Abstract
Neuroinflammation is now recognized as a major etiological factor in neurodegenerative disease. Mononuclear phagocytes are innate immune cells responsible for phagocytosis and clearance of debris and detritus. These cells include CNS-resident macrophages known as microglia, and mononuclear phagocytes infiltrating from the periphery. Light microscopy has generally been used to visualize phagocytosis in rodent or human brain specimens. However, qualitative methods have not provided definitive evidence of in vivo phagocytosis. Here, we describe quantitative 3D in silico modeling (q3DISM), a robust method allowing for true 3D quantitation of amyloid-β (Aβ) phagocytosis by mononuclear phagocytes in rodent Alzheimer's Disease (AD) models. The method involves fluorescently visualizing Aβ encapsulated within phagolysosomes in rodent brain sections. Large z-dimensional confocal datasets are then 3D reconstructed for quantitation of Aβ spatially colocalized within the phagolysosome. We demonstrate the successful application of q3DISM to mouse and rat brains, but this methodology can be extended to virtually any phagocytic event in any tissue.
Keywords: Medicine, Issue 118, q3DISM, TgF344-AD rat, APP/PS1 mouse, cerebral amyloidosis, phagocytosis, microglia, lysosome, phagolysosome, Alzheimer's disease
Introduction
Alzheimer's Disease (AD), the most common age-related dementia1, is characterized by cerebral amyloid-β (Aβ) accumulation as "senile" β-amyloid plaques, chronic low-level neuroinflammation, tauopathy, neuronal loss, and cognitive disturbance2. In AD patient brains, neuroinflammation is earmarked by reactive astrocytes and mononuclear phagocytes (referred to as microglia, although their central vs. peripheral origin remains unclear) surrounding Aβ deposits3. As the innate immune sentinels of the CNS, microglia are centrally positioned to clear brain Aβ. However, microglial recruitment to Aβ plaques is accompanied by very little, if any, Aβ phagocytosis4,5. One hypothesis is that microglia are initially neuroprotective by phagocytozing small assemblies of Aβ. However, eventually these cells become neurotoxic as overwhelming Aβ burden and/or age-related functional decline, provokes microglia into a dysfunctional proinflammatory phenotype, contributing to neurotoxicity and cognitive decline6.
Recent Genome-wide Association Studies (GWAS) have identified a cluster of AD risk alleles belonging to core innate immune pathways7 that modulate phagocytosis8-11. Consequently, the immune response to cerebral amyloid deposition has become a major area of interest, both in terms of understanding AD etiology and for developing new therapeutic approaches12-14. Yet, there is a vital need for methodology to evaluate Aβ phagocytosis in vivo. To address this unmet need, we have developed quantitative 3D in silico modeling (q3DISM) to enable true 3D quantitation of cerebral Aβ phagocytosis by mononuclear phagocytes in rodent models of Alzheimer-like disease.
Limited only by the extent to which they recapitulate disease, animal models have proven invaluable for understanding AD pathoetiology and for evaluating experimental therapeutics. Owing to the fact that mutations in the Presenilin (PS) and Amyloid Precursor Protein (APP) genes independently cause autosomal dominant AD, these mutant transgenes have been extensively used to generate transgenic rodent models. Transgenic APP/PS1 mice simultaneously coexpressing "Swedish" mutant human APP (APPswe) and Δ exon 9 mutant human presenilin 1 (PS1ΔE9) present with accelerated cerebral amyloidosis and neuroinflammation15,16. Further, we have generated bi-transgenic rats coinjected with APPswe and PS1ΔE9 constructs (line TgF344-AD, on a Fischer 344 background). Unlike transgenic mouse models of cerebral amyloidosis, TgF344-AD rats develop cerebral amyloid that precedes tauopathy, apoptotic loss of neurons, and behavioral impairment17.
In this report, we describe a protocol for immunostaining microglia, phagolysosomes and Aβ deposits in brain sections from APP/PS1 mice and TgF344-AD rats, and acquisition of large z-dimensional confocal images. We detail in silico generation and analysis of true 3D reconstructions from confocal datasets allowing quantitation of Aβ uptake into microglial phagolysosomes. More broadly, the methodology that we detail here can be used to quantify virtually any form of phagocytosis in vivo.
Protocol
Statement of research ethics: All experiments involving animals detailed herein were approved by the University of Southern California Institutional Animal Care and Use Committee (IACUC) and performed in strict accordance with National Institutes of Health guidelines and recommendations from the Association for Assessment and Accreditation of Laboratory Animal Care International.
1. Rodent Brain Isolation and Preparation for Immunostaining
DAY 1:
Place aged TgF344-AD rats (14-month-old) or APP/PS1 mice (12-month-old) under continuous deep isoflurane anesthesia (4%). Assess the depth of anesthesia by toe pinch and the absence of withdrawal reflex.
