Abstract
Angiogenesis is the process by which new blood vessels are formed from existing vessels. New vessel growth requires coordinated endothelial cell proliferation, migration, and alignment to form tubular structures followed by recruitment of pericytes to provide mural support and facilitate vessel maturation. Current in vitro cell culture approaches cannot fully reproduce the complex biological environment where endothelial cells and pericytes interact to produce functional vessels. We present a novel application of the in vivo matrix gel plug assay to study endothelial-pericyte interactions and formation of functional blood vessels using severe combined immune deficiency mutation (SCID) mice. Briefly, matrix gel is mixed with a solution containing endothelial cells with or without pericytes followed by injection into the back of anesthetized SCID mice. After 14 days, the matrix gel plugs are removed, fixed and sectioned for histological analysis. The length, number, size and extent of pericyte coverage of mature vessels (defined by the presence of red blood cells in the lumen) can be quantified and compared between experimental groups using commercial statistical platforms. Beyond its use as an angiogenesis assay, this matrix gel plug assay can be used to conduct genetic studies and as a platform for drug discovery. In conclusion, this protocol will allow researchers to complement available in vitro assays for the study of endothelial-pericyte interactions and their relevance to either systemic or pulmonary angiogenesis.
Keywords: Cellular Biology, Issue 118, angiogenesis, endothelial cells, pericytes, matrix gel plug, in vivo assay, mice
Introduction
Angiogenesis is the process by which new blood vessels are formed from a pre-existing vascular network1 and is the focus of ongoing research across many areas ranging from normal development to disease. This dynamic process involves the proliferation and migration of endothelial cells (ECs) and recruitment of pericytes to construct a vascular tube that is directed toward the site that needs oxygen and nutrient delivery2. To study this process requires an equally dynamic assay, most importantly one that can recapitulate the three-dimensional nature of tube formation. In vitro 3D matrix assays have been developed to address this need and have worked well to allow researchers to define the discrete steps in space and time in which angiogenesis takes place3,4,5,6. However, these in vitro 3D matrix models are limited to studying non-perfused vessels and therefore lack critical components pertinent to the angiogenesis process (e.g., circulating growth and inhibitory factors, unnatural tension/forces across the vascular bed) and fail to simulate the complex environment present in live tissue. To address this limitation, several in vivo angiogenesis assays have been developed7, including the matrix gel plug assay which will be the focus of our report8,9.
The matrix gel plug assay is a well-established in vivo angiogenesis assay that appeals to researchers as it provides a robust platform to test the roles of different cells and substances in angiogenesis. Matrix gel is a commercially available basement membrane solution that is secreted by the Engelbreth-Holm-Swarm (EHS) mouse sarcoma cell line that solidifies into a gel-like material at 37 °C. The matrix gel can be mixed with cells and/or substances, such as growth factors, and injected subcutaneously into the mouse. The host ECs will invade the plug over 14 days, form a vascular network, and become perfused with the host's blood. To date, matrix gel plug assays have focused exclusively on the study of endothelial cell behavior during angiogenesis, however, to the best of our knowledge no effort has yet been made to determine whether this assay can be used to co-culture endothelial cells and pericytes to study how these two cell types interact during angiogenesis. Specifically, understanding the relationship between ECs and pericytes is valuable for studying diseases where blood vessel loss is pathologic, including microvascular ischemia and peripheral vascular disease10,11,12.
Here, we describe a protocol that introduces human-derived pericytes to the matrix gel mixture along with human ECs and fibroblast growth factor (bFGF). This mixture can then be injected subcutaneously in the dorsum of SCID mice to allow formation of fully functional, pericyte coated, hybrid vessels. Our protocol describes how to prepare matrix gel plugs containing human ECs either with or without human pericytes, placement into SCID mice and how to analyze the histological sections for critical angiogenesis endpoints.
Protocol
Ethics Statement: Procedures involving animal subjects have been approved by the Institutional Animal Care and Use Committee at Stanford University School of Medicine.
NOTE: Animals are under anesthetization with 3% vaporizer isoflurane and 3% supply of O2 gas. Use of vet ointment on eyes may help to prevent dryness while under anesthesia.
1. Cell Preparation
- Grow human endothelial and pericyte cell cultures in 100 mm plates with 10 ml of the appropriate media. Replace 10 ml of fresh medium every 2 - 3 days until cells reach 80% confluency.
