SUMMARY
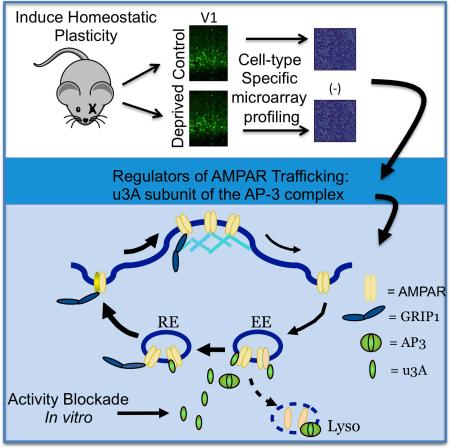
Synaptic scaling is a form of homeostatic plasticity driven by transcription-dependent changes in AMPA-type glutamate receptor (AMPAR) trafficking. To uncover the pathways involved we performed a cell-type specific screen for transcripts persistently altered during scaling, which identified the μ subunit (μ3A) of the adaptor protein complex AP-3A. Synaptic scaling increased μ3A (but not other AP-3 subunits) in pyramidal neurons, and redistributed dendritic μ3A and AMPAR to recycling endosomes (REs). Knockdown of μ3A prevented synaptic scaling and this redistribution, while overexpression (OE) of full-length or truncated μ3A (that cannot interact with the AP-3A complex) was sufficient to drive AMPAR to REs. Finally, OE of μ3A acted synergistically with GRIP1 to recruit AMPAR to the dendritic membrane. These data suggest that excess μ3A acts independently of the AP-3A complex to re-route AMPAR to RE, generating a reservoir of receptors essential for the regulated recruitment to the synaptic membrane during scaling up.
Graphical abstract
INTRODUCTION
The ability of networks to maintain stable function over time, and to efficiently store information, is thought to rely on homeostatic plasticity mechanisms that stabilize neuronal and network activity (Davis, 2013; Turrigiano and Nelson, 2004). Synaptic scaling is a form of homeostatic plasticity that scales postsynaptic strength up or down in response to perturbations in neuronal firing (Gainey et al., 2009, 2015; Goold and Nicoll, 2010; Ibata et al., 2008; Meadows et al., 2015), a process thought to contribute to the stabilization of firing rates both in vitro (Turrigiano et al., 1998) and in vivo (Hengen et al., 2013, 2016). Synaptic scaling is accomplished through changes in the abundance of postsynaptic AMPA receptors (AMPAR), but despite great effort the full set of molecular trafficking events that homeostatically adjust synaptic AMPAR abundance are poorly understood (Pozo and Goda, 2010; Turrigiano, 2012). In particular, although synaptic scaling is known to be transcription-dependent (Gainey et al., 2015; Goold and Nicoll, 2010; Ibata et al., 2008, Meadows et al., 2015), the factor or factors that are transcriptionally regulated to drive synaptic scaling are largely unknown. We thus set out to devise an unbiased screen for factors that are persistently upregulated during synaptic scaling, in the hopes of gaining deeper insight into the transcription-dependent AMPAR trafficking pathways involved in this critical form of synaptic plasticity.
Synaptic scaling up is induced within primary visual cortex (V1) by brief sensory deprivation (Desai et al., 2002; Lambo and Turrigiano, 2013). Several studies have examined the transcriptional changes within extracts of V1 following visual deprivation protocols (Lachance and Chaudhuri, 2004; Majdan and Shatz, 2006; Tropea et al., 2006). However, these earlier studies probed tissue derived from total V1, including all cell types and all layers. This is problematic because synaptic scaling is expressed in a cell-type and layer specific manner (Desai et al., 2002; Maffei and Turrigiano, 2008), so this approach does not provide the necessary sensitivity to isolate transcripts that are specifically involved in synaptic scaling. For this reason we designed a screen that would allow us to probe for transcriptional changes in a defined population of pyramidal neurons in which we know synaptic scaling is induced. Two days of visual deprivation via intraocular TTX injection induces synaptic scaling up of mEPSCs onto Layer 4 (L4) star pyramidal neurons in rodent visual cortex during early postnatal development (Desai et al., 2002). Here we generated a mouse with mCitrine expressed within these L4 star pyramids, allowing us to probe for transcriptional changes in this specific cell population (Sugino et al., 2006). This highly targeted approach revealed a relatively small number of transcripts (30) with expression changes that reached criterion (fold change > 1.5, p < 0.0034) following visual deprivation.
Surprisingly, 2 of these, Ap3m1 and Ap4m, code for μ subunits (μ3A and μ4, respectively) of the heterotetrameric clathrin adaptor protein (APC) complexes AP-3 and AP-4. The APC family is comprised of 5 members (AP-1 through 5) that sort and shuttle membrane-bound cargo between different endosomal and cell-surface compartments (Faundez et al., 1998; Hirst et al., 2013; Le Borgne et al., 1998; Nakatsu and Ohno, 2003; Newell-Litwa et al., 2007; Robinson and Bonifacino, 2001; Simpson et al., 1997); the μ subunits are critical for most cargo recognition (Bonifacino and Traub, 2003; Mardones et al., 2013; Ohno et al., 1998; Traub and Bonifacino, 2013). Although none of these complexes were previously known to have activity-regulated expression, several of them have been implicated in basal sorting and trafficking of Glutamate receptors (Margeta et al., 2009; Kastning et al., 2007; Lee et al., 2002). In particular, μ3A and μ4 can bind AMPAR indirectly through interactions with TARPS (Matsuda et al., 2008; Matsuda et al., 2013), and this interaction is important for dendritic trafficking of AMPAR (Matsuda et al., 2008) and for NMDA-induced trafficking of internalized AMPAR from early endosomes to lysosomes (AP-3, Matsuda et al., 2013). These considerations suggest that μ3A could contribute to the regulated trafficking of AMPAR that underlies synaptic scaling up (Kennedy and Ehlers, 2006; Turrigiano, 2008).
Here we show that μ3A upregulation plays an essential role in synaptic scaling up, by trafficking GluA2-containing AMPAR into the recycling pathway. AP-3 is classically known for sorting membrane proteins into the lysosomal pathway for degradation (Bonafacino and Traub, 2003), so it was surprising to find that TTX treatment re-routed μ3A from lysosomes to recycling endosomes (RE). Both TTX treatment and overexpression (OE) of μ3A were able to drive AMPAR into the RE pathway, as was OE of a truncated μ3A that cannot interact with the rest of the AP-3 complex (Mardones et al., 2013). Further, knockdown (KD) of μ3A (which blocks the normal increase in μ3A induced by activity-blockade) prevented the activity-dependent re-routing AMPAR to REs, and blocked synaptic scaling. Finally, OE of μ3A acted synergistically with the GluA2 trafficking protein GRIP1 to recruit AMPAR to the dendritic surface. Taken together these data show that activity-blockade transcriptionally upregulates μ3A to re-route AMPAR into the RE pathway, and is essential for the recruitment of AMPAR to the cell surface during synaptic scaling up (Fig. S1).
RESULTS
We set out to identify in an unbiased manner a set of candidate “scaling factors” that might participate in transcription-dependent synaptic scaling up. The ex vivo slice and profiling studies were conducted on HsCt5 mice (see below) at postnatal day (P)14-15, an age when synaptic scaling can be induced within L4 star pyramidal neurons by 2 days of optic nerve blockade using intraocular TTX (Desai et al., 2002, Fig. 1A). In vitro experiments were performed on postnatal visual cortical cultures after 7-8 days in vitro, and pyramidal neurons were targeted using standard morphological features as described (Watt et al., 2000).
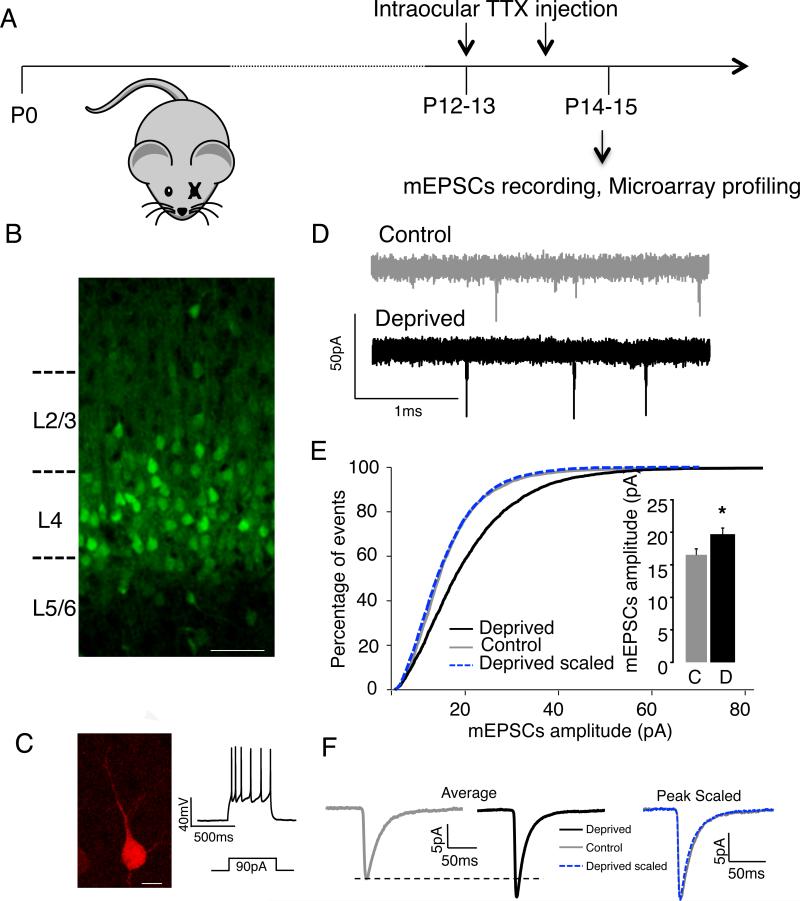
Figure 1. Characterization of synaptic scaling in L4 star pyramidal neurons in the HsCt5 mouse.
