Abstract
There is an increasing interest in using Drosophila to model human brain degenerative diseases, map neuronal circuitries in adult brains, and study the molecular and cellular basis of higher brain functions. A whole-mount preparation of adult brains with well-preserved morphology is critical for such whole brain-based studies, but can be technically challenging and time-consuming. This protocol describes an easy-to-learn, one-step dissection approach of an adult fly head in less than 10 s, while keeping the intact brain attached to the rest of the body to facilitate subsequent processing steps. The procedure helps remove most of the eye and tracheal tissues normally associated with the brain that can interfere with the later imaging step, and also places less demand on the quality of the dissecting forceps. Additionally, we describe a simple method that allows convenient flipping of the mounted brain samples on a coverslip, which is important for imaging both sides of the brains with similar signal intensity and quality. As an example of the protocol, we present an analysis of dopaminergic (DA) neurons in adult brains of WT (w1118) flies. The high efficacy of the dissection method makes it particularly useful for large-scale adult brain-based studies in Drosophila.
Keywords: Neuroscience, Issue 118, Drosophila, Adult, Brain, Dissection, Dopamine, Neurons, Morphology, Fruit fly, Method, Immunofluorescence
Introduction
The model organism Drosophila, commonly known as the fruit fly, has long been valued for its elegant genetic tools, short reproductive times, and highly conserved molecular and cellular pathways. The fruit fly has been successfully employed to dissect basic signaling pathways, the patterning mechanisms of multicellular organisms, as well as the mechanisms underlying neuronal development, functions, and diseases1,2. With recent advances in cell labeling and imaging technologies, the fruit fly brain has become especially powerful in fine mapping of neuronal circuitry and in dissecting the molecular and cellular basis of higher brain functions, such as learning and memory, and circadian rhythm1,3,4,5,6,7,8.
One particular advantage of the Drosophila system is its relatively small size, allowing whole-mount preparation and examination of the brain using a regular compound or confocal microscope. This feature enables detailed anatomic and functional analyses of neuronal circuitry, or even a single neuron, at cellular and subcellular levels, in the context of a whole brain tissue, thus providing both a holistic view of the studied subject and its exact geometry within the whole brain. However, given the rather miniature size of the brain, it also presents a technical challenge in efficiently dissecting an intact brain tissue out of the protective exoskeleton head case in an adult fly. Various effective and relatively simple dissection methods have been described in detail, which usually involve careful and step-wise removal of the head case and the associated tissues including the eyes, trachea, and fat from the brain proper9, 10. These microsurgical dissection methods often place rather stringent demands on the quality of the dissection forceps, relying on forceps with fine well-aligned tips that can be easily damaged. Moreover, as the dissected brains are often separated from the rest of the body, the brains can be easily lost during the subsequent staining and washing processes because of their small sizes and their transparency in the processing buffer. Here, we describe a relatively simple and easy-to-learn, one-step dissection protocol for adult brains that keeps the dissected brains attached to the torso. The dissection process often easily clears away most of the brain-associated tissues such as the eye and trachea and reduces the demand for good quality dissection forceps.
Additionally, when imaging the brain under the fluorescent compound microscope or confocal microscope, the side of the brain that is away from the fluorescent light source often produces a weaker signal and less clear images due to the thickness of the whole-mount brain. Here, we also describe a simple mounting method that allows easy flipping of the brain samples, enabling convenient imaging of both sides of the brain with similar signal intensity and quality.
As a proof-of-concept for the application of this method to study the adult brain, we further examined the presence of DA neurons in the brains of w1118 flies; a genotype that is often used as the parental line for generating transgenic flies and the wildtype control in many Drosophila studies.
Protocol
1. Solutions Used for Brain Dissection and Immunofluorescent Staining
Dissect the adult fly brains in artificial Cerebral Spinal Fluid (aCSF): 119 mM NaCl, 26.2 mM NaHCO3, 2.5 mM KCl, 1 mM NaH2PO4, 1.3 mM MgCl2, and 10 mM glucose. Before use, gas the aCSF with 5% CO2/95% O2 for 10 - 15 min and spike with 2.5 mM CaCl2. Sterilize the aCSF solution by filtering through a 0.22 µm membrane filter. Note: Store aCSF at 4 °C to prevent bacterial contamination. Fly brains can also be dissected in a Phosphate-buffered Saline (PBS) solution, although detailed comparisons of the effects of the two dissection solutions on the final imaging quality of the dissected brains have not been performed.
