Abstract
In the past decades, new methods for tumor staging, restaging, treatment response monitoring, and recurrence detection of a variety of cancers have emerged in conjunction with the state-of-the-art positron emission tomography with 18F-fluorodeoxyglucose ([18F]-FDG PET). 13C magnetic resonance spectroscopic imaging (13CMRSI) is a minimally invasive imaging method that enables the monitoring of metabolism in vivo and in real time. As with any other method based on 13C nuclear magnetic resonance (NMR), it faces the challenge of low thermal polarization and a subsequent low signal-to-noise ratio due to the relatively low gyromagnetic ratio of 13C and its low natural abundance in biological samples. By overcoming these limitations, dynamic nuclear polarization (DNP) with subsequent sample dissolution has recently enabled commonly used NMR and magnetic resonance imaging (MRI) systems to measure, study, and image key metabolic pathways in various biological systems. A particularly interesting and promising molecule used in 13CMRSI is [1-13C]pyruvate, which, in the last ten years, has been widely used for in vitro, preclinical, and, more recently, clinical studies to investigate the cellular energy metabolism in cancer and other diseases. In this article, we outline the technique of dissolution DNP using a 3.35 T preclinical DNP hyperpolarizer and demonstrate its usage in in vitro studies. A similar protocol for hyperpolarization may be applied for the most part in in vivo studies as well. To do so, we used lactate dehydrogenase (LDH) and catalyzed the metabolic reaction of [1-13C]pyruvate to [1-13C]lactate in a prostate carcinoma cell line, PC3, in vitro using 13CMRSI.
Keywords: Cancer Research, Issue 118, hyperpolarization, DNP, 13C, NMR, magnetic resonance spectroscopy, magnetic resonance imaging, MRI, in vitro, LDH, pyruvate, lactate, prostatic carcinoma
Introduction
Presently, the most widely used clinical method for tumor staging, restaging, treatment response monitoring, and recurrence detection of a wide variety of cancers is [18F]-FDG PET.1 However, recently, several novel and alternative approaches have emerged. One of those methods is 13CMRSI. This technique involves the introduction of the 13C-molecule into a biological sample, followed by minimally invasive MRI to assess the metabolism in vitro or in vivo in real time. Nevertheless, the biggest challenge of 13CMRSI, compared to the other methods such as [18F]-FDG PET or computed tomography, is its low signal-to-noise ratio.
The NMR signal is directly proportional to the level of polarization, a ratio of the spin ½ nuclei population difference in two energy states to the total population (Figure 1A). The polarization is a product of the gyromagnetic ratio (γ) of the nuclei and the applied magnetic field strength over the temperature. A typical polarization of 1H nuclei is in the order of 0.001% to 0.005% at 3 T, which gives a relatively poor signal-to-noise ratio. Today's state-of-the-art MRI has been a successful imaging method only due to the high abundance of 1H in biological samples and the high gyromagnetic ratio of 1H (γ1H = 42.576 MHz/T). However, observing other nuclei, such as carbon, is more demanding. The only stable, magnetically active carbon isotope, 13C, makes up only 1.1% of all carbon atoms. In addition, the gyromagnetic ratio of 13C (γ13C = 10.705 MHz/T) is four times lower than that of 1H, leading to a lower detection efficiency. In summary, the low 13C abundance and low γ13C cause thermal 13C measurements to achieve 0.0176% of the sensitivity of a 1H-NMR measurement in vivo.
Dynamic Nuclear Polarization
A method to overcome the relatively poor sensitivity of 13C measurements is DNP. It was originally described for metals in 1953 by Albert W. Overhauser. In his article, he stated: "It is shown that if the electron spin resonance of the conduction electrons is saturated, the nuclei will be polarized to the same degree they would be if their gyromagnetic ratio were that of the electron spin."2 Later that year, Carver and Slichter experimentally confirmed Overhauser's hypothesis3. In 1958, Abragam and Proctor described this effect for electrons in liquids and named it the "solid effect." At temperatures below 4 K, electron-spin polarization reaches nearly 100% and is more than three orders of magnitude higher than the nuclear-spin polarization (Figure 1B)4. This occurs because the gyromagnetic ratio of the electron (γe = 28024.944 MHz/T) is three orders of magnitude higher than the nuclear gyromagnetic ratios. The weak interactions between electrons and nuclei, such as the Overhauser effect, the solid effect, the cross effect, and the thermal mixing effect, allow the transfer of polarization from electron spins to nuclear spins using microwave irradiation with a frequency close to the corresponding electron paramagnetic resonance (EPR) frequency5,6. DNP theory has been further developed to involve more electrons and thermal mixing. Nevertheless, to date, no unified quantitative theoretical description of DNP has been published7,8.
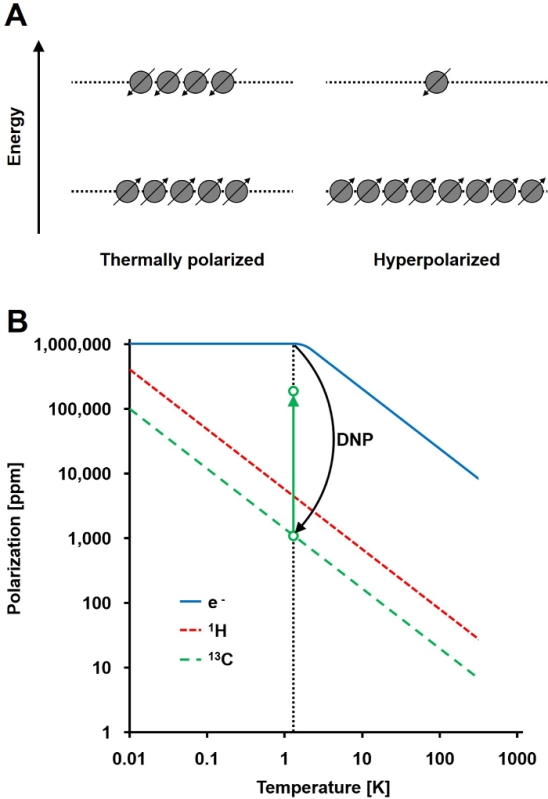 Figure 1: Understanding Dynamic Nuclear Polarization and Hyperpolarization. A) A schematic comparison of the spin population in the thermal equilibrium polarization state and the hyperpolarized state. B) The polarization is dependent upon temperature. The polarization of an electron (e-) reaches 100% below 1.4 K. The DNP allows the transfer of the polarization from the e- to the 13C nuclei, which increases their polarization up to 105-fold. Please click here to view a larger version of this figure.
Figure 1: Understanding Dynamic Nuclear Polarization and Hyperpolarization. A) A schematic comparison of the spin population in the thermal equilibrium polarization state and the hyperpolarized state. B) The polarization is dependent upon temperature. The polarization of an electron (e-) reaches 100% below 1.4 K. The DNP allows the transfer of the polarization from the e- to the 13C nuclei, which increases their polarization up to 105-fold. Please click here to view a larger version of this figure.
To introduce DNP in studies of biological systems using 13C NMR, subsequent rapid sample dissolution had to be developed. 50 years after Overhauser's hypothesis, Jan H. Ardenkjaer-Larsen et al. solved the technically challenging issue of bringing the hyperpolarized frozen sample into the liquid state with minimal hyperpolarization loss6. Dissolution DNP opened a new field of research called 13CMRSI, providing a new method to investigate and characterize various disease states9,10. As stable carriers of an unpaired electron, a trityl radical tris (8-carboxy-2,2,6,6-tetra-(hydroxyethyl)-benzo-[1,2-4,5]-bis-(1,3)-dithiole-4-yl)-methyl sodium salt (OX063) or (2,2,6,6-Tetramethylpiperidin-1-yl)oxyl (TEMPO) is usually used. These are mixed with the desired 13C-labeled molecule and exposed to microwave irradiation with a frequency close to the corresponding EPR frequency. Using this technique, the polarization of 13C nuclei can be increased up to 37%11. This results in a 105-fold polarization enhancement compared to the thermal equilibrium polarization11,12. However, as soon as the microwave irradiation is stopped and/or the 13C-molecule is transferred to the liquid state, the polarization decays with the longitudinal relaxation time (T1) of the 13C nucleus that was polarized. Thus, the invention of fast dissolution techniques or any subsequent technique shortening the time before experimental measurement (i.e., injection) is crucial for biological applications13.
There are three major requirements that the candidate molecule needs to fulfill for successful 13CMRSI studies. First, the 13C nucleus of interest has to have a sufficiently long T1 (> 10 s). The choice of the 13C-label is crucial. The best candidate nuclei are carbons with no direct contact with 1H-nuclei via a bond. It also needs to be rapidly metabolized within 2 - 3 T1 times, resulting in a downstream metabolic product with a significantly different chemical shift from the original substance. The sample mixture must also form an amorphous glass when in a solid state so that the spatial distribution decreases the distance between the electron and 13C, allowing the transfer of polarization. If the candidate molecule does not form amorphous glass naturally, it needs to be highly soluble in a glassing agent, such as glycerol or dimethyl sulfoxide14. These requirements result in a relatively small number of candidate molecules. However, even after the successful discovery of a suitable molecule, developing a working protocol for hyperpolarization can be technically challenging9,14,15.
