Abstract
Evidence has emerged suggesting that diet-induced obesity can have a negative effect on cognitive function. Here, we exploited a mouse genetic reference population to look for the linkage between these two processes on a genome-wide scale. The focus of this report is to determine whether the various BXD RI strains exhibited different behavioral performance and hippocampal function under high fat dietary (HFD) condition. We quantified genetic variation in body weight gain and consequent influences on behavioral tests in a cohort of 14 BXD strains of mice (8–12 mice/strain, n=153), for which we have matched data on gene expression and neuroanatomical changes in the hippocampus. It showed that BXD66 was the most susceptible, whereas BXD77 was the least susceptible strain to dietary influences. The performance of spatial reference memory tasks was strongly correlated with body weight gain (P<0.05). The obesity-prone strains displayed more pronounced spatial memory defects compared to the obesity-resistant strains. These abnormalities were associated with neuro inflammation, synaptic dysfunction, and neuronal loss in the hippocampus. The biological relevance of DSCAM gene polymorphism was assessed using the trait correlation analysis tool in Genenet work. Further more, a significant strain-dependent gene expression difference of DSCAM was detected in the hippocampus of obese BXD strains by real-time quantitative PCR. In conclusion, a variety of across-strain hippocampal alterations and genetic predispositions to diet-induced obesity were found in a set of BXD strains. The obesity-prone and obesity-resistant lines we have identified should be highly useful to study the molecular genetics of diet-induced cognitive decline.
Keywords: High-fat diet, Hippocampus, Spatial learning, BXD mice, DSCAM
1. Introduction
The increasing prevalence of diet-induced obesity (DIO) has become a major public health concern in modern society for the serious medical issues it causes, such as type 2 diabetes mellitus, hypertension, atherosclerosis, and stroke [1–3].Several longitudinal studies have found that weight gain has also been associated with long-term decline in cognitive performance independently of other medical conditions [4–6]. The negative effects of dietary manipulations on learning and memory performance have also been observed in rodents [7, 8].However, the influence of genetic differences amongst individuals in their susceptibility to DIO and deficits in cognitive performance is largely unknown. One important reason for these difficulties could be that there are many factors involved in weight gain, including genetic, metabolic, psychosocial, and environmental influences [9]. The interaction between genes and diet is important, but the causative neuro cognitive phenotype effect has not been precisely defined and measured. In the current study, we use BXD family of RI strains to probe the genetic architecture of quantitative traits and how they contribute to DIO and decline in cognitive function.
The BXD set of RI strains was derived by crossing common inbred mouse strains of C57BL/6J (B6) and DBA/2J (D2) and inbreeding progeny for 20 or more generations [26], each strain representing a unique mosaic of B and D alleles with fully sequenced parental strains. The sequence variation throughout the panel is exceptionally well-defined, thereby enabling extensive replication studies of the same genotype and tight experimental control [19, 27]. The families of this RI inbred strains can be used as a high power and high precision genetic reference population to assess complex interactions of gene networks and disease susceptibility [28], by which numerous genome and phenome data sets have been accumulated over the past decades. Meanwhile, the genotyping data can be queried for polymorphisms in the sequencing database of this reference population which may enable uncovering candidate genes associated with specific phenotype traits.
Although it has been argued that the high prevalence of obesity is primarily due to environmental factors, such as sedentary lifestyle and consumption of high-fat energy-dense diets [10], it has also been suggested that DIO susceptibility is strongly influenced by genetic factors. A population- and family-based study has shown that women with obese parents were more susceptible to weight gain when exposed to high dietary fat intakes [11]. A recent twin study has reported that genetic predisposition to obesity is increasingly expressed throughout childhood [12]. The differences between and within strains in response to DIO have also been well recognized in rodent studies [13–15], suggesting that genetic background not only regulates weight gain but also significantly affects the susceptibility to DIO. Meanwhile, the gene-environment correlation (GXE) on susceptibility differences are supported by the large scale genome-wide association studies (GWAS). Researchers have identified a large number of quantitative trait loci (QTLs) and genes associated with body mass index (BMI) in humans, such as FTO, MC4R, SH2B1, BDNF, etc.[16–18].However, how these genetic variations influence obesity phenotype is unclear, partially due to an inability to control environmental factors and difficulty in obtaining certain types of physiological and molecular data. Thus, an effective population-based experimental model that can simplify complex genetically admixed human populations is needed to dissect the influences of intricate GXEs [19]. In contrast, several congenic and recombinant inbred (RI) mouse strains have been tested in the field of obesity and metabolic disorders [20]. A growing number of QTLs have been identified that influence various obesity related traits, such as diet induced obesity [21], resistance to diet induced obesity[22], juvenile obesity[23], and obesity associated diseases[24]. But most of the obesity loci identified by quantitative studies do not correspond to ‘classical’ obesity mutations such as ob, tubby or fat [25], suggesting a relatively large pool of genes with allelic variations accounting for body-weight regulation.
Mouse Down Syndrome Cell Adhesion Molecule (DSCAM) gene is located on chromosome 16, a syntenic region for human chromosome band 21q22. Its allelic differences are known to be involved in regulating body weight, motor function, and motor learning [29].In parallel with this, we present the results of a trait correlation analysis in order to test the hypothesis for a functional correlation between DSCAM gene polymorphism and phenotype traits in BXD mice. Thus, the BXD RI mice strains provide an experimental model that allows us to examine the interaction between genes and diet, which seem likely to provide insight in to the biological basis of variation in DIO and behavioral traits.
2. Materials and methods
2.1. Mice and Diets
BXD RI strains (5–7 wk) were provided by Dr. Robert W. Williams and Dr. Lu Lu (University of Tennessee Health Science Center, Memphis, TN, USA). Mice were housed three to five per cage in an environmentally controlled animal facility with a 12-h light/dark cycle and given free access to food and water. A total of 14 BXD RI strains containing 153 mice were used in this study. All experimental protocols were conducted in accordance with the NIH Animal Care guidelines and were approved by the University of Tennessee Health Science Center Animal Care and Use Committee.
Standard chow diet containing (by weight) 7.2% fat was from Harlan Teklad (TD.94045). High fat diet containing 45% fat, 35% sucrose (D12451; Research Diets Inc., New Brunswick, New Jersey, USA) was fed for 4 months. Mice weights were recorded at intervals of 4 weeks or less.
2.2. Behavioral testing
To assess the differences of spatial learning and memory, and anxiety-related behavior between BXD strains, a battery of behavioral tests were performed. All mice were subjected to all behavioral tests, and the testing order was consistent across animals.
2.2.1. Spontaneous alternation behavior
Spontaneous alternation behavior was assessed by using a cross-maze. The maze was composed of 4 symmetrical arms, with each arm measuring 30 × 8 × 15 cm with a central platform of 25 cm across. The testing was conducted by placing the mice on the center platform and allowing 5 min of unimpeded exploration. The sequence of arm entries was recorded for calculation of a percent alternation score. An arm entry was recorded when all four paws entered an arm. One successful alternation was defined as any non-repetitive sequence of four arm entries. Using this procedure, possible alternation sequences were equal to the number of arm entries minus 3. The percentage alternation score is equal to the ratio of (actual alternations /possible alternations) multiplied by 100. Chance performance on this task is 22.2%. The number of arm entries was also recorded to obtain an index of spontaneous exploration and general locomotion. Mice that made fewer than 11 arm entries were excluded from the analysis.
2.2.2. Morris water maze (MWM)
Spatial learning was examined in a MWM task with hidden platform. Mouse relies on the spatial visual cues to navigate a submerged escape platform. The experimental apparatus consisted of a circular water tank, 110 cm in diameter and 60 cm in depth and filled with 22–25°C water at a depth of 30 cm. A transparent lucid platform (5.5 cm in diameter, 14.5 cm in height) was submerged 1 cm beneath the surface of the water, and placed at the midpoint in the north-west quadrant of the pool. The water was opaque by mixing with nontoxic white paint to make the platform invisible. Each mouse received 4 consecutive trials per day with an inter-trial interval of 16 s for 8 consecutive days. Four starting points were varied daily. Each trial lasted until the mice had found the platform or for a max 1 min. A video camera mounted at the height of 180 cm above the center of the maze and all data were recorded with a computerized video system. Escape latency (finding the submerged escape platform) and path length to find the hidden platform were recorded. On day 9, the probe test was performed by removing platform and allowing each mouse to swim freely for 60 s. The total length of the swim path during the testing period was recorded. The time that mice spent swimming in the target quadrant (where the platform was located during hidden platform training) was measured. For the probe trials, the number of times the mice crossed where the platform had been located was also measured and calculated.
2.2.3. Fear conditioning
The experiments were performed with conditioned freezing chambers (Coulbourn Instruments) as described previously [30].
2.2.4. Barnes maze
For the parental C57BL/6J and DBA/2J strains, a modified Barnes maze was performed to assess spatial learning as described previously[31] after HFD feeding for 4 months. Briefly, mice were trained on four-trial blocks per day for 4 days to find a target escape box. If the target box was not successfully entered within 4 min, the investigator guided the mouse in to the target box, and a latency of 240 s was assigned. Spatial learning was assessed using total and primary errors (errors committed before the first encounter with the escape hole). Escape latency and path length were also measured. Testing was digitally recorded and analyzed manually using ANY-maze v4.99 software (Stoelting Co., USA).
2.3. Trait correlation analysis
The haplotype structure and SNPs of DSCAM gene were extracted from mouse phenome database at the Jackson Laboratory (http://phenomejax.org). The genetic correlations between DSCAM gene polymorphism and phenotypic traits were analyzed using a phenotype database of over 4500 published and unpublished traits from previous studies on BXD strains at http://www.genenetwork.org. Correlation networks were constructed using on-line tools in GN.
