Abstract
Peptide-major histocompatibility complex class I (pMHC-I) tetramers have been an invaluable tool to study CD8+ T-cell responses. Because these reagents directly bind to T-cell receptors on the surface of CD8+ T-lymphocytes, fluorochrome-labeled pMHC-I tetramers enable the accurate detection of antigen (Ag)-specific CD8+ T-cells without the need for in vitro re-stimulation. Moreover, when combined with multi-color flow cytometry, pMHC-I tetramer staining can reveal key aspects of Ag-specific CD8+ T-cells, including differentiation stage, memory phenotype, and activation status. These types of analyses have been especially useful in the field of HIV immunology where CD8+ T-cells can affect progression to AIDS. Experimental infection of rhesus macaques with simian immunodeficiency virus (SIV) provides an invaluable tool to study cellular immunity against the AIDS virus. As a result, considerable progress has been made in defining and characterizing T-cell responses in this animal model. Here we present an optimized protocol for enumerating SIV-specific CD8+ T-cells in rhesus macaques by pMHC-I tetramer staining. Our assay permits the simultaneous quantification and memory phenotyping of two pMHC-I tetramer+ CD8+ T-cell populations per test, which might be useful for tracking SIV-specific CD8+ T-cell responses generated by vaccination or SIV infection. Considering the relevance of nonhuman primates in biomedical research, this methodology is applicable for studying CD8+ T-cell responses in multiple disease settings.
Keywords: Immunology, Issue 118, MHC, tetramer, vaccine, SIV, rhesus, macaque
Introduction
CD8+ T-cells comprise a crucial component of the adaptive immune system as they participate in tumor immune surveillance and contribute to the eradication of intracellular pathogens1. In simple terms, CD8+ T-cells express T-cell receptors (TCRs) that specifically recognize peptide-major histocompatibility complex class I (pMHC-I) molecules present on the plasma membrane of host cells. Since these peptides are derived from the proteolysis of endogenously synthesized proteins, cell surface pMHC-I complexes provide a window into the intracellular environment. Upon virus infection, for example, infected cells will display MHC-I molecules containing virus-derived peptides that can serve as ligands for TCRs expressed by patrolling CD8+ T-cells. In the event a virus-specific CD8+ T-cell encounters an infected cell presenting its pMHC-I ligand, TCR engagement will result in CD8+ T-cell activation and ultimately lead to target cell lysis. Given the critical nature of these TCR/pMHC-I interactions, determining the magnitude, specificity, and phenotype of responding CD8+ T-cells can often reveal important clues about human diseases.
Until the early 1990s, quantification of Ag-specific CD8+ T-cells relied on the technically demanding limiting dilution assay (LDA)2,3. Not only did the LDA require several days to be completed, it also failed to detect cells that lacked proliferative potential. As a result, the LDA vastly underestimated the actual frequency of antigen (Ag)-specific CD8+ T-cells participating in an immune response. Although the development of ELISPOT and intracellular cytokine staining assays greatly improved the ability to measure cellular immunity, these methods still required in vitro stimulation for quantifying Ag-specific T-cells4. It was not until 1996 that Altman, Davis, and colleagues published their landmark article reporting the development of the pMHC-I tetramer technology5. Critical to the success of this technique was the multimerization of pMHC-I molecules, which extended the half-life of TCR/pMHC-I interactions, thereby reducing the probability of pMHC-I tetramers falling off during the washing steps of flow cytometric assays. The main advantage of pMHC-I tetramers over the aforementioned assays is the ability to accurately detect Ag-specific CD8+ T-cells directly ex vivo without the need for in vitro re-stimulation. Moreover, the combination of pMHC-I tetramer staining with multi-color flow cytometry has allowed detailed analyses of the differentiation stage, memory phenotype, and activation status of Ag-specific CD8+ T-cells2-4. In light of recent technical advances for characterizing CD8+ T-cell repertoires by pMHC-I multimer staining6, the breadth of applications for this methodology is likely to continue expanding.
Few areas in biomedical research have benefited more from pMHC-I tetramer staining than the field of HIV immunology7. Although CD8+ T-cells had been temporally associated with the initial control of HIV viremia by the time of the publication by Altman, Davis, and colleagues8,9, the use of pMHC-I tetramers in the ensuing years significantly expanded our understanding of the HIV-specific CD8+ T-cell response. For example, pMHC-I tetramer staining helped confirm the robust size of virus-specific CD8+ T-cell responses in most HIV-infected individuals10-12. This methodology also facilitated the characterization of HIV- and SIV-specific CD8+ T-cell responses restricted by MHC-I molecules associated with spontaneous control of viral replication in the absence of antiretroviral therapy—a phenomenon known as "elite control"13-15. Furthermore, pMHC-I tetramers were instrumental in establishing the programmed death 1 (PD-1)/PD ligand 1 (PD-L1) axis as a reversible pathway for the dysfunctional phenotype of HIV-specific CD8+ T-cells in uncontrolled chronic infection16,17. Collectively, these studies underscore the utility of pMHC-I tetramers for monitoring CD8+ T-cell responses against the AIDS virus.