- Culture endothelial cells (ECs) in complete endothelial cell media (ECM) supplemented with company supplied 5% fetal bovine serum (FBS), 10% penicillin/streptomycin and 10% endothelial cell growth supplement.
- Culture pericytes in complete pericyte media (PM) supplemented with company supplied 2% FBS, 10% penicillin/streptomycin and 10% pericyte growth supplement.
- Prepare cell mixtures
- Calculate the required number of cell for the experimental and control groups. NOTE: Each plug in the experimental group contains one million (1 x 106) human ECs and two hundred thousand (2 x 105) pericytes. For the control group, each plug contains 1.2 million human ECs only. The negative control group has matrix gel only. Each group should include at least four plugs, which would require two mice as plugs can be injected into each side of a mouse's lower back. Three plugs would serve as triplicate for the experiment and the fourth could be used in case one fails. Prepare 25% extra cells to match the extra volume of gel.
- Collect and count cells from culture plates.
- To detach cells, remove media and wash once with room temperature 1x phosphate buffered saline (PBS). Remove PBS, add 1 ml 0.25% Trypsin/2.21 mM EDTA, and incubate for 1 min. Wash cells off the plate by adding 2 ml medium and collect in a 15 ml tube.
- Count cell using a hemocytometer according to the manufacturer's instructions.
- Centrifuge the appropriate number of cells into a pellet at 400 x g for 5 min. For the experimental group, centrifuge both ECs and pericytes down together into one pellet.
2. Matrix Gel Preparation
Thaw out matrix gel completely at 4 °C for 4 hr or overnight to obtain a homogenous solution. No mixing is required.
Aliquot matrix gel in 1.5 ml tubes and store at 4 °C.
Pre-chill sterile 1.5 ml tubes and 28 G 1 cc insulin syringes at 4 °C. Caution: Keep matrix gel on ice at all times.
Mix cold matrix gel with bFGF at a volume ratio of 1:100 (final concentration of bFGF= 0.5 μg/ml; bFGF stock solution= 50 μg/ml) After mixing matrix gel solution by pipetting up and down several times, incubate on ice for 30 - 60 min before mixing with the cells in step 3.1. NOTE: Thaw the stock bFGF before mixing. Calculate the number of plugs needed for injection. For example, each plug requires 200 μl matrix gel. Therefore, prepare 25% extra gel.
3. Mix Matrix Gel with Cells
For each group, resuspend the cell pellet (containing appropriate cell number) with the calculated volume of matrix gel containing bFGF. Mix gently to avoid foaming and leave in the 15 ml tube on ice until mice are prepared for injection.
4. Mouse Preparation
Anesthetize 1 SCID mice for 5 min in 3% Isoflurane plus 3% O2 in an induction chamber. Focus on one experimental group, or 2 mice, at a time.
Remove the mouse from the induction chamber, place on a heating pad ventral side down and quickly attach a snout nozzle to the mouse's face via a non-rebreathing system to supply anesthesia throughout the injection procedure. Switch the gas flow from the induction chamber to the non-rebreathing system and lower the gas pressure to 1.5% Isoflurane and 1.5% O2.
Remove hair from both lateral hind regions (around 1 cm diameter) with a shaver or hair removal cream.
Wipe the skin area with a 70% alcohol pad.
Return the mouse to the induction chamber and repeat steps 4.2 - 4.4 with the second mouse.
5. Matrix Gel Injection
Prepare one mouse for injection by attaching to the non-rebreathing system and placing on the heating pad ventral side down.
- Load the matrix gel into a pre-chilled 28 G 1 cc insulin syringe one mouse at a time.
- Load the complete 500 μl volume of matrix gel for two plugs (200 μl per plug plus 25% extra) into the syringe.
- Make sure to inject the matrix gel into the mice within 3 min of loading the cells into the syringe to prevent the cells from settling to the side of the syringe and to prevent matrix gel from solidifying inside of the syringe.
Lift the back skin to locate subcutaneous space, then inject 200 μl of the matrix gel slowly and evenly into the subcutaneous space of the back posterior of the rib cage.
Remove the injection needle slowly, being careful to prevent matrix gel from leaking out of the injection site. A bump will form at the site of injection. Wipe injection site with an alcohol pad.
