Abstract
Accurate segmentation of perimysium plays an important role in early diagnosis of many muscle diseases because many diseases contain different perimysium inflammation. However, it remains as a challenging task due to the complex appearance of the perymisum morphology and its ambiguity to the background area. The muscle perimysium also exhibits strong structure spanned in the entire tissue, which makes it difficult for current local patch-based methods to capture this long-range context information. In this paper, we propose a novel spatial clockwork recurrent neural network (spatial CW-RNN) to address those issues. Specifically, we split the entire image into a set of non-overlapping image patches, and the semantic dependencies among them are modeled by the proposed spatial CW-RNN. Our method directly takes the 2D structure of the image into consideration and is capable of encoding the context information of the entire image into the local representation of each patch. Meanwhile, we leverage on the structured regression to assign one prediction mask rather than a single class label to each local patch, which enables both efficient training and testing. We extensively test our method for perimysium segmentation using digitized muscle microscopy images. Experimental results demonstrate the superiority of the novel spatial CW-RNN over other existing state of the arts.
1 Introduction
Many important morphological properties, such as the distribution of muscle fibers and their nuclei with respect to the perimysium, are important biomarkers for early diagnosis of many muscle diseases [6]. To compute these spatial morphological parameters, accurate and efficient segmentation of perimysium is an essential prerequisite. However, muscle perimysium often shares similar appearances to other structures in the muscle, such as endomysium, epimysium, and blood vessels. The large variations in staining intensity, global structure, and morphology further complicate the automated segmentation task.
Recently, deep learning based methods have achieved great success in object detection and segmentation, among which many are mainly dominated by variations of the convolutional neural network (CNN) [10]. One popular strategy is applying sliding-window to local image patch either by classification [5] or regression [14]. This group of methods fails to exploit the global semantic information. Recently, recurrent neural (RNN) network and it’s variations (gated recurrent neural network (GRU) [3] and long short-term memory network (LSTM) [8]) have witnessed great success in modeling global semantic information in chain-structured data. Both GRU [3] and LSTM [8] are proposed to handle the exploding or vanishing gradient issue of plain RNN with the cost of a heavier computational load. Recently, the clockwork RNN (CW-RNN), which contains even a smaller number of parameters than plain RNN, has been proposed in [9] and proven effective in modeling long-term dependency. CW-RNN separates the hidden recurrent units into different groups, each runs their own computation at specific, discrete clock period. Since only a portion of the modules are active at each time step, it is more efficient than plain RNN. Furthermore, it is also shown to outperform RNN and even LSTM in various tasks [9].
Enormous efforts have been devoted to utilizing RNN on computer vision tasks. Francesco [12] applies GRU [3] to sweep the images as one chain-structured data but along four different directions to model the context information. Some pioneering works [7,2] that exploit the potentials of multi-dimensional RNN in semantic image segmentation have also achieved promising results. However, 2D plain RNN [7] suffers from the exploding or the vanishing gradient problem for large images, and 2D LSTM [2] contains much more parameters than 2D RNN, which makes it inefficient at the runtime, and sometimes over-fit can happen especially when the amount of training data is limited.
In this paper, we propose a 2D spatial clockwork RNN which extends the applicability of chain structured CW-RNN [9] to 2D image domain for efficient perimysium segmentation. Our model directly exploits the 2D structure of images and encodes the global context information among local image patches. Different from [7,2], our model contains a much smaller number of parameters, which makes it computationally efficient and suitable for medical image segmentation with limited training data. In our algorithm, instead of conducting inefficient patch-wise classification, we integrate the structured regression [14] into the proposed algorithm. This allows us to use non-overlapping stride in both training and testing stages. Extensive experimental results demonstrate the effectiveness and efficiency of our proposed model. To the best of our knowledge, this is the first work to propose a 2D spatial CW-RNN that achieves promising results on biomedical image segmentation.
2 Methodology
2.1 Recurrent Neural Network Revisited
The recurrent neural network (RNN) is one type of neural network that is equipped with recurrent connections, which enable the network to memorize past input patterns. For the simple RNN (SRNN), at each time step, it’s current hidden state ht is a non-linear transformation of the current input xt and the hidden state ht−1 from the last step. The output ot is directly connected to ht. Mathematically, those relationships can be expressed by the following equations:
| (1) |
| (2) |
where f(.), g(.) represent the nonlinear activation functions, W and U are weight matrices connecting input units to hidden units, and hidden units to themselves, respectively. V is the weight matrix connecting hidden units to the output units. bh and bo represent the bias terms for the hidden and output layer, respectively.
