Abstract
Liver metastasis is the primary cause of death for colorectal cancer (CRC) patients. To investigate the prognostic value of long non-coding RNAs (lncRNAs) on colorectal liver metastases, quantitative reverse-transcriptase PCR (quantitative RT-PCR) was performed on 15 lncRNAs in 51 stage IV CRC with liver metastases and 57 stage I/II CRC specimens. The expression levels of four lncRNAs (GAS5, H19, MEG3 and Yiya) were significantly different between liver metastases and primary tumors of stage IV CRC patients. Furthermore, the high expression levels of GAS5 and Yiya were significantly associated with future occurrence of liver metastases in early stage CRC patients. Kaplan-Meier analysis showed that the high expression levels of GAS5 or Yiya were correlated with poor prognosis of early stage CRC patients (p = 0.0206 and 0.0005 for GAS5 and Yiya, respectively). Yiya expression was proved to be an independent prognostic indicator of colorectal liver metastases in a multivariate analysis (relative risk = 10.7; p < 0.0001). Our study revealed that GAS5 and Yiya were promising prognostic biomarkers of liver metastases for early stage CRC patients.
Keywords: prognostic biomarker, long non-coding RNA, early stage colorectal cancer, liver metastasis, quantitative RT-PCR
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer worldwide, with an estimated 1.4 million cases and 693,900 deaths occurring in 2012 [1]. Metastatic disease is the primary cause of death for CRC patients [2]. According to official statistics, the five-year survival rate for localized CRC patients is up to 90% [3], while it is only 6% for stage IV CRC with distant metastases [4]. Liver is the most common site of metastatic lesions due to the portal drainage. Up to 20% of CRC patients have concurrent hepatic metastases and the five-year cumulative hepatic metastases rate is 14.5% [5, 6].
Clinically, survival time of early stage CRC patients without lymph node or distant metastases varies widely after surgical resections. Most patients do not present hepatic metastases within five years while the others show metastatic deposits in liver soon. This observation emphasizes the clinical and molecular heterogeneity in CRC [4]. Therefore, finding out the factors involved in this heterogeneity will assist clinical intervention and treatment against liver metastases for early stage CRC patients.
Long non-coding RNAs (lncRNAs), typically >200bp, are nature prognostic biomarkers in indicating the intrinsic characteristics of cancer metastasis since they are the effector molecules [7]. A growing body of evidence suggests that lncRNAs are associated with carcinogenesis, tumor metastasis and therapy [8–12]. In addition, clinical application of lncRNAs in cancer prognosis has also made inspiring progress [7, 13]. Nonetheless, little is known about the correlation between lncRNAs and colorectal liver metastases. Besides, whether lncRNAs are involved in the poor prognosis of CRC patients without metastasis has not been investigated yet. To fill up this gap, we evaluated the prognostic values of lncRNAs on liver metastases for patients with early stage CRC.
As lncRNA expression spectrums are often overlapped across different types of cancer, lncRNAs associated with at least two types of cancer are likely to be involved in colorectal liver metastases. Therefore, we collected 15 such lncRNAs (CCAT1, GAS5, H19, HOTAIR, IGF2-AS, lncRNA-LET, MALAT1, MEG3, MIR17HG, p15AS, PANDAR, PVT1, UCA1, XIST and Yiya) from public databases and literature [8, 11, 14–27]. Using quantitative RT-PCR with three non-overlapping short amplicons [28], we measured the expression levels of the 15 collected lncRNAs in stage IV CRC specimens together with liver metastases and identified four candidates for predicting colorectal liver metastases, which were GAS5, H19, MEG3 and Yiya. Furthermore, we validated these candidates by comparing their expression levels between stage I/II CRC patients who presented liver metastases within five years and who did not. Our results revealed that GAS5 and Yiya were promising prognostic biomarkers of liver metastases for patients with early stage CRC.
RESULTS
Quantification correlation of short amplicons in 15 lncRNAs
Quantification correlation of three non-overlapping short amplicons for each of the 15 collected lncRNAs and β-actin gene was determined in 51 stage IV CRC specimens and corresponding liver metastases (Supplementary Table S1). A strong correlation amongst three amplicons for every lncRNA was achieved in CRC and liver metastases (Spearman rank correlation coefficient >0.70). However, HOTAIR and PANDAR were excluded from this study due to the low expression levels (Cq value >35 in more than 20% samples). Finally, 13 lncRNAs were left for the analysis of association with colorectal liver metastases.