2.4. Quantitative real time PCR
Total RNA was purified from mouse hippocampus using Trizol reagent (Invitrogen, Carlsbad, CA) according to the protocol. RNA quality and purity was monitored by 260/280 nm OD ratios. The cDNA was synthesized from 2 µg total RNA and analyzed in use of 5 Prime Real Master Mix SYBR ROX (5 Prime) with an Eppendorf Master cyclerrealplex system. The qRT-PCR runs were performed under the following thermocycler conditions: initial denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 55°C for 15 s, and 68°C for 20 s. A melting test was conducted to verify that only one product was amplified. All tests were run in duplicate, the expression values are represented as Mean±SEM relative to GAPDH expression. Data analysis was performed using the 2-[ΔΔCT] method [32]. Primer sequences are available on request.
2.5. Western blots
To extract protein from the hippocampus, the tissues were manually homogenized in 300 µl of cold RIPA buffer containing protease and phosphatase inhibitor cocktail and 10 mM EDTA (Thermo Fisher Scientific). Lysates were centrifuged at 14,000 rpm for 30 minutes at 4°C. The supernatants were collected and assayed for protein content using a standardized BSA kit (Pierce, Rockford, IL, USA) prior to storage at −80°C. 25 µg protein samples were mixed Laemmli Sample Buffer (LSB), reduced with 50 mM dithiothreitol and denatured in a boiling water bath for 5 min. Samples were resolved via 4–20% Tris-Glycine SDS-PAGE gel electrophoresis (Invitrogen), and transferred to PVDF membranes. Non-specific protein binding to the membrane was blocked by incubating with 5% skim milk in 1X TBS-T wash buffer for 90 min at room temperature. Thereafter, the samples were reacted in blocking buffer mixed with primary antibody for β-actin (Sigma-Aldrich, St. Louis, MO, USA, at 1:10000 dilution); drebrin (Novus) and all other Abs (Cell Signaling Technology) diluted 1:1000 and incubated overnight at 4°C. Anti-mouse IgG and anti-rabbit IgG HRP-conjugated secondary Abs (Chemicon, Temecula, CA, USA) were used in 5% skim milk in TBST for 1 h at room temperature at 1:10000 dilution. Membranes were washed three times for 10 min with TBST after incubation with each antibody. Immunoblots were imaged on Kodak film using the ECL prime reagent and quantified using ImageJ software. All proteins were normalized to the loading control of β-actin.
2.6. Immunohistochemistry
Mice were perfused with 4% paraformaldehyde in PBS. Brains were rapidly removed and post-fixed for 4 h in the same fixative, and placed in 20% sucrose in PBS at 4 °C until they sank. Sections were coronally cut at 35 µm thickness on a sliding microtome. Four sets of serial sections were collected in glass vials. After blocking at room temperature for 1 hour in NGS (10% normal goat serum, 0.2% Triton X-100, and 0.02% NaN3 in TBS), free-floating sections were incubated ON at 4°C with anti-GFAP monoclonal antibody (Sigma-Aldrich, dilution 1:1000) in blocking solution. AlexaFluor 488 secondary antibody (Invitrogen, dilution 1:500) was used for 2 hours at RT. Sections were washed three times for 10 min with PBS after incubation with each antibody. The sections were then mounted in ProLong Gold Anti-Fade Reagent with DAPI (Invitrogen), and examined by fluorescence microscopy. Nissl staining and image quantification were performed as previously described [33].
2.7. Statistical analysis of strain differences and heritability estimates
All data are presented as mean±SEM. Statistical tests and graphing were done with GraphPad Prism 6.0 software. Two-way ANOVA followed by Bonferroni post hoc testing was used to analyze bodyweight, western blot for the parental strains, and behavioral test data, with strain and HFD feeding as between-subject factors. One-way ANOVA was used to compare gene expression and neuronal density data to detect significant inter-strain differences. Tukey’s post-hoc test was used for multiple comparisons. Regression analysis was used to examine correlation between body weight gain and behavioral measures. Statistical significance was defined at P<0.05. Heritability was determined as h2= VA/(VA + VE), where VA is the additive genetic variation estimated by the between-strain variance and VE is the environmental variance estimated by the within-strain variance from the ANOVA results [34].
3. Results
3.1. HFD feeding provoked behavioral impairment and hippocampal dysfunction in the parental strains B6 and D2 mice
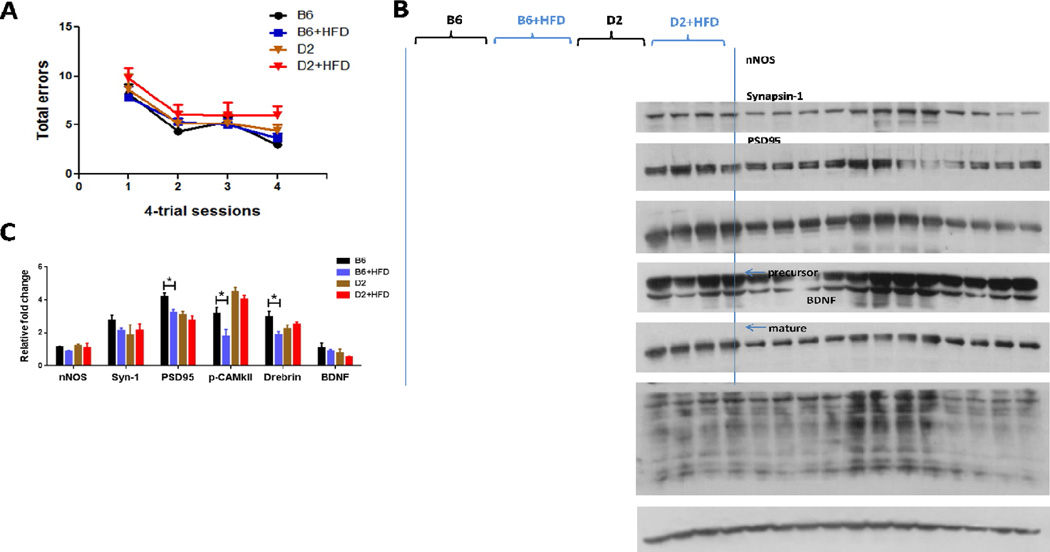
We observed a significant effect of HFD feeding on spatial pattern learning in the Barnes maze task (F(3, 128) =2.849, P<0.05) for the parental B6 and D2 mice. In line with the previous studies by others [35, 36],B6 mice did appear to out perform D2 mice although there was no significance between these two groups (Fig. 1A). Similarly, the association between hippocampus-related behavioral trait and differences in synaptic plasticity was also confirmed by examining the expression of the presynaptic marker Synapsin 1, postsynaptic marker PSD95, and other proteins involved in synaptic function such as drebrin, p-CAMKII, and BDNF (F(3, 72) =8.086, p<0.0001; Fig. 1B and C).
Figure 1. Comparison of Barnes maze test and synaptic protein expressions in hippocampus of B6 and D2 strains with high fat diet-induced obesity.
(A) The Barnes maze was used to test spatial memory acquisition and retention. Performance improved significantly in all groups over the course of training. Although experimental groups did not differ on the measures of total errors, D2+HFD mice had a higher number of visits to incorrect holes before reaching the target hole. Data are presented in mean ± SEM of four trials per day (n=8–10 per group). (B) Homogenates of the hippocampi of mice were analyzed by Western blots developed with the indicated antibodies; each lane represents an individual mouse sample. (C) Quantitative analysis of densitometrical data from western blots after normalization with the β-actin. The corresponding protein levels are presented as mean ± SEM (n = 4/group); statistical analysis is described in the results section, *p < 0.05.