A, Visual deprivation paradigm. B, Coronal section from HsCt5 mouse at P15, showing mCitrine+ neurons (green) in V1. Scale bar, 50 μm. C, Recordings from L4 mCitrine+ neurons at P15. Fill during current clamp recording (left), and examples of evoked firing (right). D, Representative mEPSC recordings from control and deprived mCitrine+ neurons in L4 V1. E, Cumulative mEPSC amplitude distributions for control and deprived conditions; dashed line shows scaled-down amplitude distribution. Inset, average mEPSC amplitudes. F, Left, Average mEPSC waveforms and Right, Peak scaled mEPSC waveforms, to illustrate average kinetics. See also Fig. S2. Here and below, * indicates different from control, p<0.05; ** p<0.01; n's are given in the text. All summary data here and below are expressed as mean ± SEM.
Generation and characterization of the HsCt5 mouse
The HsCt5 mouse was generated as part of a lentiviral enhancer trap screen (Kelsch et al., 2012; Shima et al., 2016) through random insertion of a lentiviral vector containing the tet-transactivator (tTA) driving mCitrine. Upon screening lines for restricted mCitrine expression the HsCt5 line was identified as having expression largely restricted to neocortical Layer 4 (Fig. 1B, Fig. S2A). For many labeled neurons in V1 a thin apical dendrite could be discerned (Fig. S2B), as is typical of star pyramidal neurons (Desai et al., 2002; Maffei et al., 2004). When slices were fixed and stained against GABA (Fig. S2B,C) 97.5% of mCitrine+ neurons were GABA-negative (n = 685 cells from 3 animals). Whole-cell current clamp recordings from labeled neurons in slices from V1 revealed that recorded neurons had regular-spiking properties, and fills revealed the typical star-pyramid morphology (Fig. 1C, n = 5). Thus the vast majority of labeled neurons were excitatory star pyramidal neurons.
Next, we verified that excitatory synapses onto labeled neurons undergo synaptic scaling up upon visual deprivation. We used an established protocol (Desai et al., 2002; Maffei and Turrigiano, 2008) to block optic nerve impulses with intraocular TTX injections for 2 days commencing just before eye opening (~P12, Fig. 1A). Only one optic nerve was blocked, allowing us to use the contralateral monocular V1 from the same animals as a control. Slice recordings were then obtained from mCitrine+ neurons in either the deprived or the control hemisphere, and AMPAR-mediated mEPSCs measured as described previously (Desai et al., 2002; Maffei and Turrigiano, 2008)(Fig. 1D). Consistent with our previous findings in rats (Desai et al., 2002), 2d of optic nerve block significantly increased mEPSC amplitude in mCitrine+ neurons (Fig. 1E,F, n =11 each condition, p = 0.015), without affecting mEPSC frequency (p=0.17), mEPSC kinetics (rise and decay times, p > 0.29; Fig. F, peak-scaled mEPSC waveforms), or passive cellular properties (input resistance, Rin; resting membrane potential, Vm; and membrane capacitance, Cm, p > 0.05). Further, mEPSC amplitude was scaled up multiplicatively (Fig. 1E) as is characteristic of synaptic scaling both in vitro (Turrigiano et al., 1998) and in vivo at this developmental stage (Desai et al., 2002; Goel and Lee, 2007).
Microarray screen for the synaptic-scaling associated transcriptome
To probe for candidate synaptic scaling factors with expression levels persistently and significantly altered by visual deprivation, we used the same deprivation paradigm described above (Fig. 1A), then isolated the monocular portion of V1 from both the deprived and control hemispheres. L4 mCitrine+ cells were isolated, manually sorted, and their mRNA linearly amplified and used to probe affymetrix microarrays as previously described (see Methods; Sugino et al., 2006). Expression levels were then compared between the control and deprived hemisphere in three biological replicates. We identified the top 30 differentially-expressed transcripts based on robust expression changes (fold change > 1.5), rank-ordered by p value (Table 1; these data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE56758, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56758).
Table 1.
L4 Pyramidal Neuron Genes with Significantly Altered Expression During Synaptic Scaling.
| UP-REGULATED DURING SCALING | ||||
|---|---|---|---|---|
| Gene symbol | Fold Change | P value | Gene Name (description) | Function/Pathway |
| Ube2g1 * | 60 | 0.0001 | Ubiquitin-conjugating enzyme E2G 1 | Ub |
| Ap3m1 | 2.9 | 0.0004 | Clathrin adaptor-related protein complex 3, μA subunit | A |
| Ap4m | 2.2 | 0.0005 | Clathrin adaptor-related protein complex 4, μ subunit | A |
| Usp48 | 1.9 | 0.0002 | Ubiquitin Specific Peptidase 48 | Ub |
| Sic7A | 1.8 | 0.0003 | Solute carrier family 7 (cationic amino acid transporter) | T |
| Arh | 2.1 | 0.0007 | Aryl-hydrocarbon receptor | St |
| Cacnb1 | 1.9 | 0.0011 | Calcium channel, voltage-dependent, beta 1 subunit | |
| Ptprj | 3.0 | 0.0012 | Protein tyrosine phosphatase, receptor type, J | K/P |
| Dlg1 * | 17.4 | 0.0013 | Disks, large homolog 1 (SAP97) | |
| Nyap2 | 2.8 | 0.0016 | Neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 | K/P |
| PyCard | 3.5 | 0.0017 | PYD and CARD domain containing | |
| Mtmr15 | 1.9 | 0.0018 | Myotubularin related protein 15 | K/P |
| Apbh | 1.7 | 0.0019 | Androgen-binding protein beta | St |
| Tbrg1 | 6.2 | 0.0021 | Transforming growth factor beta regulated gene 1/NIAM | |
| Pfn1 | 2.1 | 0.0022 | Profilin 1 | |
| Pggt1b | 3.3 | 0.0022 | Protein geranylgeranyltransferase type I, beta subunit | |
| Hsdl2 | 5.6 | 0.0023 | Hydroxysteroid dehydrogenase-like 2 | St |
| Abcc1 | 1.5 | 0.0023 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 | T |
| Dph3 | 2.5 | 0.0024 | DPH3 Homologue | |
| Olfr138 | 1.7 | 0.0026 | Olfactory receptor 138 | |
| Rhbdd2 | 3.8 | 0.0026 | Rhomboid domain containing 2 | |
| Arfgef1 | 20 | 0.0026 | ADP-ribosylation factor guanine nucleotide-exchange factor 1 | |
| Map3k2 | 2.8 | 0.0027 | Mitogen activated protein kinase kinase kinase 2 | K/P |
| Dbp | 2.1 | 0.0030 | D site albumin promoter binding protein | |
| DOWN-REGULATED DURING SCALING | ||||
| Dync2h1 | 1.7 | 0.0003 | Dynein cytoplasmic 2 heavy chain 1 | |
| Sgpp1 | 2.0 | 0.0007 | Sphingosine-1-phosphate phosphatase 1 | K/P |
| Atg5 * | 2.4 | 0.0011 | Autophagy related 5 | |
| V1ra8 | 2.0 | 0.0015 | Vomeronasal 1 receptor, A8 | |
| Anapc1 * | 2.1 | 0.0021 | Anaphase promoting complex 1 (E3Ubligase) | Ub |
| Tceb1 | 1.5 | 0.0034 | Transcription elongation factor B | |
Key:
intronic probe set
A, Clathrin Adaptor Protein Complex family; Ub, ubiquitin pathway; St, steroid response; K/P, kinase or phosphatase; T, transporter. See also Table S1.
None of the classic activity-regulated genes (such as c-Fos, Arc, BDNF, or Npas1-4, Flavell and Greenberg, 2008), some of which (BDNF, Arc) have previously been implicated in synaptic scaling (Shepherd et al., 2006; Real Verde et al., 2006; Rutherford et al., 1998) showed changes in expression that reached criterion, likely because the manipulation we used here (2 d of optic nerve block) is subtle relative to classic activity deprivation or enhancement paradigms such as prolonged enucleation/dark-rearing or High K+/seizure induction (Flavell and Greenberg, 2008; Nedivi et al., 1993). There was also no change in expression of AMPA receptor or other glutamate receptor subunits. Finally, transcripts coding for a number of trafficking proteins, regulatory proteins, and kinases previously implicated in synaptic scaling (including GRIP1, PICK1, Homer1, PSD-95, Narp1, Plk2, CDK5, DHHC2, CaMKIV, and MeCP2, Turrigiano 2012), did not reach criterion for altered expression (Table S1).