Experimental SIV infection of rhesus macaques (Macaca mulatta) remains the best animal model for evaluating immune interventions against HIV/AIDS18,19. In the past 25 years, substantial progress has been made in the identification and characterization of SIV-specific CD8+ T-cells in this monkey species, including the discovery of MHC-I alleles and the definition of peptide binding motifs13,20-27. As a result, pMHC-I tetramers have been developed for the analysis of SIV-specific CD8+ T-cell responses in this animal model28. Most of these reagents are made of the gene products of four rhesus macaque MHC-I alleles: Mamu-A1*001, Mamu-A1*002:01, Mamu-B*008:01, and Mamu-B*017:01. Of note, rhesus macaques do not express an MHC-C locus29. The vast majority of the pMHC-I tetramers used in the present experiments were produced at the NIH Tetramer Core Facility at Emory University. Nevertheless, some of these reagents, including Mamu-A1*001 tetramers bound to the immunodominant Gag CM9 epitope, can only be obtained from commercial sources due to licensing agreements. Using pMHC-I tetramers of the four rhesus macaque alleles listed above, we have successfully enumerated CD8+ T-cells against a total of 21 SIV epitopes (Table 1), which were induced by vaccination or primary SIV infection30,31 (Martins et al., unpublished observations).
The present manuscript provides an optimized pMHC-I tetramer staining protocol for determining the frequency and memory phenotype of SIV-specific CD8+ T-cells in rhesus macaques. The assay begins with an elective 30 min incubation with a protein kinase inhibitor (PKI; here, Dasatinib is used) in order to decrease TCR internalization and thereby improve pMHC-I tetramer staining32. As described below, this treatment is especially useful when using Mamu-B*017:01 tetramers. Instructions on how to label the cells with fluorochrome-conjugated pMHC-I tetramers and monoclonal antibodies (mAbs) are also provided. This protocol also includes a cell permeabilization step for the intracellular detection of the cytolysis-associated molecule granzyme B (Gzm B). The mAb against CD3 is added at this step as well to improve detection of this TCR signaling molecule. As a reference, all fluorochromes employed in this staining panel are listed.
Protocol
The peripheral blood mononuclear cell (PBMC) samples utilized in this manuscript were obtained from Indian rhesus macaques housed at the Wisconsin National Primate Research Center. These animals were cared for in accordance with the Weatherall Report under a protocol approved by the University of Wisconsin Graduate School Animal Care and Use Committee33. All animal procedures were performed under anesthesia and all efforts were made to minimize potential suffering.
1. PKI Treatment
NOTE: This is optional, but recommended for Mamu-B*017:01 tetramers. See Results and Discussion Sections.
Resuspend PBMC in R10 medium at a concentration of 1.6 x 107 cells/ml.
Add 50 µl of this cell suspension to the corresponding flow cytometry tubes. Add 50 µl of a 100 nM solution of PKI to each tube.
Vortex each tube. Incubate at 37 °C for 30 min. By the end of this step, each tube should have 100 µl of a cell suspension containing 8.0 x 105 PBMC and 50 nM of PKI.
Proceed to the pMHC-I tetramer staining step.
2. Staining with Fluorochrome-labeled pMHC-I Tetramers
Before preparing the pMHC-I tetramer master mix, centrifuge the pMHC-I tetramer tubes at 20,000 x g for at least 15 min at 4 °C. The goal of this step is to pellet protein aggregates in the pMHC-I tetramer solution that may increase background staining. Once the centrifugation step is done, avoid pipetting from the bottom of the tube where the protein aggregates will have accumulated.
Prepare enough pMHC-I tetramer master mix to stain all experimental tubes. Prepare an excess of 15% of the total volume of this master mix to account for pipetting error.
- Dilute pMHC-I tetramers in stain buffer (e.g., Brilliant Stain Buffer) so that 25 µl of the pMHC-I tetramer master mix are added to each test.
- Label PBMC with two pMHC-I tetramers per test; one conjugated to allophycocyanin (APC) and the other to Brilliant Violet (BV) 421.
Add 8.0 x 105 PBMC to the corresponding flow cytometry tubes in a final volume of 100 µl. If the cells were subjected to the PKI treatment described above, they should already be resuspended in 100 µl at this step.
Add 25 µl of pMHC-I tetramer master mix to the corresponding flow cytometry tubes. By the end of this step, the final volume in each tube should be 125 µl.
Vortex each tube in order to homogenize the cell suspension.
Incubate in the dark at room temp for 45 min. Prepare the surface staining mAb cocktail during this 45 min incubation.
3. Surface Staining
Prepare enough mAb master mix to stain all experimental tubes. Prepare an excess of 15% of the total volume of this master mix to account for pipetting error.
Adjust the volume of the master mix with stain buffer so that 50 µl are added per test.
For proper exclusion of non-CD8+ T-cells and delineation of memory subsets, use titrated amounts of mAbs directed against the following molecules in the surface staining master mix. NOTE: The fluorochromes conjugated to each mAbs are provided as a reference: CD14 BV 510, CD16 BV 510, CD20 BV 510, CD8α BV 785, CD28 PE Cy7, and CCR7 FITC. Note that the mAbs against CD14, CD16, and CD20 are conjugated to the same fluorochrome (i.e., BV 510) since they will be included in the "dump" gate.
Include a fixable dye for discriminating dead cells in the surface staining master mix [i.e., amine reactive dye (ARD)]. Make sure that the ARD reagent is conjugated to a fluorochrome with a similar emission spectrum as the ones used in the "dump" gate. In this case, use ARD Aqua.