3.2. Correlation between performance of hippocampus-dependent tasks and body weight gain under HFD conditions in BXD mice
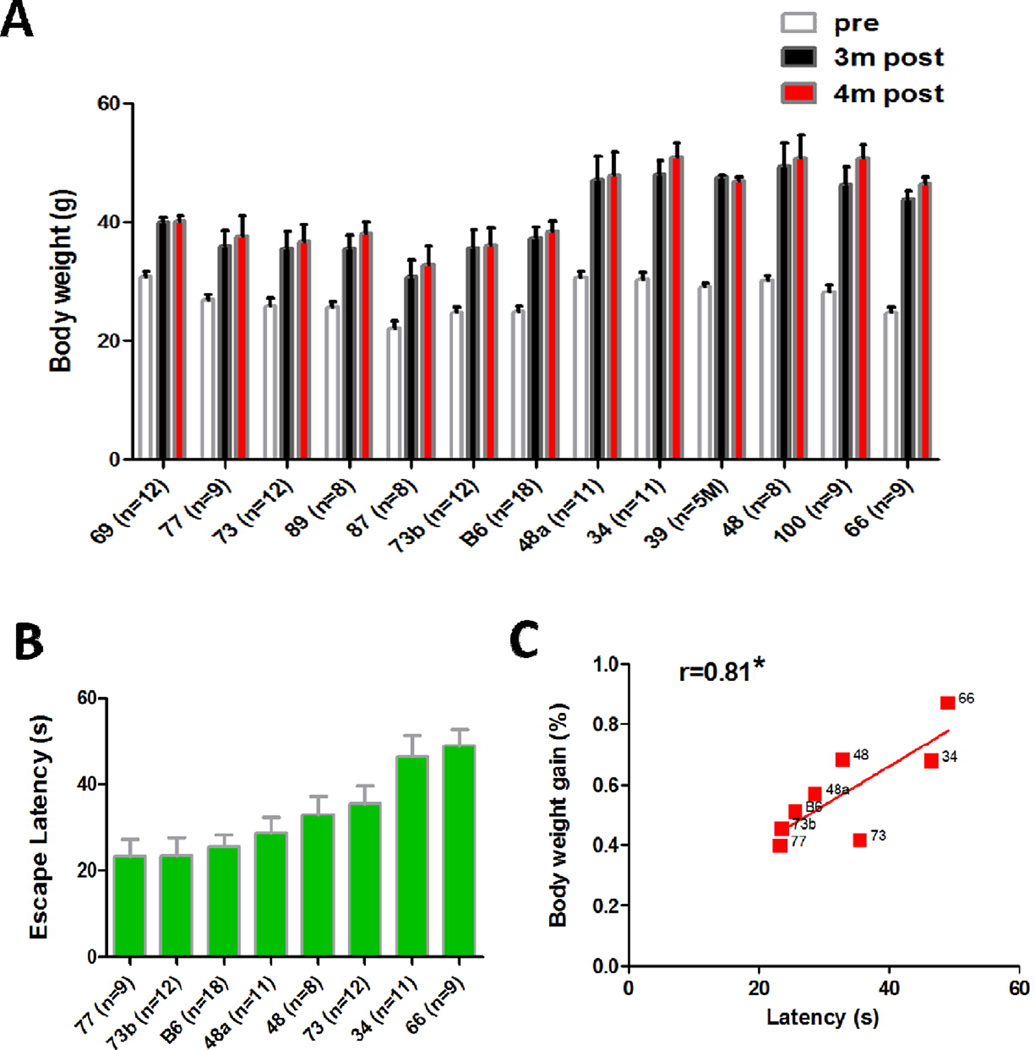
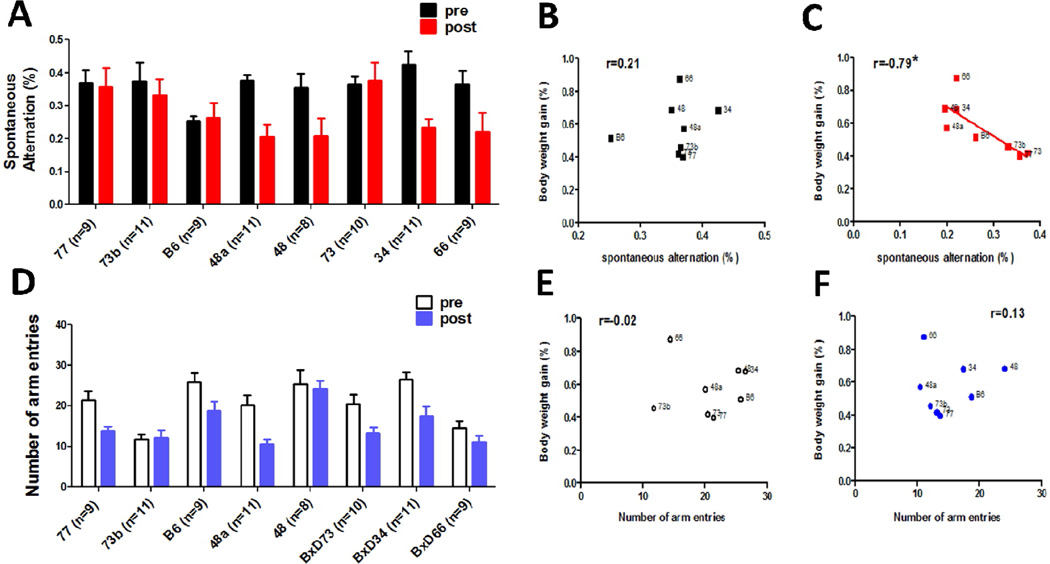
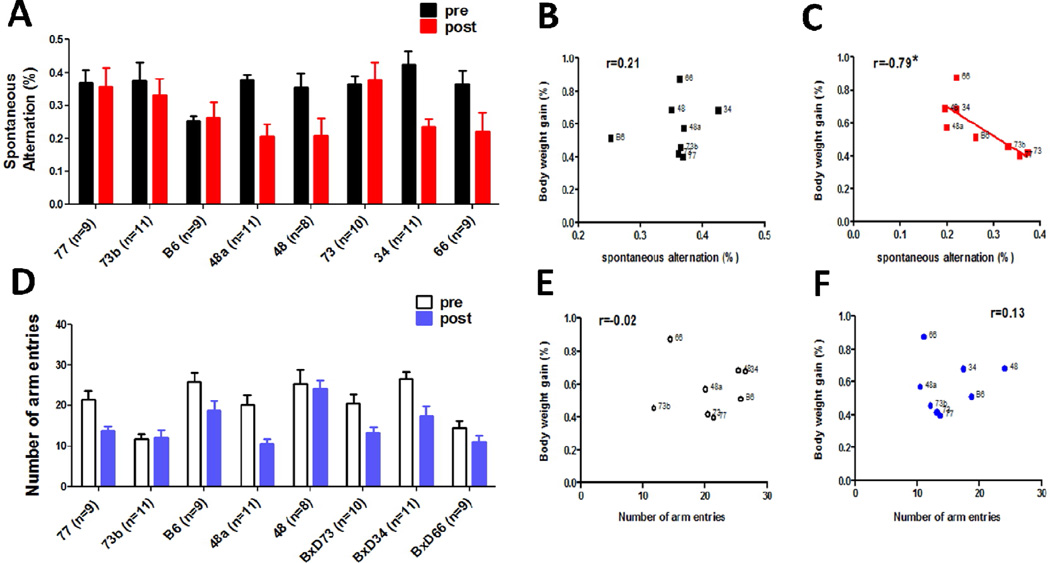
Thirteen BXD strains were tested for susceptibility to high fat diet. The body weight gain reached plateau levels by 3–4 months, and varied significantly (from 30.5% in BXD77 to 87.1% in BXD66) across the BXD strains after 4 months HFD feeding (Fig. 2A). There was a significant effect of strain (F(12, 238) =8.679, P<0.0001) on the body weight gain. The heritability (h2=0.72) was computed by dividing the between strain variance by the total variance. These results suggest that the body weight gain is a highly heritable trait. Eight representative strains were selected to perform the behavior test. The Pearson correlation between body weight gain and average escape latency (Fig. 2B) during the entire escape trial period of water maze task was 0.81 (P=0.018; Fig. 2C). For the cross maze test, the ANOVA revealed a significant effect of HFD feeding, but not strain, on spontaneous alternation (F(1,126) = 7.79, P<0.0005; Fig. 3A). The Pearson correlation between body weight gain and spontaneous alternate was −0.79 (P=0.019; Fig. 3C). The total amount of arm entries was also analyzed during each trial, which was taken as a measure of locomotor activity. There was a significant between group difference (F(1,125)= 12.13, P<0.0001; Fig. 3D), suggesting the locomotor activity was affected by HFD consumption, but no significant correlation was found between body weight gain and total amount of arm entries (Fig. 3E and F). With regard to the fear conditioning test, body weight gain was not significantly correlated with any of the variables (data not shown). Taken together, these results suggest that body weight gain can affect the spatial memory performance specifically, and this effect could not be explained by differences in motor activity.
Figure 2. Correlation between water-maze performance and body weight gain in BXD RI strains after 4-month HFD treatment.
(A) Comparison of the body weight of high fat diet induced-obesity mice after 4 months of consumption. Data are represented as mean ± SEM from each time-point (i.e. 0, 3 or 4 months of diets). Strains are arranged from smallest to largest body weight gain (left to right). (B) The average escape latency (error bar represents SEM, n=8–18/group) is shown for each BXD RI strain and the parent C57BL/6 strain. (C) A strong correlation exists between the percentage of body weight gain and the average escape latency in B BXD strains after 4 months of HFD consumption. The number next to each data point refers to the BXD strain number (P<0.05, r=0.81).
Figure 3. Correlation between spontaneous alternation behavior in the cross maze test and body weight gain in BXD RI strains after 4-month HFD treatment.
There were strain differences in both % alternation (A) and entry number (D) in n = 8–11 per BXD strain after 4 months of HFD consumption. Data are expressed as means ± SEM. There was a significant association between body weight gain and the percentage of spontaneous alternation in the cross maze test (C). The number next to each data point refers to the BXD strain number (P<0.05, r=−0.79).However, there was a weak correlation between body weight gain, entry number, and the percentage of spontaneous alternation before HFD feeding (B, E, and F).
3.3. Significant strain-differences of molecular and structural alterations in the hippocampus
3.3.1. Synaptic markers
For the levels of post-synaptic marker, PSD-95, one-way ANOVA revealed significant inter-strain differences under high fat feeding conditions (F(5,12)=5.880, P=0.0057; Fig. 4A and B). With regards to pre-synaptic marker, Synapsin-I, a very close to statistical significance (F(5,12)=2.823, P=0.0654; Fig. 4A and B) was observed. p-CAMKII level significantly decreased in DIO susceptible strains as compared to resistant strains of mice (F(5,12)=12.01, P=0.0002; Fig. 4A and B).
Figure 4. Verification of the molecular characteristics and structural alterations in the hippocampus of BXD RI strains after 4 months of HFD feeding.
(A) Homogenates of the hippocampi of mice were analyzed by Western blots developed with the indicated antibodies; each lane represents an individual mouse sample. (B) Quantitative analysis of densitometrical data from western blots after normalization with the β-actin. The corresponding protein levels are presented as mean ± SEM (n = 3/group); statistical analysis is described in the results section. (C) Representative microphotographs of GFAP-immunoreactive (IR) astrocytes in hippocampal CA1 regions of obesity-resistant strain (BXD77, left) and obesity-prone strain (BXD66, right), scale bar=50 µm. (D) Representative light microphotographs of Nissl-stained hippocampal CA1 regions of BXD77 (left) and BXD66(right) strains, scale bar=100 µm. (E) The Nissl-stained neurons were quantified using the ImageJ software. The data are presented as the mean of percent changes ± SEM (n = 3 per group), where the values of the B6 group are set as 100%; statistical analysis is described in the results section.