Use 4% paraformaldehyde (PFA) prepared in 1x PBS as the fixative solution, which is normally aliquoted and stored at -20 °C. Caution: PFA is toxic and corrosive and should be handled with care.
Use 1x PBS to rinse the PFA from the brains and after the last washing step, during the immunofluorescent staining of the dissected brains. Prepare the 1x PBS from 10x PBS stock solution (1.37 M NaCl, 27 mM KCl, 100 mM Na2HPO4, and 18 mM KH2PO4).
Use 0.3% Tween-20 in 1x PBS (1x PBT) for all the subsequent washing steps during immunofluorescent staining of the dissected brains.
2. Microsurgical Dissection of Adult Fly Heads
Note: The dissection procedure is illustrated in Figure 1.
Anesthetize the adult flies with CO2. Using forceps, pick up a fly and briefly dewax its cuticle by dipping it in 70% ethanol for 3 - 5 s. This ensures that no bubbles adhere to the fly cuticle when immersed in the aCSF or 1X PBS dissection solution.
Under a dissecting microscope with a light source at an angle that will not be blocked by hands, set the objective lens to attain a clear view of the fly (1.2X magnification). Use forceps with the nondominant hand to immobilize the animal and immerse the fly into the cold dissection solution with its abdomen facing upwards, as shown in Figure 1A.
Keep the forceps at a 160 - 170° angle with respect to the dissection plate, while exerting a small force onto the abdomen of the fly (illustrated as arrows in Figure 1A). This step will immobilize the fly and force it to move the head backwards by 15- 25° while extending the proboscis upwards. In the process, a soft and translucent region of the cuticle, underneath the proboscis, should become apparent. Increase the magnification and adjust the focal plane onto this region. Note: Do not hold the forceps too tightly, but rather just firm enough to stabilize the fly. Too much force will cause the inside organs to rupture into the solution.
- Use the dominant hand to press the dissecting forceps so that its tips are closed, which will generate a mild resistant force on the tips of the forceps, so that the forceps will spring open when pressure from the holding hand is released. Position the forceps perpendicularly to the forceps holding the fly, as shown in Figure 1B.
- Pierce the closed tips of the dissection forceps through the soft and translucent region of the cuticle underneath the proboscis, as illustrated by the red arrow in Figure 1B. It is important not to penetrate the forceps too deeply that it touches the brain. The brain proper should become visible. Maintain a stable grasp of the fly with the nondominant hand.
Quickly but steadily, release the tips of the dissection forceps by its own force and rely on the momentum from this release to tear away the exoskeleton surrounding the fly head. An intact brain proper should become visible (the white tissue immediately above the tip of the bottom forcep in Figure 1C) Follow an imaginary line to guide the momentum of the released forcep tips (illustrated as the outward red arrows shown in Figure 1C). This will gently remove the exoskeleton and most of the brain-associated eye and trachea from the brain. The proper timing and force applied to open the exoskeleton will be dependent on the investigator's experience. Note: This is the most critical step during the dissection. It is important to rely on the natural force generated from the momentum of the opening forceps to remove the exoskeleton while leaving the exposed brain remains attached to the rest of the body.
Use the forceps in the dominant hand to carefully remove the remnant accessory tissues, such as the tracheal that appears as white fiber structures remaining attached to the brain. Be careful not to pull or damage the brain proper.
3. Immunofluorescent Staining of the Brains
Fix the dissected brain samples with their associated torsos in approximately 1 mL of 4% PFA for 40 - 60 min. Note: For this and subsequent steps, keep the tubes or plates with samples on a nutator to properly mix the samples in the solution.
Rinse with 1 mL of 1x PBS by pipetting the solution up and down 3 times. Be careful not to aspirate the brains away.
Wash 5 times with 1x PBT for 5 min each. Keep the samples on the nutator during incubation.
Incubate with primary antibodies at proper dilution O/N at 4 °C.
The next day, remove the primary antibodies, and wash the samples 5 times with 1x PBT. Leave the samples for 5 - 10 min in the 1x PBT solution in between each wash. Keep the samples on the nutator during incubation.
Incubate the washed samples in appropriate secondary antibodies with the suggested dilution and time of incubation.
Wash the samples six times with 1x PBT with a 10 min incubation between each wash. This step is critical for the removal of nonspecifically bound secondary antibodies from the sample.