SRNN is usually trained with a discriminative objective function using the back propagation through time (BPTT) algorithm [13]. However, the fact that the computed gradients of SRNN are either exploding or vanishing when T becomes large hinders the SRNN from learning long-term temporal dependencies. Instead of introducing gated connections [3,8] to complicate the model, clockwork RNN (CW-RNN) [9] addresses the long-term dependency issue by using a clever trick. Specifically, the hidden units h are partitioned into M modules (hm for i = 1, …, M), each is of size k and associated with a clock (or temporal) period Ti ∈ {T1, …, TM}. The total length of the hidden units is hid = M × k. At each time step t, the neurons in module i will be updated only when t satisfies (t mod Ti) = 0. Units corresponding to slower rates are thus capable of preserving long-term information. In addition, connections between hidden units are restricted that faster modules can only receive information from slower ones and not vice-versa, this mechanism further reduces the total number of active weights.
2.2 Spatial Clockwork RNN
Since there are no existing sequences presented in static images, the aforementioned CW-RNN is not directly applicable to our application. To ameliorate this problem, we extend the CW-RNN to a two-dimensional domain, in which current state can receive information from it’s predecessors in both row and column directions. In order for the spatial CW-RNN to process the image, all image patches need to be sorted to an acyclic sequence.
Specifically, we maintain one sub-hidden state for both row and column dimension, denoted as and , which are composed together as the hidden states Denote respectively the weights matrix connecting the current hidden states to it’s row and column predecessor as Û and Ũ, which are split into four hid × hid block matrices; W connecting the input units to hidden units is partitioned into 2 input_dim × hid blocks-columns; the bias bh is also evenly separated into 2 groups: , , W = (W1 W2), and b = (b1 b2). Each block matrix Û(m,n), Ũ(m,n) are further partitioned into M × M smaller block-matrices with the same size k × k. Wm and bm is partitioned into M blocks-columns as well; , , and . Recall that each sub-hidden state and is partitioned into M module, each runs at specific temporal rate. Denote i, j ∈ {1, …, M} as the modules index, u ∈ {1, 2} matrix identifier, and (r, c) as the time-step. For brief narrative, we define the following general matrix placeholders: , , and . The updating rule for the i-th module of (similar case for ) at time step (r, c) is given as:
| (3) |
Note that the aforementioned method only considers the 4 connected neighborhood, namely, every patch only receives information from it’s left, right, up and lower adjacent patches. But it is trivial to extend our method to 8 connected neighborhood. Both of the two cases are evaluated in the experiment part.
2.3 Structured Prediction with Full Sweeping
Due to the temporal dependency property of spatial CW-RNN, each local patch only receives context information from the region spanned by it’s predecessors. However, in 2D images, each local patch is surrounded by both it’s predecessors and postdecessors, we thus want the model to be aware of such bi-directional context information. To this end, we sweep the input image (or feature map) from four different corners (upper-left, lower-left, upper-right, lower-right) to the opposite corners. For each local image patch, activations from four directional sweepings are concatenated together as the full-context representation, which is fed to the successive layers to produce the final prediction output. The illustration of this process is shown in Figure 1.
Fig. 1.
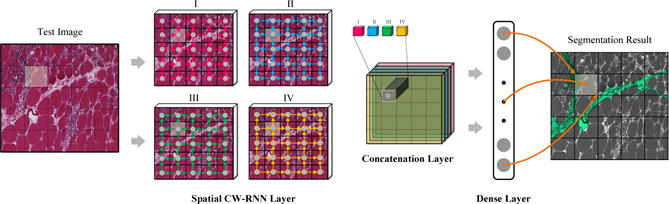
One exemplar architecture. Spatial CW-RNN and dense layer represent the proposed spatial clockwork RNN and fully connected layer, respectively. Sweepings in different directions are illustrated in spatial CW-RNN layer using colorful arrows. Activations from four sweepings are concatenated together in the concatenation layer as the global context information for each local patch. The mapping between the output of dense layer and the predicted mask (overlaid on the original image) for each local patch is illustrated using brown arrows.
Now, we omit the module index i in and define , , and as the total hidden activations (containing all the modules) for each directional sweeping at time step (r, c). The output after applying one dense layer to those concatenated features can be computed as:
| (4) |
where d′ ∈ {↘, ↙, ↗, ↖} denotes different sweeping direction. Please note that dense layer is applied individually across all the time step, and local patches corresponding to different time steps share the same weights Wd′.
Given a set of training data , where N is the total number of training data, Xi is the i-th training image and Yi is the corresponding mask label. Let Ri and Ci denote the total number of local patches in row and column dimension for the i-th pair of training data. Denote Θ as the model’s parameter, and ψ as our model. The objective function defined on {(Xi, Yi)} is given by:
| (5) |
where both of and are reshaped into a vector to computed the loss.