None of the top differentially-expressed transcripts have previously been implicated in synaptic scaling. These include Ptprj (a protein tyrosine phosphatase), Map3K2 (a member of the Map kinase pathway), and Dbp (the clock gene D site albumin promoter binding protein)(Table 1). There were several transcripts involved in ubiquitin processing, including Ube2g1 (a ubiquitin conjugating enzyme), Usp48 (a predicted but uncharacterized ubiquitin peptidase), and Anapc1 (anaphase promoting complex 1, a member of a large E3 ubiquitin ligase complex); and several with known or putative involvement in protein trafficking, including Dync2h1 (cytoplasmic dynein 2 heavy chain 1), Dlg1 (the trafficking/scaffold protein SAP97), and Ap3m1 and Ap4m (μ subunits of the tetrameric clathrin adaptor protein complex, or APC, family members AP-3 and AP-4). The two most significant hits in this group were Ap3m1 (coding for μ3A) and Ap4m (coding for μ4), which increased 2.9 and 2.2 fold, respectively (Table 1). Interestingly, none of the other AP-3 or AP-4 subunits showed altered expression, and a second isoform of Ap3m, Ap3m2, (coding for μ3B) was also unaffected.
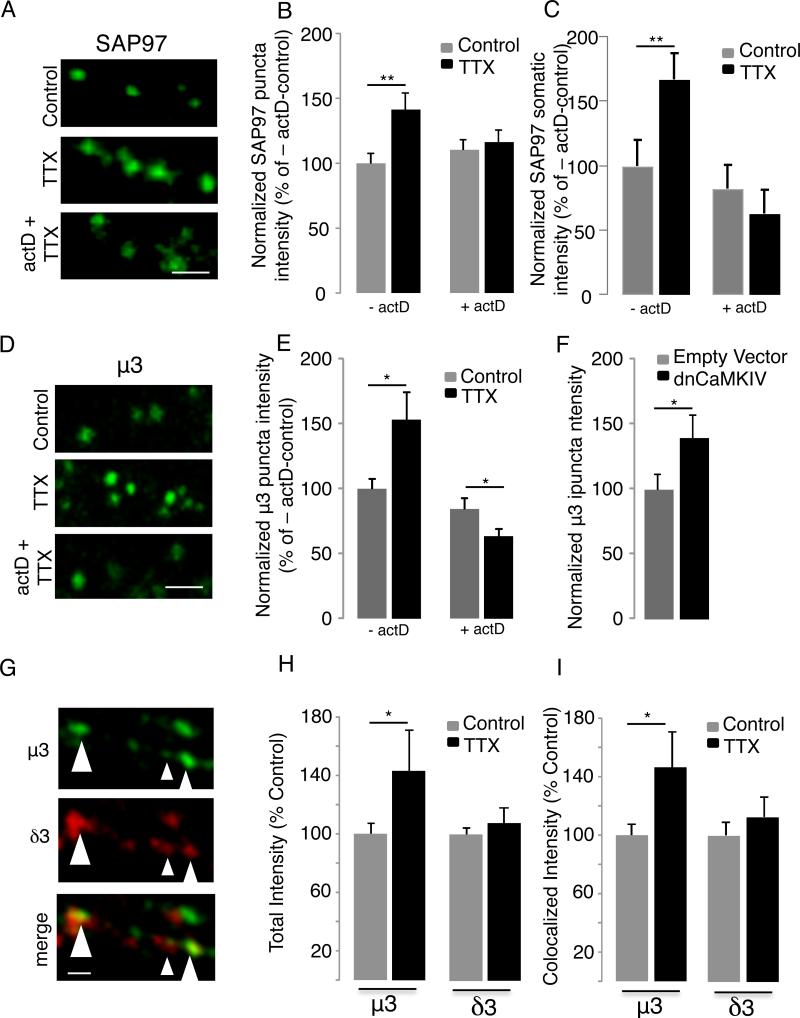
To determine whether these expression changes were predictive of increased protein levels during synaptic scaling, we analyzed a subset of these hits using quantitative immunohistochemistry in vitro (Fig 2, Fig S6A). We treated neocortical cultures with TTX for 6 hr (a period of time sufficient to induce transcription-dependent scaling up in vitro, Ibata et al., 2008, Gainey et al., 2015), then fixed, permeabilized, and stained against SAP97, μ3, or profilin 1, and quantified the intensity of the signal within the somatic and dendritic compartments of morphologically identified pyramidal neurons as described (Gainey et al., 2015, see methods)(Fig. 2). As there were no reliable commercially available antibodies against μ3 that worked for immunohistochemistry, we generated and characterized a μ3 rabbit polyclonal antibody (Fig. S3A-C, see also Methods). There is ~80% homology between μ3A and μ3B (Pevsner et al., 1994), so this antibody recognizes both μ3 subunits but does not recognize other μ subunits such as μ4 (Fig S3B). SAP97 (Fig. 2A-C; n = 10-24/condition) and μ3 (Fig. 2D,E; n = 16-23 neurons/condition) increased in a transcription-dependent manner within pyramidal neuronal dendrites (Fig. 2B,E) and somata (Fig. 2A-E) following TTX treatment. Another means of inducing synaptic scaling up is to express a dominant-negative form of CaMKIV in pyramidal neurons (Ibata et al., 2008; Pratt et al., 2011); like TTX treatment, this also significantly increased μ3 protein in pyramidal neuron dendrites (Fig. 2F, n = 11-13/condition, p < 0.04). In contrast, the profilin 1 signal was not significantly elevated by 6 hrs TTX treatment (p > 0.07); thus 2 of the 3 hits tested showed rapid upregulation at the protein level during synaptic scaling.
Figure 2. Activity-deprivation in vitro increases SAP97 and μ3 protein in pyramidal neurons.
A, Examples of pyramidal neuron dendrites stained against endogenous SAP97 in control, 6 h TTX, or 6 h TTX + ActD treated cultures; scale bar = 3 μm. B, Intensity of punctate SAP97 signal in the dendrites for conditions in A. C, Total intensity of SAP97 signal from pyramidal neuron somata. D, Examples of cultured pyramidal neuron dendrites stained against endogenous μ3 under control, 6 h TTX, or 6 h TTX + ActD; scale bar = 5 μm. E, Intensity of punctate μ3 signal for conditions in E. F, Total intensity of dendritic μ3 signal for EV and dnCaMKIV. G, Example of dendritic double-label against μ3 and δ3; scale bar = 2 μm. H,I; Quantification of puncta intensity for all μ3 and δ3 dendritic puncta (H) and for colocalized puncta (I) following TTX treatment. See also Fig. S3 for μ3 antibody characterization.
AP-3A is a heterotetrameric complex. To determine whether other subunits increased in tandem with μ3, we double-labeled against μ3 and the δ subunit δ3, which is obligatory for full AP-3 complex formation (Kantheti et al., 1998; Peden et al., 2002). Both antibodies recognized endosomal compartments within the soma and dendrites, with ~50% of δ3 dendritic puncta having detectible levels of colocalized μ3 (Fig. 2G). While TTX increased both total μ3 (Fig. 2H) and μ3 at colocalized sites (Fig. 2I, p<0.05), neither total δ3 nor colocalized δ3 were elevated by TTX (Fig. 2H, I; n = 17-20 neurons/condition). Thus, consistent with our microarray data, synaptic scaling is associated with a selective increase in the μ3 protein with no change in δ3. Further, a significant fraction of endosomal compartments contain detectible μ3 without detectible δ3 (Fig. 2G).
Synaptic scaling changes the endosomal distribution of μ3
AP-3A, which associates indirectly with AMPAR through μ3A binding to TARPs, has been implicated in the sorting and trafficking of glutamate receptors (Matsuda et al., 2013), suggesting that transcriptional upregulation of μ3A might play a critical role in synaptic scaling. As expected, immunohistochemistry localized μ3 to a number of endosomal compartments within pyramidal neurons (Fig. 3A-F), including a subset of those labeled with internalized transferrin receptor (TfR, a marker of RE), EEA1 (a marker of early endosomes, EE), and LAMP1 (a lysosomal marker).