Cut through both sides of the rib cage and lift to expose the heart. Insert a 23 G needle into the left ventricle of the heart and make a small incision into the right atrium. Proceed to exsanguination by transcardial perfusion with ice-cold phosphate-buffered saline (PBS) using a peristaltic pump (30 mL for mice; 150 - 200 mL for rats).
Make a caudal midline incision into the skin and move the skin and muscle aside. Cut through the top of the skull along the midline and between the eyes. Remove the bone plates and isolate the whole brain from the skull.
Place the brain into a coronal rodent brain matrix and slice it into quarters. Incubate posterior quarters O/N (16 h) in paraformaldehyde fixative (4% PFA in PBS) at 4 °C. Wash 3x in PBS, and then transfer to 70% ethanol. Caution: PFA is toxic and should be handled under a chemical hood with appropriate personal protection equipment.
DAY 2:
Place brain quarters into embedding cassettes and progressively dehydrate tissue in successively more concentrated 1 h ethanol baths (70%, 80%, 95%, and 100% x3).
Clear ethanol from the tissue with three successive 100% xylene baths (1 h each). Caution: Xylene is toxic and should be handled under a chemical hood with appropriate personal protection equipment.
Embed tissue in paraffin blocks after two molten paraffin wax baths (56 - 58 °C, 90 min each).
DAY 3:
Cut 10 μm-thick sections of paraffin-embedded brains using a microtome. Dip sections into a water bath (50 °C for 1 min) and apply to microscope slides. Leave the slides to dry O/N, ensuring tissue adhesion to the slide.
2. Immunostaining
Note: Different combinations of antibodies can be utilized for the staining procedure described below. Antibody cocktails compatible with brain tissue from rats and mice are listed in Table 1.
DAY 4:
Deparaffinize brain sections using 2x 100% xylene baths (12 min each).
Rehydrate brain sections in successive ethanol baths — 100% for 10 min, 95% for 5 min, 80% for 10 min, and finally 70% for 15 min — followed by 3x PBS washes [5 min at RT, with light agitation]. Meanwhile, heat antigen retrieval solution to 95 - 97 °C on a hot plate with magnetic bar stirring.
Incubate brain sections in antigen retrieval solution at 95 - 97 °C for 30 min. Then, wash 3x in PBS (5 min at RT, with light agitation).
Quickly dry slides using delicate task wipers to avoid tissue drying, and draw a hydrophobic barrier around the tissue area with a hydrophobic barrier pen. Fill the encircled tissue region with blocking buffer [PBS containing 0.3% Triton X-100 and 10% Normal Donkey Serum (NDS)], and incubate at RT for 1 h in a humidified chamber.
Replace blocking buffer with Iba1 primary antibody (diluted in blocking buffer) to label mononuclear phagocytes, and incubate O/N at 4 °C in a humidified chamber. For antibody hosts and working dilutions, see Table 1 and the Table of Materials.
DAY 5:
Rinse primary antibody with 3x PBS baths (5 min at RT with light agitation). Incubate with fluorescent secondary antibody (conjugated with a 594 nm emission fluorophore) for 1 h (in blocking buffer at RT in the dark) followed by 3x PBS baths (5 min at RT with light agitation). At this time, maintain sections in the dark to avoid fluorescent signal bleaching.
DAY 6 - 7:
Repeat steps 2.5 & 2.6 with CD68 (rat brains) or LAMP1 (mouse tissue) antibodies and appropriate secondary antibodies (coupled with a 488 nm emission fluorophore) to label phagolysosomes.
DAY 7 - 8:
Repeat steps 2.5 & 2.6 with OC (rat tissue) or 4G8 (mouse brains) antibodies and appropriate secondary antibodies (coupled to a 647 nm fluorophore) to label Aβ deposits. NOTE: Alternatively, 6E10 antibody can be used successfully both on mouse and rat tissue. For appropriate antibody combinations, see the Table of Materials.
Allow sections to completely dry O/N at RT in the dark. Then, cover specimens with a cover slip sealed by fluorescence mounting media containing DAPI.
3. Acquisition of Large Z-stack Confocal Datasets
Note: This protocol requires a fully automated laser scanning confocal microscope equipped with a 60X objective and 405 nm, 488 nm, 594 nm, and 647 nm lasers. All equipment is computer controlled by imaging and laser control software. Prior to beginning the imaging protocol, power on the computer, epifluorescent lamp, microscope, lasers and camera.
DAY 9:
Select the 60X microscope objective. Add immersion oil to the lens, and place the sample onto the microscope stage slide holder. Raise the objective until the oil makes contact with the slide. Adjust the focal plane to locate amyloid plaques in the hippocampus or cerebral cortex using epifluorescent illumination through the oculars.
