Abstract
MicroRNAs (miRNAs) are integral to the gene regulatory network. A single miRNA is capable of controlling the expression of hundreds of protein coding genes and modulate a wide spectrum of biological functions, such as proliferation, differentiation, stress responses, DNA repair, cell adhesion, motility, inflammation, cell survival, senescence and apoptosis, all of which are fundamental to tumorigenesis. Overexpression, genetic amplification and gain-of-function mutation of oncogenic miRNAs (“onco-miRs”) as well as genetic deletion and loss-of-function mutation of tumor suppressor miRNAs (“suppressor-miRs”) are linked to human cancer. In addition to the dysregulation of a specific onco-miR or suppressor-miRs, changes in global miRNA levels resulting from a defective miRNA biogenesis pathway play a role in tumorigenesis. The function of individual onco-miRs and suppressor-miRs and their target genes in cancer has been described in many different articles elsewhere. In this review we primarily focus on the recent development regarding the dysregulation of the miRNA biogenesis pathway and its contribution to cancer.
Keywords: Argonaute, microRNA, pri-miRNA, pre-miRNA, post-translational modifications, Drosha, DGCR8, Dicer, TRBP, stability, processing
Introduction
MicroRNAs (miRNAs) are small noncoding RNAs (ncRNAs) of ~22-nucleotides (nt) which mediate destabilization and/or translational suppression of target mRNAs bearing partially complementary sequences(Kim et al., 2009, Siomi and Siomi, 2010). The biogenesis of miRNAs starts by transcription of the miRNA gene encoded in the genome by RNA polymerase II (Pol II). This process generates long primary (pri-miRNA) transcripts comprising a stem-loop hairpin structure (Kim et al., 2009, Siomi and Siomi, 2010) (Figure 1). Pri-miRNAs undergo stepwise processing. The first processing takes place in the nucleus and involves the RNase III enzyme Drosha and its cofactor DiGeorge syndrome critical region gene 8 (DGCR8), which compose the “Drosha microprocessor” complex. The Drosha microprocessor complex recognizes the base of the stem-loop hairpin structure, cleaves it and releases a ~60–70-nt hairpin-shaped precursor miRNA (pre-miRNA). The pre-miRNA is then exported to the cytoplasm by exportin 5 (Xpo5) and undergoes the second processing by the RNase III enzyme Dicer and the cofactor transactivation-responsive RNA-binding protein (TRBP, also known as TARBP2), which generates a ~22-nt miRNA duplex(Kim et al., 2009, Siomi and Siomi, 2010) (Figure 1). As exceptions to this general pathway, some miRNAs are generated by Drosha-independent or Dicer-independent mechanisms, including splicing of miRNA-containing introns, which are known as miRtrons (Yang and Lai, 2011). The miRNA duplex is loaded into an Argonaute (Ago) protein, which preferentially ejects one strand (“passenger strand”) and retains the mature miRNA (“guide strand”) (Meister, 2013). Ago proteins and the GW182/TNRC6 family of proteins form the miRNA-induced silencing complex (miRISC). MiRNA/Ago complexes recognize target mRNAs by paring the 5′ end of the miRNA molecule (nts 2–8), called the “seed” region, with a partially complementary sequence in the 3′-untranslated region (3′-UTR) of target mRNAs. A specific miRNA can regulate hundreds of target mRNAs simultaneously, although the degree of regulation is only 30–50%. During the last decade, numerous regulatory pathways have been shown to modulate the biogenesis (transcription and processing), stability, and silencing activity of miRNAs. Dysregulation of these processes has been implicated in the pathogenesis of human disorders, including cancer. Several review articles summarize the involvement of individual miRNAs in cancer (Hata and Lieberman, 2015, Lin and Gregory, 2015). This review mainly focuses instead on the dysregulation of “global” miRNA levels in the context of cancer as a result of aberrant expression and/or activity of molecules that control miRNA biogenesis.
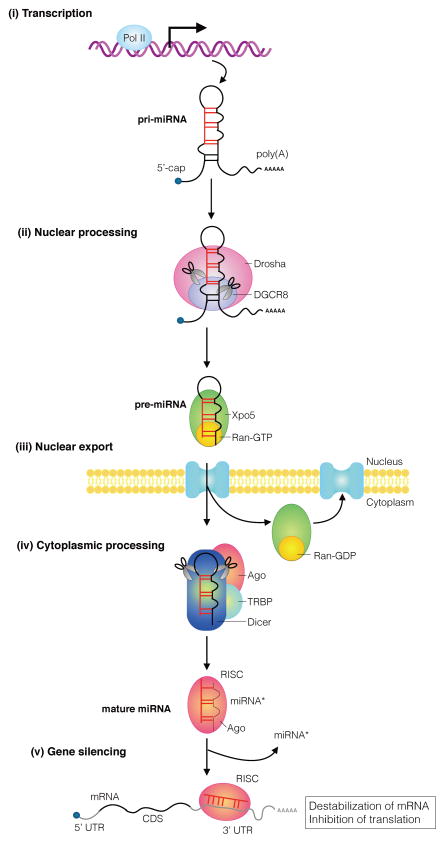
Figure 1. Overview of miRNA biogenesis pathway.
The biogenesis of an miRNA is a stepwise process that includes (i) transcription of a primary transcript (pri-miRNA), (ii) nuclear cropping to produce the pre-miRNA, (iii) export to the cytoplasm, and (iv) cytoplasmic cropping to a ds miRNA precursor. miRNA genes are generally transcribed into long, 5′-capped, and 3′-polyadenylated transcripts (pri-miRNAs) by RNA polymerase II (Pol II) and subjected to a primary processing step by a nuclear enzyme of the RNase III family, Drosha in the microprocessor complex. The primary processing products, hairpin-loop RNAs known as precursor-miRNAs (pre-miRNAs) are recognized by the Exportin-5 (Xpo5)/Ran-GTP transporter and exported to the cytoplasm, where an another enzyme of the RNase III family, Dicer, catalyzes the secondary processing to produce miRNA/miRNA* duplexes. Dicer, TRBP, and Argonaute (Ago) proteins mediate the processing of pre-miRNAs and the assembly of the RNA-induced silencing complex (RISC) in humans. In most cases, one strand of each duplex remains on the Ago protein as the mature miRNA; the other strand (miRNA*) is degraded. Ago associates with Dicer in both the cropping and RISC assembly steps. Every molecule that plays a role in miRNA biogenesis and RISC assembly can be regulated in response to environmental changes or physiological stimuli. Mutations and dysregulation of the expression of miRNA pathway molecules can contribute to human cancer. 5′ cap: 5′-7-Methylguanosine cap; poly(A): polyadenylation; TRBP: transactivation-responsive RNA-binding protein; CDS: coding sequence, UTR: untranslated region;
Global dysregulation of miRNAs and its activity on cancer
The level of expression and activity of different components of the miRNA biogenesis pathway are often found to be dysregulated in cancer. For example, the expression of Drosha and Dicer is either increased or decreased in various types of cancers where it inversely correlates with advanced stages of tumor and poor clinical outcome (Table 1). Defects in components of the miRNA biogenesis pathway produce expression changes in the large number of cellular miRNAs indicated as “altered miRNA profile (AMP)” in Table 1. Instances of global “downregulation” of miRNAs in tumors, especially in poorly differentiated ones, have been reported (Lu et al., 2005). One explanation proposed for this widespread downregulation of miRNAs in cancer cells is that the function of many miRNAs is to define lineage specific properties and, therefore, the low abundance of miRNAs promotes the undifferentiated state of tumor cells and enhances their potential of invasion and metastasis (Lu et al., 2005). In support of a general “tumor suppressor” action of global miRNAs, knockdown or haploinsufficiency of Dicer1 in mouse has been found to foster tumor formation and progression (Kumar et al., 2007, Kumar et al., 2009, Lambertz et al., 2010). On the other hand, although global “upregulation” of miRNAs is uncommon, elevated levels of the processing enzymes Drosha and Dicer have been found in tumor cells. Below, we discuss some examples of dysregulation of different components of the miRNA biogenesis pathway.
Table 1.
Dysregulation of proteins in the miRNA biogenesis pathway in various cancers.
| Dysregulated Protein | Role in tumorigenesis | Type of Cancer | Clinical Relevance | References |
|---|---|---|---|---|
| Drosha | Oncogene | BCC | NO | (Sand et al., 2010) |
| Cervical SCC | AMP; AAS | (Muralidhar et al., 2007, Muralidhar et al., 2011) | ||
| Esophageal cancer | APP | (Sugito et al., 2006) | ||
| Gastric cancer | APP | (Tchernitsa et al., 2010) | ||
| Non-small cell lung cancer | APP | (Diaz-Garcia et al., 2013) | ||
| SCC | NO | (Sand et al., 2010) | ||
| Serous ovarian carcinoma | AAS | (Vaksman et al., 2012) | ||
| Smooth muscle tumours | AAS | (Papachristou et al., 2012) | ||
| Triple-negative breast cancer | NO | (Passon et al., 2012, Avery-Kiejda et al., 2014) | ||
| Tumor suppressor | Bladder cancer | AMP | (Catto et al., 2009) | |
| Breast cancer | NO | (Yan et al., 2012) | ||
| Cutaneous melanoma | APP | (Jafarnejad et al., 2013) | ||
| Endometrial cancer | AAS | (Torres et al., 2011) | ||
| Gallbladder adenocarcinoma | APP | (Shu et al., 2012) | ||
| Nasopharyngeal carcinoma | APP | (Guo et al., 2012) | ||
| Neuroblastoma | AMP;APP | (Lin et al., 2010) | ||
| Ovarian cancer | APP | (Merritt et al., 2008) | ||
| DGCR8 | Oncogene | Bladder cancer | AMP | (Catto et al., 2009) |
| Colorectal carcinoma | NO | (Kim et al., 2014) | ||
| Esophageal cancer | APP | (Sugito et al., 2006) | ||
| Ovarian cancer | APP | (Guo et al., 2015) | ||
| Prostate cancer | AMP | (Ambs et al., 2008) | ||
| SCC and BCC | NO | (Sand et al., 2012) | ||
| Dicer1 | Oncogene | Colorectal cancer | ASS; APP | (Faber et al., 2011, Stratmann et al., 2011, Papachristou et al., 2011) |
| Cutaneous melanoma | AAS | (Ma et al., 2011) | ||
| Gastric cancer | Correlated with tumor subtype | (Tchernitsa et al., 2010) | ||
| Oral cancer | NO | (Jakymiw et al., 2010) | ||
| Precursor lesions of lung adenocarcinoma | AAS | (Chiosea et al., 2007) | ||
| Prostate cancer | AMP; AAS | (Ambs et al., 2008, Chiosea et al., 2006, Vaksman et al., 2012) | ||
| Serous ovarian carcinoma | AAS | (Vaksman et al., 2012) | ||
| Smooth muscle tumours | AAS | (Papachristou et al., 2012) | ||
| Tumor suppressor | BCC | NO | (Sand et al., 2010) | |
| Bladder cancer | AMP | (Catto et al., 2009, Wu et al., 2012) | ||
| Breast cancer | APP | (Yan et al., 2012, Khoshnaw et al., 2012) | ||
| Chronic lymphocytic leukemia | APP | (Zhu et al., 2012) | ||
| Colorectal cancer | APP | (Faggad et al., 2012) | ||
| Endometrial cancer | NO | (Torres et al., 2011) | ||
| Gallbladder adenocarcinoma | APP | (Shu et al., 2012) | ||
| Hepatocellular carcinoma | NO | (Wu et al., 2011b) | ||
| Nasopharyngeal carcinoma | APP | (Guo et al., 2012) | ||
| Neuroblastoma | AMP;APP | (Lin et al., 2010) | ||
| Non-small cell lung cancer | APP | (Diaz-Garcia et al., 2013, Karube et al., 2005) | ||
| Ovarian cancer | AAS; APP | (Merritt et al., 2008, Pampalakis et al., 2010, Faggad et al., 2010) | ||
| Triple-negative breast cancer | NO | (Avery-Kiejda et al., 2014, Dedes et al., 2011) | ||
| Xpo5 | Oncogene | AK, SCC and BCC | NO | (Sand et al., 2012) |
| Ago1 | Oncogene | Bladder cancer | AMP | (Catto et al., 2009) |
| Serous ovarian carcinoma | AAS | (Vaksman et al., 2012) | ||
| Ago2 | Oncogene | AK, SCC and BCC | NO | (Sand et al., 2012) |
| Serous ovarian carcinoma | AAS; APP | (Vaksman et al., 2012) |
AAS, associated with advanced stages; Ago, Argonaute; AK, actinic keratoses; AMP, altered miRNA profile; APP, associated with poor prognosis; BCC, basal cell carcinoma; DGCR8, DiGeorge syndrome critical region 8; miRNA, microRNA; NO, not determined or no clinical correlation; SCC, squamous cell carcinoma; Xpo5, exportin 5.