3.3.2. NO synthase, GFAP, and BDNF
Quantitative analysis revealed that nNOS expression was significantly down-regulated in DIO susceptible strains (F(5,12)=7.672, P=0.0019; Fig. 4A and B), but iNOS expression was significantly up-regulated in these strains (F(5,12)=12.33, P=0.0002; Fig. 4A and B) after HFD feeding. Meanwhile, the susceptible strain BXD66 mice displayed increased GFAP-positive cells in the CA1 field of the dorsal hippocampus as compared to DIO resistant strain BXD77 mice (Fig. 4C). The levels of BDNF protein significantly declined in hippocampus (F(5,12)=4.224, P=0.0190; Fig. 4A and B) of BXD66 mice, as compared to BXD77 mice.
3.3.3. Neuronal loss
Neuronal density based on Nissl staining was determined in the CA1 subfield of the hippocampus. A significant strain difference in neuronal cell number was detected in the pyramidal layer of the CA1 region of BXD mice (F(5,12) =12.02, P=0.0002; Fig. 4D and E). Tukey’s post-hoc test showed that there was a significant decrease between BXD66 and other BXD strains (P<0.001 compared with BXD34, P<0.01 compared with BXD73b and BXD77 mice, respectively; Fig. 4E).
These results clearly showed that BXD strains differed significantly in the hippocampal function after HFD feeding, and suggested that DIO-induced behavioral impairments are closely related to hippocampal dysfunction.
3.4. Genetic correlation analysis of metabolic, learning and memory traits with DSCAM gene from the GN phenotype database
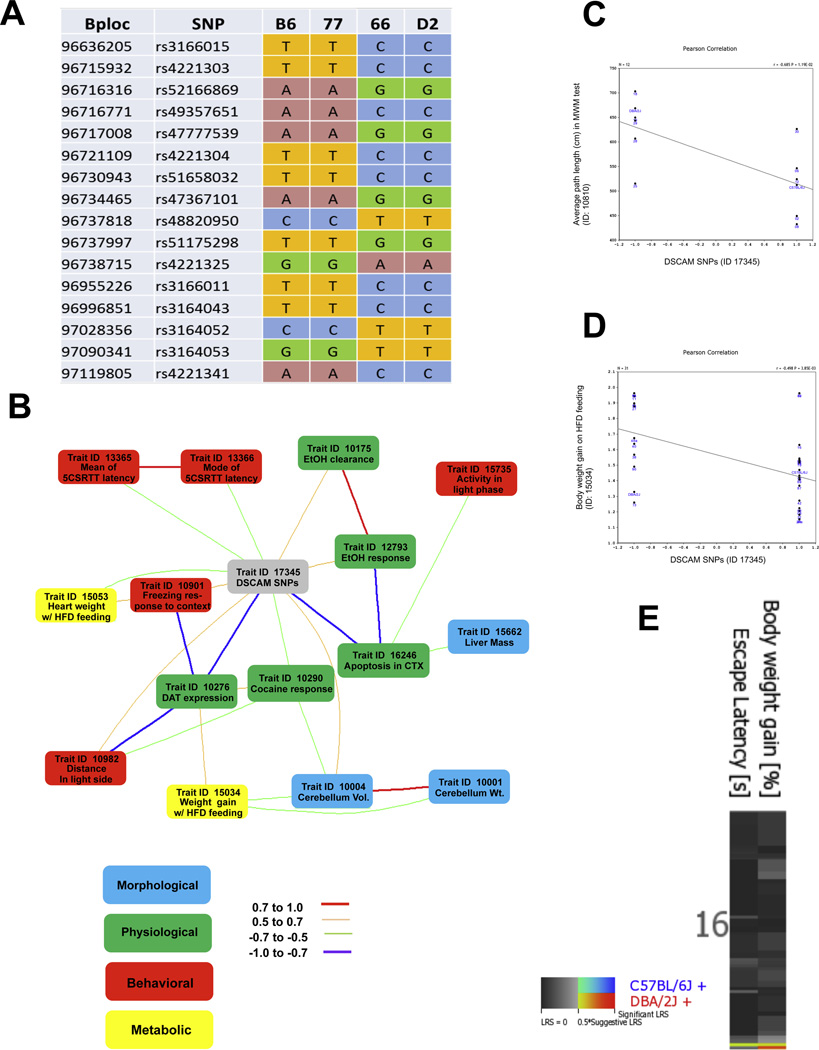
The haplotype structure of the DSCAM gene was queried through mouse phenome database at the Jackson Laboratory (http://phenome.jax.org/db/q?rtn=snp/ret1). DSCAM gene is highly polymorphic, containing at least 109 SNPs, 16 of which show strain differences. Haplotype analysis revealed that strains inheriting the paternal D2 allele exhibited a significant susceptibility to DIO and poorer behavior performance compared to those B6-like strains (Fig. 5A). To further validate the biological role of genetic variations of DSCAM gene in relation to learning behavior and obesity phenotypes, we computed genetic correlations between DSCAM gene polymorphism (GN Record ID: 17345) and over 4500 phenotypic traits of BXD RI sets from the GN database. A total of 49 anatomical, physiological and behavioral traits were found to significantly correlate with DSCAM SNPs (P<0.05). Top 15 correlation traits with a significant alpha level less than 0.005 are summarized in table 2, and Fig. 5B shows the co-variations among these traits (|0.5|≤r ≤|1|).
Figure 5. Genetic correlation analysis of DSCAM SNPs with metabolic, learning and memory traits from the BXD phenotype database.
(A) Haplotype map of DSCAM single-nucleotide polymorphism (SNP) from obesity-resistant strain (BXD77), obesity-prone strain (BXD66), and parental B6 and D2 strains. The genotype of DSCAM SNPs revealed that the B6 allele is more associated with obesity-resistance than the D2 allele. The physical position in mega bases (Mb) and SNP ID are listed in the left column, respectively. The SNPs are shown as observed nucleotides A, T, G, and C at each position among the strains. (B) GeneNetwork diagram illustrating top-ranked covariations among DSCAM SNPs (gray) and a collection of morphological (blue), physiological (green), behavioral (red), and metabolic traits (yellow). Strength of correlation between two connected traits is indicated in the legend. (C) Scatterplots illustrating correlation of DSCAM SNP genotypes with average path length to reach the platform during acquisition in the water maze test from the BXD Phenotype Database. (GN trait ID 10810, r=−0.68, P=0.01, N=12). (D) Scatterplots illustrating correlation of DSCAM SNP genotypes with body weight gain between 12 and 13 weeks under high fat diet feeding from the BXD Phenotype Database. (GN trait ID 15034, r=0.5, P<0.005, N=31). (E) Multiple QTL heat map of bodyweight gain and escape latency revealed an overlap QTL interval on the distal portion of Chr16; the candidate gene DSCAM is also located within this region.
Table 2.
Correlation of phenotypic traits in BXD mouse strains
| Record ID |
Trait description | Sample r | P-value | N cases | Compound trait |
|---|---|---|---|---|---|
| 13366 | Mode of correct response latencies in the 5- CSRT task |
−0.60718 | 1.82E-05 | 40 | Behavior/sensorimotor |
| 16246 | Post-natal neurogenesis; Apoptosis in cortex L2/3 (% caspase 3+ cells) |
−0.80841 | 0.000101 | 15 | Apoptosis |
| 13365 | Mean correct response latency in the 5- CSRT task |
−0.53897 | 0.000246 | 41 | Behavior/sensorimotor |
| 15662 | Liver mass, percentage of body | 0.49904 | 0.000621 | 42 | Morphology/Metabolism |
| 15735 | Activity (movement) during light phase | 0.49565 | 0.000688 | 42 | Behavior/movement |
| 10276 | Dopamine transporter binding maximum in membrane fragments in dorsal striatum |
−0.71582 | 0.000769 | 17 | Physiology/protein expression |
| 12793 | Dopamine (DA) level in the medial septal nucleus 72 h after ethanol vapor chamber treatment |
0.59479 | 0.000791 | 27 | Ethanol response |
| 10290 | Locomotion measured as difference from saline control after i.p injection of cocaine |
−0.63251 | 0.00156 | 21 | Cocaine response |
| 10004 | Cerebellum volume | 0.53567 | 0.001886 | 30 | Morphology |
| 15053 | Heart weight at 20 weeks on high fat diet feeding |
−0.52238 | 0.002162 | 31 | Metabolism/HFD feeding |
| 10175 | Rate of ethanol clearance | 0.56206 | 0.002293 | 26 | Ethanol response |
| 10982 | Restraint stress + ethanol treated, percent distance traveled in light side using a light- dark box,10 min session |
0.52079 | 0.003237 | 29 | Behavior/anxiety |
| 10001 | Cerebellum weight | 0.48899 | 0.003402 | 33 | Morphology |
| 15034 | Body weight gain between 12 and 13 weeks on high fat diet feeding |
−0.49769 | 0.003848 | 30 | Metabolism/HFD feeding |
| 10901 | freezing in response to context 48 hr after conditioning |
0.64072 | 0.004492 | 17 | Behavior/memory |
Data retrieved from BXD phenotype database, available at the GeneNetwork (www.genenetwork.org)
Among behavioral traits, DSCAM SNPs were highly correlated with sensorimotor-related behavior, such as: the mode and mean correct response latency (r=− 0.60, P< 0.0001, N=40, GN ID 13366; and r=− 0.54, P= 0.0002, N=40, GN ID 13365, respectively) in the 5-CSRT task; the activity during the light phase in standard housing cage (r=0.50, P< 0.001, N=42, GN ID 15735); and the locomotion from 0–5 min in the novel open field test (r=0.49, P< 0.05, N=17, GN ID 10911). It was also correlated with anxiety and memory-related behavior, such as: the freezing in response to context exposure 48 hr after conditioning in the fear conditioning test (r=0.64, P<0.005, N=17, GN ID 10901); the percent distance traveled in the light side under restraint stress and ethanol treatment in the light/dark transition test (r=0.52, P<0.005, N=29, GN ID 10982); total distance ratio and center-distance to total distance ratio in the open-field test (r=0.63, P<0.01, N=15, GN ID 12760; and r=− 0.56, P<0.05, N=15, GN ID 13536, respectively); and the average path length to reach the platform during acquisition in the Morris water maze test (r=−0.68, P=0.01, N=12, GN ID 10810; Fig. 5C).