Acquire confocal images of activated mononuclear phagocytes surrounding amyloid deposits in the hippocampus or cortex of rodents by confocal microscopy (60X magnification, z-stack steps: 0.25 μm < z < 0.40 μm, number of steps 25 < n < 35).
4. q3DISM
Note: In order to yield significant results, we suggest analyzing a minimum of 3 images per animal/region of interest. For each image, the abundance of cells to analyze may vary depending on experimental paradigms. In the representative results shown in this report, we analyzed 3 cells/condition (e.g., mononuclear phagocytes distant from or associated with plaques; see Figures 1C - D and 2C - D).
DAY 10:
- Analyze confocal datasets with scientific 3D image processing and analysis software colocalization (coloc) module for spatial proximity of Iba1/CD68 (rat tissue) or Iba1/LAMP1 (mouse tissue) staining in all z-planes simultaneously. Create Iba+/CD68+ or Iba+/LAMP1+ colocalization channels that correspond to phagolysosomes within activated mononuclear phagocytes.
- Select TRITC for channel A (corresponding to Iba1 staining coupled with 594 nm fluorophore) and FITC for channel B (corresponding to CD68 or LAMP1 staining coupled with 488 nm fluorophore). On the right-hand side of the software window for 'mode check,' select 'threshold,' and for 'coloc intensities,' select 'source channels'.
- Click 'Edit' to select 'coloc color' on the right-hand side of the software window.
- For each channel independently, adjust thresholds to include specific staining and exclude background/non-specific signals. Once adjusted, do not change thresholds between images to ensure an unbiased analysis. The colocalized voxels (pixels from all z-stacks) will appear in the color selected in step 4.1.2 in all z-stacks simultaneously.
- Click 'build coloc channel'. The colocalization channel created will appear in the display adjustment window.
- Click on the coloc channel to open channel statistics. The '% of volume/material A above threshold colocalized' represents the % of Iba1 signal (voxels corresponding to the 594 nm fluorophore) colocalized with LAMP1 or CD68 signal (voxels corresponding to the 488 nm fluorophore). More simply, this is the monocyte volume occupied by phagolysosomes (see Figures 1C and 2C). NOTE: '% of volume/material B above threshold colocalized' corresponds to % of LAMP1 or CD68 signal colocalized with Iba1. This should be close to 100%, as phagolysosomes are intracellular structures. Values are based on ratio of signal from all z-stacks above threshold colocalized.
- Using the coloc module, analyze the coloc channel created in step 4.1 for spatial proximity with OC (rat tissue) or 4G8 (mouse tissue) Aβ signals. This allows for quantitation of Aβ encapsulated within phagolysosomes.
- Select the channel A coloc dataset (corresponding to the Iba1/CD68 or Iba1/LAMP1 coloc channel created in step 4.1) and Cy5 for channel B (corresponding to OC or 4G8 staining coupled with 647 nm fluorophore).
- Build a coloc channel as described in steps 4.1.2 to 4.1.5.
- Click on the coloc channel to open channel statistics. The '% of volume/material A above threshold colocalized' represents the % of Iba1/LAMP1 or Iba1/CD68 (voxels corresponding to the coloc channel built in step 4.1.5.) colocalized with OC or 4G8 signal (voxels corresponding to the 647 nm fluorophore). This is the phagolysosomal volume occupied by Aβ (see Figures 1D and 2D). NOTE: '% of volume/material B above threshold colocalized' corresponds with % of total Aβ signal colocalized with phagolysosomes. This can be used to evaluate the fraction of total Aβ deposits encapsulated within phagolysosomes (not shown in the present representative results).
- Use the surpass module to reconstruct the confocal image stacks and generate 3D models of Aβ encapsulated within monocyte phagolysosomes.
- In the 'display adjustment' window, select TRITC (to show only Iba1 staining). In the 'volume properties' window, click 'add new surface'.
- In step '1/5 algorithm', in 'settings,' select 'surface.' Also, in 'color,' select color type in palette or RGB and adjust transparency (red is set at 60% transparency in Figures 1B and 2B). Check box 'select region of interest'. Click next.
- In Step '2/5 region of interest,' draw a window around the cell of interest by adjusting x, y and z coordinates (see white boxes in Figures 1A and 2A). Click next.
- In Step '3/5 source channel', select the TRITC source channel. Check the box 'smooth,' and set surface area detail level to 0.4 μm. For 'thresholding,' select absolute intensity. Click next.
- In Step '4/5 threshold,' adjust threshold so that the volume created overlaps perfectly with the TRITC channel signal. Click next.
- In Step '5/5 classify surfaces,' in section 'filter type,' select objects depending on their size to be included or excluded from the volumes to be created. Click finish, execute all creation steps, and exit the wizard.
- Repeat steps 4.3.1 to 4.3.6 to create a 3D surface for the FITC channel (LAMP1+ or CD68+ phagolysosomes).