(a) Dysregulation of the Drosha microprocessor
Long pri-miRNAs transcribed by Pol II undergo primary processing in the nucleus by a large complex, the Drosha microprocessor, composed of various cofactors, including DGCR8. The processing of some pri-miRNAs also requires the RNA helicases (p68 and p72) (Figure 1).
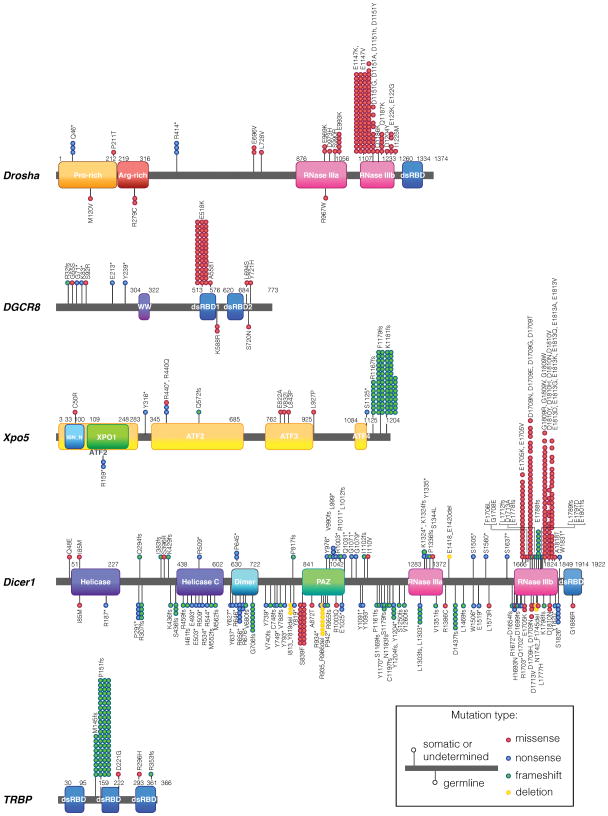
Upregulation of DGCR8 has been found in various types of cancer (Table 1). Gene mutation analyses have recently identified frequent heterozygous mutations in the Drosha gene in the rare pediatric form kidney cancer of Wilms tumors (Rakheja et al., 2014, Torrezan et al., 2014, Walz et al., 2015, Wegert et al., 2015) (Figure 2, Table 2). More than 60% of all the Drosha mutations consist of a single missense change (E to K) at amino acid (aa) 1147, located in the second RNase domain (Rakheja et al., 2014, Torrezan et al., 2014, Walz et al., 2015, Wegert et al., 2015) (Figure 2, Table 2). This E1147K mutation is thought to interfere with metal binding and negatively regulate the processing function of Drosha in a dominant fashion, which is consistent with the global downregulation of miRNAs observed in Wilms tumors harboring mutated Drosha (Rakheja et al., 2014, Torrezan et al., 2014, Walz et al., 2015, Wegert et al., 2015). In addition to the E1147K mutation, many other missense, nonsense, and splice-site mutations of Drosha have been identified in Wilms tumors (Figure 2, Table 2), but their effect has yet to be assessed.
Figure 2. The miRNA biogenesis pathway genes mutated in cancer.
Schematic representation of somatic and germline mutations of the miRNA biogenesis pathway genes (Drosha, DGCR8, Xpo5, Dicer1, and TRBP) identified in cancer. The location and the types of mutation are indicated. Somatic and germline mutations are plotted above and below the protein structure, respectively. Undermined mutations are plotted together with somatic mutations. ATF: armadillo-type fold; dsRBD,: double-stranded RNA-binding domain; IBN_N: importin-β amino-terminal domain; NLS: nuclear localization signal; PAZ: Piwi–Argonaute–Zwille domain; RNase: ribonuclease; pre-miRNA: precursor miRNA; TRBP: transactivation-responsive RNA-binding protein; WW: WW domain (also known as WWP-repeating motif); XPO1:exportin 1/importinβ-like domain; Xpo5: exportin 5.
Table 2.
Somatic and germline mutations associated with molecules in the miRNA biogenesis pathway in various cancers.
| Gene | Location | Mutation | Type of mutation | Type of cancer | References |
|---|---|---|---|---|---|
| Drosha | Q46* | Nonsense | Somatic | Wilms tumors | (Torrezan et al., 2014, Walz et al., 2015) |
| M120V | Missense | Gemline | Wilms tumors | (Rakheja et al., 2014) | |
| P211T | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| R279C | Missense | Gemline | Wilms tumors | (Wegert et al., 2015) | |
| R414* | Nonsense | Somatic | Wilms tumors | (Torrezan et al., 2014, Walz et al., 2015) | |
| E696V | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| L728V | Missense | Somatic | Wilms tumors | (Rakheja et al., 2014) | |
| R967W | Missense | Gemline | Wilms tumors | (Rakheja et al., 2014) | |
| E969K | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| D973H | Missense | Somatic | Wilms tumors | (Wegert et al., 2015) | |
| S990R | Missense | Somatic | Wilms tumors | (Wegert et al., 2015) | |
| E993K | Missense | Somatic | Wilms tumors | (Walz et al., 2015, Wegert et al., 2015) | |
| E1147K | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014, Rakheja et al., 2014, Walz et al., 2015, Wegert et al., 2015) | |
| E1147V | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| D1151Y | Missense | Somatic | Wilms tumors | (Rakheja et al., 2014) | |
| D1151A | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| D1151H | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| D1151G | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014, Walz et al., 2015) | |
| Q1186K | Missense | Somatic | Wilms tumors | (Wegert et al., 2015) | |
| Q1187K | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| D1204Y | Missense | Somatic | Wilms tumors | (Wegert et al., 2015) | |
| E1222K | Missense | Somatic | Wilms tumors | (Walz et al., 2015, Wegert et al., 2015) | |
| E1222G | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| I1225M | Missense | Somatic | Wilms tumors | (Wegert et al., 2015) | |
| DGCR8 | R32fs | Frameshift | Somatic | Wilms tumors | (Torrezan et al., 2014) |
| G55S | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| G71* | Nonsense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| K82* | Nonsense | Somatic | Wilms tumors | (Wegert et al., 2015) | |
| S92R | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| E213* | Nonsense | Somatic | Wilms tumors | (Wegert et al., 2015) | |
| Y239* | Nonsense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| E518K | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014, Wegert et al., 2015, Walz et al., 2015) | |
| A558T | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| K588R | Missense | Gemline | Wilms tumors | (Wegert et al., 2015) | |
| L694S | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| S720N | Missense | Gemline | Wilms tumors | (Rakheja et al., 2014) | |
| Y721H | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| Xpo5 | C50R | Missense | Somatic | Wilms tumors | (Walz et al., 2015) |
| R159* | Nonsense | Germline | Wilms tumors | (Walz et al., 2015) | |
| Y316* | Nonsense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| R440* | Nonsense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| R440Q | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| Q572fs | Frameshift | Somatic | Wilms tumors | (Walz et al., 2015) | |
| E822A | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| V832I | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| L843P | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| L927P | Missense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| S1125* | Nonsense | Somatic | Wilms tumors | (Walz et al., 2015) | |
| R1167fs | Frameshift | ND | Mix tumors | (Melo et al., 2010) | |
| F1179fs | Frameshift | ND | Mix tumors | (Melo et al., 2010) | |
| K1181fs | Frameshift | ND | Mix tumors | (Melo et al., 2010) | |
| TRBP | M145fs | Frameshift | ND | Mix tumors | (Melo et al., 2009) |
| P151fs | Frameshift | ND | Mix tumors | (Melo et al., 2009) | |
| D221G | Missense | Somatic | Wilms tumors | (Rakheja et al., 2014) | |
| R296H | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| R353fs | Frameshift | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| Dicer1 | R187* | Nonsense | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) |
| I383fs | Frameshift | ND | Pleuropulmonary blastoma | (Seki et al., 2014) | |
| S436fs | Frameshift | Gemline | Pleuropulmonary blastoma | (Foulkes et al., 2011) | |
| R459fs | Frameshift | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| I461fs | Frameshift | Germline | Pleuropulmonary blastoma | (Seki et al., 2014) | |
| E493* | Nonsense | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| R534* | Nonsense | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| R544* | Nonsense | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| M552fs | Frameshift | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| M562fs | Frameshift | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| Y627* | Nonsense | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| R646* | Nonsense | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| R656* | Nonsense | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| V680fs | Frameshift | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| Y739* | Nonsense | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| P740fs | Frameshift | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| T788fs | Frameshift | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| R934* | Nonsense | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| T955fs | Frameshift | Germline | Pleuropulmonary blastoma | (Seki et al., 2014) | |
| R1003* | Nonsense | ND | Pleuropulmonary blastoma | (Seki et al., 2014) | |
| Y1091* | Nonsense | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| P1161fs | Frameshift | Germline | Pleuropulmonary blastoma | (Seki et al., 2014) | |
| Y1170* | Nonsense | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| C1197fs | Frameshift | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| S1250fs | Frameshift | Germline | Pleuropulmonary blastoma | (Seki et al., 2014) | |
| L1469fs | Frameshift | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| W1506* | Nonsense | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| E1519* | Nonsense | Germline | Pleuropulmonary blastoma | (de Kock et al., 2013) | |
| L1573R | Missense | Gemline | Pleuropulmonary blastoma | (Hill et al., 2009) | |
| S1637* | Nonsense | ND | Pleuropulmonary blastoma | (Seki et al., 2014) | |
| D1654fs | Frameshift | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| E1705K | Missense | Somatic | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| E1705V | Missense | ND | Pleuropulmonary blastoma | (Seki et al., 2014) | |
| G1708E | Missense | Somatic | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| D1709N | Missense | Somatic | Pleuropulmonary blastoma | (Pugh et al., 2014, Seki et al., 2014) | |
| K1798fs | Frameshift | Germline | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| G1809R | Missense | Somatic | Pleuropulmonary blastoma | (Pugh et al., 2014, Seki et al., 2014) | |
| D1810Y | Missense | Somatic | Pleuropulmonary blastoma | (Pugh et al., 2014, Seki et al., 2014) | |
| E1813D | Missense | Somatic | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| E1813G | Missense | Somatic | Pleuropulmonary blastoma | (Pugh et al., 2014, Seki et al., 2014, de Kock et al., 2013) | |