Add 50 µl of the staining master mix described in 3.1-3.4 to the corresponding flow cytometry tubes. By the end of this step, the final volume in each tube should be 175 µl.
Vortex each tube. Incubate in the dark at room temp for 25 min.
Wash cells with wash buffer (PBS solution containing 0.1% of bovine serum albumin and 0.45 g/L NaN3). CAUTION: Sodium azide is a toxic substance. Exposure to even minute amounts can cause symptoms. Handle this substance according to the guidelines specified by the Environmental Health and Safety (EHS) Office at the institution where these experiments are being performed.
Centrifuge tubes at 510 x g for 5 min. Carefully decant supernatant into a separate waste container. Be sure not to disturb the pellet. CAUTION: Do not decant wash buffer supernatant into reservoirs containing bleach as it can react with the sodium azide present in the wash buffer and result in the formation of a toxic gas34. Contact the EHS Office at the institution where these experiments are being performed for guidelines on how to dispose of sodium azide.
After decanting, vortex the cells in the leftover liquid retained in each tube.
4. Cell Fixation
- Add 250 µl of a 2% paraformaldehyde (PFA) solution to all tubes to fix the cells. CAUTION: PFA is a toxic substance. Exposure to even minute amounts can cause symptoms. Handle this substance according to the guidelines specified by the EHS Office at the institution where these experiments are being performed.
- Since the 2% PFA solution must be isotonic to the cells, use PBS to prepare this solution. One hundred milliliters of a 2% PFA solution is enough for multiple experiments. Thoroughly vortex all tubes immediately after the addition of 2% PFA in order to prevent the formation of cell aggregates.
Incubate in the dark at 4 °C for 20 min.
Wash cells by repeating steps 3.7-3.9. Once this step is completed, the cells can be stored in the dark at 4 °C for 24-48 hr. Before proceeding to the Permeabilization phase, vortex the tubes thoroughly.
5. Permeabilization of Cells
Add 500 µl of permeabilization buffer. Vortex all tubes.
Incubate in the dark at room temp for 10 min.
Wash cells by repeating steps 3.7-3.9.
6. Intracellular Staining
Prepare enough mAb master mix to stain all experimental tubes.
Adjust the volume with PBS or stain buffer so that 50 µl of the intracellular mAb master mix are added per test.
Prepare the mAb master mix described in 6.1 and 6.2 using titrated amounts of mAbs directed against CD3 and Gzm B. NOTE: TCR engagement by pMHC-I tetramers can result in CD3 internalization, which can interfere with the detection of tetramer+ CD3+ CD8+ T-cells if the anti-CD3 mAb is added to the surface staining master mix. To avoid this, add the anti-CD3 mAb at this stage, that is, after the cells are permeabilized. The fluorochromes conjugated to each mAb are listed as a reference: CD3 PerCP Cy5.5 and Gzm B PE.
Add 50 µl of mAb cocktail to the corresponding tubes. Incubate in the dark at room temp for 30 min.
Wash cells by repeating steps 3.7-3.9. The tubes are ready to be acquired in a flow cytometer.
Representative Results
The protocol described here has been used to determine the magnitude and memory phenotype of vaccine-induced, Gag CM9-specific CD8+ T-cell responses in a Mamu-A1*001+ rhesus macaque. For this analysis, an APC-conjugated Mamu-A1*001/Gag CM9 tetramer was used in an 8-color flow cytometric staining panel. Figure 1A-F shows the gating strategy used to analyze the data, which should be applied for both tetramers present in each test. Note that the pMHC-I tetramer+ CD8+ T-cell population is well separated with minimal background (Figures 1F and G). Memory CD8+ T-cells in rhesus macaques can be classified as three subsets based on the surface expression of CD28 and CCR7: central memory (TCM; CD28+CCR7+), transitional memory (TEM1; CD28+CCR7-), and effector memory (TEM2; CD28-CCR7-)35. This protocol also includes a cell permeabilization step for intracellular detection of Gzm B and CD3. Figure 1G-I shows the delineation of memory subsets and Gzm B-expressing CD8+ T-cells within the tetramer+ gate.
Background staining can yield suboptimal results in pMHC-I tetramer labeling assays. To avoid this, a few precautions are suggested. As stated in the protocol section, the vials containing the pMHC-I tetramer reagents should always be centrifuged at 20,000 x g for at least 15 min prior to the preparation of the master mixes. The goal of this step is to pellet any protein aggregates that may be in the pMHC-I tetramer solution. Additionally, titrating each reagent prior to use is also recommended. Ideally, MHC-I-matched cells that do or do not contain the Ag-specific CD8+ T-cell population of interest should be labeled with the relevant pMHC-I tetramer. This can be accomplished by staining cryopreserved PBMC from SIV naïve (negative control) and SIV-infected (positive control) macaques side by side with various amounts of a newly obtained pMHC-I tetramer. In Figure 2, for example, 1.0 µl/test of an APC-conjugated Mamu-B*008:01/Nef RL10 tetramer yielded the best separation between tetramer positive and negative CD8+ T-cell populations. Lastly the choice of fluorochrome can also significantly impact the results obtained from pMHC-I tetramer stainings. In this regard, pMHC-I tetramers conjugated to dim molecules (e.g., fluorescein) should be avoided. In our experience, pMHC-I tetramers conjugated to bright fluorochromes, such as PE, APC, and BV 421, have yielded the best results.