- Repeat steps 4.3.1 to 4.3.6 to create a 3D surface for the Cy5 channel (OC+ or 4G8+ Aβ deposits).
Representative Results
Using the multi-stage methodology for q3DISM detailed above, we are able to quantify Aβ uptake into monocyte phagolysosomes in the brains of APP/PS1 mice (Figure 1) and TgF344-AD rats (Figure 2). Therefore, the q3DISM methodology has enabled analysis of mononuclear phagocytes in mouse and rat models of AD. Interestingly, the volume occupied by CD68+ phagolysosomes is significantly increased in Iba1+ mononuclear phagocytes associated with compared to distant from plaques both in APP/PS1 mice (Figure 1B, C) and TgF344-AD rats (Figure 2B, C). These data show that mononuclear phagocytes near plaques are poised for phagocytosis compared to cells located away from the plaque. In order to illustrate the range of data from staining different Aβ conformers, we used various antibodies in mouse and rat tissue (detailed in Table 1 and discussion)- we are not seeking to directly compare mouse and rat models of AD with regards to Aβ phagocytosis. Nonetheless, it was noteworthy that plaque associated mononuclear phagocytes had increased Aβ uptake in APP/PS1 mice compared to cells distant from plaques (Figure 1D). TgF344-AD rats had very modest uptake of Aβ fibrils overall, and no change in Aβ fibril phagocytosis as a function of distance from plaques (Figure 2D).
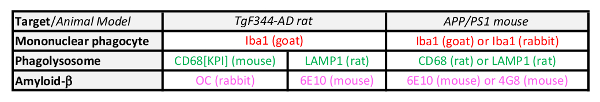 Table 1: Immunostaining Mononuclear Phagocytes, Phagolysosomes and Amyloid-β in Brain Tissue from APP/PS1 Mice or TgF344-AD Rats. Rat or mouse specific antibody cocktails are listed with their respective host in parentheses. Colors indicate the fluorophore used for the secondary antibody. Red: Alexa Fluor 594; green: Alexa Fluor 488; magenta: Alexa Fluor 647.
Table 1: Immunostaining Mononuclear Phagocytes, Phagolysosomes and Amyloid-β in Brain Tissue from APP/PS1 Mice or TgF344-AD Rats. Rat or mouse specific antibody cocktails are listed with their respective host in parentheses. Colors indicate the fluorophore used for the secondary antibody. Red: Alexa Fluor 594; green: Alexa Fluor 488; magenta: Alexa Fluor 647.
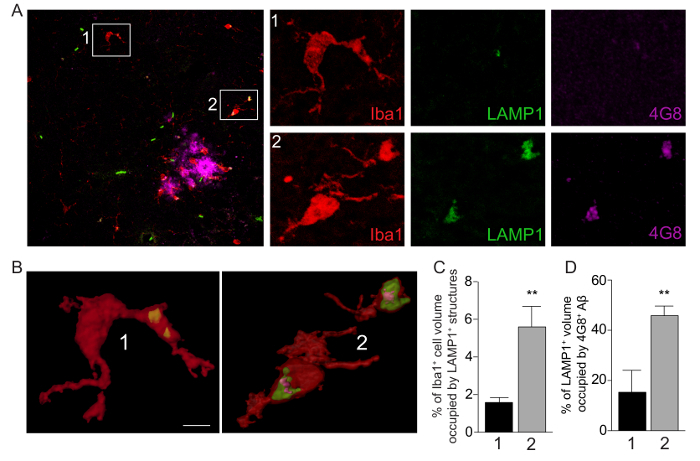 Figure 1: Visualization and Quantitation of Amyloid-β Phagocytosis in APP/PS1 Mouse Brains. (A) Confocal images of brain sections from an APP/PS1 mouse showing Iba1+ cells (red), LAMP1+ phagolysosomes (green) and 4G8+ amyloid deposits (magenta). Inset 1 highlights a mononuclear phagocyte distant from the plaque, while inset 2 shows a mononuclear phagocyte associated with the plaque. Scale bar denotes 50 μm. Smaller panels on the right are enlargements of Iba1, LAMP1 and 4G8 signals within insets 1 and 2. (B) 3D reconstruction of Iba1, LAMP1 and 4G8 signals for insets 1 and 2. Scale bar denotes 3 μm. (C) Quantitation of Iba1+ mononuclear phagocyte volume occupied by LAMP1+ phagolysosomes. (D) Quantitation of intracellular LAMP1+ phagolysosomal volume occupied by 4G8+ Aβ. Values are based on ratio of signal (voxels) above threshold that are colocalized. Data are shown as mean ±standard error of the mean (n = 6 cells) for mononuclear phagocytes distant from plaques (1), or associated with plaques (2). ** p <0.01 by student's t-test. Please click here to view a larger version of this figure.