| E1813K | Missense | Somatic | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| E1813Q | Missense | Somatic | Pleuropulmonary blastoma | (Pugh et al., 2014) | |
| Y1820* | Nonsense | Germline | Pleuropulmonary blastoma | (Seki et al., 2014) | |
| E503* | Nonsense | Germline | Nonepithelial ovarian cancers | (Schultz et al., 2011) | |
| R459fs | Frameshift | Germline | Nonepithelial ovarian cancers | (Schultz et al., 2011) | |
| C748fs | Frameshift | Germline | Nonepithelial ovarian cancers | (Schultz et al., 2011) | |
| Y819* | Nonsense | Gemline | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| P942* | Nonsense | Gemline | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| E1025* | Nonsense | Germline | Nonepithelial ovarian cancers | (Schultz et al., 2011) | |
| G1079* | Nonsense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| Y1204* | Nonsense | Gemline | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| V1260fs | Frameshift | Germline | Nonepithelial ovarian cancers | (Schultz et al., 2011) | |
| D1699fs | Frameshift | Germline | Nonepithelial ovarian cancers | (Schultz et al., 2011) | |
| R1703* | Nonsense | Gemline | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| E1705K | Missense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012, Witkowski et al., 2013) | |
| D1709N | Missense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012, Witkowski et al., 2013) | |
| D1709N | Missense | ND | Nonepithelial ovarian cancers | (Schultz et al., 2011) | |
| D1709E | Missense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| D1709G | Missense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| E1788fs | Frameshift | Somatic | Nonepithelial ovarian cancers | (Witkowski et al., 2013) | |
| D1810H | Missense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| D1810N | Missense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| D1810Y | Missense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012, Witkowski et al., 2013) | |
| D1810V | Missense | Somatic | Nonepithelial ovarian cancers | (Witkowski et al., 2013) | |
| E1813A | Missense | ND | Nonepithelial ovarian cancers | (Schultz et al., 2011) | |
| E1813D | Missense | Somatic | Nonepithelial ovarian cancers | (Witkowski et al., 2013) | |
| E1813K | Missense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012, Witkowski et al., 2013) | |
| E1813G | Missense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| E1813Q | Missense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012, Witkowski et al., 2013) | |
| W1831* | Nonsense | Somatic | Nonepithelial ovarian cancers | (Heravi-Moussavi et al., 2012) | |
| Q48E | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| I85M | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| I85M | Missense | Gemline | Wilms tumors | (Walz et al., 2015) | |
| R307fs | Frameshift | Germline | Wilms tumors | (Foulkes et al., 2011, Wu et al., 2013) | |
| S436fs | Frameshift | Germline | Wilms tumors | (Foulkes et al., 2011, Wu et al., 2013) | |
| P645* | Nonsense | Somatic | Wilms tumors | (Wu et al., 2013) | |
| G706fs | Frameshift | Germline | Wilms tumors | (Foulkes et al., 2011, Wu et al., 2013) | |
| A872T | Missense | Germline | Wilms tumors | (Wu et al., 2013) | |
| L999* | Nonsense | Somatic | Wilms tumors | (Wu et al., 2013) | |
| R1003Q | Missense | Gemline | Wilms tumors | (Walz et al., 2015) | |
| A1011* | Nonsense | Somatic | Wilms tumors | (Wu et al., 2013) | |
| R1071* | Nonsense | Somatic | Wilms tumors | (Wu et al., 2013) | |
| I1102fs | Frameshift | Somatic | Wilms tumors | (Rakheja et al., 2014) | |
| I1110V | Missense | ND | Wilms tumors | (Wu et al., 2013) | |
| K1324* | Nonsense | Somatic | Wilms tumors | (Wu et al., 2013) | |
| S1344L | Missense | Somatic | Wilms tumors | (Wu et al., 2013) | |
| R1386C | Missense | Gemline | Wilms tumors | (Walz et al., 2015) | |
| S1505* | Nonsense | Somatic | Wilms tumors | (Wu et al., 2013) | |
| A1560* | Nonsense | Somatic | Wilms tumors | (Wu et al., 2013) | |
| H1693N | Missense | Gemline | Wilms tumors | (Walz et al., 2015) | |
| E1705K | Missense | Gemline | Wilms tumors | (Walz et al., 2015) | |
| D1709N | Missense | Gemline | Wilms tumors | (Walz et al., 2015) | |
| D1713A | Missense | Somatic | Wilms tumors | (Wu et al., 2013) | |
| D1713V | Missense | Gemline | Wilms tumors | (Walz et al., 2015) | |
| L1777H | Missense | Germline | Wilms tumors | (Wu et al., 2013) | |
| E1788fs | Frameshift | Somatic | Wilms tumors | (Wu et al., 2013) | |
| G1809R | Missense | Somatic | Wilms tumors | (Rakheja et al., 2014) | |
| G1809V | Missense | Somatic | Wilms tumors | (Rakheja et al., 2014) | |
| D1810N | Missense | Somatic | Wilms tumors | (Torrezan et al., 2014) | |
| D1810N | Missense | Gemline | Wilms tumors | (Walz et al., 2015) | |
| A1818T | Missense | Somatic | Wilms tumors | (Wu et al., 2013) | |
| G1886R | Missense | Gemline | Wilms tumors | (Wegert et al., 2015) | |
| K429fs | Frameshift | Gemline | Pituitary blastoma | (de Kock et al., 2014a) | |
| R509* | Nonsense | Gemline | Pituitary blastoma | (de Kock et al., 2014a) | |
| R676* | Nonsense | Gemline | Pituitary blastoma | (de Kock et al., 2014a) | |
| Y793* | Nonsense | Gemline | Pituitary blastoma | (de Kock et al., 2014a) | |
| N1093* | Nonsense | Gemline | Pituitary blastoma | (de Kock et al., 2014a) | |
| S1179fs | Frameshift | Gemline | Pituitary blastoma | (de Kock et al., 2014a) | |
| D1437fs | Frameshift | Gemline | Pituitary blastoma | (de Kock et al., 2014a) | |
| D1709H | Missense | Gemline | Pituitary blastoma | (de Kock et al., 2014a) | |
| D1709T | Missense | Somatic | Pituitary blastoma | (de Kock et al., 2014a) | |
| D1709N | Missense | Somatic | Pituitary blastoma | (de Kock et al., 2014a) | |
| G1809W | Missense | Somatic | Pituitary blastoma | (de Kock et al., 2014a) | |
| E1813D | Missense | Somatic | Pituitary blastoma | (de Kock et al., 2014a) | |
| E1813V | Missense | Somatic | Pituitary blastoma | (de Kock et al., 2014a) | |
| E1813K | Missense | Somatic | Pituitary blastoma | (de Kock et al., 2014a) | |
| Y793* | Nonsense | Germline | Differentiated thyroid carcinoma | (de Kock et al., 2014b) | |
| S1169fs | Frameshift | Germline | Differentiated thyroid carcinoma | (de Kock et al., 2014b) | |
| N1193fs | Frameshift | Germline | Differentiated thyroid carcinoma | (de Kock et al., 2014b) | |
| E1705K | Missense | Somatic | Differentiated thyroid carcinoma | (de Kock et al., 2014b) | |
| E1813D | Missense | Somatic | Differentiated thyroid carcinoma | (de Kock et al., 2014b) | |
| E1813G | Missense | Somatic | Differentiated thyroid carcinoma | (de Kock et al., 2014b) | |
| Q249fs | Frameshift | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| S396R | Missense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| K429fs | Frameshift | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| R509* | Nonsense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| P817fs | Frameshift | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| Y976* | Nonsense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| V990fs | Frameshift | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| L1012fs | Frameshift | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| Q1031* | Nonsense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| K1324fs | Frameshift | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| Y1335* | Nonsense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| P1336fs | Frameshift | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| D1437fs | Frameshift | Germline | Familial cystic nephroma | (Bahubeshi et al., 2010) | |
| E1705K | Missense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| D1709N | Missense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| E1778fs | Frameshift | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| G1809R | Missense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| D1810H | Missense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| E1813D | Missense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| E1813G | Missense | ND | Childhood cystic nephroma | (Doros et al., 2014) | |
| E1813K | Missense | Childhood | Childhood cystic nephroma | (Doros et al., 2014) | |
| S1826* | Nonsense | Germline | Familial cystic nephroma | (Bahubeshi et al., 2010) | |
| Y637* | Nonsense | Germline | Embryonal rhabdomyosarcoma (ERMS) | (Doros et al., 2012) | |
| G706fs | Frameshift | Germline | Cervix embryonal rhabdomyosarcoma | (Foulkes et al., 2011) | |
| Y749* | Nonsense | Germline | Embryonal rhabdomyosarcoma (ERMS) | (Doros et al., 2012) | |
| S1179fs | Frameshift | Germline | Embryonal rhabdomyosarcoma (ERMS) | (Tomiak et al., 2014) | |
| Y1204fs | Frameshift | Germline | Cervix embryonal rhabdomyosarcoma | (Foulkes et al., 2011) | |
| L1303* | Nonsense | Germline | Rhabdomyosarcoma | (Heravi-Moussavi et al., 2012) | |
| L1303fs | Frameshift | Germline | Cervix embryonal rhabdomyosarcoma | (Foulkes et al., 2011) | |
| E1418_E1420del | Deletion | Somatic | Embryonal rhabdomyosarcoma (ERMS) | (Doros et al., 2012) | |
| D1437fs | Frameshift | Germline | Embryonal rhabdomyosarcoma (ERMS) | (Doros et al., 2012) | |
| Q1702* | Nonsense | Germline | Embryonal rhabdomyosarcoma (ERMS) | (Doros et al., 2012) | |
| E1705K | Missense | Somatic | Rhabdomyosarcoma | (Heravi-Moussavi et al., 2012) | |
| L1789fs | Frameshift | Somatic | Embryonal rhabdomyosarcoma (ERMS) | (Doros et al., 2012) | |
| E1813K | Missense | Somatic | Embryonal rhabdomyosarcoma (ERMS) | (Tomiak et al., 2014) | |
| P291* | Nonsense | Germline | Nontoxic multinodular goiter | (Rio Frio et al., 2011) | |