We have noticed that Mamu-B*017:01 tetramers typically yield dim staining, even when conjugated to the bright fluorochromes mentioned above. The reasons for this low fluorescence intensity are not entirely clear but might include low-affinity TCR/Mamu-B*017:01 interactions and rapid TCR internalization upon tetramer binding36,37. Along these lines, PKIs have been shown to inhibit TCR internalization on the surface of CD8+ T-cells32. As a result, PKIs can improve detection of Ag-specific CD8+ T-cells by pMHC-I tetramer staining. We have confirmed this effect in our experiments, where the quality of Mamu-B*017:01 tetramer stainings can be substantially improved by pre-treating PBMC with a PKI (Figure 3). Curiously, pre-treatment with this PKI did not significantly improve the staining of other pMHC-I tetramers tested here (data not shown), suggesting that Mamu-B*017:01-restricted CD8+ T-cells might be unique in their sensitivity to PKI treatment.
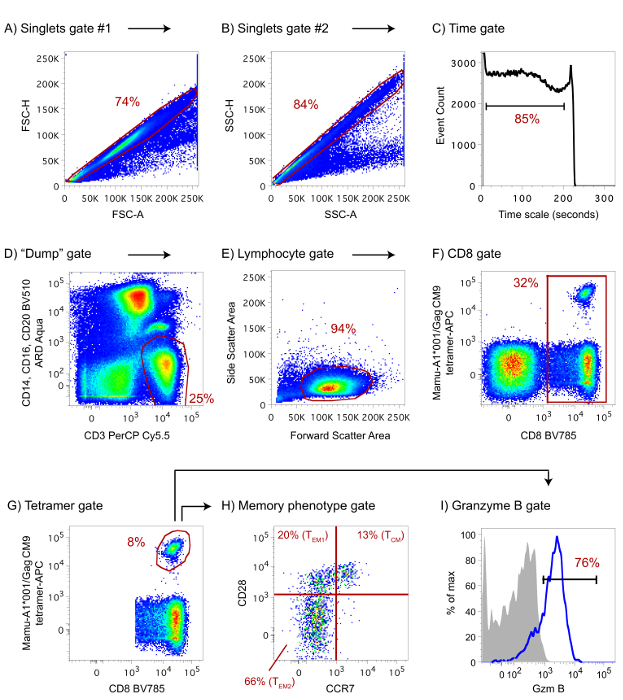 Figure 1: Gating strategy and representative results of a successful pMHC-I tetramer staining. We evaluated the magnitude and memory phenotype of vaccine-induced Gag CM9-specific CD8+ T-cells in PBMC from a Mamu-A1*001+ rhesus macaque. Here, an APC-conjugated Mamu-A1*001/Gag CM9 tetramer was used in an 8-color flow cytometric staining panel. (A-F) Gating strategy for flow cytometric analysis. First, a gate on diagonally clustered singlets was created by plotting forward scatter height (FSC-H) versus FSC area (FSC-A) and then side scatter height (SSC-H) versus SSC area (SSC-A; A and B). Next, a time gate was created that included only those events that were recorded within the 5th and 90th percentiles. The resulting cells were then gated on "dump channel" negative, CD3+ cells (C and D). At this stage, the lymphocyte population was delineated based on its FSC-A and SSC-A properties and subsequent analyses were conducted within CD8+ cells (E and F). After outlining pMHC-I tetramer+ cells (G), the memory phenotyping analysis was performed within this gate (H). Rhesus macaque memory T-cells can be classified into three subsets based on the expression of CD28 and CCR7: central memory (TCM; CD28+CCR7+), transitional memory (TEM1; CD28+CCR7-), and effector memory (TEM2; CD28-CCR7-). The present protocol also includes a cell permeabilization step for the intracellular evaluation of Granzyme B (Gzm B) expression within tetramer+ CD8+ T-cells (I). The shaded area in the histogram corresponds to tetramer+ CD8+ T-cells stained with an isotype-matched control mAb. Although the BV 421-conjugated tetramer+ population that was present in this staining is not shown in the figure, the same gating strategy should be used to analyze it. Please click here to view a larger version of this figure.
Figure 1: Gating strategy and representative results of a successful pMHC-I tetramer staining. We evaluated the magnitude and memory phenotype of vaccine-induced Gag CM9-specific CD8+ T-cells in PBMC from a Mamu-A1*001+ rhesus macaque. Here, an APC-conjugated Mamu-A1*001/Gag CM9 tetramer was used in an 8-color flow cytometric staining panel. (A-F) Gating strategy for flow cytometric analysis. First, a gate on diagonally clustered singlets was created by plotting forward scatter height (FSC-H) versus FSC area (FSC-A) and then side scatter height (SSC-H) versus SSC area (SSC-A; A and B). Next, a time gate was created that included only those events that were recorded within the 5th and 90th percentiles. The resulting cells were then gated on "dump channel" negative, CD3+ cells (C and D). At this stage, the lymphocyte population was delineated based on its FSC-A and SSC-A properties and subsequent analyses were conducted within CD8+ cells (E and F). After outlining pMHC-I tetramer+ cells (G), the memory phenotyping analysis was performed within this gate (H). Rhesus macaque memory T-cells can be classified into three subsets based on the expression of CD28 and CCR7: central memory (TCM; CD28+CCR7+), transitional memory (TEM1; CD28+CCR7-), and effector memory (TEM2; CD28-CCR7-). The present protocol also includes a cell permeabilization step for the intracellular evaluation of Granzyme B (Gzm B) expression within tetramer+ CD8+ T-cells (I). The shaded area in the histogram corresponds to tetramer+ CD8+ T-cells stained with an isotype-matched control mAb. Although the BV 421-conjugated tetramer+ population that was present in this staining is not shown in the figure, the same gating strategy should be used to analyze it. Please click here to view a larger version of this figure.