In recent years, several substrates have been successfully polarized, such as [1-13C]pyruvate12,16-36, [2-13C]pyruvate37, [1-13C]ethyl pyruvate38, [1-13C]lactate39, [1-13C]fumarate40-43, 13C-bicarbonate36,44,45, [1-13C]sodium acetate43,46-49, 13C-urea6,36,50,51, [5-13C]glutamine15,52,53, [1-13C]glutamate53,54, [1-13C]2-oxoglutarate55, [1-13C]alanine, and others14,56. A particularly interesting and commonly used substrate for hyperpolarization is [1-13C]pyruvate. It is widely used in preclinical studies to investigate the cellular energy-metabolism in various diseases14,17,22. [1-13C]pyruvate meets all the requirements for successful hyperpolarization, including a relatively long T1 and rapid transport across the cell membrane before subsequently being metabolized. Preclinical studies with [1-13C]pyruvate are currently being translated into the clinic57.
Metabolism of Pyruvate
It is well known that there is a direct link between mutations in a cancer cells' DNA and changes in their metabolic pathways. Already in the 1920s, Otto Warburg discovered that there is an increased metabolism of glucose and production of lactate in tumors compared to healthy tissue58-60. Subsequently, various alternations in other metabolic pathways, such as the pentose-phosphate pathway, the tricarboxylic acid cycle, oxidative phosphorylation, and the synthesis of nucleotides and lipids, have been described.
Pyruvate is the final product of glycolysis. In the tumor, it undergoes anaerobic glycolysis catalyzed by LDH61 and reacts with the reduced form of the coenzyme nicotinamide adenine dinucleotide(NADH), resulting in lactate and the oxidized form of the coenzyme (NAD+). Alternatively, pyruvate undergoes a transamination reaction with glutamate to form alanine, catalyzed by alanine transaminase (ALT). Both reactions are readily reversible. Pyruvate also undergoes decarboxylation catalyzed by pyruvate dehydrogenase (PDH) to carbon dioxide and acetyl-CoA, representing an irreversible reaction at this step. Alternations in these reaction rates can be linked to tumor metabolism17,21,22,25,62. The metabolic pathways are summarized in Figure 2.
 Figure 2: Diagram of the major metabolic reaction of pyruvate. Pyruvate/lactate conversion is catalyzed by LDH, and pyruvate/alanine conversion is catalyzed by ALT. Pyruvate is irreversibly converted to acetyl-CoA and CO2 by PDH, and CO2 is in a pH-dependent equilibrium with bicarbonate80. Please click here to view a larger version of this figure.
Figure 2: Diagram of the major metabolic reaction of pyruvate. Pyruvate/lactate conversion is catalyzed by LDH, and pyruvate/alanine conversion is catalyzed by ALT. Pyruvate is irreversibly converted to acetyl-CoA and CO2 by PDH, and CO2 is in a pH-dependent equilibrium with bicarbonate80. Please click here to view a larger version of this figure.
The detection of hyperpolarized [1-13C]pyruvate and its metabolites has been previously demonstrated in the rat heart37,63-65, liver66, muscle, and kidney62,67. One study demonstrated significant differences in the lactate-to-alanine ratio between the normal and fasted rat liver66 and demonstrated a highly elevated and hyperpolarized [1-13C]lactate level in liver cancer68,69. There is evidence that the tumor grade can be identified in a transgenic adenocarcinoma of mouse prostate (TRAMP) using hyperpolarized [1-13C]pyruvate22, with the hyperpolarized lactate levels showing a high correlation with the histological grade of the excised tumors. The alanine catalyzed from pyruvate by ALT has also been suggested as a useful marker in rat hepatocellular carcinoma23.
Measuring the pyruvate-lactate metabolic flux has been used for monitoring ischemia63,65,70 and as a response to treatment with cytotoxic chemotherapy17,40, targeted drugs24,25,41, or radiotherapy26 in animal models. It has also been used for the detection of the phosphatidylinositol 3-kinase (PI3K) inhibitor LY294002 response in glioblastoma and breast cancer mouse models25. Changes in pyruvate metabolism in brain tumors26 and prostate cancer24,71 have also been observed after treatment.
Prostate Carcinoma
Prostate carcinoma is the predominant cancer in elderly men and the second leading cancer related to death in men worldwide72. To date, no reliable, non-invasive methods are available for an early diagnosis and characterization of prostate cancer73,74, emphasizing the urgent need for novel metabolic imaging techniques to enable stringent detection and staging of patients. Prostate carcinoma was used as a model to demonstrate the possibilities of dissolution DNP combined with 13CMRSI in patients57. This work was continued in a first clinical trial employing [1-13C]pyruvate and 13CMRSI for the imaging of prostate cancer, and it has just recently has been completed (NCT01229618).
The motivation behind this work was to illustrate in more detail and for a wider audience the application of the 13CMRSI method in a preclinical setting with cells. Measuring the LDH-catalyzed metabolism of [1-13C]pyruvate to [1-13C]lactate in vitro in the PC3 prostate carcinoma cell line, we demonstrate the possible application of dissolution DNP in in vitro studies and address the crucial steps and challenges during experiments.
Protocol
1. Sample Stock Solution Preparation
Add gadoterate meglumine (GadM, 0.5 mol/L) to concentrated [1-13C]pyruvic acid to give a final concentration of 1-mmol/L GadM. Add trityl radical tris (8-carboxy-2,2,6,6-tetra-(hydroxyethyl)-benzo-[1,2-4,5]-bis-(1,3)-dithiole-4-yl)-methyl sodium salt (OX063) to this mixture to give a final concentration of 15 mmol/L. Vortex until complete dissolution. NOTE: This stock solution preparation is designed for usage with a 3.35-T preclinical DNP hyperpolarizer. When a 7-T clinical hyperpolarizer is used, the gadoterate meglumine is not required because, at a higher magnetic field, its benefits are negligible. The addition of a gadolinium-based contrast agent increases the achievable solid-state polarization and also the polarization rate. However, in the liquid state, the contrast agent shortens the T1 relaxation time.
2. Growing the Cell Culture
Grow PC3 cells in a culture flask with a 125-cm2 growth area. Use F-12K medium containing 10% fetal calf serum (FCS) and maintain the cells at 37 °C in a humidified atmosphere at 5% CO2. Before the dissolution step, remove the medium from the culture flask. NOTE: Each cell line requires a particular preparation protocol for cell propagation. Consult the requirements with the cell line provider.
3. Preparation of the Cells for the Experiment
Remove the cell medium and wash the cells with ~ 10 mL of phosphate-buffered saline (PBS).
Add 5 mL of trypsin to the flask and return the cell culture flasks to the incubator for 3 to 5 min.
Add ~ 5 mL of F-12K medium to deactivate the trypsin.
Count the cells using an automatic cell counter. Mix 10 µL of the cell solution with 10 µL of the stain solution. Mix well with the pipette and transfer 10 µL of the mixture into the chamber of a "counting glass".
Remove and count the cells in the flask(s). Transfer the appropriate volumes containing the desired number of cells (e.g., 5 x 106 up to 108) into plastic vials.
Centrifuge the cells at 1,200 x g for 5 min and discard the supernatant.
Re-suspend the cells in the F-12K medium containing 10% FCS to a total volume of 800 μL and transfer them into a reaction cup (2 mL). Place the reaction cup into a plastic vial filled with warm water.
4. Dissolution Agent Preparation
NOTE: The dissolution agent is a liquid that is used to dissolve the hyperpolarized sample. In biological applications, dissolution is usually performed with H2O-based or deuterium oxide (D2O)-based buffers, such as PBS or tris(hydroxymethyl)aminomethane (Tris), containing 1 g/L ethylenediaminetetraacetic acid (EDTA).
- Preparation of 20 mmol/L PBS buffer
- To prepare 100 mL of the dissolution agent, dissolve 36 mg of monosodium phosphate (NaH2PO4), 247 mg of disodium phosphate (Na2HPO4), and 10 mg of EDTA in a solution of 20 mmol/L sodium hydroxide (NaOH) in D2O. Mix properly until complete dissolution. NOTE: EDTA (1 g/L) is added to the buffer to eliminate possible ferromagnetic ions, which can spoil the hyperpolarization. The NaOH is used to neutralize the pyruvic acid in a 1:1 mol ratio to reach a pH of 7.4.
5. Variable Temperature Insert (VTI) Cooldown
In the DNP-NMR polarizer program main window, click on "Cooldown." NOTE: This switches on the vacuum pump and evacuates the VTI to approximately 5.0 mbar. Subsequently, the needle valve between the VTI and the liquid helium reservoir fully opens, allowing liquid helium to flow into the VTI. The flow rate is regulated by the needle valve to maintain the optimal amount of liquid helium in the VTI until it reaches the helium boiling temperature. Then, the VTI is evacuated to almost complete vacuum, and the temperature reaches approximately 1.4 K. The VTI is filled with liquid helium up to 65%. At this point, the instrument is ready for sample insertion.
6. Sample Preparation and Insertion
Using a micropipette, add ~ 8 µL of 13C-labeled sample stock solution into a plastic cup.
Attach the plastic cup to the insertion rod and initiate the sample insertion process by pressing "Insert sample" in the main program window. Select "Normal sample" and click "Continue." NOTE: During this process, the needle valve first closes to discontinue the flow of liquid helium into VTI, and the pressure in the VTI then increases. The sample holder inside the VTI is raised from the liquid helium, the inlet valve at the top of the VTI opens, and a gaseous helium flow is introduced from the inlet valve to prevent outside contamination by air moisture.