| R509* | Nonsense | ND | Multinodular goiter | (Darrat et al., 2013) | |
| S839F | Missense | Germline | Nontoxic multinodular goiter | (Rio Frio et al., 2011) | |
| I813_Y819del | Deletion | Germline | Nontoxic multinodular goiter | (Rio Frio et al., 2011) | |
| R935_R996del | Deletion | Germline | Nontoxic multinodular goiter | (Rio Frio et al., 2011) | |
| R1672* | Nonsense | Germline | Nontoxic multinodular goiter | (Rio Frio et al., 2011) | |
| N1742_F1745del | Deletion | Germline | Multinodular goiter | (Rath et al., 2014) | |
| R656* | Nonsense | Germline | Pulmonary sequestration | (Foulkes et al., 2011) | |
| V1351fs | Frameshift | Germline | Primitive neuroectodermal tumor | (Foulkes et al., 2011) | |
| F1706L | Missense | ND | Non-small cell lung cancers | (Kim et al., 2013) | |
| L1712fs | Frameshift | ND | Gastric carcinoma | (Kim et al., 2013) | |
| E1801fs | Frameshift | ND | Prostate carcinoma | (Kim et al., 2013) | |
| E1797D | Missense | ND | Soft tissue sarcoma | (Kim et al., 2013) |
DGCR8, DiGeorge syndrome critical region 8; ND, not determined; TRBP: transactivation-responsive RNA-binding protein; Xpo5, exportin 5
Somatic and germline mutations of DGCR8 were also found in Wilms tumors (Torrezan et al., 2014, Walz et al., 2015, Wegert et al., 2015) (Figure 2, Table 2). The E518K missense mutation in the first double strand RNA (dsRNA) binding domain (dsRBDe) of DGCR8 is clearly a “hot-spot” and makes up for more than 70% of the DGCR8 mutations in Wilms tumors (Torrezan et al., 2014, Walz et al., 2015, Wegert et al., 2015) (Figure 2, Table 2). The E518K mutation causes a reduction of critical miRNAs in tumors (Torrezan et al., 2014, Walz et al., 2015, Wegert et al., 2015), which is consistent with the observation that knockdown of DGCR8 promotes tumor growth (Kumar et al., 2007).
Besides gene mutations, the expression of alternatively spliced variants of Drosha has been reported in melanoma and teratocarcinoma cells (Grund et al., 2012). The splice variants encode a Drosha protein with a truncated carboxyl (C)-terminal RNase domain (see Figure 2). Its dsRBD fails to interact with DGCR8, and therefore is functionally compromised and potentially acts as dominant negative (Grund et al., 2012). Conversely, an increased copy number of the Drosha gene or overexpression of Drosha protein, which in turn lead to a global change in miRNA levels, have been found in more than half of the advanced cervical squamous cell carcinomas (Muralidhar et al., 2011). Further study is required to uncover why Drosha mutations are frequently found in Wilms tumors, and why Drosha levels are regulated in opposite directions depending on the tumor type.
Various post-translational modifications (PTMs) of Drosha and DGCR8 that are able to affect miRNA processing have been identified. For example, phosphorylation of serine residues by glycogen synthase kinase 3β (GSK3β) is required for nuclear localization of Drosha (Tang et al., 2011, Tang et al., 2010); acetylation of Drosha by p300, CBP and GCN5 prevents ubiquitin-mediated degradation and stabilizes Drosha (Tang et al., 2013); deacetylation of DGCR8 by histone deacetylase 1 (HDAC1) increases the affinity of DGCR8 for pri-miRNAs (Wada et al., 2012), while phosphorylation of DGCR8 by Erk stabilizes DGCR8 protein and promotes miRNA production (Herbert et al., 2013). It is not clear whether changes in miRNA production that result from these Drosha PTMs play an important role in cancer. However, GSK3β and HDAC1 are often dysregulated in cancer, therefore it is possible that different Drosha PTMs may be found in tumor cells versus non-tumor cells.
Drosha activity is regulated by different nuclear proteins, in a manner that generally affects the biogenesis of only a small subset of miRNAs. One such nuclear protein is adenosine deaminase acting on RNA (ADAR). ADAR is a RNA editing enzyme that converts adenosine (A) to inosine (I) in double-stranded RNAs. ADAR1 forms a complex with DGCR8 and inhibits Drosha activity (Nemlich et al., 2013). In metastatic melanoma in which ADAR1 level is reduced, two miRNAs (miR-17 and miR-432) are overproduced and promote tumor growth (Nemlich et al., 2013). Smad proteins, the signal transducers of the transforming growth factor-β (TGF-β)–superfamily of growth factors, also modify Drosha activity in the nucleus. Although primarily cytoplasmic at steady state, Smad proteins translocate to the nucleus upon ligand activation. Smads are bona fide transcription factors with DNA binding and transcription activating domains (Massague, 2012). However, they are also recruited to the Drosha microprocessor complex through physical interaction with the RNA helicase p68 (also known as DDX5), where they promote pri- to pre-miRNA processing of about 20 miRNAs (Davis et al., 2008), including miR-21 and miR-199. miR-21 is one of the most commonly upregulated onco-miRs in nearly all tumor samples and is known to inhibit a large group of tumor suppressor genes, including PTEN, PDCD4, TPM1, SPRY1/2 and TP53BP2(Di Leva et al., 2014). In addition, miR-21 can drive tumorigenesis by inhibiting negative regulators of the Ras/MEK/Erk pathway(Hatley et al., 2010). The specificity of R-Smad-mediated regulation of pri-miRNA processing is achieved by direct association of R-Smads with a sequence element in the stem region of pri-miR-21 (Davis et al., 2010). Like Smads, p53 induces the expression of a group of suppressor-miRs (miR-15/16, miR-143, miR-145 and miR-203) by associating with the Drosha microprocessor complex via p68 and facilitating pri-miRNA processing (Suzuki et al., 2009). Genotoxic stimuli, which induce acetylation of K120 in the DNA binding domain of p53, do not affect the transcription activity of p53, but prompt the association of p53 with the Drosha microprocessor complex and elevate miR-203 levels to promote apoptosis instead of cell cycle arrest (Chang et al., 2013). It is yet unclear how regulation of processing of a specific subset of miRNA is accomplished by p53, and how TGF-β-mediated activation of Smads can affect p53-dependent regulation of miRNA processing.
A recent study also identified Yes-associated protein (YAP), a signal transducer of the Hippo pathway that controls organ size by sensing cell density, as a regulator of Drosha microprocessor activity (Mori et al., 2014). When Hippo signaling is inactive, such as in cancer cells or in normal cells at low cell density, YAP accumulates in the nucleus and binds the RNA helicase p72 (also known as DDX17), sequestering it from the Drosha microprocessor and thus inhibiting processing of a subset of miRNAs that targets Myc (Mori et al., 2014). It is plausible that this YAP/p72-dependent inhibition of Drosha may play a significant role in the widespread downregulation of a large number of miRNAs in cancer.
The single strand RNA binding factor KH-type splicing regulatory protein (KSRP, also known as FUBP2)(Trabucchi et al., 2009) binds the terminal loop of a small subset of pri-miRNAs, including onco-miRs in the let-7 family, miR-21 and miR-125, and promotes processing by Drosha through an unknown mechanism (Trabucchi et al., 2009, Briata et al., 2012). When the PI3K/Akt signaling pathway is activated, KSRP is phosphorylated at S274 and S670, which triggers enhanced binding to pri-miRNAs and stimulates their Drosha-dependent processing (Briata et al., 2012). KSRP-enhanced miRNA biogenesis is also activated by DNA damage (Zhang et al., 2011). KSRP is highly expressed in chronic myeloid leukemia (CML) acute phase/blast crisis, compared with the chronic phase disease. It remains unclear, however, whether altered expression or function of KSRP contributes to leukemogenesis as a result of dysregulation of miRNA processing (Radich et al., 2006). A few other RNA binding proteins, such as hnRNP A1, serine/arginine-rich splicing factor 1 (SRSF1) and FUS (also known as TLS), have been found to interact with pri-miRNAs and facilitate Drosha processing, but there is yet no evidence of a direct link to tumorigenesis.
(b) Alteration of pre-miRNA export
Following Drosha processing and synthesis in the nucleus, pre-miRNAs are exported to the cytoplasm where they undergo secondary processing by Dicer (Figure 1). Xpo5 and Ran-GTP form a nuclear complex that transports and releases pre-miRNAs into the cytoplasm. Xpo5 protein is rapidly induced during cell cycle entry by a PI3K-dependent post-transcriptional mechanism, leading to an overall increase in mature miRNAs in proliferating cells (Iwasaki et al., 2013). Nuclear export of pre-miRNAs is also induced upon DNA damage. Ataxia telangiectasia mutated (ATM) activates Akt kinase to phosphorylate Nup153, a key component of the nucleopore, leading to enhanced interaction between Nup153 and Xpo5 and more efficient nuclear export of pre-miRNAs (Wan et al., 2013). The PI3K-dependent mechanism underlying increased Xpo5 protein and pre-miRNA nuclear export has not been defined, but could potentially also be mediated by Akt, which is activated by PI3K.
Somatic and germline heterozygous mutations in Xpo5 occur in gastric, endometrial, and colon tumors with microsatellite instability (Melo et al., 2010) (Figure 2, Table 2). Xpo5 mutations impair the export of pre-miRNAs, causing their accumulation in the nucleus, consequent reduction of mature miRNAs, and possibly tumorigenesis (Melo et al., 2010). Genetic and epigenetic association studies reveal a link between variations in Xpo5 and the risk of developing breast cancer (Leaderer et al., 2011), adding support to a role for dysregulation of miRNA biogenesis and transport in cancer. Upregulation of Xpo5 and dysregulation of miRNA expression profile have also been reported in bladder cancer (Table 1).