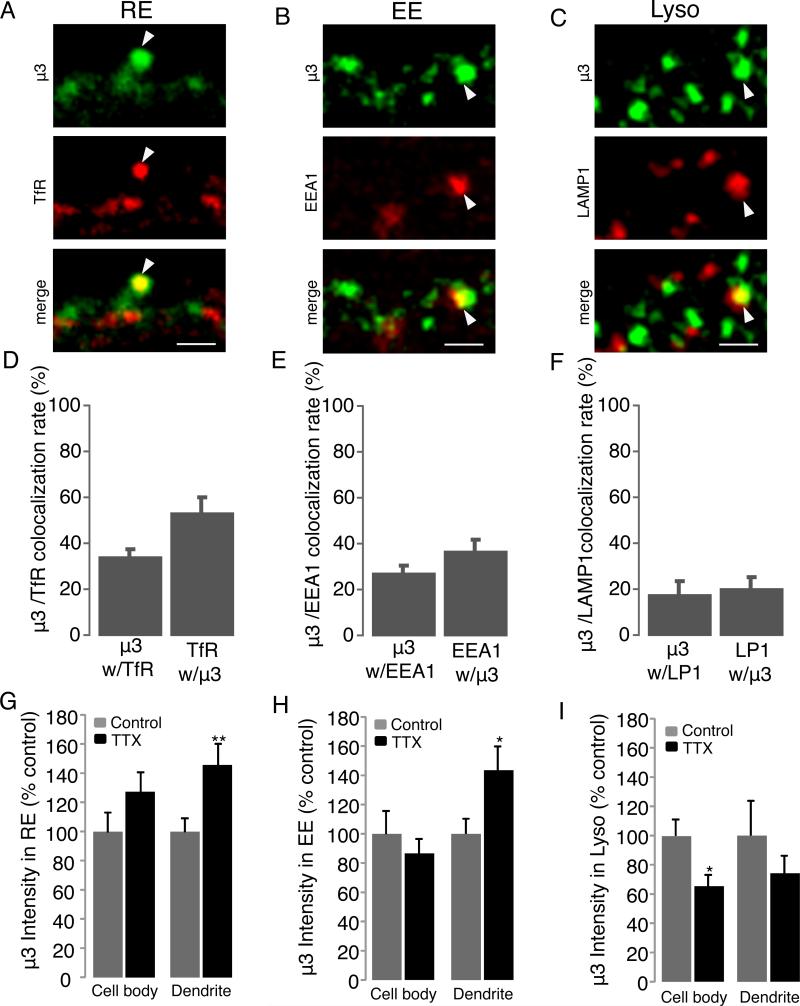
Figure 3. Activity-deprivation alters the endosomal distribution of μ3 within pyramidal neuron dendrites.
A-C, Examples of staining in pyramidal dendrites against endogenous μ3 (green) and (A) transferrin-labeled RE (TfR, red); (B) EE (EEA1, red); and (C) Lyso (LAMP1, red). Arrows show example of co-localized puncta. Scale bars = 2 μm. D-F, Quantification of colocalization rates between μ3 and TfR (D), EEA1 (E), and LAMP1 (F). G-H, Intensity of μ3 puncta in either the cell body or dendrites, for μ3 puncta colocalized with TfR in RE (G), with EEA1 in EE (H), and with LAMP1 in lysosomes (I) for control and TTX-treated neurons.
To determine whether TTX increases the accumulation of μ3 within specific endosomal compartments we performed double-labeling against μ3 and LAMP1 (lysosomes), TfR (RE), or EEA1 (EE), and quantified the intensity of the μ3 signal at colocalized sites. The μ3 signal associated with RE and EE was unchanged in the cell body, but was significantly elevated in the dendrites (Fig. 3G, H; n = 9-11 neurons/condition; p =0.007 RE, and 0.017 EE). In contrast, the μ3 signal associated with the lysosomal compartment decreased following TTX treatment in the cell body (Fig. 3I, n = 9-14 neurons/condition, p < 0.01). Thus TTX reduces the association of μ3 with lysosomes, while increasing its association with dendritic RE and EE. TTX did not significantly affect the colocalization rates between μ3 and these various endosomal markers (not shown). As a control, we verified that the experimental colocalization rate between μ3 and TfR is significantly higher than expected if the distribution of μ3 were random relative to the TfR signal, as described (Gainey et al., 2015); we ran the same control for triple colocalization between GluA2, μ3, and TfR, and found that the observed rates were many fold higher than predicted for random association (Fig. S4).
Upregulation of μ3A is necessary for synaptic scaling
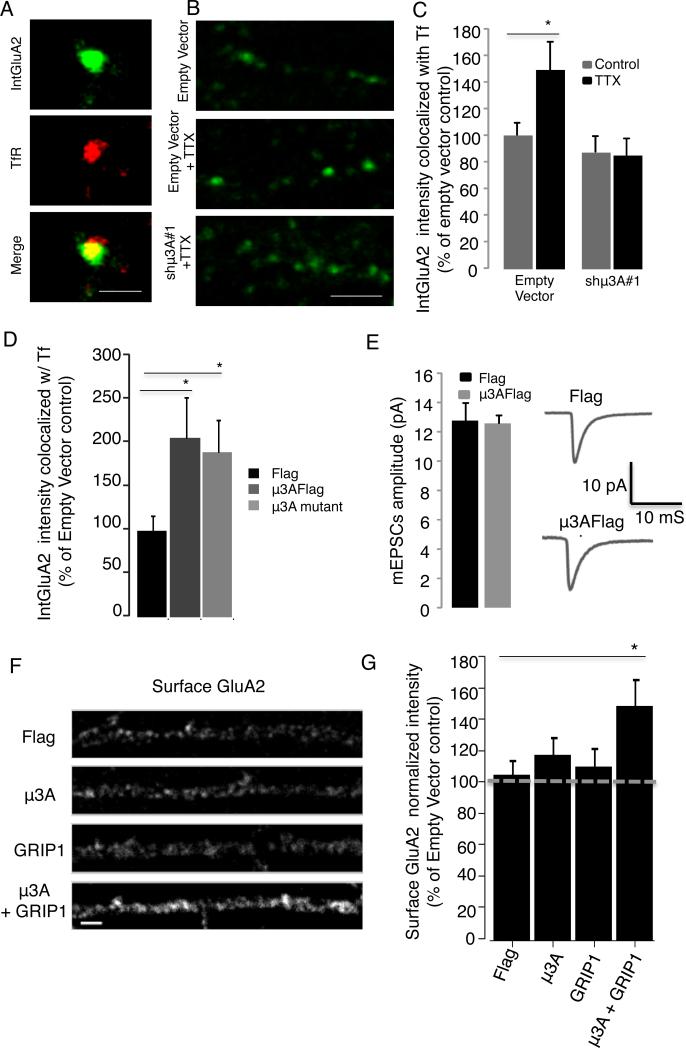
To determine whether μ3A is necessary for synaptic scaling up, we designed two small hairpin RNAs (shRNAs) targeted against two distinct and unique regions of the μ3A mRNA in order to knock down expression of μ3A, an approach that allowed us to target μ3A in an acute and cell-autonomous manner, thus avoiding circuit level and developmental defects due to prolonged loss of μ3A. Neurons were transfected with the hairpins or an empty vector (EV) along with EGFP at low efficiency (Fig. S6) for three days, then fixed, stained, and processed for immunohistochemistry against μ3. Both shRNAs reduced the intensity of μ3 staining in the somatic and dendritic compartments, to ~60% and ~40% of control, respectively (Fig. 4A-B, n = 9-13 neurons/condition, p < 0.05 soma and p < 0.01 dendrite). Note that this likely represents an underestimate of the degree of μ3A KD, as the shRNAs are specific for μ3A but the antibody recognizes both μ3A and μ3B (Fig S3B). Importantly, in KD neurons TTX treatment was not able to increase μ3 expression in either the cell body or dendrites (Fig. 4C).
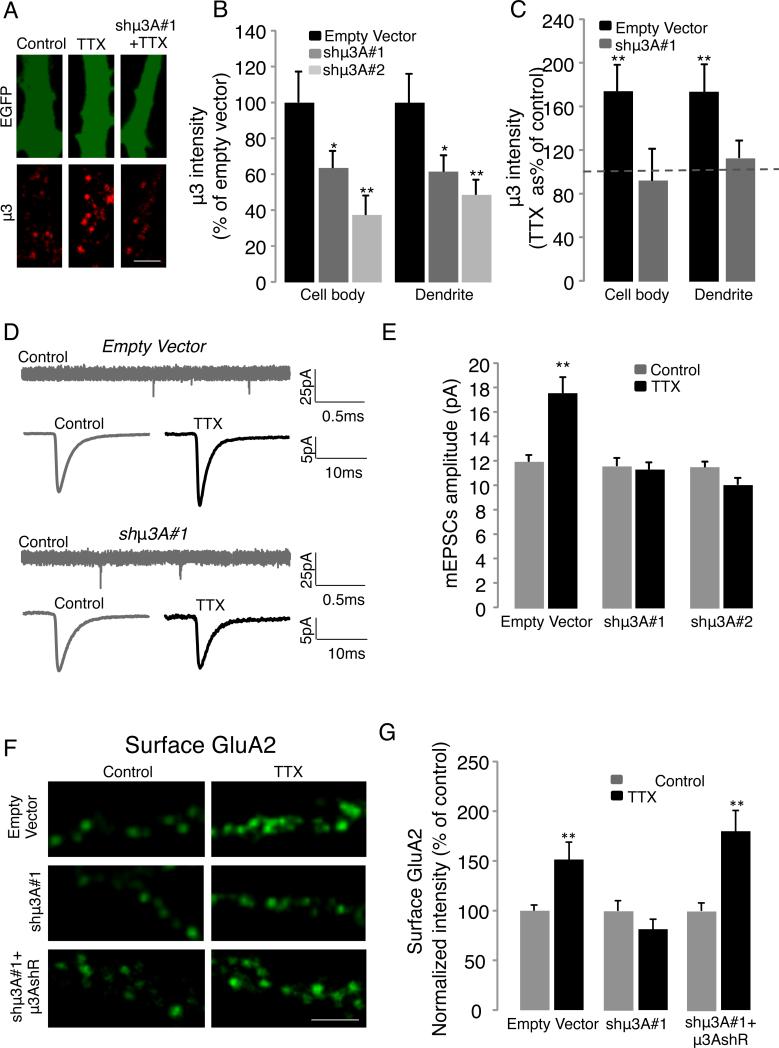
Figure 4. Knockdown of μ3A blocks synaptic scaling.