Repeat injection steps 5.2 - 5.5 on the other side of mouse's back, then leave the mouse on its back for 1 min to allow gelation of the matrix gel on the heat pad.
Outline both bumps using a permanent marker to identify the bump after 14 days. After some hair grows back, it will be easier to find the bump.
6. Matrix Gel Plug Isolation: 14 Days after Injection
Euthanize the mice humanely by exposing the animals to >5 min of carbon dioxide inhalation followed by cervical dislocation to assure death.
- Remove the matrix gel plug from the mouse's back.
- Gently remove regrown hair around injection site where the plug is located by repeating step 4.3. The marker line indicates the size of original bump under the skin.
- Excise the plug along with the surrounding skin and muscle layers attached on the top and bottom sides of the plug, respectively.
- Using fine surgical scissors, make an incision on the marked border of the plug that is perpendicular to the skin and cut through to the muscle underneath the plug. Then, carefully cut around the periphery of the plug along the marked border.
- Remove the plug, keeping it sandwiched between the skin and muscle layers. Immediately rinse in a small beaker with 1x PBS to wash away excess blood. The plug is now ready to be fixed (next step, 6.3).
- Fix the plug in 4% paraformaldehyde.
- Place the entire plug, including the skin and muscle layers, in a 50 ml tube with 25 ml of 4% paraformaldehyde for 24 hr at room temperature without rocking. The next day, transfer the plug into 25 ml of 70% ethanol for another 24 hr before paraffin embedding. NOTE: Handle paraformaldehyde with caution. Wear gloves.
7. Paraffin Process the Matrix Gel Plug
- Prepare the plug for paraffin embedding.
- Orient the plug as shown in Figure 1a; place the plug in a tissue cassette so that the side of the plug is facing up and both the skin and muscle layers are visible across the surface of the tissue block that will be cut first during sectioning.
Paraffin embed the plug according to standard paraffin embedding protocols13.
Cut 8 - 10 μm thick sections off of the tissue block and mount onto microscope slides according to standard paraffin sectioning protocols13.
Store the slides in a cool dry place at room temperature until ready for staining.
8. Stain Sections with Hematoxylin and Eosin Stain (H&E)13
De-paraffinize the tissue sections by passing the slides through 100% xylene twice, 100% ethanol, 95% ethanol, 70% ethanol and lastly 1x PBS, each for 5 min at room temperature. Leave slides in 1x PBS until the next step.
Pipette 100 μl H&E solution onto the section and incubate for 1 - 5 min, or until there is clear contrast between blue nuclei and pink cytoplasm in a cell as viewed under an upright light microscope at 10X magnification. Be careful not to let any H&E solution touch the objective.
Wash H&E solution off of the slide by dunking the slide in a large beaker of tap water, then place the slide in a rack in the beaker and flush with running water for 2 min.
Move to step 9.8 to mount the slides.
9. Stain Other Slides for Human Capillaries and Pericytes
De-paraffinize the tissue sections by passing the slides through 100% xylene twice, 100% ethanol, 95% ethanol, 70% ethanol and lastly 1x PBS, each for 5 min at room temperature. Leave slides in 1x PBS until the next step.
- Boil the sections in antigen retrieval solution.
- In a 2 L beaker, prepare boiling antigen retrieval solution using 1x citrate buffer (pH 7.0) at 95 °C on a heating block.
- Once boiling, place the slides in a metal rack and submerge in the boiling antigen retrieval solution for 20 min.
- Carefully remove the beaker from the heating block with the slides still submerged and allow the solution to slowly return to room temperature for about 30 min.
- Place the slides in 1x PBS until next step.
- Block the sections in blocking buffer (1x PBS, 5% Goat Serum, 0.3% Triton X-100).
- Draw a hydrophobic circle around the edges of each section with a PAP pen.
- Place the slides in a humidity tray with distilled water at the bottom of the tray.
- Pipette 100 μl blocking buffer onto the sections making sure all of the tissue is covered by fluid (may require more than 100 μl).
- Incubate sections with blocking buffer for 1 hr.
- Incubate sections with primary antibody.