(c) Dysregulation of the Dicer/TRBP processing complex
Pre-miRNAs exported from the nucleus via Xpo5 undergo secondary processing by Dicer and its cofactor TRBP in the cytoplasm to finally give rise to ~22-nt mature miRNAs (Figure 1). Unlike Drosha processing, which is not required for the processing of a subset of miRNAs known as “miRtrons” that are encoded in the intronic region of other genes and processed by the splicing machinery, Dicer processing is essential for the biogenesis of all miRNAs, with the sole exception miR-451, whose pre-miRNA is processed by Ago2 (Yang and Lai, 2010, Yang et al., 2010). Germline mutations in the Dicer1 gene have been identified in a broader spectrum of inherited tumors compared to Drosha mutations (Figure 2, Table 2). Heterozygous germline Dicer1 mutations were first reported in a rare pediatric lung cancer, pleuropulmonary blastoma (PPB)(Hill et al., 2009). Currently, both germline and somatic Dicer1 mutations have been associated not only with PPB (de Kock et al., 2013, Hill et al., 2009, Pugh et al., 2014, Schultz et al., 2011, Seki et al., 2014, Wagh et al., 2014) but also with other tumors, including Wilms tumor (Foulkes et al., 2014, Torrezan et al., 2014, Wu et al., 2013, Heravi-Moussavi et al., 2012, Schultz et al., 2011, Witkowski et al., 2013), nonepithelial ovarian cancer (Heravi-Moussavi et al., 2012, Schultz et al., 2011, Witkowski et al., 2013), pituitary blastoma (de Kock et al., 2013), cystic nephroma (Doros et al., 2014), and embryonal rhabdomyosarcoma (ERMS) (Doros et al., 2012). As the carriers of Dicer1 mutations exhibit a predisposition to distinctive inheritable tumors, this condition has been defined “the Dicer1 syndrome” (OMIM #601200)(de Kock et al., 2015). For many of these mutations, the predicted effect is a reduction of Dicer protein level and/or processing activity and a resulting global reduction of miRNA levels and their tumor suppressor activities (Foulkes et al., 2014). Loss of heterozygosity (LOH) of Dicer has not been found in human cancer.
Beside gene mutations, both increase and decrease of Dicer expression have been reported in different types of tumors (Table 1). For example, Dicer expression is positively regulated transcriptionally by the p53 homolog TAp63 (Su et al., 2010). The induction of Dicer is critical for the ability of TAp63 to suppress tumor formation and inhibit metastasis (Su et al., 2010). Also, tumor hypoxia causes reduction of Dicer. This occurs through an epigenetic mechanism that involves inhibition of oxygen-dependent Histone H3K27me3 KDM6A/B and silencing of the Dicer promoter (van den Beucken et al., 2014).
Expression of an alternative splice variant of Dicer1 which lacks the dsRBD is detected in neuroblastoma cells but not in normal tissues(Potenza et al., 2010). It is intriguing to speculate that this splice variant might act as a dominant negative mutant that promotes tumorigenesis by downregulating global miRNA levels.
Several proteins regulate the activity of Dicer. For example, association of KSRP to pre-miRNA promotes Xpo5-mediated export of the pre-miRNA, while its binding to Dicer facilitates pre-miRNA processing when the PI3K/Akt signaling pathway is activated (Briata et al., 2012). Also, the Dicer cofactor TRBP stabilizes Dicer and enhances pre-miRNA processing. Mutations in the TARBP2 gene encoding TRBP are found in sporadic and hereditary carcinoma with microsatellite instability (Figure 2, Table 2). The TRBP expression is reduced in the cancer stem cell (CSC) population of Ewing sarcoma family tumor (ESFT)(De Vito et al., 2012). Restoration of TARBP2 activity inhibits ESFT CSC clonogenicity and tumor growth, suggesting that Dicer/TRBP-mediated miRNA processing plays a tumor suppressor function(De Vito et al., 2012). The TRBP activity is post-translationally regulated. The MAPK/Erk pathway mediates TRBP phosphorylation, which enhances global miRNA biogenesis by increasing Dicer levels and facilitating Dicer/TRBP miRNA processing (Paroo et al., 2009). Expression of phosphomimetic TRBP enhances growth-promoting miRNAs and increases cell proliferation (Paroo et al., 2009).
(d) Post-translational modifications of Ago proteins
Ago proteins are the critical downstream effectors of miRNA-mediated gene silencing (Figure 1). There are four Ago proteins (Ago1–4) in humans. They associate with cofactors of the GW182/TNRC6 family in the RISC to guide miRNAs to target specific transcripts and mediate gene silencing (Meister, 2013). Perfectly paired siRNA or miRNA duplexes are cleaved by catalytically active Ago2, while Ago1, Ago3, and Ago4 are catalytically inactive and unable to cleave RNAs (Meister, 2013). Binding to Ago proteins stabilizes mature miRNAs, possibly for weeks to months under basal conditions. In the absence of Ago2 in the cell, miRNAs are unstable, with half-lives of approximately 8 hours, and global miRNA levels are markedly reduced (Winter and Diederichs, 2011). Thus, a pathological change in the level of Ago proteins and/or their activity could result in major impairments in global miRNA stability and/or silencing. Indeed, upregulation of Ago1 and Ago2 has been reported in various types of cancer (Table 1). Currently, there is no report of mutations in the genes encoding Ago1–4. However, different PTMs found in Ago proteins can affect their stability and/or activity and critically modulate global miRNA levels (Figure 1). Prolyl-hydroxylation of Ago proteins by type I collagen prolyl-4-hydroxylase I [C-P4H(I)] was the first of these critical PTMs to be reported (Qi et al., 2008). C-P4H(I) hydroxylates multiple human Ago proteins, and especially Ago2 at P700, which stabilizes the protein and augments siRNA-mediated silencing. Hypoxia potently induces C-P4H(I) expression and promotes Ago2 hydroxylation, resulting in a rapid rise in the silencing activity of miRNAs (Wu et al., 2011a). In response to mitogens and growth factor signaling, Ago1 and Ago2 are also phosphorylated. Phosphorylation of the serine residue S387 of Ago2, mediated by the p38 mitogen-activated protein kinase (MAPK) pathway and Akt3, promotes its association with the TNRC6 cofactor and localization to the processing bodies (P-bodies)(Zeng et al., 2008), where translational repression by miRNAs is enhanced (Horman et al., 2013). It is currently unclear whether phosphorylation of S387 in Ago2 is augmented in human tumors. Another phosphorylation event, at the Ago2 tyrosine Y393, is induced by activation of epidermal growth factor receptor (EGFR) signaling and causes a reduction of the Ago2 association with Dicer and its cofactor TRBP (Shen et al., 2013). Hypoxia, which upregulates EGFR and increases its activity, consequently also enhances P-Y393 Ago2. The P-Y393 Ago2 modification suppresses the maturation of a subset of growth-repressing pre-miRNAs with longer terminal loops and leads to increased survival in response to hypoxia. P-Y393 Ago2 is elevated in hypoxic areas of human breast tumors. Moreover, higher levels of P-Y393 Ago2 correlate with poor overall survival in breast cancer patients (Shen et al., 2013). The effect of the P-Y393 Ago2 modification on the RISC activity, miRNA stability or subcellular localization of miRNAs remains to be investigated in the future. The RISC activity of Ago proteins can also be modulated by ubiquitination and poly(ADP-ribosyl)ation (PARylation). Ubiquitination of Ago leads to its proteosomal degradation and to global downregulation of miRNAs. This occurs rapidly during T cell activation when the miRNA repertoire is suddenly altered to allow T cell clonal expansion and activated T cell effector functions (Bronevetsky et al., 2013). PARylation of Ago2 complexes occurs rapidly during infection with viruses, such as the Herpes simplex viruses, that trigger innate immune alarms and interferons via an unknown mechanism that involves PARP13 (Seo et al., 2013). PARylation inhibits RISC activity and the silencing activity of miRNAs. This has the net effect of derepressing antiviral interferon-stimulated genes, which would be otherwise inhibited by cellular miRNAs, and enhancing the cell’s ability to withstand viral infection (Seo et al., 2013). Whether the Ago2 amino acid residues targeted by PTMs are mutated in human tumors remains the subject of future investigations.
Conclusions
Considerable evidence points to a significant role of miRNAs in human disease. The evolutionary conservation of miRNAs and their biogenesis pathway, the highly tuned regulation of their tissue- and developmental stage-specific expression, and the reproducible detection of their biological activity in vitro altogether predict a key biological function. Yet, the functional significance of miRNAs, especially in the context of human development and physiology, has been questioned because the knockdown of a single miRNA, unlike a protein-coding gene, rarely gives rise to a phenotype in an organism. However, the recent discovery of germline and somatic mutations in the genes encoding core components of the miRNA biogenesis pathway in human cancer illuminate the significance of the proper production and action of miRNAs in order to maintain homeostasis. During the last decade of miRNA research, the focus of the investigators in the cancer field has yielded the identification of onco-miRs and suppressor-miRs, many of which are expressed strictly in tumor type-specific and stage-specific manner. Some of these studies led to the identification of a biomarker that can properly diagnose a specific type or stage of cancer. However, modulation by antisense oligonucleotides or miRNA mimicry of a single onco-miR or suppressor-miR, or even a few of them, has not been successful in treating cancer in humans. The recent work that we have summarized in this review raises a new hope. Future research may uncover novel gene mutations and PTMs of the components of the miRNA biogenesis pathway in cancer and help us understand the activities they disrupt, which might lead to the discovery of regulatory factors amenable to pharmacological intervention. These tools will enable us to modulate globally the expression and activity of miRNAs in tumor cells or in the tumor environment, and could be applied to cancer prevention and treatment.
Acknowledgments
We thank Dr. Lagna for critical reading of the manuscript. A.H. is supported by the LeDucq Transatlantic Network of Excellence in Cardiovascular Research Program and the National Institutes of Health (NIH) (HL108317 and HL116191).