Association of 13 lncRNAs with colorectal liver metastases
Expression levels of 13 lncRNAs were compared between primary stage IV CRC and matched liver metastases in 51 pairs of samples (Table 1). Paired samples were from the same patient to eliminate individual differences. The expression levels of GAS5, H19 and Yiya were up-regulated in liver metastases compared with primary CRC (fold change = 0.4, 1.0 and 0.6, respectively; all p < 0.0500), while MEG3 was down-regulated (fold change = −0.3; p < 0.0010). Scatter-plots showed the differential expressions of these four lncRNAs between primary CRC and liver metastases (Supplementary Figure S1). The areas under the receiver operating characteristic curve (AUC; represents discrimination accuracy) were 0.68, 0.64, 0.63 and 0.73 for GAS5, H19, Yiya and MEG3, respectively. Thus, these four lncRNAs were further assessed on their prognostic values on live metastases for early stage CRC patients.
Table 1. Expression profiles of 13 lncRNAs in stage IV CRC patients compared with matched liver metastases.
| Liver metastases / colorectal carcinoma | ||||
|---|---|---|---|---|
| Fold change | P value | AUC | 95% CI | |
| CCAT1 | 0.4 | 0.1004 | 0.59 | 0.49 - 0.69 |
| GAS5 | 0.4 | 0.0038 | 0.68 | 0.58 - 0.77 |
| H19 | 1.0 | 0.0314 | 0.64 | 0.54 - 0.73 |
| IGF2-AS | −0.5 | 0.3582 | 0.59 | 0.49 - 0.69 |
| lncRNA-LET | −0.1 | 0.5322 | 0.57 | 0.47 - 0.67 |
| MALAT1 | 0 | 0.7225 | 0.58 | 0.48 - 0.68 |
| MEG3 | −0.3 | 0.0003 | 0.73 | 0.64 - 0.82 |
| MIR17HG | −0.2 | 0.3072 | 0.57 | 0.46 - 0.67 |
| p15AS | 0.2 | 0.7194 | 0.58 | 0.48 - 0.68 |
| PVT1 | 0.2 | 0.2114 | 0.54 | 0.44 - 0.64 |
| UCA1 | 0.2 | 0.4989 | 0.52 | 0.42 - 0.62 |
| XIST | −0.6 | 0.3485 | 0.54 | 0.44 - 0.64 |
| Yiya | 0.6 | 0.0272 | 0.63 | 0.53 - 0.72 |
CRC, colorectal cancer
AUC, area under the receiver operating characteristic curve
CI, confidence interval
Association of four lncRNAs with liver metastases in early stage CRC
An independent cohort of 57 patients with stage I/II CRC was divided into two groups according to the presence (n = 21) or absence (n = 36) of liver metastases within five years after surgery (Table 2). Quantitative RT-PCR results of these two groups were shown in Table 3. Higher expression levels of the four lncRNAs were observed in the group presenting liver metastases. However, only the differential expressions of GAS5 and Yiya were statistically significant (fold change = 0.5 and 3.0 for GAS5 and Yiya, respectively; both p < 0.0500, Supplementary Figure S2). The AUC values were 0.65 and 0.70 for GAS5 and Yiya, respectively. The results indicated that the high expression levels of GAS5 and Yiya stimulated liver metastases in early stage CRC patients.
Table 2. Characteristics of study subjects.
| Variable | Selection phase Identification phase |
Validation phase | |
|---|---|---|---|
| Patient | 51 | 57 | |
| Gender | |||
| Male | 30 | 30 | |
| Female | 21 | 27 | |
| Age (year) | |||
| Median (range) | 61 (35-77) | 63 (32-81) | |
| Tissue type | |||
| Colon | 38 | 29 | |
| Rectum | 13 | 25 | |
| Colorectum | 0 | 3 | |
| Liver metastasis | 51 | 0 | |
| TNM stage | |||
| Tumor stage (T) | |||
| Tis | 0 | 0 | |
| T1 | 0 | 2 | |
| T2 | 3 | 18 | |
| T3 | 5 | 13 | |
| T4 | 43 | 24 | |
| Nodal status (N) | |||
| N0 | 15 | 57 | |
| N1 | 20 | 0 | |
| N2 | 16 | 0 | |
| Distant metastases (M) | |||
| M0 | 0 | 57 | |
| M1 | 51 | 0 | |
| Pathological stage | |||
| 0 | 0 | 0 | |
| I | 0 | 20 | |
| II | 0 | 37 | |
| III | 0 | 0 | |
| IV | 51 | 0 | |
| Histologic grade | |||
| I | 0 | 1 | |
| II | 29 | 42 | |
| III | 22 | 14 | |
| Liver metastases within five years | |||
| Presence | 51 | 21 | |
| Absence | 0 | 36 | |
Table 3. Expression profiles of candidate lncRNAs between stage I/II CRC patients with and without liver metastases.