Incubate in a 1:10,000 dilution of 1 mg/mL 4',6'-diamidino-2-phenylindole (DAPI) in 1x PBT solution for 30 min. DAPI is a DNA dye that binds to the AT regions of DNA and can be used to reveal the overall morphology of the brain.
Rinse three times with 1x PBS.
Transfer samples to a dissection dish, and separate the brains from the bodies in a dissection dish using forceps.
- After the staining and washing steps, mount the brains in between two coverslips and place the coverslips on top of a mounting slide. In this way, the brain samples can be easily flipped so that both sides of the brain can be imaged with a similar signal intensity. Note: Mounting the brain on the coverslip instead of directly on the mounting slide allows flipping of the sample's orientation freely. The mounting process is explained in more detail below.
- Place an 18 x 18 mm coverslip on top of a mounting slide. Widen the opening of a pipette tip with a blade and use it to transfer the brains in 1x PBS solution onto the 18 x 18 mm coverslip.
- Place the brains on the middle of the coverslip, remove the liquid around the brains with a fine pipette tip, and then add another 20 - 40 µL of anti-fade mounting medium (0.2% (w/v) n-propyl gallate in 99.5% ACS grade glycerol) over the brains. Note: The amount of mounting medium added depends on the number of brains examined.
- Gently spread out the brains while displacing the viscous mounting medium outwards. Place a second 18 x 18 mm coverslip on top of the brains by first lowering the slide from one side until it makes contact with the surface of the mounting slip, and then slowly lowering the other side to minimize brain contact with air bubbles. Note: There is no need to use a spacer in between the two coverslips, as the volume of the mounting medium and the brains should provide a space sufficient to separate the two coverslips.
- Allow the slides to settle down. Remove excessive liquid that come out from the sides of the slips with a laboratory tissue.
- To prevent the coverslips with the mounted brains from falling off the mounting slide during transportation, use a small piece of tape to attach the coverslips to the mounting slide. Note: Brains mounted in between the slides can be imaged immediately or preserved at 4 °C for up to one week without significant loss of fluorescent signal, without a need to use any sealant to seal the slides. For long-term preservation of the stained brains, the side of the two slips can be sealed with nail polish and stored in a -20 °C freezer, although stained brains might lose their signals over time. This is not recommended.
- Use an appropriate compound fluorescent microscope or confocal microscope to image the brains.
- First, acquire the image from the side of the brain closer to the objective, and then carefully flip the coverslips to have the samples sandwiched in the middle. This step facilitates acquiring the image of the other side of the same brain.
- Use an upright compound fluorescent microscope and a 20X objective lens to image the whole brain (examples in Figure 2) and a 40X objective to image smaller regions of the brain, such as DA neurons in the Paired Anterior Medial (PAM) cluster region (examples in Figure 3).
Representative Results
Figure 1 illustrates the main procedures for adult brain dissection, as described above. Figures 2 and 3 are representative images of 3-day-old WT (genotype: w1118) adult fly brains, which were costained with an antibody against Tyrosine Hydroxylase (TH, colored in red in Figure 2 and white in Figure 3), a marker commonly used to label DA neurons11, in addition to the DNA dye DAPI to label all the cell nuclei and an antibody against Elav protein, a marker for all differentiated neurons in the fly (colored in green), which together revealed the overall structure of the brain.
For the primary antibodies in Figures 2 and 3, we used rabbit anti-TH antibody at a 1:200 dilution and rat anti-Elav antibody at a 1:100 dilution, which were prepared in 1x PBT with 5% normal goat serum to block nonspecific binding. For secondary antibodies, we used Alexa Fluor 488-conjugated goat-anti-rat antibody and AlexaFluor596-conjugated goat anti-rabbit antibody at a 1:500 dilution in 1x PBT.
To image the brains, we used an upright compound fluorescent microscope equipped with an apotome function to acquire Z-stack images of brain slices that cover the whole depth of the region of the interest in the brain. For example, to obtain a clear visualization of the general distribution of DA neurons in the whole brain, we used a 20X objective lens to separately image the anterior and posterior sides of the same brain (Figure 2). The depth of each side of the brain imaged that can cover all the DA neurons is about 20 - 25 µm in total. To reliably visualize and quantify DA neurons in the PAM cluster region, which have smaller cell sizes and weaker TH signals (see below), we used a 40X objective lens. Additionally, in the Z-series acquisition function from the commercial software, we selected a section thickness of 0.5 - 1 µm between each imaged slice, which ensured optimal image quality in a 3D reconstruction of the PAM cluster region (Figure 3c). The thickness of PAM cluster in the brain that covers all the DA neurons is about 8 - 10 µm in total. In our experience, the apotome function can significantly reduce the noise signal in imaging thick samples with high background, such as a whole brain or the PAM cluster region.