Our proposed spatial CW-RNN is inherently capable of capturing semantic information in the entire image. Meanwhile, it is totally end-to-end trainable and can be optimized using standard BPTT algorithm [13]. It takes an input image with any size and produces the result mask with the same size as the input.
3 Experimental Results
Dataset and Implementation Details
The proposed spatial CW-RNN has been extensively evaluated using 348 H&E stained skeletal muscle microscopy images (each image roughly contains 300 × 600 pixels). All the images are manually annotated and double checked by two neuromuscular pathologists. In total 150 images are chosen for testing and the rest for training. Both qualitative and quantitative experiments are reported. The detailed architecture of our method is summarized in Table. 1. The first layer is a dense layer, and the next 4 spatial CW-RNN layers are used to sweep the input feature map in four different directions. The size of the none-overlapping patches is set to 10 × 10 × 3. The model is trained using RMSprop algorithm with a learning rate of 0.003. M and k in Section 2.2 are 4 and 48, respectively. Time period is set to exponential series: Ti = 2i−1. Our model is implemented in python with Theano [1] and Keras [4]. The experiments are performed on a PC endowed with an Intel Xeon E5-1650 CPU and an NVIDIA Quadro K4000 GPU.
Table 1.
The network architecture. Dense represents the fully connected layer applied individually to every time step. SCR represents spatial CW-RNN, where the arrow indicates the sweeping direction. The Inputs row specifies the layer ID of each layer’s inputs. Layer 7 takes the concatenation of the output from layer 3,4,5 and 6 as input.
| Layer ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| Layer | Input | Dense | SCR↘ | SCR↙ | SCR↗ | SCR↖ | Dense | Dense |
| Size | 300 | 100 | 384 | 384 | 384 | 384 | 100 | 100 |
| Inputs | – | 1 | 2 | 2 | 2 | 2 | [3,4,5,6] | 7 |
Evaluation metrics
Denote mij as the number of pixels of class i labeled as class j, ti = Σj mij as the number of pixels of class i. The following metrics are computed (IU represents region intersection over union):
Mean accuracy (MA): (1/2) Σi mii/ti.
Average IU (AIU): (1/2) Σi (mii/(ti + Σj mji − mii)).
Weighted IU (WIU): (1/Σi ti) Σi(timii/ti + Σj mji − mii)).
Precision (P), recall (R) and F1 score.
Comparison with Other Works
We compare our method with several variations of other deep learning based frameworks, e.g., multi-layer perception (MLP), convolutional neural network (CNN). The detailed performance comparison are given in Table 2. SCW-RNN(4) and SCW-RNN(8) denote the proposed method for 4 and 8 connected neighborhood, respectively. CNN-nips is the famous architecture utilized in [5] to segment neuronal membranes, which consists of 4 convolutional layers and 4 max-pooling layers followed by two fully connected layer. This network uses a large input window size (95 × 95) to capture the context information. We also compare our method with U-NET [11], an end-to-end CNN architecture. To demonstrate our method’s capability of handling spatial context information, a plain MLP network that shares similar architecture to our model, denoted as MLP-10 are considered for comparison as well. We also try a larger window size (48 × 48) for MLP network, denoted as MLP-48.
Table 2.
The quantitative comparative results of muscle perimysium segmentation results. T represents the average running time (measured in second).
| P | R | F1 score | MA | AIU | WIU | T | |
|---|---|---|---|---|---|---|---|
|
| |||||||
| MLP-10 | 0.768 | 0.803 | 0.776 | 0.883 | 0.787 | 0.447 | 7.36 |
| MLP-48 | 0.805 | 0.82 | 0.804 | 0.897 | 0.811 | 0.453 | 22.14 |
| U-NET [11] | 0.764 | 0.792 | 0.761 | 0.869 | 0.774 | 0.442 | 1.7 |
| CNN-nips [5] | 0.834 | 0.855 | 0.84 | 0.916 | 0.841 | 0.463 | 319.8 |
|
| |||||||
| SCW-RNN(4) | 0.854 | 0.843 | 0.842 | 0.909 | 0.842 | 0.462 | 2.6 |
| SCW-RNN(8) | 0.836 | 0.866 | 0.845 | 0.918 | 0.844 | 0.462 | 3.6 |
As we show in Table 2, both versions of the proposed method, SCW-RNN(4) and SCW-RNN(8), achieve the best overall performance compared with others. It is obvious that the utilization of more spatial context information in SCW-RNN(8) leads to performance improvement than SCW-RNN(4), especially in terms of recall and F1 score. MLP-10, which does not consider such spatial context information across local patches, produces a lot of false positive evidenced by the low precision and F1 score. MLP-48, which has a larger receptive field outperforms MLP-10 with a large margin. CNN-nips, which uses a really large window size (95×95), achieves comparative results as ours, but its running time is almost 100 times slower than our method. Although for certain architecture, fast scanning can be utilized to remove redundant computations of convolution operation, it is not applicable to our case, which conducts patch-wise normalization. U-NET [11], which does not invoke patch based testing, is also very efficient, but it produces a much lower F1 score and AIU than ours, one of the possible reasons is that we do not apply aggressive data augmentation in all of our experimental settings.