References
- AMBS S, PRUEITT RL, YI M, HUDSON RS, HOWE TM, PETROCCA F, WALLACE TA, LIU CG, VOLINIA S, CALIN GA, YFANTIS HG, STEPHENS RM, CROCE CM. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–70. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AVERY-KIEJDA KA, BRAYE SG, FORBES JF, SCOTT RJ. The expression of Dicer and Drosha in matched normal tissues, tumours and lymph node metastases in triple negative breast cancer. BMC Cancer. 2014;14:253. doi: 10.1186/1471-2407-14-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAHUBESHI A, BAL N, RIO FRIO T, HAMEL N, POUCHET C, YILMAZ A, BOURON-DAL SOGLIO D, WILLIAMS GM, TISCHKOWITZ M, PRIEST JR, FOULKES WD. Germline DICER1 mutations and familial cystic nephroma. J Med Genet. 2010;47:863–6. doi: 10.1136/jmg.2010.081216. [DOI] [PubMed] [Google Scholar]
- BRIATA P, LIN WJ, GIOVARELLI M, PASERO M, CHOU CF, TRABUCCHI M, ROSENFELD MG, CHEN CY, GHERZI R. PI3K/AKT signaling determines a dynamic switch between distinct KSRP functions favoring skeletal myogenesis. Cell Death Differ. 2012;19:478–87. doi: 10.1038/cdd.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRONEVETSKY Y, VILLARINO AV, EISLEY CJ, BARBEAU R, BARCZAK AJ, HEINZ GA, KREMMER E, HEISSMEYER V, MCMANUS MT, ERLE DJ, RAO A, ANSEL KM. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. 2013;210:417–32. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CATTO JW, MIAH S, OWEN HC, BRYANT H, MYERS K, DUDZIEC E, LARRE S, MILO M, REHMAN I, ROSARIO DJ, DI MARTINO E, KNOWLES MA, MEUTH M, HARRIS AL, HAMDY FC. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69:8472–81. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANG J, DAVIS-DUSENBERY BN, KASHIMA R, JIANG X, MARATHE N, SESSA R, LOUIE J, GU W, LAGNA G, HATA A. Acetylation of p53 stimulates miRNA processing and determines cell survival following genotoxic stress. EMBO J. 2013;32:3192–205. doi: 10.1038/emboj.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIOSEA S, JELEZCOVA E, CHANDRAN U, ACQUAFONDATA M, MCHALE T, SOBOL RW, DHIR R. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIOSEA S, JELEZCOVA E, CHANDRAN U, LUO J, MANTHA G, SOBOL RW, DACIC S. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–50. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- DARRAT I, BEDOYAN JK, CHEN M, SCHUETTE JL, LESPERANCE MM. Novel DICER1 mutation as cause of multinodular goiter in children. Head Neck. 2013;35:E369–71. doi: 10.1002/hed.23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS BN, HILYARD AC, LAGNA G, HATA A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS BN, HILYARD AC, NGUYEN PH, LAGNA G, HATA A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–84. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE KOCK L, DRUKER H, WEBER E, HAMEL N, TRAUBICI J, MALKIN D, ARSENEAU J, STEWART CJ, BOURON-DAL SOGLIO D, PRIEST JR, FOULKES WD. Ovarian embryonal rhabdomyosarcoma is a rare manifestation of the DICER1 syndrome. Hum Pathol. 2015;46:917–22. doi: 10.1016/j.humpath.2015.02.008. [DOI] [PubMed] [Google Scholar]
- DE KOCK L, PLOURDE F, CARTER MT, HAMEL N, SRIVASTAVA A, MEYN MS, ARSENEAU J, BOURON-DAL SOGLIO D, FOULKES WD. Germ-line and somatic DICER1 mutations in a pleuropulmonary blastoma. Pediatr Blood Cancer. 2013;60:2091–2. doi: 10.1002/pbc.24692. [DOI] [PubMed] [Google Scholar]
- DE KOCK L, SABBAGHIAN N, PLOURDE F, SRIVASTAVA A, WEBER E, BOURON-DAL SOGLIO D, HAMEL N, CHOI JH, PARK SH, DEAL CL, KELSEY MM, DISHOP MK, ESBENSHADE A, KUTTESCH JF, JACQUES TS, PERRY A, LEICHTER H, MAEDER P, BRUNDLER MA, WARNER J, NEAL J, ZACHARIN M, KORBONITS M, COLE T, TRAUNECKER H, MCLEAN TW, ROTONDO F, LEPAGE P, ALBRECHT S, HORVATH E, KOVACS K, PRIEST JR, FOULKES WD. Pituitary blastoma: a pathognomonic feature of germ-line DICER1 mutations. Acta Neuropathol. 2014a;128:111–22. doi: 10.1007/s00401-014-1285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE KOCK L, SABBAGHIAN N, SOGLIO DB, GUILLERMAN RP, PARK BK, CHAMI R, DEAL CL, PRIEST JR, FOULKES WD. Exploring the association Between DICER1 mutations and differentiated thyroid carcinoma. J Clin Endocrinol Metab. 2014b;99:E1072–7. doi: 10.1210/jc.2013-4206. [DOI] [PubMed] [Google Scholar]
- DE VITO C, RIGGI N, CORNAZ S, SUVA ML, BAUMER K, PROVERO P, STAMENKOVIC I. A TARBP2-dependent miRNA expression profile underlies cancer stem cell properties and provides candidate therapeutic reagents in Ewing sarcoma. Cancer Cell. 2012;21:807–21. doi: 10.1016/j.ccr.2012.04.023. [DOI] [PubMed] [Google Scholar]
- DEDES KJ, NATRAJAN R, LAMBROS MB, GEYER FC, LOPEZ-GARCIA MA, SAVAGE K, JONES RL, REIS-FILHO JS. Down-regulation of the miRNA master regulators Drosha and Dicer is associated with specific subgroups of breast cancer. Eur J Cancer. 2011;47:138–50. doi: 10.1016/j.ejca.2010.08.007. [DOI] [PubMed] [Google Scholar]
- DI LEVA G, GAROFALO M, CROCE CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIAZ-GARCIA CV, AGUDO-LOPEZ A, PEREZ C, LOPEZ-MARTIN JA, RODRIGUEZ-PERALTO JL, DE CASTRO J, CORTIJO A, MARTINEZ-VILLANUEVA M, IGLESIAS L, GARCIA-CARBONERO R, FRESNO VARA JA, GAMEZ-POZO A, PALACIOS J, CORTES-FUNES H, PAZ-ARES L, AGULLO-ORTUNO MT. DICER1, DROSHA and miRNAs in patients with non-small cell lung cancer: implications for outcomes and histologic classification. Carcinogenesis. 2013;34:1031–8. doi: 10.1093/carcin/bgt022. [DOI] [PubMed] [Google Scholar]
- DOROS L, YANG J, DEHNER L, ROSSI CT, SKIVER K, JARZEMBOWSKI JA, MESSINGER Y, SCHULTZ KA, WILLIAMS G, ANDRE N, HILL DA. DICER1 mutations in embryonal rhabdomyosarcomas from children with and without familial PPB-tumor predisposition syndrome. Pediatr Blood Cancer. 2012;59:558–60. doi: 10.1002/pbc.24020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOROS LA, ROSSI CT, YANG J, FIELD A, WILLIAMS GM, MESSINGER Y, CAJAIBA MM, PERLMAN EJ, KAS, CATHRO HP, LEGALLO RD, LAFORTUNE KA, CHIKWAVA KR, FARIA P, GELLER JI, DOME JS, MULLEN EA, GRATIAS EJ, DEHNER LP, HILL DA. DICER1 mutations in childhood cystic nephroma and its relationship to DICER1-renal sarcoma. Mod Pathol. 2014;27:1267–80. doi: 10.1038/modpathol.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FABER C, HORST D, HLUBEK F, KIRCHNER T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur J Cancer. 2011;47:1414–9. doi: 10.1016/j.ejca.2011.01.006. [DOI] [PubMed] [Google Scholar]
- FAGGAD A, BUDCZIES J, TCHERNITSA O, DARB-ESFAHANI S, SEHOULI J, MULLER BM, WIRTZ R, CHEKEROV R, WEICHERT W, SINN B, MUCHA C, ELWALI NE, SCHAFER R, DIETEL M, DENKERT C. Prognostic significance of Dicer expression in ovarian cancer-link to global microRNA changes and oestrogen receptor expression. J Pathol. 2010;220:382–91. doi: 10.1002/path.2658. [DOI] [PubMed] [Google Scholar]
- FAGGAD A, KASAJIMA A, WEICHERT W, STENZINGER A, ELWALI NE, DIETEL M, DENKERT C. Down-regulation of the microRNA processing enzyme Dicer is a prognostic factor in human colorectal cancer. Histopathology. 2012;61:552–61. doi: 10.1111/j.1365-2559.2011.04110.x. [DOI] [PubMed] [Google Scholar]
- FOULKES WD, BAHUBESHI A, HAMEL N, PASINI B, ASIOLI S, BAYNAM G, CHOONG CS, CHARLES A, FRIEDER RP, DISHOP MK, GRAF N, EKIM M, BOURON-DAL SOGLIO D, ARSENEAU J, YOUNG RH, SABBAGHIAN N, SRIVASTAVA A, TISCHKOWITZ MD, PRIEST JR. Extending the phenotypes associated with DICER1 mutations. Hum Mutat. 2011;32:1381–4. doi: 10.1002/humu.21600. [DOI] [PubMed] [Google Scholar]
- FOULKES WD, PRIEST JR, DUCHAINE TF. DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer. 2014;14:662–72. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- GRUND SE, POLYCARPOU-SCHWARZ M, LUO C, EICHMULLER SB, DIEDERICHS S. Rare Drosha splice variants are deficient in microRNA processing but do not affect general microRNA expression in cancer cells. Neoplasia. 2012;14:238–48. doi: 10.1593/neo.111586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO X, LIAO Q, CHEN P, LI X, XIONG W, MA J, LUO Z, TANG H, DENG M, ZHENG Y, WANG R, ZHANG W, LI G. The microRNA-processing enzymes: Drosha and Dicer can predict prognosis of nasopharyngeal carcinoma. J Cancer Res Clin Oncol. 2012;138:49–56. doi: 10.1007/s00432-011-1058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUO Y, TIAN P, YANG C, LIANG Z, LI M, SIMS M, LU L, ZHANG Z, LI H, PFEFFER LM, YUE J. Silencing the double-stranded RNA binding protein DGCR8 inhibits ovarian cancer cell proliferation, migration, and invasion. Pharm Res. 2015;32:769–78. doi: 10.1007/s11095-013-1219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HATA A, LIEBERMAN J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal. 2015;8:re3. doi: 10.1126/scisignal.2005825. [DOI] [PubMed] [Google Scholar]
- HATLEY ME, PATRICK DM, GARCIA MR, RICHARDSON JA, BASSEL-DUBY R, VAN ROOIJ E, OLSON EN. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERAVI-MOUSSAVI A, ANGLESIO MS, CHENG SW, SENZ J, YANG W, PRENTICE L, FEJES AP, CHOW C, TONE A, KALLOGER SE, HAMEL N, ROTH A, HA G, WAN AN, MAINES-BANDIERA S, SALAMANCA C, PASINI B, CLARKE BA, LEE AF, LEE CH, ZHAO C, YOUNG RH, APARICIO SA, SORENSEN PH, WOO MM, BOYD N, JONES SJ, HIRST M, MARRA MA, GILKS B, SHAH SP, FOULKES WD, MORIN GB, HUNTSMAN DG. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med. 2012;366:234–42. doi: 10.1056/NEJMoa1102903. [DOI] [PubMed] [Google Scholar]
