomson K, Seppänen J, Vuopio J. Human cases of methicillin-resistant Staphylococcus aureus CC398, Finland. Emerg Infect Dis. 2010;16:1626–1629. doi: 10.3201/eid1610.091571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz FJ, Hofmann B, Hansen B, Scheuring S, Lückefahr M, Klootwijk M, Verhoef J, Fluit A, Heinz HP, Köhrer K, Jones ME. Relationship between ciprofloxacin, ofloxacin, levofloxacin, sparfloxacin and moxifloxacin (BAY 12-8039) MICs and mutations in grlA, grlB, gyrA and gyrB in 116 unrelated clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 1998;41:481–484. doi: 10.1093/jac/41.4.481. [DOI] [PubMed] [Google Scholar]
- Shearer JE, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, LaMarre J, Lindsay JA, Bayles K, Nicholson A, O’Brien F, Jensen SO, Firth N, Skurray RA, Summers AO. Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci. G3 (Bethesda) 2011;1:581–591. doi: 10.1534/g3.111.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore AC, Deasy EC, Slickers P, Brennan G, O’Connell B, Monecke S, Ehricht R, Coleman DC. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:3765–3773. doi: 10.1128/AAC.00187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore AC, Rossney AS, O’Connell B, Herra CM, Sullivan DJ, Humphreys H, Coleman DC. Detection of staphylococcal cassette chromosome mec-associated DNA segments in multiresistant methicillin-susceptible Staphylococcus aureus (MSSA) and identification of Staphylococcus epidermidis ccrAB4 in both methicillin-resistant S. aureus and MSSA. Antimicrob Agents Chemother. 2008;52:4407–4419. doi: 10.1128/AAC.00447-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieradzki K, Leski T, Dick J, Borio L, Tomasz A. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: Multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J Clin Microbiol. 2003;41:1687–1693. doi: 10.1128/JCM.41.4.1687-1693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Green LE, Medley GF, Bird HE, Fox LK, Schukken YH, Kruze JV, Bradley AJ, Zadoks RN, Dowson CG. Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J Clin Microbiol. 2005;43:4737–4743. doi: 10.1128/JCM.43.9.4737-4743.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana D, Blanco J, Tormo-Más MÁ, Selva L, Guinane CM, Baselga R, Corpa JM, Lasa Í, Novick RP, Fitzgerald JR, Penadés JR. Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol Microbiol. 2010;77:1583–1594. doi: 10.1111/j.1365-2958.2010.07312.x. [DOI] [PubMed] [Google Scholar]
- Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. 2005;11:1965–1966. doi: 10.3201/eid1112.050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waness A. Revisiting methicillin-resistant Staphylococcus aureus infections. J Glob Infect Dis. 2010;2:49–56. doi: 10.4103/0974-777X.59251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Archer GL. Roles of CcrA and CcrB in excision and integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J Bacteriol. 2010;192:3204–3212. doi: 10.1128/JB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H, Louie L, Lo RY, Simor AE. Characterization of Staphylococcus aureus isolates with a partial or complete absence of staphylococcal cassette chromosome elements. J Clin Microbiol. 2010;48:3525–3531. doi: 10.1128/JCM.00775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulf MW, Sørum M, van Nes A, Skov R, Melchers WJ, Klaassen CH, Voss A. Prevalence of methicillin-resistant Staphylococcus aureus among veterinarians: An international study. Clin Microbiol Infect. 2008;14:29–34. doi: 10.1111/j.1469-0691.2007.01873.x. [DOI] [PubMed] [Google Scholar]