For quantitative comparison, some challenging images with segmentation results overlaid on the original image are shown in Figure 2. It can be observed that our method produces the most accurate results with much better global consistency. This further provides evidences that our proposed spatial CW-RNN has strong capability to learn the global context information, which is the key to differentiate perimysium from endomysium, epimysium, and blood vessels.
Fig. 2.
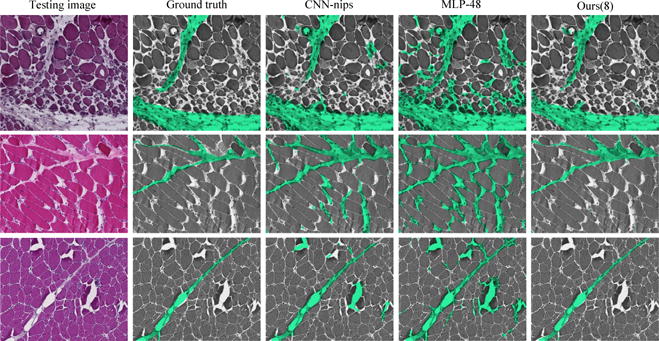
Perymisum segmentation results on three challenging skeleton muscle images which show strong global structure and demonstrates a lot of appearance similarity between perimysium (true positive) and endo/epimysium (false positive). Comparing with other methods, our results show much better global consistency because it can capture global spatial configurations.
4 Conclusion
In this paper, we propose a formulation of the novel 2D spatial clock-work recurrent neural network and pave the way to utilize RNN architecture to process 2D biomedical image data. Our spatial CW-RNN is totally end-to-end trainable and capable of encoding the global context information into the features of each local image patch, which tremendously improves the performance. In addition, we utilize the structured output for each local image patch, making it efficient for both training and testing.
References
- 1.Bergstra J, Breuleux O, Bastien F, Lamblin P, Pascanu R, Desjardins G, Turian J, Warde-Farley D, Bengio Y. Theano: a CPU and GPU math expression compiler. SciPy. 2010 Jun; [Google Scholar]
- 2.Byeon W, Breuel TM, Raue F, Liwicki M. Scene labeling with LSTM recurrent neural networks. CVPR. 2015:3547–3555. [Google Scholar]
- 3.Cho K, van Merrienboer B, Bahdanau D, Bengio Y. On the properties of neural machine translation: Encoder-decoder approaches. 2014 vol. abs/1409.1259. [Google Scholar]
- 4.Chollet F. keras. GitHub. 2015 https://github.com/fchollet/keras.
- 5.Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J. Deep neural networks segment neuronal membranes in electron microscopy images. NIPS. 2012:2852–2860. [Google Scholar]
- 6.Dalakas MC, Hohlfeld R. Polymyositis and dermatomyositis. 2003;362:971–982. doi: 10.1016/S0140-6736(03)14368-1. [DOI] [PubMed] [Google Scholar]
- 7.Graves A, Fernández S, Schmidhuber J. Multi-dimensional recurrent neural networks. ICANN. 2007:549–558. [Google Scholar]
- 8.Hochreiter S, Schmidhuber J. Long short-term memory. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 9.Koutník J, Greff K, Gomez F, Schmidhuber J. A clockwork rnn. ICML. 2014;32:1863–1871. [Google Scholar]
- 10.Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. 1998;86:2278–2324. [Google Scholar]
- 11.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. MICCAI. 2015:234–241. [Google Scholar]
- 12.Visin F, Kastner K, Courville AC, Bengio Y, Matteucci M, Cho K. Reseg: A recurrent neural network for object segmentation. 2015 vol. abs/1511.07053. [Google Scholar]
- 13.Werbos PJ. Backpropagation through time: What it does and how to do it. 1990;78:1550–1560. [Google Scholar]
- 14.Xie Y, Xing F, Kong X, Su H, Yang L. Beyond classification: Structured regression for robust cell detection using convolutional neural network. MICCAI. 2015;9351:358–365. doi: 10.1007/978-3-319-24574-4_43. [DOI] [PMC free article] [PubMed] [Google Scholar]