When prompted, push the insertion rod with the attached plastic cup down into the VTI. Make sure to reach the sample holder at the bottom of the VTI. Otherwise, the gaseous helium can push the sample out of the VTI.
Detach and remove the insertion rod.
Finish the procedure by clicking "Next" in the dialog window. The sample insertion procedure should not take longer than 10 s. NOTE: The inlet valve then closes, the gaseous helium flow is discontinued, the sample holder with the sample cup is submerged into liquid helium, and the needle valve is opened to allow the liquid helium to flow into the VTI. After 5 - 10 min, the VTI is cooled below 1.4 K, allowing all of the free electrons to be polarized.
Confirm that the plastic cup with the sample was introduced correctly into the VTI by checking that it is not attached to the insertion rod or pushed out from the VTI by helium gas. Then click "Finish."
7. Microwave Sweep (optional)
NOTE: A microwave sweep allows the determination of the optimal microwave frequency to maximize the hyperpolarization rate of the 13C nuclei in the target compound.
To measure the microwave sweep, start the RINMR program, type "HYPERSENSENMR," and click "Select Config" and "Do Microsweep."
To initiate the process, select the "calibrate" tab on the main program window.
Click "Generate" and choose the beginning and ending frequency (e.g., 94.100 GHz-94.200 GHz), the frequency step size (e.g., 20 MHz), the power (100 mW), and the time (60 s). Click "Continue," "Enable," and "Start." NOTE: With these settings, the hyperpolarizer first polarizes the sample for 60 s using a microwave frequency of 94.100 GHz and a power of 100 mW. Then, it applies a 90° radio-frequency (RF) pulse and acquires the hyperpolarized 13C signal using the built-in spectrometer. These steps are repeated for each step in specified frequency range. For subsequent hyperpolarization, choose the microwave frequency with the maximal signal amplitude measured.
8. Polarization
To measure the polarization build-up, start the RINMR program, type "HYPERSENSENMR," and click "Select Config" and "Solid Build-up."
In the DNP-NMR polarizer program main window, click "Polarization" to initiate the hyperpolarization process.
Choose the optimal microwave frequency (obtained during the microwave sweep) and the power (e.g., 100 mW) for the sample and click "Next."
Enable "Polarization build-up monitoring" and click "Finish."
Polarize the sample to > 95% (~ 60 min for [1-13C]pyruvate). NOTE: During the polarization, microwaves are guided into the VTI and to the sample, causing the 13C spins to align with the hyperpolarized unpaired electron spins. To measure the hyperpolarization buildup, RF pulses with a flip angle (FA) of 5° are applied periodically (e.g., every 300 s), and the resulting signal is plotted as a polarization build-up curve.
9. Dissolution
When the polarization reaches > 95%, initiate the dissolution process by clicking "Dissolution" in the DNP-NMR polarizer program main window.
Choose the dissolution process from the drop-down menu and click "Next." NOTE: The polarizer allows one to define the desired dissolution process by choosing the timing of the chasing gas.
Load ~ 5 mL of the dissolution agent through the top valve into a heated vessel in the dissolution part of the polarizer. Calculate the exact volume of the dissolution agent needed using following equation:
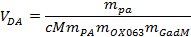 where VDA is the wanted volume of dissolution agent, mPA, mOX063, and mGad are the masses of the pyruvate, OX063 and gadoterate meglumine, respectively, added to the sample stock solution.
where VDA is the wanted volume of dissolution agent, mPA, mOX063, and mGad are the masses of the pyruvate, OX063 and gadoterate meglumine, respectively, added to the sample stock solution.Place the dissolution stick in the active position above the inlet valve. NOTE: This allows the instrument to connect its dissolution instrumentation to the sample cup in the VTI.
Click "Finish" to start the dissolution process. NOTE: The dissolution agent is pressurized to 3 bar by helium gas and is subsequently heated up to 200 °C, causing an increase in pressure. When the pressure reaches 10 bar, the needle valve closes to discontinue the flow of liquid helium into the VTI. The sample holder raises the cup from the liquid helium. The dissolution stick is lowered into the VTI and connected to the sample cup. The conditioned dissolution agent is pushed by the pressure, which results from the heating vessel containing the dissolution buffer and helium gas, through the dissolution stick to the cup, causing a rapid dissolution of the sample. The solution then flows out into the collection flask via plastic tubing. The dissolution stick with the attached cup is then raised from the VTI.
Move the dissolution stick with the attached cup to the "cleaning" position and finish the process by clicking "Finish."
10. Detection of the 13C Hyperpolarized Signal
- 13C metabolic magnetic resonance spectroscopy in vitro
- Mix 200 µL of the 20-mmol/L dissolved hyperpolarized sample from the collection flask with 800 µL of the cell solution. NOTE: The resulting final concentration of [1-13C]pyruvate is 4 mmol/L.
- Mix the suspension well using a micropipette and transfer ~ 600 µL into a 5-mm NMR tube.
- Insert the 5-mm NMR tube into the 1-T NMR spectrometer. In the main window of the software, click "Run" to start the measurement, applying series of one hundred 10° RF pulses every 3 s. NOTE: Measure the time between the initial mixing of the hyperpolarized sample with the cells and the start of the spectroscopic acquisition. Ensure that the mixing procedure does not exceed 30 s to minimize polarization loss.
- 13C metabolic magnetic resonance imaging
- To build a container for the in vitro experiments using the MRI spectrometer, take a 5-mL syringe and connect it to a catheter (d = 1.2 mm) that is long enough to reach from the spectrometer's iso-center to the approachable area of the spectrometer.
- Fill the in vitro container with the cell solution of the desired concentration for the experiment (e.g., 108) or with an enzymatic solution.
- Place an in vitro container at the isocenter of the MRI magnet. Place a 13C-tuned radio frequency receiver coil on the container. Place a concentrated 13C-labeled calibration phantom (e.g., 10-mol/L 13C-urea) nearby.
- Insert the "in vitro container" near the iso-center of the NMR scanner.
- Run the scanner's standard 3-plane localization sequence and adjust the in vitro container's position to the iso-center, as needed.
- Run a 1H T2-weighted "anatomical" sequence covering the in vitro container localization. Use the following settings: 2D spin echo with axial orientation, repetition time (TR) = 2,000 ms, echo time (TE) = 20 ms, slice thickness = 1 mm, field of view covering the in vitro container, and 16 echoes per excitation. Ensure that field shimming is done on protons during this step.
- In the anatomical images, select 5 contiguous slices centered on the region of interest. Prescribe a 13C spectroscopic calibration acquisition covering the selected anatomical slices. Use the following settings: 2D Block-Siegert calibration sequence with axial orientation 12 x 12 centric encoded, TR = 1,000 ms, slice thickness = 5 mm, field of view matching anatomical images, number of scans (NS) = 64, bandwidth = 5,000 Hz, and FA = 90°.
- Select the 13C spectroscopic calibration sequence (for more information, see Schulte et al. 2011)75 from the pulse sequence library. Download the pulse sequence to the scanner from the computer by clicking "Download." Click on "Spectra Prescan" to run the spectroscopic prescan. In the spectrum magnitude plot, adjust the peak from the 13C calibration phantom to the center of the scanner frequency. Set the receiver gains to the maximum. Click "Start" to run the 13C spectroscopic calibration sequence. Note the reported transmit gain and centric frequency.
- Set a 13C chemical shift imaging (CSI) acquisition covering the selected anatomical slices. Use the following settings: 2D echo-planar spectroscopic imaging (EPSI) with axial orientation 12 x 12 centric encoded, TR = 400 ms, slice thickness = 5 mm, field of view matching anatomical images, NS = 300, and bandwidth = 5,000 Hz. NOTE: EPSI samples a single line in k-space repeatedly after one RF excitation to acquire both spatial and spectral information simultaneously. For more information about the acquisition techniques, see the article by Durst et al. 201576.
- Download the 13C CSI sequence and run the spectroscopic prescan. Adjust the scanner frequency and transmit the gain as specified by the calibration sequence output.
- After the hyperpolarized solution is deposited in the collection flask, draw up ~ 3 mL into a syringe and then inject it into the catheter connected to the in vitro container. Start the acquisition. After the acquisition is complete, save the raw data file for subsequent reconstruction.
11. Data Reconstruction
- Apply one of the two described kinetic models to analyze the acquired data.
- In the first method for describing the LDH kinetics, kinetic value (k), compare the sum of the lactate signal (MLAC) to the signal of all hyperpolarized molecules (Mx)21,77.
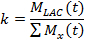
- In the other method, measure the lactate and pyruvate signals over time and fit these to a kinetic model17,25,71. To solve the metabolic exchange rate, kPA→LAC, and the effective signal decay rate of lactate, rLAC, use the following linear differential equations using the two-site exchange differential model, yielding for lactate:
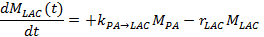
Note: The effective lactate signal decay rate rLAC is dependent upon the lactate longitudinal relaxation time (T1,LAC), the opposite metabolic exchange rate from lactate to pyruvate kLAC→PA, the applied FA and TR, and the signal intensity of pyruvate (MPA) and lactate (MLAC), taking into account the irreversible signal reduction after each successive excitation: ![]()
Therefore, rLAC results in a single, inseparable term of signal decay. Since it is possible to correct for the flip angle and the repetition time, and even though there is a flux LAC→PA, we assume that the exchange rate from lactate to pyruvate (kLAC→PA) does not need to be included in the calculation, based on the results of Harrison et al. 201278. Their results show that the kLAC→PA does not play as crucial a role as one would assume. This mode allows the T1 relaxation time of lactate to be quantified. This model is independent of pyruvate administration to the measurement, which, in the case of in vitro experiments, is not crucial and can be neglected. It does, however, play an important role for in vivo measurements79.