Figures 2A-E show the anterior view of a brain, whereas Figures 2F-J show the posterior view of the same brain after flipping the coverslips that hold the brain samples, which display a similar level of signal intensity between two sides of the brain for all the three imaged channels. The DA neurons were grouped into different clusters following the early designation11, although here we use the name Paired Posterior Medial (PPM) to include the PPM1, PPM2, and PPM3 clusters at the anterior side of the brain, and the name Paired Posterior Lateral (PPL) to cover the PPL1 and PPL2 clusters from the posterior side of the brain, as depicted in Figures 2E and 2J. Figures 2E and 2J in white show anterior and posterior views of the same brain in high-magnification, revealing the different clusters of DA neurons on both sides of the brain. The white dashed lines highlight the prominent PAM and Paired Anterior Lateral (PAL) clusters in the anterior side of the brain (Figure 2E) as well as the PPL and the PPM clusters in the posterior side of the brain (Figure 2J).
Figures 3A and 3B are summaries of the quantification results for some of the DA clusters in w1118 flies. We counted DA neurons slice by slice and also from 3D reconstructed images, which should minimize imprecise counting. We estimate that the 3-day-old w1118 flies have an average of 27 DA neurons overall in the PPM cluster per brain, 16 DA neurons in the PPL cluster per hemisphere, 5 DA neurons in PAL cluster per hemisphere (Figure 3A), and 97 DA neurons in each of the PAM clusters (Figure 3B). Figure 3C is a representative 3D reconstruction of DA neurons in the PAL and PAM clusters, projected from slices of high-magnification images covering all the DA neurons in this region. It is apparent that compared to other clusters such as the PAL, DA neurons in the PAM cluster have relatively smaller cell sizes and weaker TH staining signals.
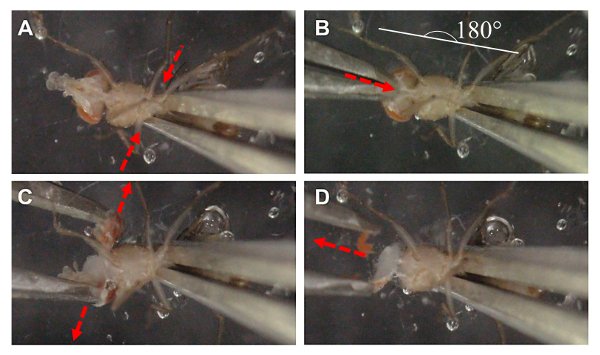 Figure 1: Graphical Illustration of the Dissection Protocol for Adult Drosophila Brain. (A) Flies are positioned with the ventral side upwards, and held by the thorax using the non-dominant hand. Force was gently applied (red arrows) to the forceps to induce backward tilting of the fly head and slight outward extension of the proboscis, exposing the white transparent region below. (B) Forceps from the dominant hand are inserted in the transparent region beneath the proboscis, creating a shallow incision (red arrow). Note that the forceps we use do not have very fine tip ends. (C) Forceps are allowed to come apart through its own force, as indicated by the red arrows. The momentum generated by opening the forceps removes the eyes and exoskeleton, exposing a relatively clean Drosophila brain. Most of the trachea are removed in the process. (D) Excess accessory tissues surrounding the brain are carefully trimmed away for full exposure of the brain proper. Please click here to view a larger version of this figure.
Figure 1: Graphical Illustration of the Dissection Protocol for Adult Drosophila Brain. (A) Flies are positioned with the ventral side upwards, and held by the thorax using the non-dominant hand. Force was gently applied (red arrows) to the forceps to induce backward tilting of the fly head and slight outward extension of the proboscis, exposing the white transparent region below. (B) Forceps from the dominant hand are inserted in the transparent region beneath the proboscis, creating a shallow incision (red arrow). Note that the forceps we use do not have very fine tip ends. (C) Forceps are allowed to come apart through its own force, as indicated by the red arrows. The momentum generated by opening the forceps removes the eyes and exoskeleton, exposing a relatively clean Drosophila brain. Most of the trachea are removed in the process. (D) Excess accessory tissues surrounding the brain are carefully trimmed away for full exposure of the brain proper. Please click here to view a larger version of this figure.