For metabolic traits, there were significant correlations with body weight gain between 12 and 13 weeks and heart weight at 20 weeks under high fat diet feeding (r=−0.50, P<0.005, N=30, GN ID 15034, Fig. 5D; and r=− 0.52, P< 0.005, N=31, GN ID 15053, respectively). DSCAM SNPs were also correlated with ethanol clearance rate (r=0.56, P< 0.005, N=26, GN ID 10175); Iron level in dorsal striatum of females (r=−0.68, P= 0.006, N=14, GN ID 10242), ventral midbrain (r=−0.56, P< 0.05, N=14, GN ID 10246), dorsal striatum of males (r=−0.56, P< 0.05, N=14, GN ID 10241), and nucleus accumbens (r=−0.56, P<0.05, N=14, GN ID 10244); and copper level in medial prefrontal cortex (r=−0.65, P<0.01, N=14, GN ID 10733).
Among morphological traits, DSCAM SNPs were highly correlated with liver mass (% of body, r=0.50, P= 0.0006, N=42, GN ID 15662); and cerebellum volume and weight (r=0.53, P=0.002, N=30, GN ID 10004; and r=0.48, P= 0.003, N=33, GN ID 10001, respectively); It was also correlated with striatum cholinergic neurons (r=0.50, P<0.01, N=26, GN ID 10109) and septal nuclei and cochlear nuclei volume (r=0.72, P=0.01, N=11, GN ID 10893; and r=0.68, P=0.02, N=11, GN ID 10938, respectively).
DSCAM SNPs were also associated with many other physiological traits, such as apoptosis in cortex L2/3(r=−0.80, P=0.0001, N=15, GN ID 16246); longevity of females (r=−0.55, P<0.01, N=22, GN ID 10148); and dopamine transporter binding capacity in dorsal striatum (r=−0.58, P=0.01, N=17, GN ID 10278); In addition, there were correlations between DSCAM SNPs and a set of traits (Trait IDs: 10064, 10141, 10287, 10290, 10291, 12002) which measured ethanol response and the locomotion response to cocaine administration (P<0.01).
These results provide further evidence supporting that there are significant strain differences on the susceptibility to DIO-induced spatial memory impairment, and also add information concerning the contribution of allelic variants of DSCAM gene family to specific phenotypic variations.
3.5. Validation of HFD-responsive genes by real-time PCR
To assess HFD-induced gene expression changes in the hippocampus, we measured the mRNA levels of several selected neuroinflammation cytokines, synaptic plasticity markers, and obesity and metabolic factors in obesity-resistant strain BXD77, obesity-prone strain BXD66, and parental B6 and D2 strains.
3.5.1. Neuroinflammatory response factors
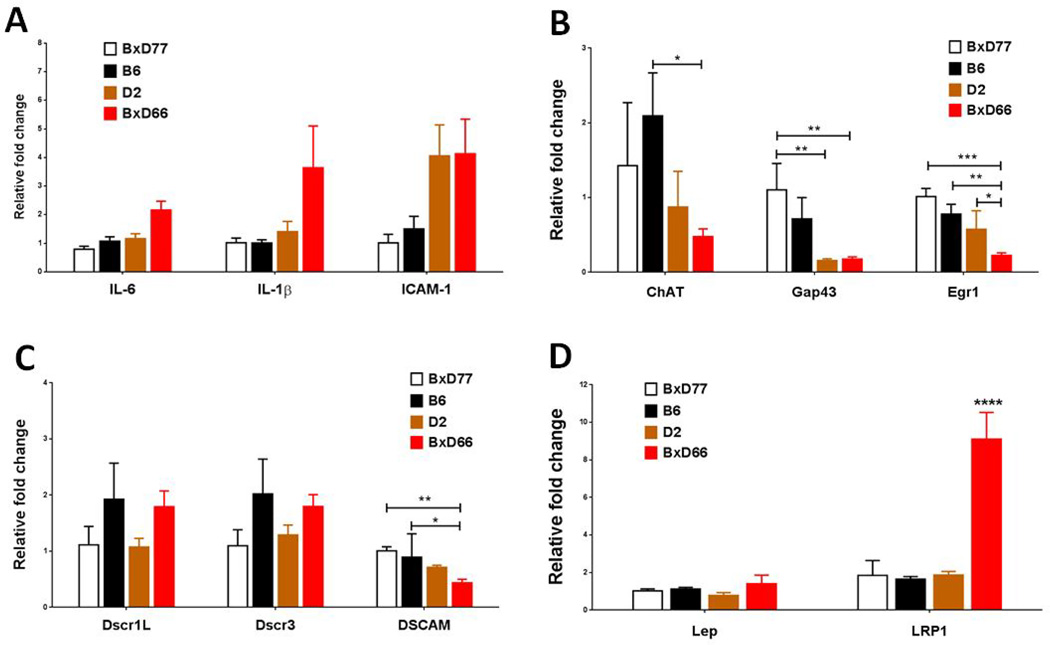
IL-6 is a cytokine that having both pro- and anti-inflammatory properties [37]. A one-way ANOVA showed no strain difference for IL-6 mRNA expression (P>0.05). For the expression of inflammatory cytokine IL-1β and cellular adhesion molecules ICAM1, BXD66 mice showed the trend of increasing but did not reach significance (F(3,19) = 2.699, P=0.0747; and F(3,20) = 2.847, P=0.0634, respectively, Fig. 6A).
Figure 6. HFD-induced gene expression changes in the hippocampus of obesity-resistant strain (BXD77), obesity-prone strain (BXD66), and parental B6 and D2 strains.
(A) Changes in hippocampal mRNA expression of neuro inflammation cytokines, from left to right, interleukin (IL)-6, IL-1β, and ICAM-1. (B) Synaptic plasticity markers ChAT, GAP43, and Egr-1. (C) Down Syndrome-related genes Dscr1L, Dscr3, and DSCAM. (D) Obesity related genes Lep and LRP1. Data are expressed as means ± SEM (n = 3 per group, duplicate experiments were performed).The relative transcript levels were normalized to the expression level of GAPDH mRNA. *, p<0.05, **, p<0.01, ***, p<0.001, and ****, p<0.0001.
3.5.2. Cognitive function-related genes
For the levels of ChAT, there was a statistically significant difference between groups as determined by one-way ANOVA (F(3,18) = 3.388, P<0.05). Tukey’s post-hoc test showed that there was a significant decrease in BXD66 and D2 strains (P<0.001), as compared to B6 strain. Gap43 expression significantly decreased in BXD66 and D2 mice (F(3,18)=12.08, P<0.0001). Egr-1 expression also significantly decreased in BXD66 mice (F(3,18)=12.38, P<0.0001; Fig. 6B).
3.5.3. Neuroendocrine factors
Leptin plays a critical role in the body weight regulation. We found the expression of leptin was slightly increased in the hippocampus of BXD66 mice. Whereas, a robust increase in the expression of leptin receptor was observed in the hippocampus after HFD feeding (F(3,18)=17.19, P<0.0001; Fig. 6D).
3.5.4. DSCR gene and DSCAM gene
The mRNA expression levels of DSCR3 and DSCAM were measured in the hippocampus of BXD lines. We found BXD66 mice displayed lower levels of DSCAM mRNA expression than other strains (F(3,18)= 7.926, P=0.0014), and that the expression level was closely related with the behavioral phenotype. Whereas, there was no expression difference was found in the levels of DSCR expression among the BXD strains (Fig. 6C).
We also examined pre-existing transcriptome data to assess the normative expression levels of these responsive genes. The BXD hippocampal expression database is publicly available in GeneNetwork. No difference in transcription level of the corresponding genes was observed among BXD mice, as summarized in Table 3. Taken together, these results suggest that the alterations of cognitive function-related gene expression in the hippocampus of obese-prone strain are associated with the cis-acting genetic variation in BXD mice.
Table 3.