Representative Results
The results of the “microwave sweep” are illustrated in Figure 3. It shows that the optimal microwave frequency for the [1-13C]pyruvate sample is at 94.156 GHz for the local 3.35-T hyperpolarizer. All following hyperpolarization experiment (n = 14) were performed using this microwave frequency with a power of 100 mW. The microwave irradiation was applied for 60 to 80 min, leading to a solid-state hyperpolarization higher than 90%. The results are presented in Figure 4. The hyperpolarized [13C]pyruvate was mixed with 5 × 106 (n = 2), 107 (n = 2), 2 × 107 (n = 1), 3 × 107 (n = 2), 4 × 107 (n = 1), 6 × 107 (n = 2), 8 × 107 (n = 2), and 108 (n = 1) of the prostate cancer cell line PC3.
The resulting data are summarized in Figure 5 and Figure 6. Acquired data with spectral and temporal resolution are shown in Figure 5A-D and Figure 6A-D, with only a temporal resolution for each molecule observed (Figure 5E-H and Figure 6E-H), and with only a spectral resolution (Figure 5I-L and Figure 5I-L). We have observed three major hyperpolarized signals representing [1-13C]pyruvate, [1-13C]pyruvate hydrate, and [1-13C]lactate, with chemical shifts at 173 ppm, 181 ppm, and 185 ppm, approximately relative to the trimethylsilyl propanoic acid (TMSP) at pH 7.4 and temperature 20 °C. The signal ratios between the three metabolites are summarized in Table 1. The data show a clear correlation between the lactate signal and the number of cells present in the sample (Figure 7). However, the results from the experiments with less than 2 × 107 cells exhibit significant deviation, likely due to a low signal-to-noise ratio. Therefore, we suggest using more cells than this for further experiments. When the relative lactate signal (kinetic value) is normalized by the number of cells (Figure 8), it clearly demonstrates similar uptake and metabolism throughout all of the cells. However, there is a trend of decreasing lactate production per cell with an increasing number of cells. We believe that one of the causes of reduced cell metabolic activity is a very high concentration of cells in a very small volume, resulting in the increased viscosity of the sample. The results of the two-site exchange differential model are summarized in Table 2 and shown in Figure 9. The data follow a trend similar to the previous model: increasing kPA→LAC with an increasing number of cells. However, this model results in a steeper increase of the kinetics with the number of cells. When the metabolic exchange rate kPA→LAC is normalized to the number of cells, we can again see a clear trend of decreasing kPA→LAC with an increasing number of cells (Figure 10).
Figure 11 demonstrates the possibility of the addition of spatial localization to the experiment. It shows a phantom injected with 80 mmol/L hyperpolarized [1-13C]pyruvate next to a 10 mol/L 13C-urea phantom. The technique allows the attainment of a spectrum with temporal and special resolution (Figure 11A) or of the signal decay of the chosen metabolite signals in time (Figure 11B). The spectra in the time domain can also be summed to receive a better signal-to-noise ratio (Figure 11C). The special resolution allows the choice of the desired frequency region of the 13C spectrum belonging to certain metabolites, such as [1-13C]pyruvate (Figure 11D), [1-13C]pyruvate hydrate (Figure 11E), or reference 13C-urea (Figure 11F). It can be co-registered with a 1H image. The pulse sequence used (EPSI) allows the acquisition of an image of the whole slice every 4.9 s. In summary, this technique can provide data with a spatial, temporal, and spectral resolution for any metabolite.
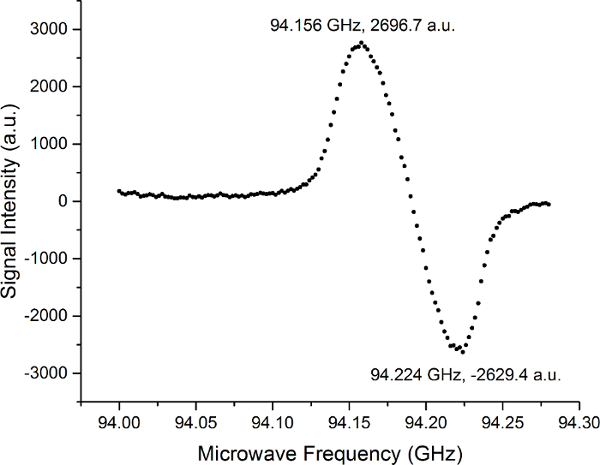 Figure 3: Results of a Microwave Sweep with [1-13C]pyruvate at the Local 3.35-T Hyperpolarizer. The result of the measurements determining the optimal microwave frequency to maximize the hyperpolarization rate of 13C nuclei in the target compound of [1-13C]pyruvate. The microwave sweep has a shape of an EPR absorption spectrum. The shape and separation of the peaks are based on the radical used (in this case, trityl radical), and the biggest influence have a solid effect and thermal mixing. Please click here to view a larger version of this figure.
Figure 3: Results of a Microwave Sweep with [1-13C]pyruvate at the Local 3.35-T Hyperpolarizer. The result of the measurements determining the optimal microwave frequency to maximize the hyperpolarization rate of 13C nuclei in the target compound of [1-13C]pyruvate. The microwave sweep has a shape of an EPR absorption spectrum. The shape and separation of the peaks are based on the radical used (in this case, trityl radical), and the biggest influence have a solid effect and thermal mixing. Please click here to view a larger version of this figure.
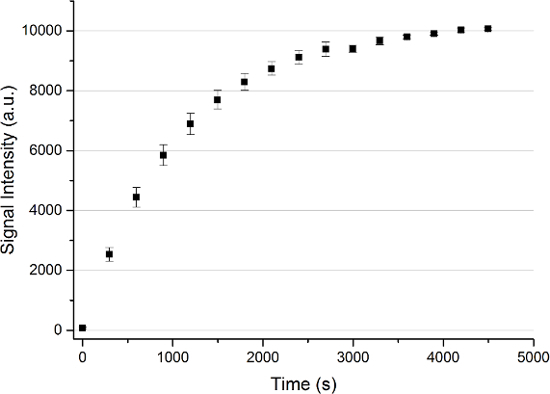 Figure 4: Solid State Polarization Buildup of a [1-13C]pyruvate Sample. An average of n = 13 solid-state polarization buildups with the error represented by the standard deviation measured every 300 s for up to 4,500 s. Please click here to view a larger version of this figure.
Figure 4: Solid State Polarization Buildup of a [1-13C]pyruvate Sample. An average of n = 13 solid-state polarization buildups with the error represented by the standard deviation measured every 300 s for up to 4,500 s. Please click here to view a larger version of this figure.
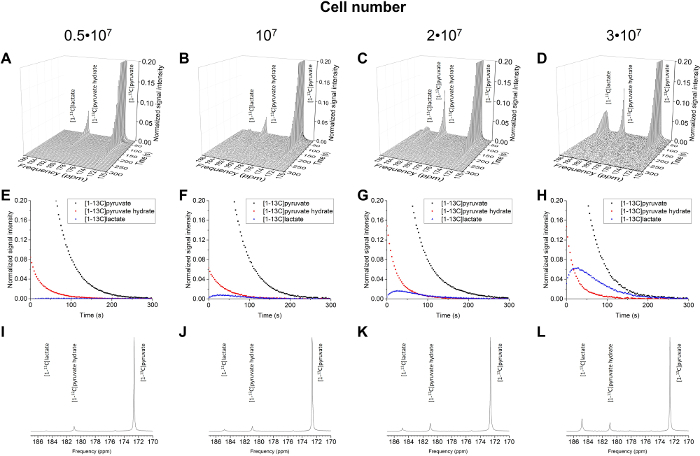 Figure 5: Results of the 13C NMR Spectroscopy for the Number of Cells (5 x 106 to 3 x 107 cells). The acquired data plotted with spectral and temporal resolution (A-D), plotted with temporal resolution only for [1-13C]pyruvate, [1-13C]pyruvate hydrate, and [1-13C]lactate (E-H), and plotted with spectral resolution only, summing all time steps (I-L). Please click here to view a larger version of this figure.
Figure 5: Results of the 13C NMR Spectroscopy for the Number of Cells (5 x 106 to 3 x 107 cells). The acquired data plotted with spectral and temporal resolution (A-D), plotted with temporal resolution only for [1-13C]pyruvate, [1-13C]pyruvate hydrate, and [1-13C]lactate (E-H), and plotted with spectral resolution only, summing all time steps (I-L). Please click here to view a larger version of this figure.