- Prepare one solution with two primary antibodies to probe each section for CD31 (ECs) and smooth muscle actin (pericytes) diluted in dilution buffer (1x PBS, 1%BSA, 0.3% Triton X-100). NOTE: Here, anti-CD31 concentration is 1:50 and anti-smooth muscle actin concentration is 1:300.
- Incubate the sections overnight at 4 °C with 100 μl primary antibodies in a humidity tray containing distilled water.
Wash the slides three times in 1x PBST (1x PBS + 0.1% Tween20) for 5 min each wash.
- Incubate sections with secondary antibody.
- Prepare a solution with a fluorophore conjugated goat anti-rabbit secondary antibody to recognize the anti-CD31 rabbit antibody diluted in dilution buffer.
- NOTE: The smooth muscle actin primary antibody is already conjugated with a CY3 fluorophore, so no secondary antibody staining is needed. Goat-anti rabbit secondary antibody concentration is 1:250.
- Incubate the sections for 1 hr with 100 μl secondary antibody in the humidity tray containing distilled water.
Wash the slides three times in 1x PBST for 5 min each wash.
- Mount the slides.
- Pipette 2 - 3 drops (~ 40 - 60 μl) of antifade mounting solution with DAPI nuclear staining onto each section.
- Gently place a no. 1.5 cover slip over the sections and mounting solution, allowing the fluid to spread out over the sections and to the edges of the cover slip. Gently use a finger to press out air bubbles that may be located over the section.
- Let the slides dry for at least 1 hr before handling.
10. Quantify Capillary Density and Structure14
- Count the number of capillaries in H&E stained sections, expressed as #/mm2.
- Acquire photos of four different fields on an upright light microscope under 40X magnification. NOTE: Capillaries are identified as tubes filled with red blood cells.
- Alternatively, instead of manually counting capillaries, use image analysis software (e.g., Wimasis Image Analysis) to quantify various angiogenesis parameters such as vessel number, average vessel length and diameter, and branch points.
- Analyze capillary structure in immunofluorescent stained sections.
- Acquire photos of four different fields on an inverted fluorescent microscope with FITC and TxRed filter cube.
- Use a FITC cube to image ECs by excitation at 488 nm wavelength and use a TxRed cube to image pericytes by excitation at 594 nm wavelength. p.p1 {margin: 0.0px 0.0px 0.0px 0.0px; font: 8.0px Arial} p.p1 {margin: 0.0px 0.0px 0.0px 0.0px; font: 8.0px Arial} span.s1 {letter-spacing: -0.1px} p.p1 {margin: 0.0px 0.0px 0.0px 0.0px; font: 8.0px Arial} span.s1 {letter-spacing: -0.1px} p.p1 {margin: 0.0px 0.0px 0.0px 0.0px; font: 8.0px Arial} span.s1 {letter-spacing: -0.1px}
Representative Results
Representative H&E and immunofluorescent staining of matrix gel plug sections are shown in Figure 2. Sections from EC only plugs display some vessels that are mostly not perfused with blood (Figure 2 top left, black arrows) whereas plugs containing both ECs and pericytes display several perfused vessels with larger diameters and complete pericyte coverage, as evidenced by positive SMA staining immediately adjacent to CD31-positive ECs. These results suggest that pericytes play a substantial role in nascent blood vessel formation and that this dynamic process can be recapitulated and easily analyzed in an in vivo model.
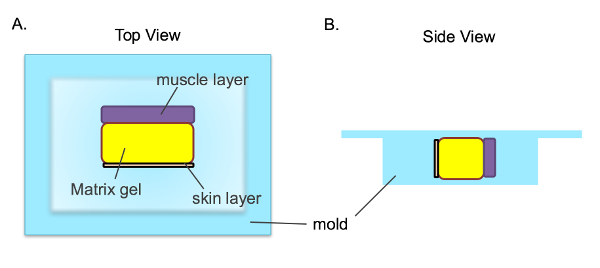 Figure 1. Tissue Orientation when Embedded in a Tissue Cassette. After the matrix gel plug (yellow) is isolated, it is in between the mouse's muscle (purple) and skin (green) layers. The plug is placed in the tissue cassette as shown so that all three layers can be seen across the surface that will be cut first during sectioning. A) View from the top of the cassette, B) View from the side of the cassette. Please click here to view a larger version of this figure.