A, Example staining against endogenous μ3 for control and TTX-treated neurons expressing EV or the shRNA shμ3A #1 in pyramidal dendrites (scale bar = 5 μm). B, Quantification of μ3 signal for EV, shμ3A#1, and shμ3A#2. C, Quantification of μ3 signal for EV or shμ3A#1, ± TTX for 6 h. D, Representative mEPSC recording (top trace) and average mEPSC waveforms ± TTX (bottom traces) from EV or shμ3A#1- transfected pyramidal neurons. E, Average mEPSC amplitude for EV, shμ3A#1, and shμ3A#2-expressing neurons, ± TTX for 6h (TTX). F, Examples of staining against endogenous surface GluA2 for neurons expressing either EV, shRNA shμ3A #1, or co-expressing shμ3A #1 and RNAi-resistant μ3A (μ3AshR), ± TTX for 6 h (scale bar = 5 μm). G, Quantification of surface GluA2 signal in dendrites for the conditions in (F).
Pyramidal neurons transfected with the EV showed normal synaptic scaling up of mEPSC amplitude following 6 h TTX (Fig. 4D,E; n = 16-12/condition, p = 0.01). In contrast, neurons in which μ3A had been knocked down using either shRNA (which target unique and distinct sequences in the μ3A mRNA) failed to show synaptic scaling up (Fig. 4D, E, n = 11-9 neurons/condition). Interestingly, neither shRNA affected baseline mEPSC amplitude (Fig. 4E, ANOVA, p = 0.61) or frequency (ANOVA, p = 0.79).
Synaptic scaling in neocortical pyramidal neurons operates primarily through changes in synaptic accumulation of AMPAR (Turrigiano, 2008), so next we examined the ability of μ3A KD to prevent the TTX-induced enhancement of surface AMPAR. The intensity of the surface punctate GluA2 signal was increased by TTX treatment as expected (Fig. 4F, G, n = 9-12 neurons/condition, p = 0.04), and this increase was completely blocked by μ3A KD (Fig. 4G, shμ3A#1); similar results were obtained with GluA1 (not shown). Finally, coexpression of an RNAi-insentitive μ3A (μ3AshR) was able to rescue the affects of endogenous μ3A KD (4G, shμ3A#1 + μ3AshR); taken together with our data above using two distinct hairpins, this shows that the inability of neurons to undergo synaptic scaling is not the result of off-target effects of the shRNA.
μ3A is necessary to traffic GluA2 to RE during synaptic scaling up
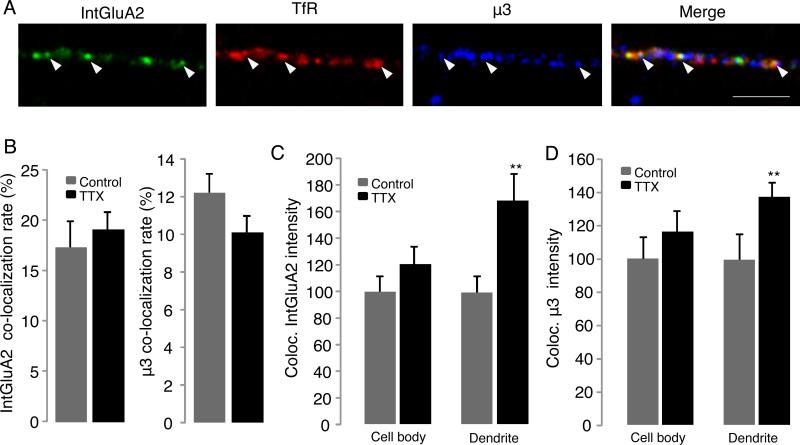
The data above suggest that enhanced expression of μ3A is critical for the regulated delivery of AMPAR to synapses during synaptic scaling. AMPAR are thought to be mobilized to the dendritic surface from RE and/or EE (Park et al., 2004), and we found that dendritic μ3A increases in both RE and EE during synaptic scaling (Fig. 3G,H). We thus wondered whether μ3A might play an important role in trafficking AMPAR to these compartments during scaling. To test this we first asked whether μ3A and GluA2 are colocalized within RE, and whether the amount of GluA2 at such colocalized sites is affected by TTX treatment. RE were labeled with TfR, and internal GluA2 was labeled by applying antibody to the surface, waiting 45 minutes for receptor internalization, stripping surface antibody, and then permeabilizing and staining for μ3 (Fig. 5A; Tatavarty et al., 2013; Gainey et al., 2015). A significant fraction of internalized GluA2 puncta were colocalized with μ3 in RE (Fig. 5B, left panel, ~18% in dendritic compartments), many fold higher than expected for random colocalization (Fig. S4). While TTX did not affect the fraction of RE with detectible GluA2 and μ3, there was a significant increase in the intensity of both the GluA2 and the μ3 signal within this population of dendritic RE following TTX treatment (Fig. 5C,D; n = 12-13/condition). Thus, TTX treatment simultaneously enhances the accumulation of GluA2 and μ3 within a subset of dendritic RE.
Figure 5. Activity-blockade increases the abundance of GluA2 in μ3-containing RE.
A, Example images of dendritic labeling for endogenous internalized GluA2 (IntGluA2, green), TfR-labeled RE (red) and endogenous μ3 (blue). Arrows indicates example triple-colocalized puncta. Scale bar = 5 μm. B, quantification of colocalization rates for Internalized GluA2 with μ3 and TfR (left), and for μ3 with IntGluA2 and TfR (right), ± TTX. C, Quantification of internalized GluA2 intensity at triple co-localized sites, in the cell body or dendrites of pyramidal neurons, ± TTX. D, Quantification of μ3 intensity at triple co-localized sites, in the cell body or dendrites of pyramidal neurons, ± TTX. See also Fig. S4.
Next, we asked whether this TTX-induced enhancement of GluA2 in RE depends on μ3A. Neurons were transfected with either EV or the shμ3A, and internal GluA2 in RE was labeled as above (Fig. 6A). In the EV condition TTX treatment increased the intensity of the GluA2 signal within the RE compartment, and this increase was completely blocked by KD of μ3A (Fig. 6B,C). This demonstrates that the deprivation-induced increase in μ3A is necessary for enhanced trafficking of GluA2-containing AMPAR to RE. Interestingly, KD of μ3A did not significantly affect basal levels of internal GluA2 (Fig. 6C, n = 12/condition, p = 0.40, compare EV to shμ3A), suggesting that the ability of μ3A KD to prevent the TTX-induced enhancement of surface GluA2 does not result from a gross defect in basal trafficking of GluA2 to RE.
Fig. 6. μ3 is necessary and sufficient to recruit GluA2 to RE, and enhances the ability of GRIP1 to recruit GluA2 to the dendritic surface.
A, Example of internalized GluA2 signal (green) within RE (TfR, red). Scale bar = 1 μm. B, dendritic internalized GluA2 signal for EV, EV + TTX, or shμ3A#1 +TTX conditions. Scale bar = 5. C, Quantification of intensity of internalized GluA2 co-localized with TfR in pyramidal dendrites transfected with EV or shμ3A#1, ± 6 h TTX. D, Quantification of intensity of internalized GluA2 co-localized with TfR in pyramidal dendrites transfected with EV, μ3A-Flag, or truncated μ3A-Flag. E, Average mEPSC amplitude for Flag or μ3AFlag-transfected pyramidal neurons. F, Example dendritic staining for surface GluA2 after transfection with Flag alone, μ3A-Flag, GRIP1, or μ3A-Flag + GRIP1. G, quantification of conditions in (F). See also Fig. S5 and S6.
To determine if upregulation of μ3A was sufficient to drive GluA2 to RE, we OE flag-tagged μ3A and quantified the GluA2 signal within RE as above. OE of μ3A indeed significantly increased the intensity of the GluA2 signal within RE, (Fig. 6D, EV, n = 46; μ3A, n = 43, p = 0.014). In contrast, μ3A OE did not affect the intensity of the GluA2 signal within other endosomal compartments (Fig. S6B,C). Next, we asked whether μ3A must interact with the AP-3 complex in order to traffic GluA2 to RE. The μ3A subunit interacts with the AP-3A complex through its N terminal domain, and interacts with TARPS (and thus AMPAR) through its C terminal domain (Aguilar et al., 1997; Mardones et al., 2013; Matsuda et al., 2013). Because TTX selectively increases μ3A without increasing expression of the other AP-3 subunits, and OE of μ3A alone is sufficient to recruit AMPAR to RE (Fig. 6D), we reasoned that μ3A might be acting independently of the full AP-3 complex during synaptic scaling. To test this idea further, we OE a truncated form of μ3A that lacks the N terminal domain but can still recognize and bind cargo (Mardones et al., 2013). Importantly, this truncated μ3A was still able to traffic to RE (Fig. S5), and, like full-length μ3A, was also able to recruit GluA2 to RE (Fig. 6D, n = 22, p= 0.03).