Figure 1: Visualization and Quantitation of Amyloid-β Phagocytosis in APP/PS1 Mouse Brains. (A) Confocal images of brain sections from an APP/PS1 mouse showing Iba1+ cells (red), LAMP1+ phagolysosomes (green) and 4G8+ amyloid deposits (magenta). Inset 1 highlights a mononuclear phagocyte distant from the plaque, while inset 2 shows a mononuclear phagocyte associated with the plaque. Scale bar denotes 50 μm. Smaller panels on the right are enlargements of Iba1, LAMP1 and 4G8 signals within insets 1 and 2. (B) 3D reconstruction of Iba1, LAMP1 and 4G8 signals for insets 1 and 2. Scale bar denotes 3 μm. (C) Quantitation of Iba1+ mononuclear phagocyte volume occupied by LAMP1+ phagolysosomes. (D) Quantitation of intracellular LAMP1+ phagolysosomal volume occupied by 4G8+ Aβ. Values are based on ratio of signal (voxels) above threshold that are colocalized. Data are shown as mean ±standard error of the mean (n = 6 cells) for mononuclear phagocytes distant from plaques (1), or associated with plaques (2). ** p <0.01 by student's t-test. Please click here to view a larger version of this figure.
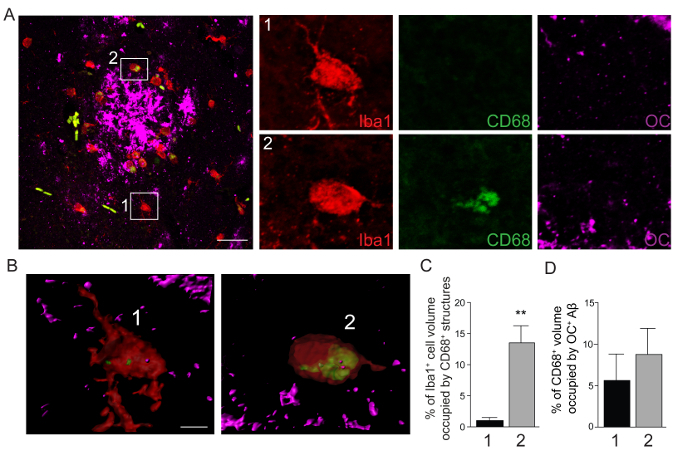 Figure 2: Visualization and Quantitation of Amyloid-β Phagocytosis in TgF344-AD Rat Brains. (A) Confocal images of brain sections from a TgF344-AD rat showing Iba1+ cells (red), CD68+ phagolysosomes (green), and OC+ soluble fibrillar Aβ (magenta). Inset 1 highlights a mononuclear phagocyte distant from the plaque, while inset 2 shows a mononuclear phagocyte associated with the plaque. Scale bar denotes 50 μm. Smaller panels on the right are enlargements of Iba1, CD68 and OC signals within insets 1 and 2. (B) 3D reconstruction of Iba1, CD68 and OC signals for insets 1 and 2. Scale bar denotes 3 μm. (C) Quantitation of Iba1+ mononuclear phagocyte volume occupied by CD68+ phagolysosomes. (D) Quantitation of intracellular CD68+ phagolysosomal volume occupied by OC+ Aβ. Values are based on ratio of signal (voxels) above threshold that are colocalized. Data are shown as mean ±standard error of the mean (n = 6 cells) for mononuclear phagocytes distant from plaques (1), or associated with plaques (2). ** p <0.01 by student's t-test. Please click here to view a larger version of this figure.
Figure 2: Visualization and Quantitation of Amyloid-β Phagocytosis in TgF344-AD Rat Brains. (A) Confocal images of brain sections from a TgF344-AD rat showing Iba1+ cells (red), CD68+ phagolysosomes (green), and OC+ soluble fibrillar Aβ (magenta). Inset 1 highlights a mononuclear phagocyte distant from the plaque, while inset 2 shows a mononuclear phagocyte associated with the plaque. Scale bar denotes 50 μm. Smaller panels on the right are enlargements of Iba1, CD68 and OC signals within insets 1 and 2. (B) 3D reconstruction of Iba1, CD68 and OC signals for insets 1 and 2. Scale bar denotes 3 μm. (C) Quantitation of Iba1+ mononuclear phagocyte volume occupied by CD68+ phagolysosomes. (D) Quantitation of intracellular CD68+ phagolysosomal volume occupied by OC+ Aβ. Values are based on ratio of signal (voxels) above threshold that are colocalized. Data are shown as mean ±standard error of the mean (n = 6 cells) for mononuclear phagocytes distant from plaques (1), or associated with plaques (2). ** p <0.01 by student's t-test. Please click here to view a larger version of this figure.