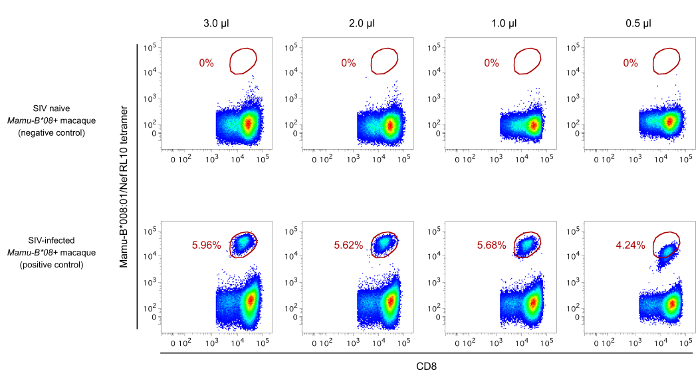 Figure 2: Representative results of the titration of an APC-conjugated Mamu-B*008:01/Nef RL10 tetramer. PBMC from two Mamu-B*008:01+ rhesus macaques were used for this experiment: one animal was SIV naïve and served as the negative control whereas the other was SIV-infected and served as the positive control. To determine the amount of Mamu-B*008:01/Nef RL10 tetramer that yields the best separation between tetramer+ and tetramer- CD8+ T-cells, PBMC from each animal were incubated with the indicated amounts of the pMHC-I tetramer reagent. Based on these results, 1.0 µl/test was chosen as the optimal amount to be used in future assays. The gating strategy utilized to analyze the data is described in Figure 1. Please click here to view a larger version of this figure.
Figure 2: Representative results of the titration of an APC-conjugated Mamu-B*008:01/Nef RL10 tetramer. PBMC from two Mamu-B*008:01+ rhesus macaques were used for this experiment: one animal was SIV naïve and served as the negative control whereas the other was SIV-infected and served as the positive control. To determine the amount of Mamu-B*008:01/Nef RL10 tetramer that yields the best separation between tetramer+ and tetramer- CD8+ T-cells, PBMC from each animal were incubated with the indicated amounts of the pMHC-I tetramer reagent. Based on these results, 1.0 µl/test was chosen as the optimal amount to be used in future assays. The gating strategy utilized to analyze the data is described in Figure 1. Please click here to view a larger version of this figure.
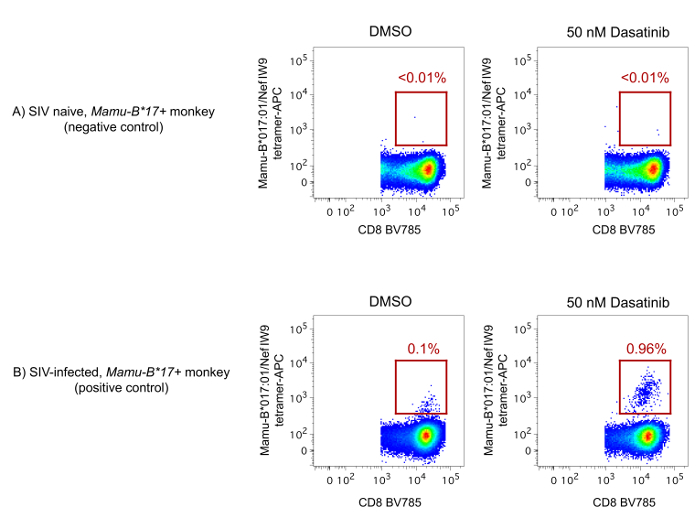 Figure 3: Effects of PKI treatment on the fluorescence intensity of Mamu-B*017:01 tetramers. The PKI inhibitor inhibits internalization of TCR complexes and thereby improves detection of Ag-specific CD8+ T-cells by fluorochrome-labeled pMHC-I tetramers32. (A-B) PBMC from two Mamu-B*017:01+ rhesus macaques were used in this experiment: one monkey was SIV naïve and served as the negative control (A), while the other was infected with SIV and served as the positive control (B). PBMC from both animals were incubated at 37 °C in the presence of PKI at a concentration of 50 nM for 30 min prior to staining with an APC-conjugated Mamu-B*017:01/Nef IW9 tetramer, as described in the Protocol section. As a reference, another set of tubes was subjected to the same incubation and staining conditions, except that dimethyl sulfoxide (DMSO) was added instead of PKI. The gating strategy utilized to analyze the data is described in Figure 1. Please click here to view a larger version of this figure.