The transcripts levels of HFD-responsive genes in the hippocampus of BXD RI strains
| Gene | ID | Location (Chr: Mb) |
BXD77 | B6 | D2 | BXD66 |
|---|---|---|---|---|---|---|
| BDNF | 12064 | 2: 109.514857 | 8.06±0.02 | 7.90±0.17 | 7.99±0.09 | 7.97±0.39 |
| LepR | 16847 | 4: 101.390044 | 6.38±0.13 | 6.80±0.21 | 6.77±0.18 | 6.80±0.69 |
| nNOS | 18125 | 5: 118.292195 | 10.15±0.12 | 9.79±0.42 | 10.39±0.13 | 9.90±0.55 |
| Leptin | 16846 | 6: 29.010221 | 6.37±0.16 | 6.23±0.19 | 6.25±0.08 | 6.27±0.18 |
| ICAM1 | 15894 | 9: 20.820404 | 8.37±0.02 | 8.47±0.11 | 8.21±0.36 | 8.39±0.04 |
| PSD95 | 13385 | 11: 69.830727 | 11.88±0.04 | 12.13±0.17 | 11.93±0.07 | 11.93±0.37 |
| iNOS | 18126 | 11: 78.734289 | 7.11±0.04 | 7.13±0.16 | 7.02±0.30 | 7.29±0.33 |
| GFAP | 14580 | 11: 102.74866 | 9.94±0.03 | 10.40±0.42 | 10.37±0.24 | 10.02±0.29 |
| ChAT | 12647 | 14: 33.221389 | 6.75±0.03 | 6.66±0.05 | 6.76±0.08 | 6.85±0.07 |
| Gap43 | 14432 | 16: 42.248665 | 10.19±0.05 | 10.26±0.16 | 10.22±0.09 | 10.55±0.31 |
| Dscr3 | 13185 | 16: 94.719395 | 8.84±0.07 | 9.05±0.06 | 9.12±0.12 | 9.07±0.22 |
| DSCAM | 13508 | 16: 96.814339 | 10.47±0.07 | 10.58±0.09 | 10.61±0.19 | 10.62±0.22 |
| Egr1 | 13653 | 18: 35.019477 | 10.01±0.06 | 9.93±0.01 | 9.91±0.36 | 10.14±0.03 |
| CamKII | 12322 | 18: 61.085286 | 13.99±0.05 | 13.96±0.20 | 14.05±0.08 | 13.94±0.42 |
| Syn-1 | 20964 | X: 20.437637 | 11.90±0.13 | 11.84±0.34 | 12.13±0.05 | 11.91±0.48 |
Data retrieved from the BXD hippocampal microarray database, available at the GeneNetwork (www.genenetwork.org)
4. Discussion
The results of the present study demonstrated that genetic background has a significant impact on the susceptibility to dietary induced obesity. We found that body weight gain significantly correlated with a number of behavioral, morphological, and gene expression phenotype variances. Our data revealed that the relationship between body weight gain and spatial memory is subject to genetic control in the BXD genetic reference panel.
This genotype-phenotype relationship was further clarified through using trait correlation analysis of DSCAM gene in GN database. The results showed that DSCAM SNPs are significantly correlated with a number of neuroanatomical, physiological, and behavioral phenotypes that reflect these functions, suggesting DSCAM as a promising positional candidate gene for the susceptibility to DIO-induced cognitive impairment. DSCAM has conserved basic functions in neural development and has been directly implicated in Down syndrome [38]. In the adult brain, DSCAM is expressed in pyramidal cells in the cortex and in Purkinje cells of the cerebellum. This expression pattern indicates a functional role of DSCAM in the development and function of motor neurons. Indeed, a recent study reported that DSCAMdel17 mutant mice have impaired motor coordination [29].It is noteworthy that the traits of sensorimotor-related behavior were also among the top correlations with DSCAM SNPs in our study (table 2). The significant negative correlation suggested that D2-like BXD strains have slower sensory-motor-processing speed compared with B6-like strains, which is in concordance with previously published studies [39, 40].
In another instance, we found highly positive correlation with cerebellum volume and weight, and highly negative correlation with apoptosis in cortex Layer 2/3. Specifically, BXD strains with B6 haplotype had larger cerebellum volume and weight, and less apoptosis in cortex compared with D2-like strains. In our experiment the BXD mice showed large variations in the degree of weight gain, spatial memory performance, and gene expression pattern in hippocampus after HFD consumption. In general, DBA/2 mice are typically considered poor learners specifically in hippocampal dependent tasks [41]. More recent reports showed different learning strategy selection between D2 and B6 mice, which is closely related to synaptic plasticity in hippocampus [42–44]. Here, we conducted DSCAM SNPs and haplotype analysis in BXD strains and the B6 and D2 parental strains. As expected, we found that BXD66 inheriting the D allele at this genetic interval exhibited more susceptibility to DIO, whereas those strains inheriting the B6 haplotype showed less susceptibility. We hypothesized that gene expression differences may be attributed to the haplotype diversity. Thus, haplotype analysis could be used to predict learning and memory performance. This hypothesis will be addressed by further population genetics studies.
From a metabolic perspective, we also found highly significant correlations between DSCAM and many metabolic traits, such as liver mass and body weight gain after HFD feeding (table 2). This correlation is of interest in view of a recent systemic genetic study indicating that DSCAM is associated with individual daily feed intake in a population of Duroc pigs[45]. In this context, DSCAM represents a plausible candidate gene for co-regulations of both the behavior trait and the body weight. Currently, DSCAM is being targeted by the Knock-Out Mouse Project (www.komp.org), and further functional studies of DSCAM gene on these knockout or conditional knockout mice can be pursued in the near future.
The QTL mapping approach is an advanced tool to identify candidate genes responsible for the variations in quantitative phenotypes. Although the small number of strains used in our analysis undermines the reliability of such an analysis, we obtained one suggestive QTL region overlapped for groups of traits at 96.64 to 98.18 Mb on chromosome 16 including the candidate gene DSCAM in a preliminary analysis (Fig. 5E). In fact, many QTL-influenced responses to body weight, body fat, glucose level, and lipid level have been mapped within DSCAM region in humans (table 1). Thus, this suggestive QTL further suggests that the candidate genes in this region may contribute to DIO-induced cognitive decline, but will require additional verification by testing in a bigger BXD cohort containing many more of the BXD strains.
Table 1.
QTLs search result for DSCAM gene of Homo sapiens
| RGD ID | Symbol | LOD | P Value | Trait | Start | Stop |
|---|---|---|---|---|---|---|
| 1357321 | AASTH53_H | 0.0096 | Reversible airflow obstruction | 26141707 | 48129895 | |
| 1298478 | BP12_H | 2.82 | Blood pressure | 26141707 | 48129895 | |
| 1643280 | BW202_H | 4.27 | 0.00001 | Body fat amount | 35037695 | 48129895 |
| 1643522 | BW296_H | 1.19 | Body weight | 41191420 | 44160967 | |
| 2289198 | BW369_H | 0.0303 | Body fat amount | 34328246 | 48129895 | |
| 2289305 | BW395_H | 1.19 | Body weight | 41191420 | 44160967 | |
| 2315822 | GLUCO185_H | 1.67 | Glucose level | 29684648 | 48129895 | |
| 1643546 | GLUCO33_H | 0.042 | Glucose level | 26141707 | 48129895 | |
| 2314742 | GLUCO94_H | 0.83 | 0.005 | Glucose level | 24827002 | 48129895 |
| 1358833 | MULTSCL25_H | Multiple sclerosis susceptibility | 35037695 | 48129895 | ||
| 2292819 | MYI37_H | Myocardial infarction | 35037695 | 48129895 | ||
| 2292955 | PRSTS145_H | 5.15 | 0.000019 | Prostate tumor susceptibility | 29684648 | 48129895 |
| 2293457 | PRSTS308_H | 0.013 | Prostate tumor susceptibility | 26141707 | 48129895 | |
| 2293481 | PRSTS311_H | 0.002 | Prostate tumor susceptibility | 28191515 | 48129895 | |
| 1559112 | SCL36_H | 2.66 | Lipid level | 40421533 | 47968032 | |
| 1559108 | SCL37_H | 2.74 | Lipid level | 40421533 | 47968032 | |
| 1559244 | SCL56_H | 2 | 0.001 | Lipid level | 41356608 | 47968217 |
Data retrieved from the Rat Genome Database (http://rgd.mcw.edu/rgdweb/search/qtls.html?term=DSCAM%5Bgene%5D&speciesType=1)
Although a strong correlation between DIO and impaired hippocampal dependent performance was found in this study, it remains unclear whether obesity is a cause or a consequence of the cognitive deficits. Several longitudinal birth cohort studies indicate that lower levels of cognitive function in early life can increase the risk and/or predict the development of later obesity [46–48]. A recent animal study showed that a short-term feeding (1–3 wks) of HFD can induce anxiety-like behaviors and learning/memory impairments prior to the onset of weight gain and/or pre-diabetes in juvenile mice [49]. In our study, the susceptible BXD66 strain with DBA/2-derived alleles at DSCAM on chromosome 16 may already have subtle impairment in hippocampal function. With this in mind, poor neurological function in these mice could have implications for body weight increase over time which establishes a bidirectional link between DIO and cognitive function.
Obesity is associated with cognitive impairments. The Long-term mechanisms underlying this association include chronic neuro inflammation [see review of 50], neuroendocrine dysregulation such as leptin and insulin resistance [see review of 51], oxidative stress [52], and synaptic dysfunction[53]. The indirect mechanisms include the vascular damage and BBB breakdown [54], and likely a combination of neuroanatomical changes and biochemical alterations. It should be noted that these alterations have synergistic effects and are not restricted to the hippocampus. Recent data found that synaptic dysfunction in hippocampus is mediated by IL1 in db/db mice [53]. Another report also indicated that DIO-induced cerebro-micro vascular damage and BBB disruption can promote neuroinflammation and oxidative stress in the hippocampus of aged mice [55]. Consistent with these findings, we observed an exacerbated neuroinflammatory response as shown by the increased number of activated astrocytes, and synaptic dysfunction as shown by significantly reduced synaptic marker —SYN-1 and PSD95 level in the hippocampus of the susceptible BXD66 strain (Fig. 3A). In addition, we found significantly reduced levels of GAP43, egr-1, and p-CAMKII in BXD66 mice. GAP-43 is a neuron-specific expressed protein that control neuronal development and synaptic plasticity; a learning-dependent increase in GAP-43 expression has been reported previously [56]. CaMKII is one of the most abundant protein kinases in the brain, and the pivotal role of CaMKII/NMDAR complex as a molecular memory is well documented. Egr-1 belongs to the immediate early gene family and has been demonstrated to be involved in synapse remodeling and memory formation, with egr-1 mutant mice showing severe deficits in long-term spatial memory. It has been recognized that both CaMKII and egr-1 expression were up-regulated in the hippocampus during spatial learning in the Morris water maze training [57, 58].