Discussion
The protocol that we describe in this report for true 3D quantitation of Aβ phagocytosis in vivo by mononuclear phagocytes relies on specific labeling of cellular and subcellular compartments as well as Aβ deposits. Specifically, we use Iba1 (Ionized-calcium Binding Adaptor molecule 1), a protein that is involved in membrane ruffling and phagocytosis upon cell activation18,19, to stain cerebral mononuclear phagocytes. While Iba1+ cells are generally regarded as brain-resident microglia, it remains possible that peripherally infiltrating mononuclear phagocytes comprise a subset of immunolabeled cells. Intracellular phagolysosomes are revealed with antibodies recognizing members of the Lysosomal/endosomal-associated membrane glycoprotein (LAMP) family: LAMP120 or CD6821. The latter is primarily localized to lysosomes and endosomes, with a smaller fraction circulating to the cell surface. LAMP1 and CD68 antibodies have both been successfully used in rat and mouse tissue, and the decision on which to use is primarily dependent on the host of other antibodies used for co-staining (see Table 1). It is important to be aware that various antibodies can be used for detection of Aβ. In mouse tissue, 4G8 (recognizing human and mouse amino acid residues 17 - 24 of Aβ peptide22, Figure 1) and 6E10 (recognizing human amino acid residues 1 - 1723, data not shown) have been used successfully, allowing detection and quantitation of Aβ content in LAMP1+ or CD68+ phagolysosomes present in Iba1+ mononuclear phagocytes. Using our q3DISM methodology, we observed increased Aβ load in Iba1+ cells associated with global reduction of Aβ load in brains of APPPS1/IL-10-/- mice16, whereas others showed decreased Aβ uptake into Iba1+ cells accompanied by increased Aβ load and plaque volume in brains of Rag-5xfAD mice24. These results suggest that the phenomenon observed is a "snapshot" of active Aβ phagocytosis, although our methodology cannot be used to predict if the phagocytosed material is efficiently digested and cleared.
In rat tissue, we detected Aβ deposits using OC (recognizing soluble Aβ42 oligomers/fibrils25,26, Figure 2) or 6E1017 (not shown). Interestingly, very little OC+ fibrils were present in monocyte phagolysosomes in TgF344-AD rat brains. This phenomenon has previously been reported in APP23 transgenic mice using immunogold staining and 3D reconstruction27. Note: a variety of antibodies for detection of different Aβ species/conformers are available and can potentially be tested and employed at the user's discretion.
Our q3DISM technique has been adopted by us and others to quantify Aβ phagocytosis in the context of AD16,24,28. However, the method can be adapted to evaluate virtually any form of phagocytosis. It may also be applied to tissues other than the brain, and to other animal species (including humans). The procedures described in this protocol rely on standard immunostaining, detected with fluorescence and imaged with a confocal microscope. In this report, we have used paraffin-embedded tissue according to guidelines provided by the antibody manufacturers. If desired, the present protocol could also be optimized for fresh tissue (embedded in 2% agarose in PBS). The protocol can also be optimized to detect phagocytosis of coaggregating proteins. This is made possible by staining and acquiring images composed of more than 4 fluorescent colors29. In this case, the creation of the first colocalization channel (monocyte/phagolysosome) would be followed by creation of a second colocalization channel (aggregated protein 1/co-aggregated protein 2). Then, the two channels would be used for 3D analysis and modeling.
The q3DISM technique is limited principally by the specificity and sensitivity of immunostaining, quality of antibodies, and colocalization approach. This multistep procedure has a few potential pitfalls that can affect quality of the staining and, therefore, 3D quantitation of Aβ uptake. The first critical step of the procedure is the isolation of rodent brains. Indeed, careful tissue handling is fundamental to avoid damage to the brain and to guarantee that brain structures remain intact when sectioned. Second, tissue fixation time is quite important since over-fixation (more than 16 h in 4% PFA) limits detection of subcellular phagolysosomes and Aβ content. Importantly, the successive staining of 1) monocytes, 2) phagolysosomes and 3) Aβ is critical for the success of the method. It is essential to use the antibodies consecutively- not concomitantly. Additionally, acquisition of the z-stack confocal images is of utmost importance, since 3D modeling of cells, phagolysosomes and Aβ peptide as well as colocalization analyses depend on the quality of imaging of cellular and subcellular structures through the thickness of the tissue section. Finally, adjustment of thresholds in the software is crucial for 3D modeling and for colocalization analysis. Indeed, thresholds for the different channels need to be carefully chosen, since they will determine which signal is included (specific) or excluded (background) from the analysis. It is also fundamental that the thresholds chosen remain consistent between samples in the case of the comparison of different animals/specimens. So far, classical immunohistostaining, magnetic resonance imaging and positron emission tomography have been the imaging methods of choice for in vivo visualization and quantitation of Aβ30. Although highly useful for large-scale quantitation of amyloid burden in brains of rodents or AD patients, these methods are inefficient for visualization of intracellular Aβ. Other techniques using confocal microscopy, fluorescence spectroscopy and flow cytometry exist and have been used by us and others to discriminate and quantify intracellular Aβ content in vitro16,31,32. Immunogold staining coupled with 3D reconstruction was previously used to highlight the absence of amyloid fibrils within microglia in vivo in APP23 transgenic mice27, and another study successfully used confocal imaging and 3D surface reconstruction of microglia-associated with β-amyloid plaques to show limited Aβ interaction and uptake33. However, these techniques do not allow for quantitation of intra-phagolysosomal Aβ species. Others have recently developed a fluorescence-based in vivo assay allowing the measurement of phagocytic activity by peripheral macrophages in the rat34. This method ingeniously allows for quantitation of fluorescent bioprobes in phagolysosomes in vivo, however, data available so far are limited to the periphery.