Figure 1. Tissue Orientation when Embedded in a Tissue Cassette. After the matrix gel plug (yellow) is isolated, it is in between the mouse's muscle (purple) and skin (green) layers. The plug is placed in the tissue cassette as shown so that all three layers can be seen across the surface that will be cut first during sectioning. A) View from the top of the cassette, B) View from the side of the cassette. Please click here to view a larger version of this figure.
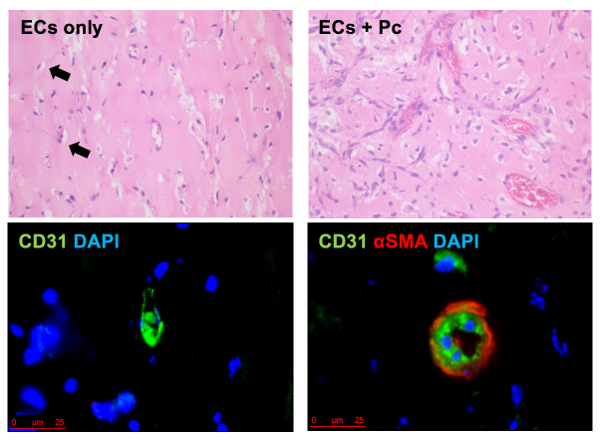 Figure 2. Representative H & E (top row) and Immunofluorescence (bottom row) of Matrix Gel. Images show the appearance of ECs alone (left images) and ECs plus healthy pericytes (Pc) (right images). Human and murine ECs are labeled in green (bottom images) and pericytes (α smooth muscle actin (αSMA)) are labeled in red (bottom right), and nuclei are stained blue with DAPI stain. Scale is 25 µm. This figure has been modified from a previous publication8. Please click here to view a larger version of this figure.
Figure 2. Representative H & E (top row) and Immunofluorescence (bottom row) of Matrix Gel. Images show the appearance of ECs alone (left images) and ECs plus healthy pericytes (Pc) (right images). Human and murine ECs are labeled in green (bottom images) and pericytes (α smooth muscle actin (αSMA)) are labeled in red (bottom right), and nuclei are stained blue with DAPI stain. Scale is 25 µm. This figure has been modified from a previous publication8. Please click here to view a larger version of this figure.
Discussion
The matrix gel plug assay has proven to be a convenient and powerful method to evaluate gene regulation in angiogenesis, angiogenic and antiangiogenic compounds in vivo, and to supplement in vitro tests. Here, we describe in detail a novel matrix gel plug assay of human angiogenesis that investigates the interaction between endothelial cells and pericytes during vessel formation.
There are a few novel and critical steps in this protocol. Cells in low passage (passage 1 - 4) are preferable because young cells will be more viable during the 14-day experimental setting. The number of endothelial cells used is minimum one million and the ratio of EC to pericyte is 5:1; however, increased pericyte cell number will enhance its coverage on capillaries. For the experimental group of endothelial cells alone, the total number of cells injected should be the sum of endothelial cells and pericytes, which is 1.2 million, therefore, the total cell number per plug is consistent. It is important to calculate the appropriate amounts of cell numbers and matrix gel volume. The basic formula is 1.2 million cells in 200 μl of matrix gel. In addition, before injection, keep pipet tips, syringes, matrix gel, and cell pellets on ice at all times. Since matrix gel is viscous and in order to avoid air bubbles forming during cell suspension, cut about 1 cm off of the tip of 1 ml pipet tips to allow for better flow. Do not mix matrix gel with cells until mice are ready to be injected. Each plug injection should take less than one minute. One mouse can carry two plugs, one on each side of its back. When extracting the plugs make sure to keep the plug sandwiched between the muscle and skin layers, then the matrix gel plug will be well collected after isolation.
The drawbacks of the matrix gel plug assay are that it is time consuming, costly, and involves tedious and delicate steps such as injection and plug isolation. If these steps were to fail, the results would be ruined and the process would have to be repeated again with new mice and materials.
However, the data is reproducible and provides flexibility in terms of experimental design. For example, growth factors or inhibitors could be administered to the plug at different stages of vascular development, which can be used for drug validation. Furthermore, incorporation of other cell types, such as growth-arrested cells, transfected cell lines or tumor cell lines, into the matrix gel plug is another possibility for manipulating the endothelial cell phenotype during angiogenesis. Our protocol utilizes this flexibility and introduces human-derived pericytes along with ECs to allow formation of fully functional, pericyte covered, hybrid vessels in vivo. Our protocol also describes how to analyze the histological sections from these plugs for critical angiogenesis endpoints, including tube number and tube length, with these two cell types present.