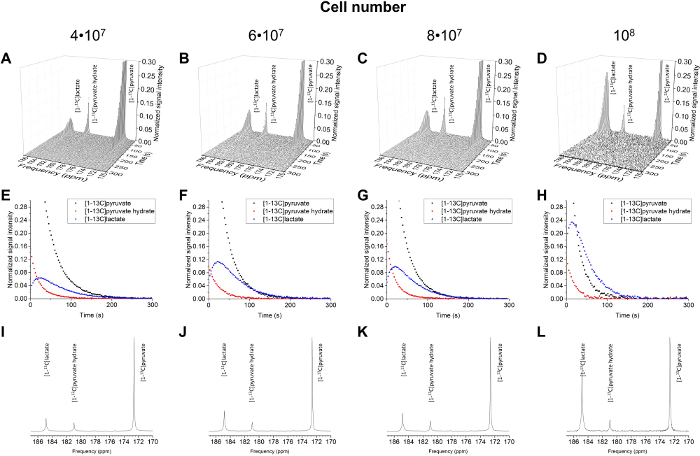 Figure 6: Results of the 13C NMR Spectroscopy for the Number of Cells (4 x 107 to 1 x 108 cells). The acquired data plotted with spectral and temporal resolution (A-D), plotted with temporal resolution only for [1-13C]pyruvate, [1-13C]pyruvate hydrate, and [1-13C]lactate (E-H), and plotted with spectral resolution only, summing all time steps (I-L). Please click here to view a larger version of this figure.
Figure 6: Results of the 13C NMR Spectroscopy for the Number of Cells (4 x 107 to 1 x 108 cells). The acquired data plotted with spectral and temporal resolution (A-D), plotted with temporal resolution only for [1-13C]pyruvate, [1-13C]pyruvate hydrate, and [1-13C]lactate (E-H), and plotted with spectral resolution only, summing all time steps (I-L). Please click here to view a larger version of this figure.
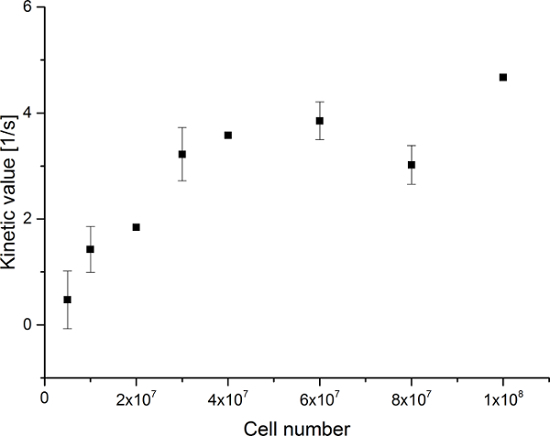 Figure 7: Results of the Simple Metabolite Ratio Kinetic Modeling. Data represents the ratio of the [1-13C]lactate signal to the sum of [1-13C]pyruvate, [1-13C]pyruvate hydrate, and [1-13C]lactate versus the number of cells in the experiments. The error represents the standard deviation. Please click here to view a larger version of this figure.
Figure 7: Results of the Simple Metabolite Ratio Kinetic Modeling. Data represents the ratio of the [1-13C]lactate signal to the sum of [1-13C]pyruvate, [1-13C]pyruvate hydrate, and [1-13C]lactate versus the number of cells in the experiments. The error represents the standard deviation. Please click here to view a larger version of this figure.
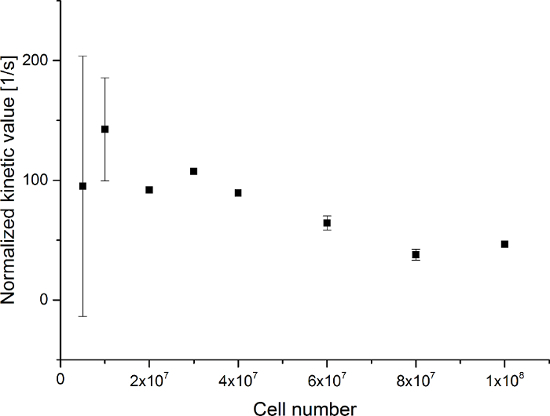 Figure 8: Results of the Simple Metabolite Ratio Kinetic Modeling Normalized to the Number of Cells. The data represent the ratio of the [1-13C]lactate signal to the sum of [1-13C]pyruvate, [1-13C]pyruvate hydrate, and [1-13C]lactate normalized to the number of cells versus the number of cells in the experiments. The error represents the standard deviation. Please click here to view a larger version of this figure.
Figure 8: Results of the Simple Metabolite Ratio Kinetic Modeling Normalized to the Number of Cells. The data represent the ratio of the [1-13C]lactate signal to the sum of [1-13C]pyruvate, [1-13C]pyruvate hydrate, and [1-13C]lactate normalized to the number of cells versus the number of cells in the experiments. The error represents the standard deviation. Please click here to view a larger version of this figure.
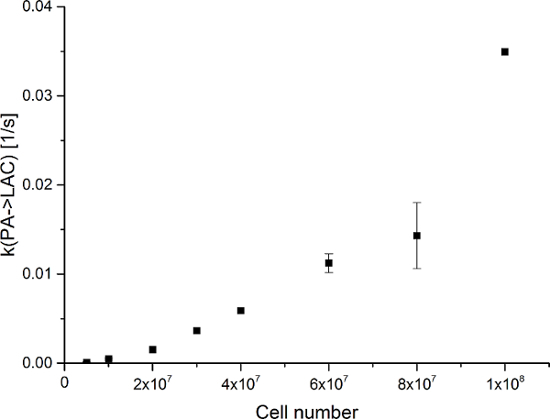 Figure 9: Results of the Two-site Exchange Differential Model. The data represent the metabolic exchange rate (kPA→LAC) versus the number of cells in the experiments. The error represents the standard deviation. Please click here to view a larger version of this figure.
Figure 9: Results of the Two-site Exchange Differential Model. The data represent the metabolic exchange rate (kPA→LAC) versus the number of cells in the experiments. The error represents the standard deviation. Please click here to view a larger version of this figure.
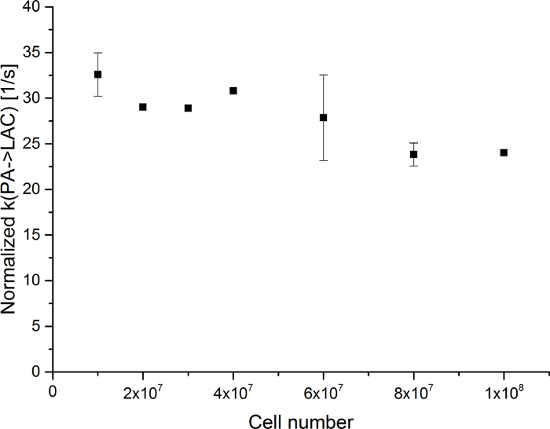 Figure 10: Results of the Two-site Exchange Differential ModelNormalized to the Number of Cells. The data represent the metabolic exchange rate (kPA→LAC) normalized to the number of cells versus the number of cells in the experiments. The error represents the standard deviation. Please click here to view a larger version of this figure.
Figure 10: Results of the Two-site Exchange Differential ModelNormalized to the Number of Cells. The data represent the metabolic exchange rate (kPA→LAC) normalized to the number of cells versus the number of cells in the experiments. The error represents the standard deviation. Please click here to view a larger version of this figure.
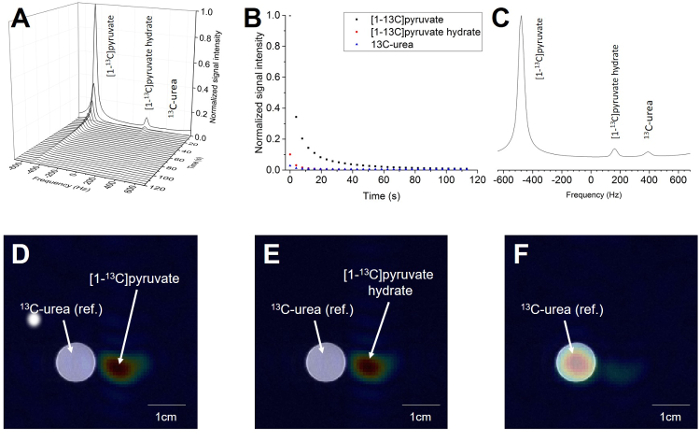 Figure 11: Result of Magnetic Resonance Imaging of the Hyperpolarized [1-13C]pyruvate Probe. A) The spectrum acquired over the whole slice and all time steps. B) The decay of the [1-13C]pyruvate and [1-13C]pyruvate hydrate signal over time. The third signal is the 10 M 13C-urea localization reference. C) The spectrum acquired from the whole spatial and temporal resolution. D) The 1H image overlaid with the 13C image of the summed [1-13C]pyruvate signal over all time steps. E) The 1H image overlaid with the 13C image of the summed [1-13C]pyruvate hydrate signal over all time steps. F) The 1H image overlaid with the 13C image of the summed 13C-urea signal over all time steps (reference). The 13C-signal in C-E is normalized to the maximum of the signal of the specific metabolite. Please click here to view a larger version of this figure.