Leptin is another neuroendocrine hormone of interest that functions in weight regulation. We found a slight increase of leptin mRNA expression in hippocampus, whereas a robust increase in leptin receptor (LepR) mRNA level was found in the obesity-prone BXD66 strain compared with the resistant BXD77 strain (Fig. 6D). Although Leptin is produced primarily in the adipocytes, there is also evidence that leptin mRNA is expressed in a number of brain regions, including the hippocampus [59], where a prominent astrocytic expression of LepR was also observed [60]. A recently study has shown that the astrocytic LepR expression was enhanced in DIO mice [61]. Another report showed that the LepRs functions as a proinflammatory factor during leptin resistance in db/db mice [62]. In response to HFD, the astrocytic leptin-receptor knockout mice did not show worsening of obesity but instead showed partial rescue of leptin resistance [63]. Similarly, we found significant increases in LepR expression level paralleled by increased astrocyte numbers in the hippocampus of the susceptible BXD strains; both of these changes are associated with cognitive deficits. However, higher leptin may enhance cognition, as proved by direct administration in to the brain [64]. Leptin receptor-deficient animals also showed impaired hippocampal LTP and poor spatial memory[65]. The mechanisms for these disparities are unclear yet necessary to gain more detailed information on the interactions between leptin and other neuronal pathways regulating cognition in response to HFD feeding. From our observation, although leptin has a protective effect in the brain, it is still not enough to ameliorate the cognitive impairment induced by DIO.
Taken together, the present study identified a population of BXD strains susceptible to DIO with severe spatial memory deficits. These findings provide a firm starting point in unraveling the genetic background of differential susceptibility to DIO-induced cognitive decline.
Highlights.
Obesity-prone strains display more pronounced memory deficits than controls
Diet-induced hippocampal dysfunction is largely dependent on genetics in BXD strains
DSCAM is implicated in body weight and diet-induced hippocampal dysfunction
Acknowledgments
We thank Dr. Lubin Lan and Mr. Bin Wang for their invaluable technical assistance in animal perfusion and tissue collection. We also thank Ms. Xiaohan Xue for her efforts in editing this paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Savini I, Catani MV, Evangelista D, Gasperi V, Avigliano L. Obesity-associated oxidative stress: strategies finalized to improve redox state. International journal of molecular sciences. 2013;14:10497–10538. doi: 10.3390/ijms140510497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeMarco VG, Aroor AR, Sowers JR. The pathophysiology of hypertension in patients with obesity. Nature reviews Endocrinology. 2014;10:364–376. doi: 10.1038/nrendo.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao GH, Thethi I, Fareed J. Vascular disease: obesity and excess weight as modulators of risk. Expert review of cardiovascular therapy. 2011;9:525–534. doi: 10.1586/erc.11.43. [DOI] [PubMed] [Google Scholar]
- 4.Lo AH, Pachana NA, Byrne GJ, Sachdev PS, Woodman RJ. Relationship between changes in body weight and cognitive function in middle-aged and older women. International journal of geriatric psychiatry. 2012;27:863–872. doi: 10.1002/gps.2797. [DOI] [PubMed] [Google Scholar]
- 5.Fagundo AB, de la Torre R, Jimenez-Murcia S, Aguera Z, Granero R, Tarrega S, et al. Executive functions profile in extreme eating/weight conditions: from anorexia nervosa to obesity. PloS one. 2012;7:e43382. doi: 10.1371/journal.pone.0043382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith E, Hay P, Campbell L, Trollor JN. A review of the association between obesity and cognitive function across the lifespan: implications for novel approaches to prevention and treatment. Obesity reviews : an official journal of the International Association for the Study of Obesity. 2011;12:740–755. doi: 10.1111/j.1467-789X.2011.00920.x. [DOI] [PubMed] [Google Scholar]
- 7.Arnold SE, Lucki I, Brookshire BR, Carlson GC, Browne CA, Kazi H, et al. High fat diet produces brain insulin resistance, synapto dendritic abnormalities and altered behavior in mice. Neurobiology of disease. 2014;67:79–87. doi: 10.1016/j.nbd.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winocur G, Greenwood CE. Studies of the effects of high fat diets on cognitive function in a rat model. Neurobiology of aging. 2005;26(Suppl 1):46–49. doi: 10.1016/j.neurobiolaging.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 9.NIH. Clinical Guidelines on the Identification, Evaluation, and Treatment of Overweight and Obesity in Adults--The Evidence Report. National Institutes of Health. Obesity research. 1998;6(Suppl 2):51S–209S. [PubMed] [Google Scholar]
- 10.Poston WS, 2nd, Foreyt JP. Obesity is an environmental issue. Atherosclerosis. 1999;146:201–209. doi: 10.1016/s0021-9150(99)00258-0. [DOI] [PubMed] [Google Scholar]
- 11.Heitmann BL, Lissner L, Sorensen TI, Bengtsson C. Dietary fat intake and weight gain in women genetically predisposed for obesity. The American journal of clinical nutrition. 1995;61:1213–1217. doi: 10.1093/ajcn/61.6.1213. [DOI] [PubMed] [Google Scholar]
- 12.Llewellyn CH, Trzaskowski M, Plomin R, Wardle J. From modeling to measurement: Developmental trends in genetic influence on adiposity in childhood. Obesity. 2014;22:1756–1761. doi: 10.1002/oby.20756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almind K, Kahn CR. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes. 2004;53:3274–3285. doi: 10.2337/diabetes.53.12.3274. [DOI] [PubMed] [Google Scholar]
- 14.Levin BE, Sullivan AC. Glucose-induced sympathetic activation in obesity-prone and resistant rats. Int J Obes. 1989;13:235–246. [PubMed] [Google Scholar]
- 15.Svenson KL, Von Smith R, Magnani PA, Suetin HR, Paigen B, Naggert JK, et al. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. Journal of applied physiology. 2007;102:2369–2378. doi: 10.1152/japplphysiol.01077.2006. [DOI] [PubMed] [Google Scholar]
- 16.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature genetics. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nature genetics. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 18.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature genetics. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andreux PA, Williams EG, Koutnikova H, Houtkooper RH, Champy MF, Henry H, et al. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150:1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dogan A, Lasch P, Neuschl C, Millrose MK, Alberts R, Schughart K, et al. ATR-FTIR spectroscopy reveals genomic loci regulating the tissue response in high fat diet fed BXD recombinant inbred mouse strains. BMC genomics. 2013;14:386. doi: 10.1186/1471-2164-14-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West DB, Waguespack J, York B, Goudey-Lefevre J, Price RA. Genetics of dietary obesity in AKR/J x SWR/J mice: segregation of the trait and identification of a linked locus on chromosome 4. Mammalian genome : official journal of the International Mammalian Genome Society. 1994;5:546–552. doi: 10.1007/BF00354928. [DOI] [PubMed] [Google Scholar]
- 22.Millward CA, Burrage LC, Shao H, Sinasac DS, Kawasoe JH, Hill-Baskin AE, et al. Genetic factors for resistance to diet-induced obesity and associated metabolic traits on mouse chromosome 17. Mammalian genome : official journal of the International Mammalian Genome Society. 2009;20:71–82. doi: 10.1007/s00335-008-9165-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuschl C, Hantschel C, Wagener A, Schmitt AO, Illig T, Brockmann GA. A unique genetic defect on chromosome 3 is responsible for juvenile obesity in the Berlin Fat Mouse. International journal of obesity. 2010;34:1706–1714. doi: 10.1038/ijo.2010.97. [DOI] [PubMed] [Google Scholar]
- 24.Vogel H, Nestler M, Ruschendorf F, Block MD, Tischer S, Kluge R, et al. Characterization of Nob3, a major quantitative trait locus for obesity and hyperglycemia on mouse chromosome 1. Physiological genomics. 2009;38:226–232. doi: 10.1152/physiolgenomics.00011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barsh GS, Farooqi IS, O’Rahilly S. Genetics of body-weight regulation. Nature. 2000;404:644–651. doi: 10.1038/35007519. [DOI] [PubMed] [Google Scholar]