| Presence / absence of liver metastases | ||||
|---|---|---|---|---|
| Fold change | P value | AUC | 95% CI | |
| GAS5 | 0.5 | 0.0486 | 0.65 | 0.51 - 0.77 |
| H19 | 0.4 | 0.4178 | 0.56 | 0.43 - 0.69 |
| MEG3 | 0.5 | 0.1276 | 0.62 | 0.48 - 0.74 |
| Yiya | 3.0 | 0.0103 | 0.70 | 0.56 - 0.81 |
CRC, colorectal cancer
AUC, area under the receiver operating characteristic curve
CI, confidence interval
Prognostic value of four lncRNAs in early stage CRC
Figure 1 shows the Kaplan-Meier curves of liver metastases for 57 patients with early stage CRC classified according to lncRNAs expression levels. Remarkably, patients with high GAS5 or Yiya expression level had a high risk of liver metastases (p = 0.0206 and 0.0005 for GAS5 and Yiya, respectively). However, no significant correlation was found between the risk of liver metastases and the expression levels of H19 and MEG3.
Figure 1. Kaplan-Meier curves of liver metastases on four candidate lncRNAs in patients with stage I/II CRC.
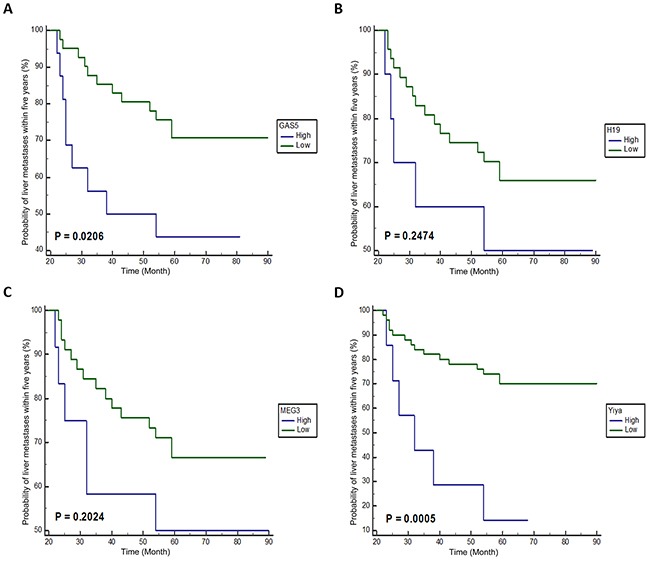
A-D. Kaplan-Meier curve of GAS5, H19, MEG3 and Yiya, respectively. High, high expression level ≥ mean 2ΔCq; Low, low expression level < mean 2ΔCq.
In the univariate analysis, three other clinicopathological characteristics (age at diagnosis, depth and histologic grade of primary tumor, Table 4) were found significantly associated with colorectal liver metastases. Among all these factors, Cox proportional-hazards regression analysis showed that the expression level of Yiya was an independent indicator of liver metastases for early stage patients (relative risk = 10.7; p < 0.0001).