Figure 11: Result of Magnetic Resonance Imaging of the Hyperpolarized [1-13C]pyruvate Probe. A) The spectrum acquired over the whole slice and all time steps. B) The decay of the [1-13C]pyruvate and [1-13C]pyruvate hydrate signal over time. The third signal is the 10 M 13C-urea localization reference. C) The spectrum acquired from the whole spatial and temporal resolution. D) The 1H image overlaid with the 13C image of the summed [1-13C]pyruvate signal over all time steps. E) The 1H image overlaid with the 13C image of the summed [1-13C]pyruvate hydrate signal over all time steps. F) The 1H image overlaid with the 13C image of the summed 13C-urea signal over all time steps (reference). The 13C-signal in C-E is normalized to the maximum of the signal of the specific metabolite. Please click here to view a larger version of this figure.
| Cell number | ||||||||
| 5×106 (n=2) | 107 (n=2) | 2×107 (n=1) | 3×107 (n=2) | 4×107 (n=1) | 6×107 (n=2) | 8×107 (n=2) | 108 (n=1) | |
| [1-13C] pyruvate | 92.9 ± 1.4 | 91.7 ± 1.0 | 86.7 | 77.5 ± 2.7 | 76 | 69.7 ± 0.5 | 65.9 ± 3.7 | 42.9 |
| [1-13C] pyruvate hydrate | 6.8 ± 1.2 | 6.7 ± 1.6 | 9.5 | 10.1 ± 1.8 | 8.9 | 7.7 ± 1.5 | 10.4 ± 0.2 | 13.4 |
| [1-13C] lactate | 0.3 ± 0.3 | 1.6 ± 0.6 | 3.8 | 12.4 ± 4.5 | 15.1 | 22.5 ± 1.1 | 23.7 ± 3.5 | 43.7 |
Table 1: The Relative Ratio of Hyperpolarized Metabolites with Respect to the Different Number of Cells.
| Cell number | ||||||||
| 5×106 (n=2) | 107 (n=2) | 2×107 (n=1) | 3×107 (n=1) | 4×107 (n=1) | 6×107 (n=2) | 8×107 (n=2) | 108 (n=1) | |
| kPA → LAC [*10-4] | 0.924±0.870 | 4.984±1.19 | 15.135 | 36.289 | 58.904 | 112.174±10.491 | 114.3±37.059 | 349.234 |
Table 2: Results of the Two-site Exchange Differential Model.
Discussion
13CMRSI with hyperpolarized probes is a promising method to monitor metabolism in real time in vitro and in vivo. One very important aspect when employing this experimental process is the proper standardization, especially regarding in vitro experiments. First, the preparation of the sample needs to be done properly and consistently to achieve the same concentration of hyperpolarized material in each experiment. This requires a precise weighing of both the sample to be hyperpolarized and the buffer. If the concentration is not correct, the final pH of the solution is not precise, which can have an influence on T1 and the cells' responses. It is also crucial to handle the cells as uniformly as possible. The cells should always be prepared in such a way that there is a minimal delay between cell harvest and the subsequent experiment in order to minimize the duration of time the cells are kept at a very high concentration and low volume. Variation in the cell preparation protocol, such as a different preparation times or temperatures, could result in substantial variations in the obtained data. The mixing of the sample with the cells should also be standardized. It is important to measure the time between the additions of the tracer to the cell suspension and the beginning of the measurement, because this can vary; during the data analysis, this should be considered.
The correct choice of the data analysis and kinetic modeling is crucial in the interpretation of the acquired data. The simple model is suitable for a linear one-way reaction with a constant exchange rate of two metabolites. As described in the introduction, pyruvate undergoes several enzymatic reactions and, more importantly, it also undergoes a non-enzymatic reversible-exchange reaction with pyruvate hydrate. This reaction played a crucial role in the experiments, and its effect is well demonstrated in the experiment with 8 × 107 cells. Although Table 1 indicates that the pyruvate hydrate relative concentration is similar to other experiments, when closely investigated in Figure 6D, it shows a much higher pyruvate hydrate signal at the beginning of the experiment compared to the other experiments. However, when the temporal resolution is summed up, this important information is lost and causes an error in the reconstruction of the data. On the other hand, the two-site exchange differential model is a more robust and precise description of the kinetics because it includes the temporal resolution in the calculation. Thus, it includes the non-enzymatic exchange with pyruvate hydrate, even if it rapidly exchanges with pyruvate during the measurement.
There are various imaging strategies to choose between to observe the hyperpolarized signal or to track the metabolism of a hyperpolarized molecule in preclinical and clinical studies. Durst et al. demonstrated the advantages and disadvantages of different pulse sequnces76. The free induction decay chemical shift imaging (FIDCSI) sequence is relatively robust but has limited use for multi-slice and temporally resolved imaging. Echo-planar spectroscopic imaging (EPSI) is robust for gradient issues and off-resonance effects but, it is prone to reconstruction artifacts. The iterative decomposition of water and fat with echo asymmetric and least-squares estimation (IDEAL)81, spiral chemical shift imaging (ISPCSI), pulse sequence35, and spiral chemical shift imaging (SPCSI) have high encoding efficiencies but are sensitive to B0 inhomogeneity. The choice of the sequence will depend on the scanner characteristics, the biological question, and the system being investigated.
There are many requirements that need to be fulfilled for successful hyperpolarization. However, there are also several limitations that the hyperpolarized 13CMRSI technique is nowadays facing. The primary and unchangeable limitation is the T1 relaxation time of the 13C nucleus in the molecule, which defines the amount of detectable signal available at the specific time of measurement. The signal is lowered by each RF excitation that causes a loss of the hyperpolarization signal repeatedly during data acquisition. Another limitation is the relatively long time period that is required to hyperpolarize a molecule. This typically takes from 30 to 90 min.
In comparison to other techniques of molecule imaging, such as [18F]-FDG PET, hyperpolarized 13CMRSI does not require tumors with increased glycolytic metabolic pathways and therefore, increased glucose consumption. The technique shows a real metabolic flux in real time. On the other hand, [18F]-FDG PET does not give direct information about metabolism but only indirect information about accumulation in the metabolically active area. This could cause a false negative result, where the tumor seems to be metabolically inactive but is actually using different metabolic pathways, such as glutaminolysis, as the carbon source for proliferation.
In conclusion, dissolution DNP can be used in a variety of applications to study an unlimited list of diseases (such as diabetes)82, measure pH15,36,45, or monitor metabolic changes in various types of cancer. These measurements can be accomplished on different levels, from in vitro cell experiments, through preclinical studies using animal models (such as mice, rats, rabbits, pigs, and dogs), to recent human clinical studies57. The future clinical applications will feature a very powerful and noninvasive diagnostic tool that could not only detect and localize the disease but also allow the observation of the treatment response in real time83.
Disclosures
Rolf F. Schulte and Marion I. Menzel are employed with GE Global Research.
Acknowledgments
E.K. gratefully acknowledges the support of the Graduate School of Bioengineering (GSB) at Technische Universität München. This work was supported by the German Research Foundation (DFG) within the SFB Collaborative Research Center 824, "Imaging for Selection, Monitoring, and Individualization of Cancer Therapies."