- 26.Peirce JL, Lu L, Gu J, Silver LM, Williams RW. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC genetics. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laughlin RE, Grant TL, Williams RW, Jentsch JD. Genetic dissection of behavioral flexibility: reversal learning in mice. Biological psychiatry. 2011;69:1109–1116. doi: 10.1016/j.biopsych.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosen GD, Chesler EJ, Manly KF, Williams RW. An informatics approach to systems neurogenetics. Methods in molecular biology. 2007;401:287–303. doi: 10.1007/978-1-59745-520-6_16. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Ye H, Shen Y, Xu Q, Zhu L, Liu J, et al. Dscam mutation leads to hydrocephalus and decreased motor function. Protein & cell. 2011;2:647–655. doi: 10.1007/s13238-011-1072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Wang B, Liu D, Li JJ, Xue Y, Sakata K, et al. Hsp90 chaperone inhibitor 17-AAG attenuates A beta-induced synaptic toxicity and memory impairment. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:2464–2470. doi: 10.1523/JNEUROSCI.0151-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison FE, Reiserer RS, Tomarken AJ, McDonald MP. Spatial and nonspatial escape strategies in the Barnes maze. Learning & memory. 2006;13:809–819. doi: 10.1101/lm.334306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome research. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 33.Barr AM, Wu CH, Wong C, Hercher C, Topfer E, Boyda HN, et al. Effects of chronic exercise and treatment with the antipsychotic drug olanzapine on hippocampal volume in adult female rats. Neuroscience. 2013;255:147–157. doi: 10.1016/j.neuroscience.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 34.Nair HK, Hain H, Quock RM, Philip VM, Chesler EJ, Belknap JK, et al. Genomic loci and candidate genes underlying inflammatory nociception. Pain. 2011;152:599–606. doi: 10.1016/j.pain.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crusio WE, Schwegler H, Lipp HP. Radial-maze performance and structural variation of the hippocampus in mice: a correlation with mossy fibre distribution. Brain research. 1987;425:182–185. doi: 10.1016/0006-8993(87)90498-7. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen PV, Abel T, Kandel ER, Bourtchouladze R. Strain-dependent differences in LTP and hippocampus-dependent memory in inbred mice. Learning & memory. 2000;7:170–179. doi: 10.1101/lm.7.3.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochimica et biophysica acta. 2011;1813:878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 38.Cvetkovska V, Hibbert AD, Emran F, Chen BE. Overexpression of Down syndrome cell adhesion molecule impairs precise synaptic targeting. Nat Neurosci. 2013;16:677–682. doi: 10.1038/nn.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loos M, Staal J, Schoffelmeer AN, Smit AB, Spijker S, Pattij T. Inhibitory control and response latency differences between C57BL/6J and DBA/2J mice in a Go/No-Go and 5-choice serial reaction time task and strain-specific responsivity to amphetamine. Behavioural brain research. 2010;214:216–224. doi: 10.1016/j.bbr.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 40.Loos M, Staal J, Pattij T, Neuro BMPC, Smit AB, Spijker S. Independent genetic loci for sensorimotor gating and attentional performance in BXD recombinant inbred strains. Genes, brain, and behavior. 2012;11:147–156. doi: 10.1111/j.1601-183X.2011.00754.x. [DOI] [PubMed] [Google Scholar]
- 41.Paylor R, Baskall-Baldini L, Yuva L, Wehner JM. Developmental differences in place-learning performance between C57BL/6 and DBA/2 mice parallel the ontogeny of hippocampal protein kinase C. Behavioral neuroscience. 1996;110:1415–1425. doi: 10.1037//0735-7044.110.6.1415. [DOI] [PubMed] [Google Scholar]
- 42.Hwang YK, Song JC, Han SH, Cho J, Smith DR, Gallagher M, et al. Differences in hippocampal CREB phosphorylation in trace fear conditioning of two inbred mouse strains. Brain research. 2010;1345:156–163. doi: 10.1016/j.brainres.2010.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sung JY, Goo JS, Lee DE, Jin DQ, Bizon JL, Gallagher M, et al. Learning strategy selection in the water maze and hippocampal CREB phosphorylation differ in two inbred strains of mice. Learning & memory. 2008;15:183–188. doi: 10.1101/lm.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Passino E, Middei S, Restivo L, Bertaina-Anglade V, Ammassari-Teule M. Genetic approach to variability of memory systems: analysis of place vs. response learning and fos-related expression in hippocampal and striatal areas of C57BL/6 and DBA/2 mice. Hippocampus. 2002;12:63–75. doi: 10.1002/hipo.10007. [DOI] [PubMed] [Google Scholar]
- 45.Do DN, Ostersen T, Strathe AB, Mark T, Jensen J, Kadarmideen HN. Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs. BMC genetics. 2014;15:27. doi: 10.1186/1471-2156-15-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luppino FS, de Wit LM, Bouvy PF, Stijnen T, Cuijpers P, Penninx BW, et al. Overweight, obesity, and depression: a systematic review and meta-analysis of longitudinal studies. Archives of general psychiatry. 2010;67:220–229. doi: 10.1001/archgenpsychiatry.2010.2. [DOI] [PubMed] [Google Scholar]
- 47.Guxens M, Mendez MA, Julvez J, Plana E, Forns J, Basagana X, et al. Cognitive function and overweight in preschool children. American journal of epidemiology. 2009;170:438–446. doi: 10.1093/aje/kwp140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osika W, Montgomery SM Longitudinal Birth Cohort S. Physical control and coordination in childhood and adult obesity: Longitudinal Birth Cohort Study. Bmj. 2008;337:a699. doi: 10.1136/bmj.a699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaczmarczyk MM, Machaj AS, Chiu GS, Lawson MA, Gainey SJ, York JM, et al. Methylphenidate prevents high-fat diet (HFD)-induced learning/memory impairment in juvenile mice. Psychoneuroendocrinology. 2013;38:1553–1564. doi: 10.1016/j.psyneuen.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castanon N, Lasselin J, Capuron L. Neuropsychiatric comorbidity in obesity: role of inflammatory processes. Frontiers in endocrinology. 2014;5:74. doi: 10.3389/fendo.2014.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fadel JR, Jolivalt CG, Reagan LP. Food for thought: the role of appetitive peptides in age-related cognitive decline. Ageing research reviews. 2013;12:764–776. doi: 10.1016/j.arr.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinha JK, Ghosh S, Swain U, Giridharan NV, Raghunath M. Increased macromolecular damage due to oxidative stress in the neocortex and hippocampus of WNIN/Ob, a novel rat model of premature aging. Neuroscience. 2014;269:256–264. doi: 10.1016/j.neuroscience.2014.03.040. [DOI] [PubMed] [Google Scholar]
- 53.Erion JR, Wosiski-Kuhn M, Dey A, Hao S, Davis CL, Pollock NK, et al. Obesity elicits interleukin 1-mediated deficits in hippocampal synaptic plasticity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:2618–2631. doi: 10.1523/JNEUROSCI.4200-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davidson TL, Monnot A, Neal AU, Martin AA, Horton JJ, Zheng W. The effects of a high-energy diet on hippocampal-dependent discrimination performance and blood-brain barrier integrity differ for diet-induced obese and diet-resistant rats. Physiology & behavior. 2012;107:26–33. doi: 10.1016/j.physbeh.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tucsek Z, Toth P, Sosnowska D, Gautam T, Mitschelen M, Koller A, et al. Obesity in Aging Exacerbates Blood-Brain Barrier Disruption, Neuroinflammation, and Oxidative Stress in the Mouse Hippocampus: Effects on Expression of Genes Involved in Beta-Amyloid Generation and Alzheimer’s Disease. The journals of gerontology Series A, Biological sciences and medical sciences. 2013 doi: 10.1093/gerona/glt177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pascale A, Gusev PA, Amadio M, Dottorini T, Govoni S, Alkon DL, et al. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1217–1222. doi: 10.1073/pnas.0307674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zoladz PR, Park CR, Halonen JD, Salim S, Alzoubi KH, Srivareerat M, et al. Differential expression of molecular markers of synaptic plasticity in the hippocampus, prefrontal cortex, and amygdala in response to spatial learning, predator exposure, and stress-induced amnesia. Hippocampus. 2012;22:577–589. doi: 10.1002/hipo.20922. [DOI] [PubMed] [Google Scholar]
- 58.Pollak DD, Herkner K, Hoeger H, Lubec G. Behavioral testing upregulates pCaMKII, BDNF, PSD-95 and egr-1 in hippocampus of FVB/N mice. Behavioural brain research. 2005;163:128–135. doi: 10.1016/j.bbr.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 59.Morash B, Li A, Murphy PR, Wilkinson M, Ur E. Leptin gene expression in the brain and pituitary gland. Endocrinology. 1999;140:5995–5998. doi: 10.1210/endo.140.12.7288. [DOI] [PubMed] [Google Scholar]
- 60.Hsuchou H, He Y, Kastin AJ, Tu H, Markadakis EN, Rogers RC, et al. Obesity induces functional astrocytic leptin receptors in hypothalamus. Brain : a journal of neurology. 2009;132:889–902. doi: 10.1093/brain/awp029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koga S, Kojima A, Kuwabara S, Yoshiyama Y. Immunohistochemical analysis of tau phosphorylation and astroglial activation with enhanced leptin receptor expression in diet-induced obesity mouse hippocampus. Neuroscience letters. 2014;571:11–16. doi: 10.1016/j.neulet.2014.04.028. [DOI] [PubMed] [Google Scholar]
- 62.Leung JC, Chan LY, Lam MF, Tang SC, Chow CW, Lim AI, et al. The role of leptin and its short-form receptor in inflammation in db/db mice infused with peritoneal dialysis fluid. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2012;27:3119–329. doi: 10.1093/ndt/gfr774. [DOI] [PubMed] [Google Scholar]
- 63.Jayaram B, Pan W, Wang Y, Hsuchou H, Mace A, Cornelissen-Guillaume GG, et al. Astrocytic leptin-receptor knockout mice show partial rescue of leptin resistance in diet-induced obesity. Journal of applied physiology. 2013;114:734–741. doi: 10.1152/japplphysiol.01499.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Figlewicz DP, Bennett J, Evans SB, Kaiyala K, Sipols AJ, Benoit SC. Intraventricular insulin and leptin reverse place preference conditioned with high-fat diet in rats. Behavioral neuroscience. 2004;118:479–487. doi: 10.1037/0735-7044.118.3.479. [DOI] [PubMed] [Google Scholar]
- 65.Li XL, Aou S, Oomura Y, Hori N, Fukunaga K, Hori T. Impairment of long-term potentiation and spatial memory in leptin receptor-deficient rodents. Neuroscience. 2002;113:607–615. doi: 10.1016/s0306-4522(02)00162-8. [DOI] [PubMed] [Google Scholar]
- 66.Kogelman LJ, Pant SD, Fredholm M, Kadarmideen HN. Systems genetics of obesity in an F2 pig model by genome-wide association, genetic network, and pathway analyses. Frontiers in genetics. 2014;5:214. doi: 10.3389/fgene.2014.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]