If the recruitment of GluA2 to RE were sufficient to drive an increase in synaptic strength during synaptic scaling, then OE μ3A should increase mEPSC amplitude. To test this, neurons were transfected with EV (n=8), or flag-tagged (n=8) or untagged μ3A (n = 10) constructs; neither μ3A construct was sufficient to scale up mEPSC amplitude (Fig. 6E). Thus increased μ3A expression is necessary (Fig. 4D,E) but not sufficient for the regulated recruitment of AMPAR to the synaptic membrane during synaptic scaling, suggesting that an additional trafficking step is required to bring internal AMPAR to the synaptic membrane. GRIP1 accumulates at sites of exocytosis as well as at synapses, and is essential to recruit AMPAR to synapses during synaptic scaling up (Gainey et al, 2015; Tan et al., 2015). To determine whether the μ3A-dependent trafficking of GluA2 into the recycling pathway enhances the ability of GRIP1 to recruit GluA2 to the membrane, we compared surface levels of endogenous GluA2 after OE of μ3A alone, GRIP1 alone, or μ3A + GRIP1 (Fig. 6F). While neither alone was sufficient to significantly increase surface GluA2, the two together produced a robust recruitment of GluA2 to the membrane (Fig. 6G, n = 25-33 neurons/condition, ANOVA followed by posthoc Tukey, test, p=0.024). Taken together, our data suggest a novel role for μ3A, in which selective activity-dependent upregulation of the μ3A (cargo-recognition) subunit reroutes AMPARs into the recycling pathway, where they can then be recruited to the membrane during a second trafficking step (Fig. S1).
DISCUSSION
To generate insight into the transcription-dependent processes that induce synaptic scaling up, we devised a cell-type specific screen to identify a set of candidate “scaling factors” with altered expression during scaling. This screen identified μ3A, the cargo-recognition subunit of the APC family member AP-3, important for sorting and trafficking of membrane-bound cargo between endosomal compartments. Because of the known association of μ3A with AMPAR, we set out to determine whether μ3A has a role in the regulated changes in AMPAR trafficking that drive synaptic scaling. We found that activity-blockade re-routes both μ3A and GluA2 into RE, without affecting the δ3 subunit (which is obligatory for complex formation). KD of μ3A prevented synaptic scaling and the redistribution of GluA2 into RE, while OE of either full-length μ3A or a truncated form that cannot interact with the AP-3A complex was sufficient to drive GluA2 to REs. Finally, OE of μ3A acted synergistically with GRIP1 to recruit GluA2 to the cell surface. Taken together these data support a model in which excess μ3A acts independently of the AP-3A complex to re-route AMPAR to RE, from whence they are recruited to the synapse to enhance synaptic strength during scaling up (Fig. S1).
We used an unbiased cell-type specific profiling approach (Sugino et al., 2006) that allowed us to probe for persistent transcriptional changes in a population of neurons (L4 star pyramidal neurons from V1) known to undergo synaptic scaling in response to visual deprivation during early postnatal development (Desai et al., 2002). None of the small number of candidates identified (Table 1) had previously been associated with synaptic scaling. Of note, there is no overlap between this candidate set and transcripts previously found to be regulated by visual deprivation in V1 of rodents or primates (Nedivi et al., 1996; Lachance and Chaudhuri, 2004; Majdan and Shatz, 2006; Tropea et al., 2006). This is likely due to large methodological differences in approach; most importantly, these previous studies probed whole V1 extracts, while here we probe changes in a specific cell type as these neurons are undergoing synaptic scaling. Because of the complexity of neocortical circuits and cell types and the diversity of plasticity mechanisms present (Feldman, 2009; Nelson and Turrigiano, 2008), this cell-type specific approach is likely critical for identifying candidates that are tied to particular forms of neocortical plasticity, rather than general circuit-wide responses to deprivation or other activity paradigms.
Interestingly, none of the signaling and trafficking proteins that have previously been linked to synaptic scaling through a candidate approach came up in our screen (see Table S1). As an example, the essential scaling factor GRIP1 is not transcriptionally regulated, consistent with the observation that, although GRIP1 increases at synapses during scaling, this is not accompanied by an increase in total GRIP1 protein (Gainey et al., 2015). This underscores the point that synaptic scaling involves the activity-dependent regulation of several AMPAR trafficking steps, only some of which are regulated at the level of transcription.
We chose to focus on μ3A because AP-3 plays a role in trafficking of receptors into dendrites and/or to the neuronal surface (Matsuda et al., 2008; Bendor et al., 2010), and more recently AP-3A has been shown to interact with AMPAR through an indirect association involving μ3A binding to the TARP stargazin (STG), an association that is important for the induction of LTD (Matsuda et al., 2013). None of the tetrameric APC family member subunits were previously known to exhibit activity-dependent transcriptional regulation, whereas here we find that both μ3A and μ4 (but not any of the other subunits of either APC) were upregulated by 2d of visual deprivation. AP-3A is primarily known for a role in sorting cargo from the Golgi or EE to lysosomes or LROs (Dell'Angelica, 1997; 2009), and during LTD the association of μ3A with stargazin enhances the trafficking of AMPAR to lysosomes (Matsuda et al., 2013). Thus it was a surprise to find that during synaptic scaling the upregulation of μ3A leads to enhanced accumulation of μ3A and AMPAR within the RE compartment, and a reduction in the association of μ3A with lysosomes. These data show that the localization of μ3A is dynamic and can be regulated by activity to redirect cargo (in particular, AMPAR) into the recycling endocytic pathway.
The RE compartment has been shown to be critical for supplying AMPAR during LTP (Hanley, 2010; Park et al., 2004), but whether they also supply AMPAR for synaptic scaling has been less clear (Gainey et al., 2015, Tan et al., 2015). Interestingly, although an increase in μ3A is necessary for scaling, and is necessary and sufficient to drive AMPAR to RE, it is not sufficient to increase mEPSC amplitude. This suggests that, once AMPAR are rerouted into the recycling pathway by μ3A, they must still be recruited to the synaptic membrane in a second activity-dependent trafficking step. The GluA2-interacting protein GRIP1 plays a critical role in recruiting AMPAR from internal endosomal compartments to the synaptic membrane during synaptic scaling (Gainey et al., 2015; Tan et al., 2015). Here we show that when μ3A and GRIP1 are OE together, they can act synergistically to enhance surface AMPAR accumulation, providing direct experimental support for a two-step trafficking model in which μ3A recruits AMPAR into the recycling pathway, where they can then be recruited to and stabilized by GRIP1 at the synaptic membrane (Fig. S1).
Systemic loss of functional AP-3 causes endosomal trafficking defects, Hermansky-Pudlak syndrome, and neurological symptoms (Kantheti et al., 1998; Lane and Deol, 1974; Peden et al., 2002; Seong et al., 2005; Sirkis et al., 2013; Swank et al., 2000; Yang et al., 2000). These have mainly been ascribed to defects in lysosomal trafficking and/or biogenesis of LROs. Interestingly, our data suggest that the μ3A subunit traffics AMPAR away from lysosomes and into the recycling pathway by acting independently of the full AP-3 complex. There are three main pieces of evidence that support this idea. First, while μ3A is upregulated during synaptic scaling, the δ3 subunit is not. Since the δ3 subunit is obligatory for formation of both the AP-3A and AP-3B complexes (Kantheti et al., 1998; Peden et al., 2002), this suggests that either μ3A is limiting for complex formation, or that activity-induced μ3A is not acting as part of the complex. The later interpretation is favored by the observation that the intensity of the μ3A signal increases even in endosomal compartments with no detectible δ3. Second, we find that OE of μ3A alone is sufficient to recruit AMPAR to RE, and to enhance the ability of GRIP1 to recruit AMPAR to the dendritic membrane. Finally, OE of a truncated μ3A that cannot interact with the AP-3 complex is also able to recruit AMPAR to RE. These data show that selectively increasing μ3A is able redirect AMPAR into the recycling pathway, possibly by protecting AMPAR from association with the full AP-3 complex. Once in the recycling pathway they can be recruited to the synapse during a subsequent trafficking step, likely involving GRIP1 (Fig. S1).
Taken together, our data identify μ3A as an essential transcription-dependent switch point that can redirect AMPAR to RE during synaptic scaling. Interestingly, these data show that both scaling up and LTP share a common reliance on AMPAR trafficking to the RE compartment (Ehlers, 2000; Park et al., 2004). Further, AP-3 is important for trafficking AMPAR to lysosomes during LTD (Matsudo et al., 2013). This raises the possibility that competition between AP-3 and μ3A for the binding and sorting of AMPAR is a key mechanism underlying several distinct forms of synaptic plasticity.