To our knowledge, q3DISM is the first method that affords 3D visualization and quantitation of Aβ phagocytosis in brains of rodent AD models. This formidable tool will undoubtedly pave the way to a deeper understanding of cerebral Aβ phagocytosis, and may also prove useful in other contexts.
Disclosures
The authors have nothing to disclose.
Acknowledgments
M-V.G-S. is supported by a BrightFocus Foundation Alzheimer's Disease Research Fellowship Award (A2015309F) and an Alzheimer's Association, California Southland Chapter Young Investigator Award. T.M.W. is supported by an ARCS Foundation and John Douglas French Alzheimer's Foundation Maggie McKnight Russell-JDFAF Memorial Postdoctoral Fellowship. This work was supported by the National Institute on Neurologic Disorders and Stroke (1R01NS076794-01, to T.T.), an Alzheimer's Association Zenith Fellows Award (ZEN-10-174633, to T.T.), and an American Federation of Aging Research/Ellison Medical Foundation Julie Martin Mid-Career Award in Aging Research (M11472, to T.T.). We are grateful for startup funds from the Zilkha Neurogenetic Institute, which helped to make this work possible.
References
- Brookmeyer R, et al. National estimates of the prevalence of Alzheimer's disease in the United States. Alzheimers Dement. 2011;7(1):61–73. doi: 10.1016/j.jalz.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease. Cold Spring Harb Perspect Biol. 2011;3(7) doi: 10.1101/cshperspect.a004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka MT, Golenbock DT, Latz E. Innate immunity in Alzheimer's disease. Nat Immunol. 2015;16(3):229–236. doi: 10.1038/ni.3102. [DOI] [PubMed] [Google Scholar]
- Mawuenyega , et al. Decreased clearance of CNS beta-amyloid in Alzheimer's disease. Science. 2010;330(6012):1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer's disease mice. J Neurosci. 2008;28(33):8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston H, Boutin H, Allan SM. Assessing the contribution of inflammation in models of Alzheimer's disease. Biochem Soc Trans. 2011;39(4):886–890. doi: 10.1042/BST0390886. [DOI] [PubMed] [Google Scholar]
- Gjoneska E, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer's disease. Nature. 2015;518(7539):365–369. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitz C, Mayeux R. Alzheimer disease: epidemiology, diagnostic criteria, risk factors and biomarkers. Biochem Pharmacol. 2014;88(4):640–651. doi: 10.1016/j.bcp.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazrati L-N, et al. Genetic association of CR1 with Alzheimer's disease: a tentative disease mechanism. Neurobiol Aging. 2012;33(12):2949. doi: 10.1016/j.neurobiolaging.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Griciuc A, et al. Alzheimer's Disease Risk Gene CD33 Inhibits Microglial Uptake of Amyloid Beta. Neuron. 2013. pp. 1–13. [DOI] [PMC free article] [PubMed]
- Li X, Long J, He T, Belshaw R, Scott J. Integrated genomic approaches identify major pathways and upstream regulators in late onset Alzheimer's disease. Scientific reports. 2015;5:12393. doi: 10.1038/srep12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz TM, Town T. Microglia in Alzheimers Disease: "Its All About Context". Int J Alzheimers Dis. 2012. p. 314185. [DOI] [PMC free article] [PubMed]
- Guillot-Sestier M-V, Doty KR, Town T. Innate Immunity Fights Alzheimer's Disease. Trends Neurosci. 2015;38(11):674–681. doi: 10.1016/j.tins.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot-Sestier M-V, Town T. Innate immunity in Alzheimer's disease: a complex affair. CNS Neurol Disord Drug Targets. 2013;12(5):593–607. doi: 10.2174/1871527311312050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky JL, Slunt HH, Ratovitski T, Jenkins NA, Copeland NG, Borchelt DR. Co-expression of multiple transgenes in mouse CNS: a comparison of strategies. Biomol Eng. 2001;17(6):157–165. doi: 10.1016/s1389-0344(01)00067-3. [DOI] [PubMed] [Google Scholar]