Figure 3: Effects of PKI treatment on the fluorescence intensity of Mamu-B*017:01 tetramers. The PKI inhibitor inhibits internalization of TCR complexes and thereby improves detection of Ag-specific CD8+ T-cells by fluorochrome-labeled pMHC-I tetramers32. (A-B) PBMC from two Mamu-B*017:01+ rhesus macaques were used in this experiment: one monkey was SIV naïve and served as the negative control (A), while the other was infected with SIV and served as the positive control (B). PBMC from both animals were incubated at 37 °C in the presence of PKI at a concentration of 50 nM for 30 min prior to staining with an APC-conjugated Mamu-B*017:01/Nef IW9 tetramer, as described in the Protocol section. As a reference, another set of tubes was subjected to the same incubation and staining conditions, except that dimethyl sulfoxide (DMSO) was added instead of PKI. The gating strategy utilized to analyze the data is described in Figure 1. Please click here to view a larger version of this figure.
| SIV protein | Amino acid position | Amino Acid Sequence | MHC Class I Molecule (new nomenclature) | MHC Class I Molecule (old nomenclature) | Epitope Short Name |
| Gag | 181-189 | CTPYDINQM | Mamu-A1*001 | Mamu-A*01 | Gag CM9 |
| Gag | 254-262 | QNPIPVGNI | Mamu-A1*001 | Mamu-A*01 | Gag QI9 |
| Gag | 72-79 | GSENLKSLY | Mamu-A1*001 | Mamu-A*01 | Gag GY9 |
| Env | 830-838 | FHEAVQAVW | Mamu-B*017:01 | Mamu-B*17 | Env FW9 |
| Env | 233-241 | CAPPGYALL | Mamu-A1*001 | Mamu-A*01 | Env CL9 |
| Env | 620-628 | TVPWPNASL | Mamu-A1*001 | Mamu-A*01 | Env TL9 |
| Env | 788-795 | RTLLSRVY | Mamu-A1*002:01 | Mamu-A*02 | Env RY8 |
| Env | 296-304 | RTIISLNKY | Mamu-A1*002:01 | Mamu-A*02 | Env RY9 |
| Vif | 123-131 | RRAIRGEQL | Mamu-B*008:01 | Mamu-B*08 | Vif RL9 |
| Vif | 173-181 | RRDNRRGL | Mamu-B*008:01 | Mamu-B*08 | Vif RL8 |
| Vif | 66–73 | HLEVQGYW | Mamu-B*017:01 | Mamu-B*17 | Vif HW8 |
| Vif | 100–109 | VTPNYADILL | Mamu-A1*001 | Mamu-A*01 | Vif VL10 |
| Vif | 97-104 | WTDVTPNY | Mamu-A1*002:01 | Mamu-A*02 | Vif WY8 |
| Rev | 13-22 | KRLRLIHLL | Mamu-B*008:01 | Mamu-B*08 | Rev KL9 |
| Tat | 28-35 | STPESAML | Mamu-A1*001 | Mamu-A*01 | Tat SL8 |
| Nef | 137-146 | RRHRILDIYL | Mamu-B*008:01 | Mamu-B*08 | Nef RL10 |
| Nef | 246-254 | RRLTARGLL | Mamu-B*008:01 | Mamu-B*08 | Nef RL9b |
| Nef | 8-16 | RRSRPSGDL | Mamu-B*008:01 | Mamu-B*08 | Nef RL9a |
| Nef | 165–173 | IRYPKTFGW | Mamu-B*017:01 | Mamu-B*17 | Nef IW9 |
| Nef | 195–203 | MHPAQTSQW | Mamu-B*017:01 | Mamu-B*17 | Nef MW9 |
| Nef | 159-167 | YTSGPGIRY | Mamu-A1*002:01 | Mamu-A*02 | Nef YY9 |
Table 1: List of pMHC-I tetramers available for monitoring SIV-specific CD8+ T-cell responses in rhesus macaques.
Discussion
A few steps in this procedure merit discussion as they are crucial for yielding optimal results. First, since the quality of the biological specimens is a strong predictor of the success of any flow cytometric assay38, all care must be taken to ensure that the cells are viable and in suspension during the staining procedure. This is particularly relevant when working with cryopreserved samples since they are more prone to clumping and typically contain higher numbers of dead cells. In these cases, passing the cell suspension through a 70 µm cell strainer prior to the pMHC-I tetramer labeling procedure might substantially improve the results. Second, proper compensation of the fluorochromes employed in the assay is critical for accurate data analysis, especially when multiple parameters are being evaluated in the same experiment. In this regard, it is recommended that fresh single-color compensation controls be prepared for every pMHC-I tetramer staining assay. We favor the use of compensation beads because of their broad anti-Ig reactivity and the fact that they yield clear bimodal distributions of positive and negative populations. Since these beads do not bind to MHC-I molecules, antibodies conjugated to the same fluorophores as the pMHC-I tetramers (e.g., APC and BV 421) should be used as compensation controls. Several of such compensation beads are available from multiple vendors and should be used according to the manufacturer's instructions. Third, since the phenotyping strategy described here requires the delineation of CD8+ T-cell subsets based on the graded expression of three molecules (i.e., CD28, CCR7, and Gzm B), gating controls, such as fluorescence minus one (FMO) tests or isotype-matched antibodies, should be used to facilitate these types of analyses. In our experiments, for instance, separate tubes containing the pMHC-I tetramer+ CD8+ T-cell population of interest are always stained with isotype control mAbs conjugated to the same fluorochromes as the mAbs against CD28, CCR7, and Gzm B. Importantly, although the aforementioned guidelines might help improve the overall quality of pMHC-I tetramer stainings, they are not the only variables that can affect the performance of this assay. Considering the uniqueness of every laboratory setting, the entire workflow described here should be performed at least once on mock samples to allow for practice and relevant adjustments.