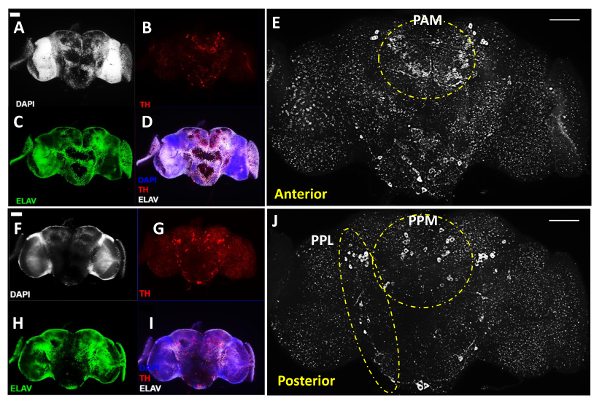 Figure 2: DA Neurons Revealed with anti-Tyrosine Hydroxylase (TH) Antibody in Adult Male Drosophila Brain (Acquired with a 20X Objective). This figure includes representative images of dissected adult brains, reconstructed using Z-stacked images of the brain region of interest obtained with the compound fluorescent microscope. (A-E) Anterior and (F-J) Posterior views of the same adult brain; (A & F) Cell nuclei were imaged by DAPI staining (white) and (C & H) neurons were labeled by pan-neuronal marker anti-Elav antibodies (green); (B & G) DA neurons are revealed by anti-TH antibodies (red). (D & I) Overlay of images for DAPI, TH and Elav treatment. (E & J) Enlarged view of the brain regions with DA neurons shown in white. Dashed lines highlight the PAL and PAM clusters in the anterior of the brain and PPL and PPM clusters in the posterior of the brain. The scale bar represents 50 µm in all of the panels. Please click here to view a larger version of this figure.
Figure 2: DA Neurons Revealed with anti-Tyrosine Hydroxylase (TH) Antibody in Adult Male Drosophila Brain (Acquired with a 20X Objective). This figure includes representative images of dissected adult brains, reconstructed using Z-stacked images of the brain region of interest obtained with the compound fluorescent microscope. (A-E) Anterior and (F-J) Posterior views of the same adult brain; (A & F) Cell nuclei were imaged by DAPI staining (white) and (C & H) neurons were labeled by pan-neuronal marker anti-Elav antibodies (green); (B & G) DA neurons are revealed by anti-TH antibodies (red). (D & I) Overlay of images for DAPI, TH and Elav treatment. (E & J) Enlarged view of the brain regions with DA neurons shown in white. Dashed lines highlight the PAL and PAM clusters in the anterior of the brain and PPL and PPM clusters in the posterior of the brain. The scale bar represents 50 µm in all of the panels. Please click here to view a larger version of this figure.
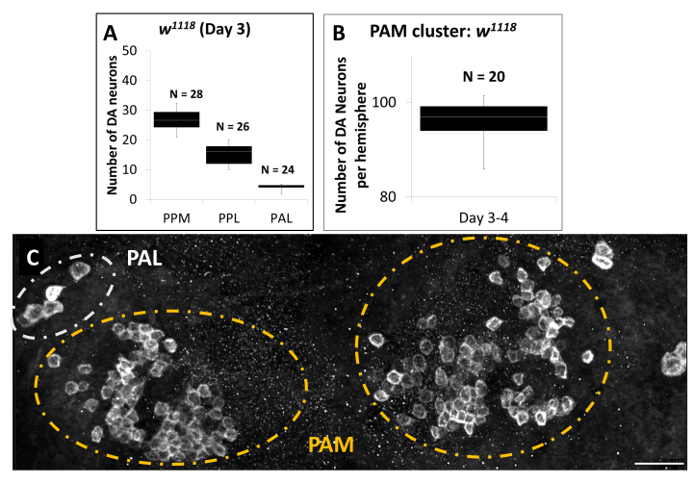 Figure 3. Quantification of DA Neurons in Brains of Male adult w1118 flies (Acquired with a 40X Objective). (A & B) Quantification of DA neurons of day 3 male w1118 flies, revealed by anti-TH staining. Data are represented by whisker box plots showing minimum and maximum values as the outliers; as well as the first (Q1), second (Q2) and third (Q3) quartiles. The second quartile is represented as the mean value. (A) PPM {(n = 28) Min: 21, Q1: 24, Average: 27, Q2: 30, Max: 32}, PPL {(n = 26) Min: 10, Q1: 12, Average: 16, Q2: 18, Max: 25}, and PAL {(n = 24), Min: 2, Q1: 4, Average: 5, Q2: 5, Max: 10} clusters; (B) PAM cluster {(n = 20) Min: 86, Q1: 94, Average: 97, Q2: 97, Max: 99}. (C) A representative projection image showing DA neurons in the PAM and PAL clusters of male w1118flies at 3 d. Scale bar represents 20 µm. Please click here to view a larger version of this figure.