References
- Rohren EM, Turkington TG, Coleman RE. Clinical applications of PET in oncology. Radiology. 2004;231(2):305–332. doi: 10.1148/radiol.2312021185. [DOI] [PubMed] [Google Scholar]
- Overhauser AW. Polarization of Nuclei in Metals. Phys. Rev. 1953;92(2):411–415. [Google Scholar]
- Carver TR, Slichter CP. Polarization of Nuclear Spins in Metals. Phys. Rev. 1953;92(1):212–213. [Google Scholar]
- Abragam A, Proctor WG. Spin Temperature. Phys. Rev. 1958;109(5):1441–1458. [Google Scholar]
- Abragam a, Goldman M. Principles of dynamic nuclear polarisation. Reports Prog. Phys. 2001;41(3):395–467. [Google Scholar]
- Ardenkjaer-Larsen JH, Fridlund B, et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. U. S. A. 2003;100(18):10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimon D, Hovav Y, Feintuch A, Goldfarb D, Vega S. Dynamic nuclear polarization in the solid state: a transition between the cross effect and the solid effect. Phys. Chem. Chem. Phys. 2012;14(16):5729–5743. doi: 10.1039/c2cp23915a. [DOI] [PubMed] [Google Scholar]
- Serra SC, Rosso A, Tedoldi F. Electron and nuclear spin dynamics in the thermal mixing model of dynamic nuclear polarization. Phys. Chem. Chem. Phys. 2012;14(38):13299–13308. doi: 10.1039/c2cp41947e. [DOI] [PubMed] [Google Scholar]
- Gallagher FA, Kettunen MI, Brindle KM. Biomedical applications of hyperpolarized 13C magnetic resonance imaging. Prog. Nucl. Magn. Reson. Spectrosc. 2009;55(4):285–295. [Google Scholar]
- Hurd RE, Yen Y-F, Chen A, Ardenkjaer-Larsen JH. Hyperpolarized 13C metabolic imaging using dissolution dynamic nuclear polarization. J. Magn. Reson. Imaging. 2012;36(6):1314–1328. doi: 10.1002/jmri.23753. [DOI] [PubMed] [Google Scholar]
- Ardenkjaer-Larsen JH, Fridlund B, et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc. Natl. Acad. Sci. U. S. A. 2003;100(18):10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golman K, in't Zandt R, Thaning M. Real-time metabolic imaging. Proc. Natl. Acad. Sci. 2006;103(30):11270–11275. doi: 10.1073/pnas.0601319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comment A, Rentsch J, et al. Producing over 100 ml of highly concentrated hyperpolarized solution by means of dissolution DNP. J. Magn. Reson. 2008;194(1):152–155. doi: 10.1016/j.jmr.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindle KM, Bohndiek SE, Gallagher FA, Kettunen MI. Tumor imaging using hyperpolarized 13C magnetic resonance spectroscopy. Magn. Reson. Med. 2011;66(2):505–519. doi: 10.1002/mrm.22999. [DOI] [PubMed] [Google Scholar]
- Gallagher FA, Kettunen MI, Day SE, Lerche M, Brindle KM. 13C MR spectroscopy measurements of glutaminase activity in human hepatocellular carcinoma cells using hyperpolarized 13C-labeled glutamine. Magn. Reson. Med. 2008;60(2):253–257. doi: 10.1002/mrm.21650. [DOI] [PubMed] [Google Scholar]
- Chen AP, Albers MJ, et al. Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3T-initial experience. Magn. Reson. Med. 2007;58(6):1099–1106. doi: 10.1002/mrm.21256. [DOI] [PubMed] [Google Scholar]
- Day SE, Kettunen MI, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat. Med. 2007;13(11):1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- Schroeder MA, Swietach P, et al. Measuring intracellular pH in the heart using hyperpolarized carbon dioxide and bicarbonate: a 13C and 31P magnetic resonance spectroscopy study. Cardiovasc. Res. 2010;86(1):82–91. doi: 10.1093/cvr/cvp396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd RE, Yen Y-F, Tropp J, Pfefferbaum A, Spielman DM, Mayer D. Cerebral dynamics and metabolism of hyperpolarized [1-(13)C]pyruvate using time-resolved MR spectroscopic imaging. J. Cereb. Blood Flow Metab. 2010;30(13):1734–1741. doi: 10.1038/jcbfm.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golman K, Zandt RI, Lerche M, Pehrson R, Ardenkjaer-Larsen JH. Metabolic imaging by hyperpolarized 13C magnetic resonance imaging for in vivo tumor diagnosis. Cancer Res. 2006;66(22):10855–10860. doi: 10.1158/0008-5472.CAN-06-2564. [DOI] [PubMed] [Google Scholar]
- Park I, Larson PEZ, et al. Hyperpolarized 13C magnetic resonance metabolic imaging: application to brain tumors. Neuro. Oncol. 2010;12(2):133–144. doi: 10.1093/neuonc/nop043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers MJ, Bok R, et al. Hyperpolarized 13C lactate, pyruvate, and alanine: noninvasive biomarkers for prostate cancer detection and grading. Cancer Res. 2008;68(20):8607–8615. doi: 10.1158/0008-5472.CAN-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen Y-F, Le Roux P, et al. T(2) relaxation times of (13)C metabolites in a rat hepatocellular carcinoma model measured in vivo using (13)C-MRS of hyperpolarized [1-(13)C]pyruvate. NMR Biomed. 2010;23(4):414–423. doi: 10.1002/nbm.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafni H, Larson PEZ, et al. Hyperpolarized 13C spectroscopic imaging informs on hypoxia-inducible factor-1 and myc activity downstream of platelet-derived growth factor receptor. Cancer Res. 2010;70(19):7400–7410. doi: 10.1158/0008-5472.CAN-10-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward CS, Venkatesh HS, et al. Noninvasive detection of target modulation following phosphatidylinositol 3-kinase inhibition using hyperpolarized 13C magnetic resonance spectroscopy. Cancer Res. 2010;70(4):1296–1305. doi: 10.1158/0008-5472.CAN-09-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day SE, Kettunen MI, et al. Detecting response of rat C6 glioma tumors to radiotherapy using hyperpolarized [1- 13C]pyruvate and 13C magnetic resonance spectroscopic imaging. Magn. Reson. Med. 2011;65(2):557–563. doi: 10.1002/mrm.22698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson H, Macholl S, Ardenkjaer-Larsen JH. Dynamic Nuclear Polarization of [1-13C]pyruvic acid at 4.6 tesla. J. Magn. Reson. 2009;197(2):167–175. doi: 10.1016/j.jmr.2008.12.016. [DOI] [PubMed] [Google Scholar]
- Durst M, Koellisch U, et al. Bolus tracking for improved metabolic imaging of hyperpolarised compounds. J. Magn. Reson. 2014;243:40–46. doi: 10.1016/j.jmr.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Khegai O, Schulte RF, et al. Apparent rate constant mapping using hyperpolarized [1-(13)C]pyruvate. NMR Biomed. 2014;27(10):1256–1265. doi: 10.1002/nbm.3174. [DOI] [PubMed] [Google Scholar]
- Sogaard LV, Schilling F, Janich MA, Menzel MI, Ardenkjaer-Larsen JH. In vivo measurement of apparent diffusion coefficients of hyperpolarized (1)(3)C-labeled metabolites. NMR Biomed. 2014;27(5):561–569. doi: 10.1002/nbm.3093. [DOI] [PubMed] [Google Scholar]
- Aquaro GD, Frijia F, et al. 3D CMR mapping of metabolism by hyperpolarized 13C-pyruvate in ischemia-reperfusion. JACC. Cardiovasc. Imaging. 2013;6(6):743–744. doi: 10.1016/j.jcmg.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel MI, Farrell EV, et al. Multimodal assessment of in vivo metabolism with hyperpolarized [1-13C]MR spectroscopy and 18F-FDG PET imaging in hepatocellular carcinoma tumor-bearing rats. J. Nucl. Med. 2013;54(7):1113–1119. doi: 10.2967/jnumed.112.110825. [DOI] [PubMed] [Google Scholar]
- Schilling F, Duwel S, et al. Diffusion of hyperpolarized (13) C-metabolites in tumor cell spheroids using real-time NMR spectroscopy. NMR Biomed. 2013;26(5):557–568. doi: 10.1002/nbm.2892. [DOI] [PubMed] [Google Scholar]
- Schulte RF, Sperl JI, et al. Saturation-recovery metabolic-exchange rate imaging with hyperpolarized [1-13C] pyruvate using spectral-spatial excitation. Magn. Reson. Med. 2013;69(5):1209–1216. doi: 10.1002/mrm.24353. [DOI] [PubMed] [Google Scholar]
- Wiesinger F, Weidl E, et al. IDEAL spiral CSI for dynamic metabolic MR imaging of hyperpolarized [1-13C]pyruvate. Magn. Reson. Med. 2012;68(1):8–16. doi: 10.1002/mrm.23212. [DOI] [PubMed] [Google Scholar]
- Wilson DM, Keshari KR, et al. Multi-compound polarization by DNP allows simultaneous assessment of multiple enzymatic activities in vivo. J. Magn. Reson. 2010;205(1):141–147. doi: 10.1016/j.jmr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MA, Atherton HJ, et al. Real-time assessment of Krebs cycle metabolism using hyperpolarized 13C magnetic resonance spectroscopy. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2009;23(8):2529–2538. doi: 10.1096/fj.09-129171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd RE, Yen Y-F, et al. Metabolic imaging in the anesthetized rat brain using hyperpolarized [1-13C] pyruvate and [1-13C] ethyl pyruvate. Magn. Reson. Med. 2010;63(5):1137–1143. doi: 10.1002/mrm.22364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AP, Kurhanewicz J, et al. Feasibility of using hyperpolarized [1-13C]lactate as a substrate for in vivo metabolic 13C MRSI studies. Magn. Reson. Imaging. 2008;26(6):721–726. doi: 10.1016/j.mri.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witney TH, Kettunen MI, et al. Detecting treatment response in a model of human breast adenocarcinoma using hyperpolarised [1-(13)C]pyruvate and. Br. J. Cancer. 2010;103(9):1400–1406. doi: 10.1038/sj.bjc.6605945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohndiek SE, Kettunen MI, et al. Detecting tumor response to a vascular disrupting agent using hyperpolarized (13)C magnetic resonance spectroscopy. Mol. Cancer Ther. 2010;9(12):3278–3288. doi: 10.1158/1535-7163.MCT-10-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher FA, Kettunen MI, et al. Production of hyperpolarized [1,4-13C2]malate from [1,4-13C2]fumarate is a marker of cell necrosis and treatment response in tumors. Proc. Natl. Acad. Sci. U. S. A. 2009;106(47):19801–19806. doi: 10.1073/pnas.0911447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR, Peitersen T, et al. Tissue-specific short chain fatty acid metabolism and slow metabolic recovery after ischemia from hyperpolarized NMR in vivo. J. Biol. Chem. 2009;284(52):36077–36082. doi: 10.1074/jbc.M109.066407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher FA, Kettunen MI, et al. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature. 2008;453(7197):940–943. doi: 10.1038/nature07017. [DOI] [PubMed] [Google Scholar]