Table 4. Candidate lncRNAs and clinicopathological factors for predicting liver metastases in patients with stage I/II CRC.
| Factors | Subset | Univariate | Multivariate | ||
|---|---|---|---|---|---|
| P value | Relative risk | 95% CI | P value | ||
| Age (year) | ≤ 50 / > 50 | 0.0399 | 0.3 | 0.10 - 0.86 | 0.0257 |
| Gender | Male / Female | 0.3334 | - | - | - |
| Organ | Colon / Rectum / C&R | 0.1455 | - | - | - |
| Deptha | T1 / T2 / T3 / T4 | 0.0137 | 2.2 | 1.16 - 4.29 | 0.0173 |
| LVI | Positive / Negative | 0.6198 | - | - | - |
| Histologic grade | I / II / III | 0.0317 | 2.6 | 0.95 - 7.24 | 0.0644 |
| GAS5 | High / Low | 0.0206 | 3.3 | 0.77 - 14.55 | 0.1105 |
| H19 | High / Low | 0.2474 | - | - | - |
| MEG3 | High / Low | 0.2024 | - | - | - |
| Yiya | High / Low | 0.0005 | 10.7 | 3.40 - 33.52 | 0.0001 |
According to Tumor-Node-Metastasis (TNM) Classification of Colorectal Carcinoma
High, high expression level ≥ mean 2ΔCq
Low, low expression level < mean 2ΔCq
CRC, colorectal cancer
C, colon; R, rectum; LVI, lymphatic vessel invasion; CI, confidence interval
DISCUSSION
Liver metastasis is the primary cause of death for CRC patients [2]. A favorable prognosis should be expected for early stage individuals without lymph node and distant metastases. Still, 26% - 40% of CRC patients in stage I and II develop distant metastases and finally died of it within five years after surgery [3, 4]. Therefore, finding out predictive biomarkers of live metastases is meaningful to improve the prognosis and reduce the mortality of CRC patients.
Colorectal carcinomas tend to be enriched for the subclones that are adept at survival, growth, invasion and metastasis during progression [29]. Thus, compared with primary CRC, the metastatic deposits theoretically contain higher proportion of subclones with strong invasion and metastases ability. By comparing the expression levels of 15 lncRNAs between stage IV CRC and corresponding liver metastases, we identified four potential biomarkers, which are GAS5, H19, MEG3 and Yiya. We further validated that the high expression level of GAS5 or Yiya was highly correlated with the poor prognosis of early stage CRC patients in a cohort of 57 stage I/II samples. Finally, Yiya was proved to be an independent prognostic biomarker of colorectal liver metastases.
So far, investigations on prognostic biomarkers for CRC liver metastases have been reported in previous studies. MicroRNA-214 was identified as a negative regulator of colorectal liver metastases by regulating fibroblast growth factor receptor 1 (FGFR1) expression [30]. Serum microRNA-29a expression was reported to be higher in CRC with liver metastases than that without liver metastases, which has strong potential as a novel noninvasive biomarker for early detection of colorectal liver metastases [31]. With regard to lncRNA, high expression level of HOTAIR was revealed to be correlated with the presence of liver metastasis in CRC [32].
To our best knowledge, we are the first to report the prognostic values of GAS5 and Yiya in early prediction of colorectal liver metastases. Previously, the well-documented GAS5 was generally considered as a tumor suppressor. Down-regulation of GAS5 was found in multiple cancers, including CRC, breast cancer and hepatocellular carcinoma [20, 33, 34]. GAS5 expression was also suggested to be an indicator of overall survival in CRC and hepatocellular carcinoma [33, 34]. However, how GAS5 was involved in colorectal liver metastases has not been fully understood. Little is known about Yiya as it was discovered recently. Identified in a cancer susceptibility region, Yiya was found to be overexpressed in breast, hepatocellular, ovary, and esophageal cancers [15]. Interestingly, the study also suggested that there located a transcription factor Prospero-related homeobox 1 (PROX1) downstream of Yiya, which might be involved in cancer metastases.
Further functional studies of GAS5 and Yiya will enrich our knowledge to understand the underlying mechanisms of colorectal liver metastases. On the other hand, as colorectal liver metastases generally depend on portal drainage, GAS5 and Yiya have a good chance to be correlated with hematogenous spread. As such, the identification of these two lncRNAs would confer great benefit on patients with other types of cancers which tend to metastasize via hematogenous pathway.
In conclusion, our study reveals that GAS5 and Yiya are novel prognostic biomarkers to predict the risk of liver metastases for early stage CRC patients. These two lncRNAs have considerable clinical values in the early prediction and timely clinical intervention of CRC liver metastases.
MATERIALS AND METHODS
Ethics statement
Investigation has been conducted in accordance with the ethical standards and according to the Declaration of Helsinki and according to national and international guidelines and has been approved by the Institutional Review Board of Shanghai Medical College in Fudan University, with written informed consent obtained from all patients.