Disclosures
No competing financial interests enclosed.
Acknowledgments
Dr. K. Yuan was supported by an American Heart Association Scientist Development Grant (15SDG25710448) and the Pulmonary Hypertension Association Proof of Concept Award (SPO121940). Dr. V. de Jesus Perez was supported by a career development award from the Robert Wood Johnson Foundation, an NIH K08 HL105884-01 award, a Pulmonary Hypertension Association Award, a Biomedical Research Award from the American Lung Association and a Translational Research and Applied Medicine award from Stanford University.
References
- Risau W. Mechanisms of angiogenesis. Nature. 1997;386(6626):671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- Ribatti D, Nico B, Crivellato E. The role of pericytes in angiogenesis. Int J Dev Biol. 2011;55(3):261–268. doi: 10.1387/ijdb.103167dr. [DOI] [PubMed] [Google Scholar]
- Koh W, Stratman AN, Sacharidou A, Davis GE. In vitro three dimensional collagen matrix models of endothelial lumen formation during vasculogenesis and angiogenesis. Methods Enzymol. 2008;443:83–101. doi: 10.1016/S0076-6879(08)02005-3. [DOI] [PubMed] [Google Scholar]
- Liu Y, Senger DR. Matrix-specific activation of Src and Rho initiates capillary morphogenesis of endothelial cells. FASEB J. 2004;18(3):457–468. doi: 10.1096/fj.03-0948com. [DOI] [PubMed] [Google Scholar]
- Grant DS, et al. Interaction of endothelial cells with a laminin A chain peptide (SIKVAV) in vitro and induction of angiogenic behavior in vivo. J Cell Physiol. 1992;153(3):614–625. doi: 10.1002/jcp.1041530324. [DOI] [PubMed] [Google Scholar]
- Madri JA, Pratt BM, Tucker AM. Phenotypic modulation of endothelial cells by transforming growth factor-beta depends upon the composition and organization of the extracellular matrix. J Cell Biol. 1988;106(4):1375–1384. doi: 10.1083/jcb.106.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks PC, Montgomery AM, Cheresh DA. Use of the 10-day-old chick embryo model for studying angiogenesis. Methods Mol Biol. 1999;129:257–269. doi: 10.1385/1-59259-249-X:257. [DOI] [PubMed] [Google Scholar]
- Yuan K, et al. Activation of the Wnt/planar cell polarity pathway is required for pericyte recruitment during pulmonary angiogenesis. Am J Pathol. 2015;185(1):69–84. doi: 10.1016/j.ajpath.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns A, Freay AD, Fraser W, Korach KS, Rubanyi GM. Disruption of estrogen receptor gene prevents 17 beta estradiol-induced angiogenesis in transgenic mice. Endocrinology. 1996;137(10):4511–4513. doi: 10.1210/endo.137.10.8828515. [DOI] [PubMed] [Google Scholar]
- Chen CW, et al. Human pericytes for ischemic heart repair. Stem Cells. 2013;31(2):305–316. doi: 10.1002/stem.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards OC, Raines SM, Attie AD. The role of blood vessels, endothelial cells, and vascular pericytes in insulin secretion and peripheral insulin action. Endocr Rev. 2010;31(3):343–363. doi: 10.1210/er.2009-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard N, et al. Increased pericyte coverage mediated by endothelial-derived fibroblast growth factor-2 and interleukin-6 is a source of smooth muscle-like cells in pulmonary hypertension. Circulation. 2014;129(15):1586–1597. doi: 10.1161/CIRCULATIONAHA.113.007469. [DOI] [PubMed] [Google Scholar]
- Buchwalow IB, Bocker W. Immunohistochemistry: Basics and Methods. 2010. pp. 1–153.
- Tata DA, Anderson BJ. A new method for the investigation of capillary structure. J Neurosci Methods. 2002;113(2):199–206. doi: 10.1016/s0165-0270(01)00494-0. [DOI] [PubMed] [Google Scholar]