- Scholz DJ, Janich MA, et al. Quantified pH imaging with hyperpolarized 13C-bicarbonate. Magn. Reson. Med. 2015;73(6):2274–2282. doi: 10.1002/mrm.25357. [DOI] [PubMed] [Google Scholar]
- Koellisch U, Gringeri CV, et al. Metabolic imaging of hyperpolarized [1-(13) C]acetate and [1-(13) C]acetylcarnitine - investigation of the influence of dobutamine induced stress. Magn. Reson. Med. 2015;74(4):1011–1018. doi: 10.1002/mrm.25485. [DOI] [PubMed] [Google Scholar]
- Koellisch U, Laustsen C, et al. Investigation of metabolic changes in STZ-induced diabetic rats with hyperpolarized [1-13C]acetate. Physiol. Rep. 2015;3(8) doi: 10.14814/phy2.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR, Meier S, Ardenkjaer-Larsen JH, Duus JO, Karlsson M, Lerche MH. Detection of low-populated reaction intermediates with hyperpolarized NMR. Chem. Commun. 2009. pp. 5168–5170. [DOI] [PubMed]
- Koelsch BL, Keshari KR, Peeters TH, Larson PEZ, Wilson DM, Kurhanewicz J. Diffusion MR of hyperpolarized 13C molecules in solution. Analyst. 2013;138(4):1011–1014. doi: 10.1039/c2an36715g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golman K, Ardenkjaer-Larsen JH, Petersson JS, Mansson S, Leunbach I. Molecular imaging with endogenous substances. Proc. Natl. Acad. Sci. U. S. A. 2003;100(18):10435–10439. doi: 10.1073/pnas.1733836100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Morze C, Larson PEZ, et al. Imaging of Blood Flow Using Hyperpolarized [(13)C]Urea in Preclinical Cancer Models. J. Magn. Reson. Imaging. 2011;33(3):692–697. doi: 10.1002/jmri.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiavazza E, Kubala E, et al. Earth's magnetic field enabled scalar coupling relaxation of 13C nuclei bound to fast-relaxing quadrupolar 14N in amide groups. J. Magn. Reson. 2013;227:35–38. doi: 10.1016/j.jmr.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Karlsson M, Meier S, Duus J, Lerche MH. Hyperpolarized amino acids for in vivo assays of transaminase activity. Chem. - A Eur. J. 2009;15(39):10010–10012. doi: 10.1002/chem.200901042. [DOI] [PubMed] [Google Scholar]
- Gallagher FA, Kettunen MI, et al. Detection of tumor glutamate metabolism in vivo using 13C magnetic resonance spectroscopy and hyperpolarized [1-13C]glutamate. Magn. Reson. Med. 2011;66(1):18–23. doi: 10.1002/mrm.22851. [DOI] [PubMed] [Google Scholar]
- Chaumeil MM, Larson PEZ, et al. Hyperpolarized [1-13C] glutamate: a metabolic imaging biomarker of IDH1 mutational status in glioma. Cancer Res. 2014;74(16):4247–4257. doi: 10.1158/0008-5472.CAN-14-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshari KR, Wilson DM. Chemistry and biochemistry of 13C hyperpolarized magnetic resonance using dynamic nuclear polarization. Chem. Soc. Rev. 2014;43(5):1627–1659. doi: 10.1039/c3cs60124b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SJ, Kurhanewicz J, et al. Metabolic Imaging of Patients with Prostate Cancer Using Hyperpolarized [1-13C]Pyruvate. Sci. Transl. Med. 2013;5(198) doi: 10.1126/scitranslmed.3006070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Warburg O, Wind F, Negelein E. {Ü}ber den Stoffwechsel von Tumoren im K{ö}rper. Klin. Wochenschr. 1926;5(19):829–832. [Google Scholar]
- Barnes AB, De Paepe G, et al. High-Field Dynamic Nuclear Polarization for Solid and Solution. Biological NMR. Appl. Magn. Reson. 2008;34(3-4):237–263. doi: 10.1007/s00723-008-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Sivridis E, Gatter KC, Harris AL. Pyruvate dehydrogenase and pyruvate dehydrogenase kinase expression in non small cell lung cancer and tumor-associated stroma. Neoplasia. 2005;7(1):1–6. doi: 10.1593/neo.04373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golman K, Petersson JS. Metabolic Imaging and Other Applications of Hyperpolarized 13C1. Acad. Radiol. 2016;13(8):932–942. doi: 10.1016/j.acra.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Golman K, Petersson JS, et al. Cardiac metabolism measured noninvasively by hyperpolarized 13C MRI. Magn. Reson. Med. 2008;59(5):1005–1013. doi: 10.1002/mrm.21460. [DOI] [PubMed] [Google Scholar]
- Merritt ME, Harrison C, Storey C, Jeffrey FM, Sherry AD, Malloy CR. Hyperpolarized 13C allows a direct measure of flux through a single enzyme-catalyzed step by NMR. Proc. Natl. Acad. Sci. U. S. A. 2007;104(50):19773–19777. doi: 10.1073/pnas.0706235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt ME, Harrison C, Storey C, Sherry AD, Malloy CR. Inhibition of carbohydrate oxidation during the first minute of reperfusion after brief ischemia NMR detection of hyperpolarized 13CO2 and H13CO3- Magn. Reson. Med. 2008;60(5):1029–1036. doi: 10.1002/mrm.21760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Chen AP, et al. In vivo carbon-13 dynamic MRS and MRSI of normal and fasted rat liver with hyperpolarized 13C-pyruvate. Mol. imaging Biol. MIB Off. Publ. Acad. Mol. Imaging. 2009;11(6):399–407. doi: 10.1007/s11307-009-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler SJ, Yen Y, et al. In vivo 13 carbon metabolic imaging at 3T with hyperpolarized 13C-1-pyruvate. Magn. Reson. Med. 2007;58(1):65–69. doi: 10.1002/mrm.21253. [DOI] [PubMed] [Google Scholar]
- Hu S, Lustig M, et al. 3D compressed sensing for highly accelerated hyperpolarized (13)C MRSI with in vivo applications to transgenic mouse models of cancer. Magn. Reson. Med. 2010;63(2):312–321. doi: 10.1002/mrm.22233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurhanewicz J, Vigneron DB, et al. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 2011;13(2):81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MA, Cochlin LE, Heather LC, Clarke K, Radda GK, Tyler DJ. In vivo assessment of pyruvate dehydrogenase flux in the heart using hyperpolarized carbon-13 magnetic resonance. Proc. Natl. Acad. Sci. U. S. A. 2008;105(33):12051–12056. doi: 10.1073/pnas.0805953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut ML, Yen Y-F, et al. Kinetic modeling of hyperpolarized 13C1-pyruvate metabolism in normal rats and TRAMP mice. J. Magn. Reson. 2010;202(1):85–92. doi: 10.1016/j.jmr.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis LK, Resnick MI. Analysis of recent trends in prostate cancer incidence and mortality. Prostate. 2000;42(4):247–252. doi: 10.1002/(sici)1097-0045(20000301)42:4<247::aid-pros1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Jambor I, Borra R, et al. Functional imaging of localized prostate cancer aggressiveness using 11C-acetate PET/CT and 1H-MR spectroscopy. J. Nucl. Med. 2010;51(11):1676–1683. doi: 10.2967/jnumed.110.078667. [DOI] [PubMed] [Google Scholar]
- Presti JCJ, Hricak H, Narayan PA, Shinohara K, White S, Carroll PR. Local staging of prostatic carcinoma: comparison of transrectal sonography and endorectal MR imaging. AJR. Am. J. Roentgenol. 1996;166(1):103–108. doi: 10.2214/ajr.166.1.8571856. [DOI] [PubMed] [Google Scholar]
- Schulte RF, Sacolick L, et al. Transmit gain calibration for nonproton MR using the Bloch-Siegert shift. NMR Biomed. 2011;24(9):1068–1072. doi: 10.1002/nbm.1657. [DOI] [PubMed] [Google Scholar]
- Durst M, Koellisch U, et al. Comparison of acquisition schemes for hyperpolarised (1)(3)C imaging. NMR Biomed. 2015;28(6):715–725. doi: 10.1002/nbm.3301. [DOI] [PubMed] [Google Scholar]
- Janich MA, Menzel MI, et al. Effects of pyruvate dose on in vivo metabolism and quantification of hyperpolarized (1)(3)C spectra. NMR Biomed. 2012;25(1):142–151. doi: 10.1002/nbm.1726. [DOI] [PubMed] [Google Scholar]
- Harrison C, Yang C, et al. Comparison of kinetic models for analysis of pyruvate-to-lactate exchange by hyperpolarized 13 C NMR. NMR Biomed. 2012;25(11):1286–1294. doi: 10.1002/nbm.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez Damián PA, Sperl JI, et al. Multisite Kinetic Modeling of (13)C Metabolic MR Using [1-(13)C]Pyruvate. Radiol. Res. Pract. 2014;2014 doi: 10.1155/2014/871619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J-N, Gutman F, et al. PET/CT in patients with hepatocellular carcinoma using [(18)F]fluorocholine: preliminary comparison with [(18)F]FDG PET/CT. Eur. J. Nucl. Med. Mol. Imaging. 2006;33(11):1285–1289. doi: 10.1007/s00259-006-0164-9. [DOI] [PubMed] [Google Scholar]
- Reeder SB, Pineda AR, et al. Iterative decomposition of water and fat with echo asymmetry and least-squares estimation (IDEAL): application with fast spin-echo imaging. Magn. Reson. Med. 2005;54(3):636–644. doi: 10.1002/mrm.20624. [DOI] [PubMed] [Google Scholar]
- Laustsen C, Ostergaard JA, et al. Assessment of early diabetic renal changes with hyperpolarized [1-(13) C]pyruvate. Diabetes. Metab. Res. Rev. 2013;29(2):125–129. doi: 10.1002/dmrr.2370. [DOI] [PubMed] [Google Scholar]
- Serrao EM, Brindle KM. Potential Clinical Roles for Metabolic Imaging with Hyperpolarized [1-(13)C]Pyruvate. Front. Oncol. 2016;6:59. doi: 10.3389/fonc.2016.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]