- HERBERT KM, PIMIENTA G, DEGREGORIO SJ, ALEXANDROV A, STEITZ JA. Phosphorylation of DGCR8 increases its intracellular stability and induces a progrowth miRNA profile. Cell Rep. 2013;5:1070–81. doi: 10.1016/j.celrep.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HILL DA, IV, ANOVICH J, PRIEST JR, GURNETT CA, DEHNER LP, DESRUISSEAU D, JARZEMBOWSKI JA, WIKENHEISER-BROKAMP KA, SUAREZ BK, WHELAN AJ, WILLIAMS G, BRACAMONTES D, MESSINGER Y, GOODFELLOW PJ. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HORMAN SR, JANAS MM, LITTERST C, WANG B, MACRAE IJ, SEVER MJ, MORRISSEY DV, GRAVES P, LUO B, UMESALMA S, QI HH, MIRAGLIA LJ, NOVINA CD, ORTH AP. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol Cell. 2013;50:356–67. doi: 10.1016/j.molcel.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IWASAKI YW, KIGA K, KAYO H, FUKUDA-YUZAWA Y, WEISE J, INADA T, TOMITA M, ISHIHAMA Y, FUKAO T. Global microRNA elevation by inducible Exportin 5 regulates cell cycle entry. RNA. 2013;19:490–7. doi: 10.1261/rna.036608.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAFARNEJAD SM, SJOESTROEM C, MARTINKA M, LI G. Expression of the RNase III enzyme DROSHA is reduced during progression of human cutaneous melanoma. Mod Pathol. 2013;26:902–10. doi: 10.1038/modpathol.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAKYMIW A, PATEL RS, DEMING N, BHATTACHARYYA I, SHAH P, LAMONT RJ, STEWART CM, COHEN DM, CHAN EK. Overexpression of dicer as a result of reduced let-7 MicroRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes Cancer. 2010;49:549–59. doi: 10.1002/gcc.20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KARUBE Y, TANAKA H, OSADA H, TOMIDA S, TATEMATSU Y, YANAGISAWA K, YATABE Y, TAKAMIZAWA J, MIYOSHI S, MITSUDOMI T, TAKAHASHI T. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHOSHNAW SM, RAKHA EA, ABDEL-FATAH TM, NOLAN CC, HODI Z, MACMILLAN DR, ELLIS IO, GREEN AR. Loss of Dicer expression is associated with breast cancer progression and recurrence. Breast Cancer Res Treat. 2012;135:403–13. doi: 10.1007/s10549-012-2169-3. [DOI] [PubMed] [Google Scholar]
- KIM B, LEE JH, PARK JW, KWON TK, BAEK SK, HWANG I, KIM S. An essential microRNA maturing microprocessor complex component DGCR8 is up-regulated in colorectal carcinomas. Clin Exp Med. 2014;14:331–6. doi: 10.1007/s10238-013-0243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM MS, LEE SH, YOO NJ. DICER1 exons 25 and 26 mutations are rare in common human tumours besides Sertoli-Leydig cell tumour. Histopathology. 2013;63:436–8. doi: 10.1111/his.12161. [DOI] [PubMed] [Google Scholar]
- KIM VN, HAN J, SIOMI MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- KUMAR MS, LU J, MERCER KL, GOLUB TR, JACKS T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- KUMAR MS, PESTER RE, CHEN CY, LANE K, CHIN C, LU J, KIRSCH DG, GOLUB TR, JACKS T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–4. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAMBERTZ I, NITTNER D, MESTDAGH P, DENECKER G, VANDESOMPELE J, DYER MA, MARINE JC. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17:633–41. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEADERER D, HOFFMAN AE, ZHENG T, FU A, WEIDHAAS J, PARANJAPE T, ZHU Y. Genetic and epigenetic association studies suggest a role of microRNA biogenesis gene exportin-5 (XPO5) in breast tumorigenesis. Int J Mol Epidemiol Genet. 2011;2:9–18. [PMC free article] [PubMed] [Google Scholar]
- LIN RJ, LIN YC, CHEN J, KUO HH, CHEN YY, DICCIANNI MB, LONDON WB, CHANG CH, YU AL. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–50. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN S, GREGORY RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–33. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LU J, GETZ G, MISKA EA, ALVAREZ-SAAVEDRA E, LAMB J, PECK D, SWEET-CORDERO A, EBERT BL, MAK RH, FERRANDO AA, DOWNING JR, JACKS T, HORVITZ HR, GOLUB TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- MA Z, SWEDE H, CASSARINO D, FLEMING E, FIRE A, DADRAS SS. Up-regulated Dicer expression in patients with cutaneous melanoma. PLoS One. 2011;6:e20494. doi: 10.1371/journal.pone.0020494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASSAGUE J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEISTER G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–59. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- MELO SA, MOUTINHO C, ROPERO S, CALIN GA, ROSSI S, SPIZZO R, FERNANDEZ AF, DAVALOS V, VILLANUEVA A, MONTOYA G, YAMAMOTO H, SCHWARTZ S, JR, ESTELLER M. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- MELO SA, ROPERO S, MOUTINHO C, AALTONEN LA, YAMAMOTO H, CALIN GA, ROSSI S, FERNANDEZ AF, CARNEIRO F, OLIVEIRA C, FERREIRA B, LIU CG, VILLANUEVA A, CAPELLA G, SCHWARTZ S, JR, SHIEKHATTAR R, ESTELLER M. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–70. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- MERRITT WM, LIN YG, HAN LY, KAMAT AA, SPANNUTH WA, SCHMANDT R, URBAUER D, PENNACCHIO LA, CHENG JF, NICK AM, DEAVERS MT, MOURAD-ZEIDAN A, WANG H, MUELLER P, LENBURG ME, GRAY JW, MOK S, BIRRER MJ, LOPEZ-BERESTEIN G, COLEMAN RL, BAR-ELI M, SOOD AK. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–50. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORI M, TRIBOULET R, MOHSENI M, SCHLEGELMILCH K, SHRESTHA K, CAMARGO FD, GREGORY RI. Hippo Signaling Regulates Microprocessor and Links Cell-Density-Dependent miRNA Biogenesis to Cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURALIDHAR B, GOLDSTEIN LD, NG G, WINDER DM, PALMER RD, GOODING EL, BARBOSA-MORAIS NL, MUKHERJEE G, THORNE NP, ROBERTS I, PETT MR, COLEMAN N. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212:368–77. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- MURALIDHAR B, WINDER D, MURRAY M, PALMER R, BARBOSA-MORAIS N, SAINI H, ROBERTS I, PETT M, COLEMAN N. Functional evidence that Drosha overexpression in cervical squamous cell carcinoma affects cell phenotype and microRNA profiles. J Pathol. 2011;224:496–507. doi: 10.1002/path.2898. [DOI] [PubMed] [Google Scholar]
- NEMLICH Y, GREENBERG E, ORTENBERG R, BESSER MJ, BARSHACK I, JACOB-HIRSCH J, JACOBY E, EYAL E, RIVKIN L, PRIETO VG, CHAKRAVARTI N, DUNCAN LM, KALLENBERG DM, GALUN E, BENNETT DC, AMARIGLIO N, BAR-ELI M, SCHACHTER J, RECHAVI G, MARKEL G. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J Clin Invest. 2013;123:2703–18. doi: 10.1172/JCI62980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAMPALAKIS G, DIAMANDIS EP, KATSAROS D, SOTIROPOULOU G. Down-regulation of dicer expression in ovarian cancer tissues. Clin Biochem. 2010;43:324–7. doi: 10.1016/j.clinbiochem.2009.09.014. [DOI] [PubMed] [Google Scholar]
- PAPACHRISTOU DJ, KORPETINOU A, GIANNOPOULOU E, ANTONACOPOULOU AG, PAPADAKI H, GRIVAS P, SCOPA CD, KALOFONOS HP. Expression of the ribonucleases Drosha, Dicer, and Ago2 in colorectal carcinomas. Virchows Arch. 2011;459:431–40. doi: 10.1007/s00428-011-1119-5. [DOI] [PubMed] [Google Scholar]
- PAPACHRISTOU DJ, SKLIROU E, CORRADI D, GRASSANI C, KONTOGEORGAKOS V, RAO UN. Immunohistochemical analysis of the endoribonucleases Drosha, Dicer and Ago2 in smooth muscle tumours of soft tissues. Histopathology. 2012;60:E28–36. doi: 10.1111/j.1365-2559.2012.04192.x. [DOI] [PubMed] [Google Scholar]
- PAROO Z, YE X, CHEN S, LIU Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–22. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PASSON N, GEROMETTA A, PUPPIN C, LAVARONE E, PUGLISI F, TELL G, DI LORETO C, DAMANTE G. Expression of Dicer and Drosha in triple-negative breast cancer. J Clin Pathol. 2012;65:320–6. doi: 10.1136/jclinpath-2011-200496. [DOI] [PubMed] [Google Scholar]
- POTENZA N, PAPA U, SCARUFFI P, MOSCA N, TONINI GP, RUSSO A. A novel splice variant of the human dicer gene is expressed in neuroblastoma cells. FEBS Lett. 2010;584:3452–7. doi: 10.1016/j.febslet.2010.06.045. [DOI] [PubMed] [Google Scholar]
- PUGH TJ, YU W, YANG J, FIELD AL, AMBROGIO L, CARTER SL, CIBULSKIS K, GIANNIKOPOULOS P, KIEZUN A, KIM J, MCKENNA A, NICKERSON E, GETZ G, HOFFHER S, MESSINGER YH, DEHNER LP, ROBERTS CW, RODRIGUEZ-GALINDO C, WILLIAMS GM, ROSSI CT, MEYERSON M, HILL DA. Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene. 2014;33:5295–302. doi: 10.1038/onc.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- QI HH, ONGUSAHA PP, MYLLYHARJU J, CHENG D, PAKKANEN O, SHI Y, LEE SW, PENG J, SHI Y. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–4. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RADICH JP, DAI H, MAO M, OEHLER V, SCHELTER J, DRUKER B, SAWYERS C, SHAH N, STOCK W, WILLMAN CL, FRIEND S, LINSLEY PS. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103:2794–9. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAKHEJA D, CHEN KS, LIU Y, SHUKLA AA, SCHMID V, CHANG TC, KHOKHAR S, WICKISER JE, KARANDIKAR NJ, MALTER JS, MENDELL JT, AMATRUDA JF. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun. 2014;2:4802. doi: 10.1038/ncomms5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RATH SR, BARTLEY A, CHARLES A, POWERS N, BAYNAM G, JONES T, PRIEST JR, FOULKES WD, CHOONG CS. Multinodular Goiter in children: an important pointer to a germline DICER1 mutation. J Clin Endocrinol Metab. 2014;99:1947–8. doi: 10.1210/jc.2013-3932. [DOI] [PubMed] [Google Scholar]