- Guillot-Sestier M-V, et al. Il10 deficiency rebalances innate immunity to mitigate Alzheimer-like pathology. Neuron. 2015;85(3):534–548. doi: 10.1016/j.neuron.2014.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen RM, et al. A transgenic Alzheimer rat with plaques, tau pathology, behavioral impairment, oligomeric aβ, and frank neuronal loss. J Neurosci. 2013;33(15):6245–6256. doi: 10.1523/JNEUROSCI.3672-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Ibata I, Ito D, Ohsawa K, Kohsaka S. A novel gene iba1 in the major histocompatibility complex class III region encoding an EF hand protein expressed in a monocytic lineage. Biochem. Biophys. Res. Commun. 1996;224(3):855–862. doi: 10.1006/bbrc.1996.1112. [DOI] [PubMed] [Google Scholar]
- Ohsawa K, Imai Y, Sasaki Y, Kohsaka S. Microglia/macrophage-specific protein Iba1 binds to fimbrin and enhances its actin-bundling activity. J Neurochem. 2004;88(4):844–856. doi: 10.1046/j.1471-4159.2003.02213.x. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay U, Nagy M, Fenton WA, Horwich AL. Absence of lipofuscin in motor neurons of SOD1-linked ALS mice. Proc Natl Acad Sci U S A. 2014;111(30):11055–11060. doi: 10.1073/pnas.1409314111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holness CL, Simmons DL. Molecular cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood. 1993;81(6):1607–1613. [PubMed] [Google Scholar]
- Connor T, et al. Phosphorylation of the translation initiation factor eIF2alpha increases BACE1 levels and promotes amyloidogenesis. Neuron. 2008;60(6):988–1009. doi: 10.1016/j.neuron.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, et al. Phospholipase D1 corrects impaired betaAPP trafficking and neurite outgrowth in familial Alzheimer's disease-linked presenilin-1 mutant neurons. Proc Natl Acad Sci U S A. 2006;103(6):1936–1940. doi: 10.1073/pnas.0510710103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh SE, et al. The adaptive immune system restrains Alzheimer's disease pathogenesis by modulating microglial function. Proc Natl Acad Sci U S A. 2016;113(9):1316–1325. doi: 10.1073/pnas.1525466113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterov I, et al. Apolipoprotein A-I deficiency increases cerebral amyloid angiopathy and cognitive deficits in APP/PS1DeltaE9 mice. J Biol. Chem. 2010;285(47):36945–36957. doi: 10.1074/jbc.M110.127738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blurton-Jones M, et al. Neural stem cells improve cognition via BDNF in a transgenic model of Alzheimer disease. Proc Natl Acad Sci U S A. 2009;106(32):13594–13599. doi: 10.1073/pnas.0901402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder M, Deller T, Staufenbiel M, Jucker M. 3D-Reconstruction of microglia and amyloid in APP23 transgenic mice: no evidence of intracellular amyloid. Neurobiol Aging. 2001;22(3):427–434. doi: 10.1016/s0197-4580(01)00209-3. [DOI] [PubMed] [Google Scholar]
- Leinenga G, Götz J. Scanning ultrasound removes amyloid-β and restores memory in an Alzheimer's disease mouse model. Sci Transl Med. 2015;7(278):33. doi: 10.1126/scitranslmed.aaa2512. [DOI] [PubMed] [Google Scholar]
- Liarski VM, et al. Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci Transl Med. 2014;6(230):46. doi: 10.1126/scitranslmed.3008146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols L, Pike VW, Cai L, Innis RB. Imaging and in vivo quantitation of beta-amyloid: an exemplary biomarker for Alzheimer's disease. Biol Psychiatry. 2006;59(10):940–947. doi: 10.1016/j.biopsych.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Skovronsky DM, Zhang B, Kung MP, Kung HF, Trojanowski JQ, Lee VM. In vivo detection of amyloid plaques in a mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2000;97(13):7609–7614. doi: 10.1073/pnas.97.13.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H, Litvinchuk A, Chiang AC-A, Aithmitti N, Jankowsky JL, Zheng H. Astrocyte-Microglia Cross Talk through Complement Activation Modulates Amyloid Pathology in Mouse Models of Alzheimer's Disease. J Neurosci. 2016;36(2):577–589. doi: 10.1523/JNEUROSCI.2117-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny R, et al. Conversion of Synthetic Aβ to In Vivo Active Seeds and Amyloid Plaque Formation in a Hippocampal Slice Culture Model. J Neurosci. 2016;36(18):5084–5093. doi: 10.1523/JNEUROSCI.0258-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaro K, et al. Development of a fluorescence-based in vivo phagocytosis assay to measure mononuclear phagocyte system function in the rat. J Immunotoxicol. 2015;12(3):239–246. doi: 10.3109/1547691X.2014.934976. [DOI] [PubMed] [Google Scholar]