Clinical specimens and study design
FFPE resection specimens of 51 stage IV colorectal carcinomas with matched liver metastases and 57 stage I/II colorectal carcinomas were collected from Zhongshan and Huashan Hospitals in Fudan University between March 2006 and July 2013. Routine histological classification according to the WHO criteria [35] was used to screen CRC samples. All cases were diagnosed by two pathologists and independently reviewed by an expert CRC pathologist. Patients who received preoperative radiotherapy or chemotherapy were excluded. Furthermore, stage I/II CRC patients who presented liver metastases within 12 months after surgery were also excluded. The clinical characteristics of patients and tumors in the study are presented in Table 2.
Our experiments mainly consisted of three different phases (Figure 2). In Selection phase, 15 lncRNAs associated with at least two types of cancer were collected from public databases and literature (Supplementary Table S2). Quantitative RT-PCR with three non-overlapping short amplicons [28] for each RNA was performed in 51 pairs of FFPE surgical tissues from stage IV CRC and matched liver metastases. Thirteen lncRNAs passing the quality control (Supplementary Table S1) were chosen for further analysis.
Figure 2. Study design.
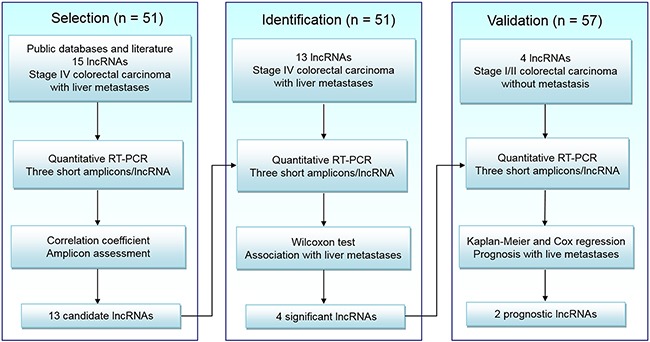
LncRNA, long non-coding RNA; RT-PCR, reverse transcriptase polymerase chain reaction.
In Identification phase, quantitative RT-PCR was performed on the chosen 13 lncRNAs in the same 51 pairs of specimens. Four candidate lncRNAs were found correlated with colorectal liver metastases.
In Validation phase, based on follow-up data, an independent cohort of 57 stage I/II CRC patients was divided into two groups: 21 patients present liver metastases within five years and 36 do not. Quantitative RT-PCR was performed between two groups to assess the prognostic value of four candidate lncRNAs on colorectal liver metastases.
Macrodissection
Macrodissection was necessary as described in our previous studies [28, 36, 37]. Hematoxylin-Eosin (H&E)-stained sections for each FFPE tissue block were prepared and reviewed by two expert CRC pathologists. If the proportion of neoplastic cells was larger than 75%, the corresponding sample was deemed suitable for experiments without purification.
If, however, the proportion is less than 75%, neoplastic cells area would be marked on the H&E sections as reference. The same area of corresponding sample was isolated with block trimming method or target tissue dissection method depending on the distribution of marked area.
RNA isolation
Total RNA of manually macrodissected FFPE samples was isolated using RecoverAll Total Nucleic Acid Isolation Kit as instructed (Ambion, Austin, Texas, USA). DNase digestion was performed on the nucleic acid samples to eliminate the genomic DNA contamination before the final extraction of RNA. The concentration of RNA was measured by the NanoDrop 2000 Spectrophotometers (Thermo Fisher Scientific, Waltham, MA, USA). RNA samples were removed from this study, if the OD 260/280 ratio was less than 1.8.
Quantitative RT-PCR
Three non-overlapping short amplicons (~ 60bp) were designed for each of 15 lncRNAs and β-actin gene in accordance with the general principles. The specificity of primer sets was assured by BLAST using the human genomic plus transcript database (Human G+T) and verified via melting curves obtained from quantitative RT-PCR. The expression of β-actin mRNA was used as an endogenous control. The primers were synthetized and purified by Sangon Biotech (Shanghai, China). Supplementary Table S3 presents the primer sequences and the amplification efficiency.
Short amplicons are more efficient and more sensitive than long amplicons in quantitative RT-PCR. And three of such short amplicons can address the problem that random fragmentation of lncRNA to different extent in different tissue types, which gives the assurance of quantification accuracy and reliability [28].