EXPERIMENTAL PROCEDURES
All experiments were approved by the Brandeis Animal Care and Use Committee and were in accordance with the National Institutes of Health guidelines. Experiments were performed on animals of both sexes. Detailed methods are provided in Supplemental Methods. Briefly, cultures were prepared and transfected 72 hr prior to recording or staining as described (Pratt et al., 2003). For culture physiology mEPSC recordings were obtained from visually identified pyramidal neurons and analyzed as described previously (Turrigiano et al., 1998; Wierenga et al., 2005). At P12-13, HsCt5 mice were subjected to monocular deprivation by intraocular injection of TTX (1 mM) as described (Desai et al., 2002; Maffei and Turrigiano, 2008); injections were performed twice (at P12 or 13 and then again at P13 or14) to maintain block for 48 h. For slice electrophysiology coronal brain slices from HsCt5 mice (300 μm) containing V1m were prepared from control and deprived hemispheres (P14-15) and recordings obtained from labeled neurons as described (Maffei et al., 2006; Loebrich et al., 2013).
Microarray screen
Isolation of labeled neurons, RNA prep, and microarray screening from L4 V1 from the HsCt5 line was performed as described previously (Sugino et al., 2006; Hempel et al., 2007), using 30 to 50 labeled cells from either the deprived or control hemisphere for each of three replicates. To stabilize the probe effects (i.e. probe-specific sources of variance) we used an approach similar to the “frozen robust multiarray analysis (fRMA)” method developed by McCall et al. (2010); details are given in supplemental methods. Data were filtered by fold change > 1.5 and the top 30 probes (ranked by p-value) annotated to RefSeq genes were included in Table 1.
Antibody generation and characterization
A rabbit anti- μ3 polyclonal antibody was raised against the peptide DMYGEKYKPFKGVKY (LifeTein, residues: 393-407) by Cocalico Biologicals. Bleeds were examined and specificity confirmed by western blot analysis against cortical extracts, and 293 lysates (ATCC) transfected with Ap3m1, Ap3m2 or Ap4m1-mCherry expression vectors. Immunohistochemical staining was abolished by preabsorption with the peptide used to generate the antibody (Fig. S3C), and was decreased when mu3A was knocked down by RNAi (Fig. 4A-C).
Statistical Analysis
Data are presented as mean ± SEM for the number of neurons indicated. Each experiment was repeated on at least three separate animals or dissociations, and the n values represent number of neurons. To determine statistical significance, unpaired two-tailed Student's t tests, or for multiple comparisons single factor ANOVAs followed by a Tukey test, or a Kruskal-Wallis test followed by a Dunn-Bonferroni post hoc test, were run as appropriate. P values ≤ 0.05 were considered significant.
Supplementary Material
Highlights.
Cell-type specific screen to identify transcripts involved in homeostatic synaptic scaling
μ3A identified as a critical switch-point in AMPAR trafficking
Upregulation of μ3A reroutes AMPAR into the recycling pathway to induce plasticity
Acknowledgments
We thank Lirong Wang, Saori Kato, Alina Chima, Dan Acker, Aram Raissi, and Zhe Meng for technical assistance. Supported by R37 NS092635 (GGT), R01EY022360 (SBN), and PO1 NS079419 (GGT and SBN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: All authors designed or performed experiments. CCS, VT, KS, HL, ML, AJ, SBN, and GGT analyzed data. GGT, CCS, and SBN wrote the manuscript.
Accession number for microarray data: GEO Series accession number GSE56758.
Literature cited
- Aguilar RC, Ohno H, Roche KW, Bonifacino JS. Functional domain mapping of the clathrin-associated adaptor medium chains mu1 and mu2. J. Bio. Chem. 1997;271:27160–6. doi: 10.1074/jbc.272.43.27160. [DOI] [PubMed] [Google Scholar]
- Bendor J, Lizardi-Ortiz JE, Westphalen RI, Brandstetter M, Hemmings HC, Jr., Sulzer D, Flajolet M, Greengard P. AGAP1/AP-3-dependent endocytic recycling of M5 muscarinic receptors promotes dopamine release. EMBO. 2010;29:2813–2826. doi: 10.1038/emboj.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Ann Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Davis GW. Homeostatic signaling and the stabilization of neural function. Neuron. 2013;80:718–728. doi: 10.1016/j.neuron.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica EC. AP-3-dependent trafficking and disease: the first decade. Curr Opin Neurobiol. 2009;21:552–559. doi: 10.1016/j.ceb.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. AP-3: an adaptor-like protein complex with ubiquitous expression. EMBO. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai NS, Cudmore RH, Nelson SB, Turrigiano GG. Critical periods for experience-dependent synaptic scaling in visual cortex. Nat. neurosci. 2002;5:783–789. doi: 10.1038/nn878. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28:511–525. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- Faundez V, Horng JT, Kelly RB. A function for the AP3 coat complex in synaptic vesicle formation from endosomes. Cell. 1998;93:423–432. doi: 10.1016/s0092-8674(00)81170-8. [DOI] [PubMed] [Google Scholar]
- Feldman DE. Synaptic mechanisms for plasticity in neocortex. Ann revi neurosci. 2009;32:33–55. doi: 10.1146/annurev.neuro.051508.135516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Ann Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainey MA, Hurvitz-Wolff JR, Lambo ME, Turrigiano GG. Synaptic scaling requires the GluR2 subunit of the AMPA receptor. J. Neurosci. 2009;29:6479–6489. doi: 10.1523/JNEUROSCI.3753-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gainey M, Tatavarty V, Nahmani M, Lin H, Turrigiano GG. Activity-dependent Synaptic GRIP1 Accumulation Drives Synaptic Scaling Up in Response to Action Potential Blockade. PNAS. 2015;112:E3590–9. doi: 10.1073/pnas.1510754112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Lee HK. Persistence of experience-induced homeostatic synaptic plasticity through adulthood in superficial layers of mouse visual cortex. J. Neurosci. 2007;27:6692–6700. doi: 10.1523/JNEUROSCI.5038-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold CP, Nicoll RA. Single-cell optogenetic excitation drives homeostatic synaptic depression. Neuron. 2010;68:512–528. doi: 10.1016/j.neuron.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JG. Endosomal sorting of AMPA receptors in hippocampal neurons. Biochem. Soc. Trans. 2010;38:460–465. doi: 10.1042/BST0380460. [DOI] [PubMed] [Google Scholar]
- Hempel CM, Sugino K, Nelson SB. A manual method for the purification of fluorescently labeled neurons from the mammalian brain. Nature protocols. 2007;2:2924–2929. doi: 10.1038/nprot.2007.416. [DOI] [PubMed] [Google Scholar]
- Hengen KB, Lambo ME, Van Hooser SD, Katz DB, Turrigiano GG. Firing rate homeostasis in visual cortex of freely behaving rodents. Neuron. 2013;80:335–342. doi: 10.1016/j.neuron.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengen KB, Torrado Pacheco A, McGregor JN, Van Hooser SD, Turrigiano GG. Neuronal firing rate homeostasis is inhibited by sleep and promoted by wake. Cell. 2016;165:180–91. doi: 10.1016/j.cell.2016.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Irving C, Borner GH. Adaptor protein complexes AP-4 and AP-5: new players in endosomal trafficking and progressive spastic paraplegia. Traffic. 2013;14:153–164. doi: 10.1111/tra.12028. [DOI] [PubMed] [Google Scholar]
- Ibata K, Sun Q, Turrigiano GG. Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron. 2008;57:819–826. doi: 10.1016/j.neuron.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Kantheti P, Qiao X, Diaz ME, Peden AA, Meyer GE, Carskadon SL, Kapfhamer D, Sufalko D, Robinson MS, Noebels JL, et al. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- Kastning K, Kukhtina V, Kittler JT, Chen G, Pechstein A, Enders S, Lee SH, Sheng M, Yan Z, Haucke V. Molecular determinants for the interaction between AMPA receptors and the clathrin adaptor complex AP-2. PNAS. 2007;104:2991–2996. doi: 10.1073/pnas.0611170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsch W, Stolfi A, Lois C. Genetic labeling of neuronal subsets through enhancer trapping in mice. PloS one. 2012;7:e38593. doi: 10.1371/journal.pone.0038593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MJ, Ehlers MD. Organelles and trafficking machinery for postsynaptic plasticity. Ann Rev Neurosci. 2006;29:325–362. doi: 10.1146/annurev.neuro.29.051605.112808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance PED, Chaudhuri A. Microarray analysis of developmental plasticity in monkey primary visual cortex. J Neurochem. 2004;88:1455–1469. doi: 10.1046/j.1471-4159.2003.02274.x. [DOI] [PubMed] [Google Scholar]
- Lambo ME, Turrigiano GG. Synaptic and Intrinsic Homeostatic Mechanisms Cooperate to Increase L2/3 Pyramidal Neuron Excitability during a Late Phase of Critical Period Plasticity. J. Neurosci. 2013;33:8810–8819. doi: 10.1523/JNEUROSCI.4502-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane PW, Deol MS. Mocha, a new coat color and behavior mutation on chromosome 10 of the mouse. J Hered. 1974;65:362–4. doi: 10.1093/oxfordjournals.jhered.a108551. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Alconada A, Bauer U, Hoflack B. The mammalian AP-3 adaptor-like complex mediates the intracellular transport of lysosomal membrane glycoproteins. J. Bio. Chem. 1998;273:29451–29461. doi: 10.1074/jbc.273.45.29451. [DOI] [PubMed] [Google Scholar]