Figure 3. Quantification of DA Neurons in Brains of Male adult w1118 flies (Acquired with a 40X Objective). (A & B) Quantification of DA neurons of day 3 male w1118 flies, revealed by anti-TH staining. Data are represented by whisker box plots showing minimum and maximum values as the outliers; as well as the first (Q1), second (Q2) and third (Q3) quartiles. The second quartile is represented as the mean value. (A) PPM {(n = 28) Min: 21, Q1: 24, Average: 27, Q2: 30, Max: 32}, PPL {(n = 26) Min: 10, Q1: 12, Average: 16, Q2: 18, Max: 25}, and PAL {(n = 24), Min: 2, Q1: 4, Average: 5, Q2: 5, Max: 10} clusters; (B) PAM cluster {(n = 20) Min: 86, Q1: 94, Average: 97, Q2: 97, Max: 99}. (C) A representative projection image showing DA neurons in the PAM and PAL clusters of male w1118flies at 3 d. Scale bar represents 20 µm. Please click here to view a larger version of this figure.
Discussion
With an increasing interest in using adult Drosophila brain to study human brain diseases, neuronal circuitry, and higher brain functions, it is necessary to develop simple and quick methods to obtain intact fly brains for whole-mount analyses, which is especially important for large-scale brain-based screens. Our method provides a simple and easy-to-learn approach to dissect out a fly head (often in less than 10 s with experience) with well-preserved morphology that is largely cleared of associated tissues. As the dissected brains are still attached to the rest of the fly bodies, they usually sink to the bottom of the sample tube quickly and are easy to recognize and collect during the washing and staining steps. This significantly minimizes sample loss that can be a prominent issue in processing the brain samples of tiny physical sizes. This issue can be especially important for large-scale studies. The brain can be easily detached from the body after the completion of the staining process, before the mounting.
During the dissection, it is important to position the fly correctly, maintain the proper angles when removing the exoskeleton from the fly head, and gently exert the proper force in performing each step. It is recommended that the forceps are placed perpendicularly to each other in order to prevent decapitation of the fly (Figure 1B). As shown in the representative images in Figure 2, the brains prepared using this protocol produce satisfactory results. In this example, we focused on the DA neurons in the PAM cluster of adult brains. A Drosophila brain in average has total of about 280 DA neurons per protocerebrum, which are distributed in distinct clusters in different regions of the brain with distinctive projection patterns and functional output11, 12. Previous characterization of DA neurons in adult Drosophila brains, including fly models of human disease genes such as in parkin mutants, have largely focused on the PAL, PPL, and PPM clusters that have relatively large cell sizes and prominent expression of TH, the standard maker used for labeling DA neurons13-18.
However, the majority of DA neurons in the fly brain are found in the PAM clusters, with about 100 DA neurons per cluster. Compared to cells in other DA clusters, DA neurons in the PAM cluster normally show a smaller level of tyrosine hydroxylase expression and smaller cell sizes. In samples prepared from our dissection and staining protocol, the DA neurons in the PAM cluster can be clearly visualized and imaged in high-quality using a compound fluorescent microscope, without confocal imaging. Given its large number, quantification of DA neurons in the PAM cluster of different genetic backgrounds might produce more reliable and reproducible results. We suggest that DA neurons in the PAM cluster represent another useful system for studying DA biology and modeling DA-related disorders, such as Parkinson's disease.
In our experience, the apotome function from the microscope we used can significantly reduce the background signal, especially in examining samples with significant thickness, such as the whole adult fly brain. Although confocal microscope can produce images with more details, higher magnification and better quality, it is often time-consuming and costly. This is especially true for imaging a large number of thick samples such as adult brains that require series of Z-stack section for clear 3D reconstruction and deconvolution analysis. In this perspective, the apotome function represents a fast and cost-effective alternative to produce images of sufficient quality (e.g., images in Figures 2 and 3).