Following the guidelines on the minimal information for publication of quantitative real-time PCR experiments (MIQE) [38], quantitative RT-PCR was performed on 7900HT Fast Real-Time PCR System (Applied Biosystems) with High Capacity cDNA Reverse Transcription Kit (Invitrogen, Foster City, California, USA) and Power SYBR Green PCR Master Mix (Applied Biosystems, Warrington, UK). For reverse transcription, 500 ng of total RNA sample was reverse transcripted into 50 μl of cDNA solution with random primers. For real-time PCR, 6 μl of the cDNA solution was amplified with 16 μl 2x SYBR Green PCR Master mix and 2 μl target-specific primers (5 μM/L) in a final volume of 32 μl. All assays were carried out in triplicate. The Cq values were determined during 40 cycles of amplifications.
Statistical analysis
The delta Cq (ΔCq) value was used to represent the expression level of lncRNA in quantitative RT-PCR. For each amplicon designed, the ΔCq value was normalized using the equation: ΔCq = Cq (target) – Cq (β-actin). The mean normalized ΔCq value of three short amplicons was also calculated.
Spearman rank correlation coefficient was used to measure the correlation of three non-overlapping short amplicons. The correlation was considered to be strong when the coefficient was between 0.70 and 1.00. The expression levels of lncRNA between stage IV CRC specimens and matched liver metastases were compared by Wilcoxon test for paired samples. Mann-Whitney unpaired test was used to evaluate the difference of lncRNA expression levels between stage I/II CRC patients presenting liver metastases within five years or not. Receiver operating characteristic (ROC) curve analysis was performed to assess the predictive performance of target lncRNAs. The area under the curve (AUC) with 95% confidence interval (CI) was used as an accuracy index of the prediction. The average expression level of each lncRNA was used as the cutoff. Low group of lncRNA expression was classified as values below the corresponding cutoff, while high group was classified as values at or above the corresponding cutoff. Kaplan-Meier methods are performed between low and high groups of four lncRNAs to analyze the correlation between lncRNAs expression levels and prognosis on liver metastases of stage I/II CRC patients. A Cox proportional hazard model was used for multivariate analysis.
MedCalc software (version 10.4.7.0; MedCalc, Mariakerke, Belgium) was used for statistical analysis. All p values were two-tailed and the difference was considered to be significant when p value was less than 0.0500.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
Acknowledgments
We thank Tengfang Zhu, Qi Chen for technical assistance and Jinyu Xu for critical revision of manuscript.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interests.
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 3.National Cancer Institute: Surveillance, Epidemiology, and End Results (SEER 18) Program. SEER Stat Fact Sheets: Colon and Rectum Cancer. 2015 Mar; http://seer.cancer.gov/statfacts/html/colorect.html
- 4.Turner JR, Lingen MW. Kumar V, Abbas AK, Aster JC. Robbins Basic Pathology. Philadelphia, PA: ELSEVIER SAUNDERS; 2013. Oral Cavity and Gastrointestinal Track; pp. 596–600. [Google Scholar]
- 5.Siriwardena AK, Mason JM, Mullamitha S, Hancock HC, Jegatheeswaran S. Management of colorectal cancer presenting with synchronous liver metastases. Nat Rev Clin Oncol. 2014;11:446–59. doi: 10.1038/nrclinonc.2014.90. [DOI] [PubMed] [Google Scholar]
- 6.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244:254–9. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2012;26:155–65. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 8.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G, Zhang H, Wan X, Yang X, Zhu C, Wang A, He L, Miao R, Chen S, Zhao H. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014:780521. doi: 10.1155/2014/780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Im JH, Muschel RJ. New evidence of lncRNA role in tumor progression and metastasis. Hepatobiliary Surg Nutr. 2012;1:55–6. doi: 10.3978/j.issn.2304-3881.2012.10.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutschner T, Hämmerle M, Eißmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Groß M. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, Laurent GS, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase expression. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–69. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He S, Yang S, Deng G, Liu M, Zhu H, Zhang W, Yan S, Quan L, Bai J, Xu N. Aurora kinase A induces miR-17-92 cluster through regulation of E2F1 transcription factor. Cell Mol Life Sci. 2010;67:2069–76. doi: 10.1007/s00018-010-0340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F, Yi F, Zheng Z, Ling Z, Ding J, Guo J, Mao W, Wang X, Wang X, Ding X, Liang Z, Du Q. Characterization of a carcinogenesis-associated long non-coding RNA. RNA Biol. 2012;9:110–6. doi: 10.4161/rna.9.1.18332. [DOI] [PubMed] [Google Scholar]