Curiously, pMHC-I tetramer positive CD8+ T-cells are occasionally observed in PBMC from SIV naïve (uninfected and unvaccinated) rhesus macaques. These cases do not appear to be the result of background staining based on visual inspection of the data. Rather, they might reflect interactions between pMHC-I molecules and killer immunoglobulin-like receptors (KIRs) expressed on the surface of rhesus macaque NK and CD8+ T-cells39,40. Indeed, Mamu-A1*002:01 tetramers folded with peptides corresponding to the Gag GY9 and Env RY8 epitopes have been shown to bind to Mamu-KIRDL0539. Along these lines, Mamu-A1*002:01/Gag GY9 and Mamu-A1*002:01/Env RY8 are more prone to exhibiting the Ag-independent pMHC-I tetramer staining described above. Given this phenomenon, it is important to control for this baseline reactivity in monkey experiments involving longitudinal assessments of SIV-specific CD8+ T-cell responses following infection or vaccination. To achieve this, it is advisable to perform pMHC-I tetramer staining on samples obtained on the first day of treatment. It might also be pertinent to freeze PBMC in several small-size aliquots at these initial time points in case comparisons with baseline samples are needed in future experiments.
One limitation of pMHC-I tetramer staining is that only CD8+ T-cells of known specificity and MHC-I restriction can be studied by this approach. This is particularly relevant in rhesus macaques given the complex organization of their MHC loci. Indeed, a single rhesus macaque haplotype can have dozens of MHC-I genes, not all of which encode molecules that are stably expressed on the cell surface29. As a result, it can be difficult to determine the set of functional MHC-I molecules expressed by a given animal. Nevertheless, this caveat can be overcome by designing experiments that include monkeys that are positive for known MHC-I alleles, such as Mamu-A1*001, Mamu-A1*002:01, Mamu-B*008:01, and Mamu-B*017:01.
In conclusion, this manuscript provides an optimized pMHC-I tetramer staining protocol for determining the frequency and memory phenotype of SIV-specific CD8+ T-cells in rhesus macaques. Since pMHC-I tetramer staining is more sensitive than IFN-γ ELISPOT and ICS assays for the detection of Ag-specific CD8+ T-cells and does not require in vitro Ag stimulation, the methodology presented here is especially useful for studying the impact of CD8+ T-cell responses in the outcome of immunodeficiency virus infection. The practical knowledge acquired from mastering this technique might also be useful for enriching Ag-specific CD8+ T-cells as part of functional studies and for monitoring CD8+ T-cell responses in various disease settings6.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We would like to thank David Watkins for supporting the experiments that enabled the optimization of the present methodology. Research reported in this publication was supported in part by a pilot grant provided by the Miami Center for AIDS Research of the National Institutes of Health under award number P30AI073961. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Parham P. Antigen Recognition by T Lymphocytes. In: Foltin J, Masson S, Ghezzi K, Engels A, Lawrence E, Jeffcock E, editors. The Immune System, 3rd ed. New York, NY: Garland Science, Taylor & Francis Group, LLC; 2009. pp. 125–154. [Google Scholar]
- Doherty PC. The tetramer transformation. J Immunol. 2011;187(1):5–6. doi: 10.4049/jimmunol.1101297. [DOI] [PubMed] [Google Scholar]
- Masopust D, Vezys V, Wherry EJ, Ahmed R. A brief history of CD8 T cells. Eur J Immunol. 2007;37:103–110. doi: 10.1002/eji.200737584. Suppl 1. [DOI] [PubMed] [Google Scholar]
- Murali-Krishna K, et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8(2):177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- Altman JD, et al. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274(5284):94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Davis MM, Altman JD, Newell EW. Interrogating the repertoire: broadening the scope of peptide-MHC multimer analysis. Nat Rev Immunol. 2011;11(8):551–558. doi: 10.1038/nri3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B, McMichael A. The T-cell response to HIV. Cold Spring Harb Perspect Med. 2012;2(11) doi: 10.1101/cshperspect.a007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68(9):6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koup RA, et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68(7):4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray CM, et al. Frequency of class I HLA-restricted anti-HIV CD8+ T cells in individuals receiving highly active antiretroviral therapy (HAART) J Immunol. 1999;162(3):1780–1788. [PubMed] [Google Scholar]
- Papagno L, et al. Comparison between HIV- and CMV-specific T cell responses in long-term HIV infected donors. Clin Exp Immunol. 2002;130(3):509–517. doi: 10.1046/j.1365-2249.2002.02005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel HM, et al. Human immunodeficiency virus type 1- and cytomegalovirus-specific cytotoxic T lymphocytes can persist at high frequency for prolonged periods in the absence of circulating peripheral CD4(+) T cells. J Virol. 2000;74(2):1018–1022. doi: 10.1128/jvi.74.2.1018-1022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo JT, et al. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J Virol. 2008;82(4):1723–1738. doi: 10.1128/JVI.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97(6):2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine LE, et al. Infection with "escaped" virus variants impairs control of simian immunodeficiency virus SIVmac239 replication in Mamu-B*08-positive macaques. J Virol. 2009;83(22):11514–11527. doi: 10.1128/JVI.01298-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CL, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443(7109):350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- Trautmann L, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006;12(10):1198–1202. doi: 10.1038/nm1482. [DOI] [PubMed] [Google Scholar]