- Lee SH, Liu L, Wang YT, Sheng M. Clathrin adaptor AP2 and NSF interact with overlapping sites of GluR2 and play distinct roles in AMPA receptor trafficking and hippocampal LTD. Neuron. 2002;36:661–674. doi: 10.1016/s0896-6273(02)01024-3. [DOI] [PubMed] [Google Scholar]
- Loebrich S, Djukic B, Tong ZJ, Cottrell JR, Turrigiano GG, Nedivi E. Regulation of glutamate receptor internalization by the spine cytoskeleton is mediated by its PKA-dependent association with CPG2. PNAS. 2013;110:E4548–4556. doi: 10.1073/pnas.1318860110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei A, Nelson SB, Turrigiano GG. Selective reconfiguration of layer 4 visual cortical circuitry by visual deprivation. Nat Neurosci. 2004;7:1353–1359. doi: 10.1038/nn1351. [DOI] [PubMed] [Google Scholar]
- Maffei A, Turrigiano GG. Multiple modes of network homeostasis in visual cortical layer 2/3. J. Neurosci. 2008;28:4377–4384. doi: 10.1523/JNEUROSCI.5298-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdan M, Shatz CJ. Effects of visual experience on activity-dependent gene regulation in cortex. Nat Neurosci. 2006;9:650–659. doi: 10.1038/nn1674. [DOI] [PubMed] [Google Scholar]
- Mardones GA, Burgos PV, Lin Y, Kloer DP, Magadan JG, Hurley JH, Bonifacino JS. Structural basis for the recognition of tyrosine-based sorting signals by the mu3A subunit of the AP-3 adaptor complex. J. Biol. Chem. 2013;288:9563–9571. doi: 10.1074/jbc.M113.450775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeta MA, Wang GJ, Shen K. Clathrin adaptor AP-1 complex excludes multiple postsynaptic receptors from axons in C. elegans. PNAS. 2009;106:1632–1637. doi: 10.1073/pnas.0812078106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Kakegawa W, Budisantoso T, Nomura T, Kohda K, Yuzaki M. Stargazin regulates AMPA receptor trafficking through adaptor protein complexes during long-term depression. Nat commun. 2013;4:2759. doi: 10.1038/ncomms3759. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Miura E, Matsuda K, Kakegawa W, Kohda K, Watanabe M, Yuzaki M. Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron. 2008;57:730–745. doi: 10.1016/j.neuron.2008.02.012. [DOI] [PubMed] [Google Scholar]
- McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA). Biostatistics. 2010;11(2):242–253. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JP, Guzman-Karlsson MC, Phillips S, Holleman C, Posey JL, Day JJ, Hablitz JJ, Sweatt JD. DNA methylation retulates neuronal glutamatergic synaptic scaling. Sci. Signal. 2015;382:ra61. doi: 10.1126/scisignal.aab0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F, Ohno H. Adaptor protein complexes as the key regulators of protein sorting in the post-Golgi network. Cell struct funct. 2003;28:419–429. doi: 10.1247/csf.28.419. [DOI] [PubMed] [Google Scholar]
- Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- Nedivi E, Fieldust S, Theill LE, Hevron D. A set of genes expressed in response to light in the adult cerebral cortex and regulated during development. PNAS. 1996;93:2048–2053. doi: 10.1073/pnas.93.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SB, Turrigiano GG. Strength through diversity. Neuron. 2008;60:477–482. doi: 10.1016/j.neuron.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K, Seong E, Burmeister M, Faundez V. Neuronal and non-neuronal functions of the AP-3 sorting machinery. J cell sci. 2007;120:531–541. doi: 10.1242/jcs.03365. [DOI] [PubMed] [Google Scholar]
- Ohno H, Aguilar RC, Yeh D, Taura D, Saito T, Bonifacino JS. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J. biol. chem. 1998;273:25915–25921. doi: 10.1074/jbc.273.40.25915. [DOI] [PubMed] [Google Scholar]
- Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD. Recycling endosomes supply AMPA receptors for LTP. Science. 2004;305:1972–1975. doi: 10.1126/science.1102026. [DOI] [PubMed] [Google Scholar]
- Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J Cell Biol. 2004;164:1065–1076. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden AA, Rudge RE, Lui WW, Robinson MS. Assembly and function of AP-3 complexes in cells expressing mutant subunits. The Journal of cell biology. 2002;156:327–336. doi: 10.1083/jcb.200107140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner J, Volknandt W, Wong BR, Scheller RH. Two rat homologs of clathrin-associated adaptor proteins. Gene. 1994;146:279–283. doi: 10.1016/0378-1119(94)90306-9. [DOI] [PubMed] [Google Scholar]
- Pozo K, Goda Y. Unraveling mechanisms of homeostatic synaptic plasticity. Neuron. 2010;66:337–351. doi: 10.1016/j.neuron.2010.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt KG, Watt AJ, Griffith LC, Nelson SB, Turrigiano GG. Activity-Dependent Remodeling of Presynaptic Inputs by Postsynaptic Expression of Activated CaMKII. Neuron. 2003;39:269–281. doi: 10.1016/s0896-6273(03)00422-7. [DOI] [PubMed] [Google Scholar]
- Pratt KG, Zimmerman EC, Cook DG, Sullivan JM. Presenilin 1 regulated homeostatic synaptic scaling through Akt signaling. Nat. Neurosci. 2011;14:1112–14. doi: 10.1038/nn.2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao VR, Pintchovski SA, Chin J, Peebles CL, Mitra S, Finkbeiner S. AMPA receptors regulate transcription of the plasticity-related immediate-early gene Arc. Nat neurosci. 2006;9:887–895. doi: 10.1038/nn1708. [DOI] [PubMed] [Google Scholar]
- Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS, Bonifacino JS. Adaptor-related proteins. Curr Opin Cell Biol. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- Rutherford LC, Nelson SB, Turrigiano GG. BDNF has opposite effects on the quantal amplitude of pyramidal neuron and interneuron excitatory synapses. Neuron. 1998;21:521–530. doi: 10.1016/s0896-6273(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Seong E, Wainer BH, Hughes ED, Saunders TL, Burmeister M, Faundez V. Genetic analysis of the neuronal and ubiquitous AP-3 adaptor complexes reveals divergent functions in brain. Mol Biol Cell. 2005;16:128–140. doi: 10.1091/mbc.E04-10-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y, Sugino K, Hempel C, Shima M, Taneja P, Bullis JB, Mehta S, Lois C, Nelson SB. A mammalian enhancer trap resource for discovering and manipulating neuronal cell types. eLife 2016. 2016 doi: 10.7554/eLife.13503. 10.7554/eLife.13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson F, Peden AA, Christopoulou L, Robinson MS. Characterization of the adaptor-related protein complex, AP-3. J Cell Biol. 1997;137:835–845. doi: 10.1083/jcb.137.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirkis DW, Edwards RH, Asensio CS. Widespread dysregulation of peptide hormone release in mice lacking adaptor protein AP-3. PLoS Genet. 2013;2013:e1003812. doi: 10.1371/journal.pgen.1003812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Swank RT, Novak EK, McGarry MP, Zhang Y, Li W, Zhang Q, Feng L. Abnormal vesicular trafficking in mouse models of Hermansky-Pudlak syndrome. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society. 2000;13(Suppl 8):59–67. doi: 10.1034/j.1600-0749.13.s8.12.x. [DOI] [PubMed] [Google Scholar]
- Tan HL, Queenan BN, Huganir RL. GRIP1 is required for homeostatic regulation of AMPAR trafficking. PNAS. 2015;112:10026–31. doi: 10.1073/pnas.1512786112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatavarty V, Sun Q, Turrigiano GG. How to Scale Down Postsynaptic Strength. J Neurosci. 2013;33:13179–89. doi: 10.1523/JNEUROSCI.1676-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM, Bonifacino JS. Cargo recognition in clathrin-mediated endocytosis. CSHPB. 2013;5:a016790. doi: 10.1101/cshperspect.a016790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropea D, Kreiman G, Lyckman A, Mukherjee S, Yu H, Horng S, Sur M. Gene expression changes and molecular pathways mediating activity-dependent plasticity in visual cortex. Nat neurosci. 2006;9:660–668. doi: 10.1038/nn1689. [DOI] [PubMed] [Google Scholar]
- Turrigiano G. Homeostatic synaptic plasticity: local and global mechanisms for stabilizing neuronal function. CSHPB. 2012;4:a005736. doi: 10.1101/cshperspect.a005736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- Watt AJ, van Rossum MC, MacLeod KM, Nelson SB, Turrigiano GG. Activity coregulates quantal AMPA and NMDA currents at neocortical synapses. Neuron. 2000;26:659–670. doi: 10.1016/s0896-6273(00)81202-7. [DOI] [PubMed] [Google Scholar]
- Wierenga CJ, Ibata K, Turrigiano GG. Postsynaptic expression of homeostatic plasticity at neocortical synapses. J. Neurosci. 2005;25:2895–2905. doi: 10.1523/JNEUROSCI.5217-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Li C, Ward DM, Kaplan J, Mansour SL. Defective organellar membrane protein trafficking in Ap3b1-deficient cells. J Cell Sci. 2000;113(Pt 22):4077–4086. doi: 10.1242/jcs.113.22.4077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.