In brain studies, it is often necessary to image cells on both sides of the same brain. For example, DA neurons are present in clusters from both the anterior and posterior sides of the brain. However, due to its thickness, after a brain is mounted onto the slide, the side away from the light source (i.e., the side facing the mounting slide) usually gives rise to weaker signals and less clear images when imaged using regular compound and confocal microscopes. By mounting the samples in between the two cover slides, it allows convenient flipping of the samples on the mounted slide and subsequent imaging of both sides of the same brain with similar signal intensities. Figure 2 is an example of imaging DA neurons from both sides of fly brains using this method, which showed relatively comparable signaling intensity and imaging quality.
Several easy-to-follow approaches for dissecting adult fly brains have been nicely described9, 10. Our approach provides an alternative method that can further simplify the dissection and staining processes of adult fly brains. With more large-scale studies being conducted to map out the whole brain neuronal circuitry as well as its molecular and cellular constituents, it is foreseeable that automated dissection and imaging approaches can be developed for adult Drosophila brains to realize high-throughput genetic and drug screens in vivo in this classical genetic model.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We acknowledge Mr. Enes Mehmet, Ms. Kiara Andrade, Ms. Pilar Rodriguez, Chris Kwok, and Ms. Danna Ghafir for their immense support to the project.
References
- Wangler MF, Yamamoto S, Bellen HJ. Fruit flies in biomedical research. Genetics. 2015;199:639–653. doi: 10.1534/genetics.114.171785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen HJ, Yamamoto S. Morgan's legacy: fruit flies and the functional annotation of conserved genes. Cell. 2015;163:12–14. doi: 10.1016/j.cell.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, et al. The neuronal architecture of the mushroom body provides a logic for associative learning. Elife. 2014;3:04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001;11:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata N, et al. Distinct dopamine neurons mediate reward signals for short- and long-term memories. Proc Natl Acad Sci U S A. 2015;112:578–583. doi: 10.1073/pnas.1421930112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nern A, Pfeiffer BD, Rubin GM. Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc Natl Acad Sci U S A. 2015;112:2967–2976. doi: 10.1073/pnas.1506763112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell S. Neural Plasticity: Dopamine Tunes the Mushroom Body Output Network. Curr Biol. 2016;26:109–112. doi: 10.1016/j.cub.2015.12.023. [DOI] [PubMed] [Google Scholar]
- Wolff T, Iyer NA, Rubin GM. Neuroarchitecture and neuroanatomy of the Drosophila central complex: A GAL4-based dissection of protocerebral bridge neurons and circuits. J Comp Neurol. 2015;523:997–1037. doi: 10.1002/cne.23705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney ST, Hidalgo A, de Belle JS, Keshishian H. Dissection of adult Drosophila brains. Cold Spring Harb Protoc. 2011;2011:1472–1474. doi: 10.1101/pdb.prot066878. [DOI] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for dissecting Drosophila melanogaster brains for live imaging or immunostaining. Nat Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- Mao Z, Davis RL. Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits. 2009;3:5. doi: 10.3389/neuro.04.005.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KE, Humphrey DM, Hirth F. The dopaminergic system in the aging brain of Drosophila. Front Neurosci. 2010;4:205. doi: 10.3389/fnins.2010.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci U S A. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene JC, et al. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100:4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth AJ, et al. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease. Proc Natl Acad Sci U S A. 2005;102:8024–8029. doi: 10.1073/pnas.0501078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesah Y, et al. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development. 2004;131:2183–2194. doi: 10.1242/dev.01095. [DOI] [PubMed] [Google Scholar]
- Trinh K, et al. Decaffeinated coffee and nicotine-free tobacco provide neuroprotection in Drosophila models of Parkinson's disease through an NRF2-dependent mechanism. J Neurosci. 2010;30:5525–5532. doi: 10.1523/JNEUROSCI.4777-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Kim SH, Kim J, Kim H, Yim J. Glutathione s-transferase omega 1 activity is sufficient to suppress neurodegeneration in a Drosophila model of Parkinson disease. J Biol Chem. 2012;287:6628–6641. doi: 10.1074/jbc.M111.291179. [DOI] [PMC free article] [PubMed] [Google Scholar]