- Mudd PA, Watkins DI. Understanding animal models of elite control: windows on effective immune responses against immunodeficiency viruses. Curr Opin HIV AIDS. 2011;6(3):197–201. doi: 10.1097/COH.0b013e3283453e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine LE, Watkins DI. Relevance of studying T cell responses in SIV-infected rhesus macaques. Trends Microbiol. 2008;16(12):605–611. doi: 10.1016/j.tim.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TM, et al. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998;160(12):6062–6071. [PubMed] [Google Scholar]
- Allen TM, et al. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J Virol. 2001;75(2):738–749. doi: 10.1128/JVI.75.2.738-749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizu M, et al. Molecular typing of major histocompatibility complex class I alleles in the Indian rhesus macaque which restrict SIV CD8+ T cell epitopes. Immunogenetics. 2007;59(9):693–703. doi: 10.1007/s00251-007-0233-7. [DOI] [PubMed] [Google Scholar]
- Loffredo JT, et al. Identification of seventeen new simian immunodeficiency virus-derived CD8+ T cell epitopes restricted by the high frequency molecule, Mamu-A*02, and potential escape from CTL recognition. J Immunol. 2004;173(8):5064–5076. doi: 10.4049/jimmunol.173.8.5064. [DOI] [PubMed] [Google Scholar]
- Loffredo JT, Valentine LE, Watkins DI. Beyond Mamu-A*01+ Indian Rhesus Macaques: Continued Discovery of New MHC Class I Molecules that Bind Epitopes from the Simian AIDS Viruses. HIV mol immunol. 2006;2007:29–51. [Google Scholar]
- Loffredo JT, et al. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182(12):7763–7775. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothe BR, et al. Characterization of the peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. J Immunol. 2002;169(1):210–219. doi: 10.4049/jimmunol.169.1.210. [DOI] [PubMed] [Google Scholar]
- Miller MD, Yamamoto H, Hughes AL, Watkins DI, Letvin NL. Definition of an epitope and MHC class I molecule recognized by gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J Immunol. 1991;147(1):320–329. [PubMed] [Google Scholar]
- Kuroda MJ, et al. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med. 1998;187(9):1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting N, et al. Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci U S A. 2005;102(5):1626–1631. doi: 10.1073/pnas.0409084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins MA, et al. Vaccine-Induced Simian Immunodeficiency Virus-Specific CD8+ T-Cell Responses Focused on a Single Nef Epitope Select for Escape Variants Shortly after Infection. J Virol. 2015;89(21):10802–10820. doi: 10.1128/JVI.01440-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd PA, et al. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491(7422):129–133. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissina A, et al. Protein kinase inhibitors substantially improve the physical detection of T-cells with peptide-MHC tetramers. J Immunol Methods. 2009;340(1):11–24. doi: 10.1016/j.jim.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherall D. The use of non-human primates in research. A working group report. 2006. p. 152.
- Betterton EA, Lowry J, Ingamells R, Venner B. Kinetics and mechanism of the reaction of sodium azide with hypochlorite in aqueous solution. J Hazard Mater. 2010;182(1-3):716–722. doi: 10.1016/j.jhazmat.2010.06.093. [DOI] [PubMed] [Google Scholar]
- Picker LJ, et al. IL-15 induces CD4 effector memory T cell production and tissue emigration in nonhuman primates. J Clin Invest. 2006;116(6):1514–1524. doi: 10.1172/JCI27564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolton G, et al. More tricks with tetramers: a practical guide to staining T cells with peptide-MHC multimers. Immunology. 2015;146(1):11–22. doi: 10.1111/imm.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge L, Lissina A, Cole DK, vanden Berg HA, Price DA, Sewell AK. Tricks with tetramers: how to get the most from multimeric peptide-MHC. Immunology. 2009;126(2):147–164. doi: 10.1111/j.1365-2567.2008.02848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M, et al. FACS Analysis of Leukocyes. In: Herzenberg LA, Weir DM, Blackwell C, editors. Weir's Handbook of Experimental Immunology, 5th ed. Boston: Blackwell Science; 1996. pp. 1–49. [Google Scholar]
- Colantonio AD, et al. KIR polymorphisms modulate peptide-dependent binding to an MHC class I ligand with a Bw6 motif. PLoS Pathog. 2011;7(3):e1001316. doi: 10.1371/journal.ppat.1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer JL, et al. Suppression of a Natural Killer Cell Response by Simian Immunodeficiency Virus Peptides. PLoS Pathog. 2015;11(9):1005145. doi: 10.1371/journal.ppat.1005145. [DOI] [PMC free article] [PubMed] [Google Scholar]