- 16.Kam Y, Rubinstein A, Naik S, Djavsarov I, Halle D, Ariel I, Gure AO, Stojadinovic A, Pan H, Tsivin V, Nissan A, Yavin E. Detection of a long non-coding RNA (CCAT1) in living cells and human adenocarcinoma of colon tissues using FIT-PNA molecular beacons. Cancer Lett. 2014;352:90–6. doi: 10.1016/j.canlet.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 17.Yang JM, Chen WS, Liu ZP, Luo YH, Liu WW. Effects of insulin-like growth factors-IR and -IIR antisense gene transfection on the biological behaviors of SMMC-7721 human hepatoma cells. J Gastroenterol Hepatol. 2003;18:296–301. doi: 10.1046/j.1440-1746.2003.02961.x. [DOI] [PubMed] [Google Scholar]
- 18.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Wang Y, Kong B, Langerod A, et al. Extensive and coordinated transcription of noncoding RNAs within cell cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 21.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–80. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Zhang P, Wang L, Piao H-l, Ma L. Long non-coding RNA HOTAIR in carcinogenesis and metastasis. Acta Biochim Biophys Sin. 2014;46:1–5. doi: 10.1093/abbs/gmt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shtivelman E, Bishop JM. The PVT gene frequently amplifies with MYC in tumor cells. Mol Cell Biol. 1989;9:1148–54. doi: 10.1128/mcb.9.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, Zhang Y, Xin DQ, Na YQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851–8. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 26.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–96. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Vincent-Salomon A, Ganem-Elbaz C, Manie E, Raynal V, Sastre-Garau X, Stoppa-Lyonnet D, Stern MH, Heard E. X inactive-specific transcript RNA coating and genetic instability of the X chromosome in BRCA1 breast tumors. Cancer Res. 2007;67:5134–40. doi: 10.1158/0008-5472.CAN-07-0465. [DOI] [PubMed] [Google Scholar]
- 28.Kong H, Zhu M, Cui F, Wang S, Gao X, Lu S, Wu Y, Zhu H. Quantitative assessment of short amplicons in FFPE-derived long-chain RNA. Sci Rep. 2014;4:7246. doi: 10.1038/srep07246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar V, Abbas AK, Aster JC. Kumar V, Abbas AK, Aster JC. Robbins Basic Pathology. Philadelphia, PA: ELSEVIER SAUNDERS; 2013. Neoplasia; p. 177. [Google Scholar]
- 30.Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, Ren C, Wang FH, Chiao LJ, Pelicano H, et al. Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology. 2014;60:598–609. doi: 10.1002/hep.27118. [DOI] [PubMed] [Google Scholar]
- 31.Wang L-g, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 36:e61–e7. doi: 10.1016/j.canep.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 33.Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303–9. [PMC free article] [PubMed] [Google Scholar]
- 34.Yin D, He X, Zhang E, Kong R, De W, Zhang Z. Long noncoding RNA GAS5 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31:1–8. doi: 10.1007/s12032-014-0253-8. [DOI] [PubMed] [Google Scholar]
- 35.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO Classification of Tumours of the Digestive System. Lyon: IARC Press; 2010. [Google Scholar]
- 36.Wang S, Wang L, Bayaxi N, Li J, Verhaegh W, Janevski A, Varadan V, Ren Y, Merkle D, Meng X. A microRNA panel to discriminate carcinomas from high-grade intraepithelial neoplasms in colonoscopy biopsy tissue. Gut. 2013;62:280–9. doi: 10.1136/gutjnl-2011-301554. [DOI] [PubMed] [Google Scholar]
- 37.Huang W, Hu J, Yang DW, Fan XT, Jin Y, Hou YY, Wang JP, Yuan YF, Tan YS, Zhu XZ, Bai CX, Wu Y, Zhu HG, et al. Two microRNA panels to discriminate three subtypes of lung carcinoma in bronchial brushing specimens. Am J Respir Crit Care Med. 2012;186:1160–7. doi: 10.1164/rccm.201203-0534OC. [DOI] [PubMed] [Google Scholar]
- 38.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