- RIO FRIO T, BAHUBESHI A, KANELLOPOULOU C, HAMEL N, NIEDZIELA M, SABBAGHIAN N, POUCHET C, GILBERT L, O’BRIEN PK, SERFAS K, BRODERICK P, HOULSTON RS, LESUEUR F, BONORA E, MULJO S, SCHIMKE RN, BOURON-DAL SOGLIO D, ARSENEAU J, SCHULTZ KA, PRIEST JR, NGUYEN VH, HARACH HR, LIVINGSTON DM, FOULKES WD, TISCHKOWITZ M. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA. 2011;305:68–77. doi: 10.1001/jama.2010.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAND M, GAMBICHLER T, SKRYGAN M, SAND D, SCOLA N, ALTMEYER P, BECHARA FG. Expression levels of the microRNA processing enzymes Drosha and dicer in epithelial skin cancer. Cancer Invest. 2010;28:649–53. doi: 10.3109/07357901003630918. [DOI] [PubMed] [Google Scholar]
- SAND M, SKRYGAN M, GEORGAS D, ARENZ C, GAMBICHLER T, SAND D, ALTMEYER P, BECHARA FG. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol Carcinog. 2012;51:916–22. doi: 10.1002/mc.20861. [DOI] [PubMed] [Google Scholar]
- SCHULTZ KA, PACHECO MC, YANG J, WILLIAMS GM, MESSINGER Y, HILL DA, DEHNER LP, PRIEST JR. Ovarian sex cord-stromal tumors, pleuropulmonary blastoma and DICER1 mutations: a report from the International Pleuropulmonary Blastoma Registry. Gynecol Oncol. 2011;122:246–50. doi: 10.1016/j.ygyno.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEKI M, YOSHIDA K, SHIRAISHI Y, SHIMAMURA T, SATO Y, NISHIMURA R, OKUNO Y, CHIBA K, TANAKA H, KATO K, KATO M, HANADA R, NOMURA Y, PARK MJ, ISHIDA T, OKA A, IGARASHI T, MIYANO S, HAYASHI Y, OGAWA S, TAKITA J. Biallelic DICER1 mutations in sporadic pleuropulmonary blastoma. Cancer Res. 2014;74:2742–9. doi: 10.1158/0008-5472.CAN-13-2470. [DOI] [PubMed] [Google Scholar]
- SEO GJ, KINCAID RP, PHANAKSRI T, BURKE JM, PARE JM, COX JE, HSIANG TY, KRUG RM, SULLIVAN CS. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe. 2013;14:435–45. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHEN J, XIA W, KHOTSKAYA YB, HUO L, NAKANISHI K, LIM SO, DU Y, WANG Y, CHANG WC, CHEN CH, HSU JL, WU Y, LAM YC, JAMES BP, LIU X, LIU CG, PATEL DJ, HUNG MC. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–7. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHU GS, YANG ZL, LIU DC. Immunohistochemical study of Dicer and Drosha expression in the benign and malignant lesions of gallbladder and their clinicopathological significances. Pathol Res Pract. 2012;208:392–7. doi: 10.1016/j.prp.2012.05.001. [DOI] [PubMed] [Google Scholar]
- SIOMI H, SIOMI MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–32. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- STRATMANN J, WANG CJ, GNOSA S, WALLIN A, HINSELWOOD D, SUN XF, ZHANG H. Dicer and miRNA in relation to clinicopathological variables in colorectal cancer patients. BMC Cancer. 2011;11:345. doi: 10.1186/1471-2407-11-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SU X, CHAKRAVARTI D, CHO MS, LIU L, GI YJ, LIN YL, LEUNG ML, EL-NAGGAR A, CREIGHTON CJ, SURAOKAR MB, WISTUBA I, FLORES ER. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–90. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUGITO N, ISHIGURO H, KUWABARA Y, KIMURA M, MITSUI A, KUREHARA H, ANDO T, MORI R, TAKASHIMA N, OGAWA R, FUJII Y. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin Cancer Res. 2006;12:7322–8. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- SUZUKI HI, YAMAGATA K, SUGIMOTO K, IWAMOTO T, KATO S, MIYAZONO K. Modulation of microRNA processing by p53. Nature. 2009;460:529–33. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- TANG X, LI M, TUCKER L, RAMRATNAM B. Glycogen synthase kinase 3 beta (GSK3beta) phosphorylates the RNAase III enzyme Drosha at S300 and S302. PLoS One. 2011;6:e20391. doi: 10.1371/journal.pone.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANG X, WEN S, ZHENG D, TUCKER L, CAO L, PANTAZATOS D, MOSS SF, RAMRATNAM B. Acetylation of drosha on the N-terminus inhibits its degradation by ubiquitination. PLoS One. 2013;8:e72503. doi: 10.1371/journal.pone.0072503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TANG X, ZHANG Y, TUCKER L, RAMRATNAM B. Phosphorylation of the RNase III enzyme Drosha at Serine300 or Serine302 is required for its nuclear localization. Nucleic Acids Res. 2010;38:6610–9. doi: 10.1093/nar/gkq547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCHERNITSA O, KASAJIMA A, SCHAFER R, KUBAN RJ, UNGETHUM U, GYORFFY B, NEUMANN U, SIMON E, WEICHERT W, EBERT MP, ROCKEN C. Systematic evaluation of the miRNA-ome and its downstream effects on mRNA expression identifies gastric cancer progression. J Pathol. 2010;222:310–9. doi: 10.1002/path.2759. [DOI] [PubMed] [Google Scholar]
- TOMIAK E, DE KOCK L, GRYNSPAN D, RAMPHAL R, FOULKES WD. DICER1 mutations in an adolescent with cervical embryonal rhabdomyosarcoma (cERMS) Pediatr Blood Cancer. 2014;61:568–9. doi: 10.1002/pbc.24826. [DOI] [PubMed] [Google Scholar]
- TORRES A, TORRES K, PASZKOWSKI T, JODLOWSKA-JEDRYCH B, RADOMANSKI T, KSIAZEK A, MACIEJEWSKI R. Major regulators of microRNAs biogenesis Dicer and Drosha are down-regulated in endometrial cancer. Tumour Biol. 2011;32:769–76. doi: 10.1007/s13277-011-0179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TORREZAN GT, FERREIRA EN, NAKAHATA AM, BARROS BD, CASTRO MT, CORREA BR, KREPISCHI AC, OLIVIERI EH, CUNHA IW, TABORI U, GRUNDY PE, COSTA CM, DE CAMARGO B, GALANTE PA, CARRARO DM. Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat Commun. 2014;5:4039. doi: 10.1038/ncomms5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRABUCCHI M, BRIATA P, GARCIA-MAYORAL M, HAASE AD, FILIPOWICZ W, RAMOS A, GHERZI R, ROSENFELD MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAKSMAN O, HETLAND TE, TROPE CG, REICH R, DAVIDSON B. Argonaute, Dicer, and Drosha are up-regulated along tumor progression in serous ovarian carcinoma. Hum Pathol. 2012;43:2062–9. doi: 10.1016/j.humpath.2012.02.016. [DOI] [PubMed] [Google Scholar]
- VAN DEN BEUCKEN T, KOCH E, CHU K, RUPAIMOOLE R, PRICKAERTS P, ADRIAENS M, VONCKEN JW, HARRIS AL, BUFFA FM, HAIDER S, STARMANS MH, YAO CQ, IV, AN M, IV, AN C, PECOT CV, BOUTROS PC, SOOD AK, KORITZINSKY M, WOUTERS BG. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:5203. doi: 10.1038/ncomms6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WADA T, KIKUCHI J, FURUKAWA Y. Histone deacetylase 1 enhances microRNA processing via deacetylation of DGCR8. EMBO Rep. 2012;13:142–9. doi: 10.1038/embor.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAGH V, POMORSKI A, WILSCHUT KJ, PIOMBO S, BERNSTEIN HS. MicroRNA-363 negatively regulates the left ventricular determining transcription factor HAND1 in human embryonic stem cell-derived cardiomyocytes. Stem Cell Res Ther. 2014;5:75. doi: 10.1186/scrt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALZ AL, OOMS A, GADD S, GERHARD DS, SMITH MA, GUIDRY AUVIL JM, MEERZAMAN D, CHEN QR, HSU CH, YAN C, NGUYEN C, HU Y, BOWLBY R, BROOKS D, MA Y, MUNGALL AJ, MOORE RA, SCHEIN J, MARRA MA, HUFF V, DOME JS, CHI YY, MULLIGHAN CG, MA J, WHEELER DA, HAMPTON OA, JAFARI N, ROSS N, GASTIER-FOSTER JM, PERLMAN EJ. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell. 2015;27:286–97. doi: 10.1016/j.ccell.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAN G, ZHANG X, LANGLEY RR, LIU Y, HU X, HAN C, PENG G, ELLIS LM, JONES SN, LU X. DNA-damage-induced nuclear export of precursor microRNAs is regulated by the ATM-AKT pathway. Cell Rep. 2013;3:2100–12. doi: 10.1016/j.celrep.2013.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEGERT J, ISHAQUE N, VARDAPOUR R, GEORG C, GU Z, BIEG M, ZIEGLER B, BAUSENWEIN S, NOURKAMI N, LUDWIG N, KELLER A, GRIMM C, KNEITZ S, WILLIAMS RD, CHAGTAI T, PRITCHARD-JONES K, VAN SLUIS P, VOLCKMANN R, KOSTER J, VERSTEEG R, ACHA T, O’SULLIVAN MJ, BODE PK, NIGGLI F, TYTGAT GA, VAN TINTEREN H, VAN DEN HEUVEL-EIBRINK MM, MEESE E, VOKUHL C, LEUSCHNER I, GRAF N, EILS R, PFISTER SM, KOOL M, GESSLER M. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell. 2015;27:298–311. doi: 10.1016/j.ccell.2015.01.002. [DOI] [PubMed] [Google Scholar]
- WINTER J, DIEDERICHS S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8:1149–57. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- WITKOWSKI L, MATTINA J, SCHONBERGER S, MURRAY MJ, CHOONG CS, HUNTSMAN DG, REIS-FILHO JS, MCCLUGGAGE WG, NICHOLSON JC, COLEMAN N, CALAMINUS G, SCHNEIDER DT, ARSENEAU J, STEWART CJ, FOULKES WD. DICER1 hotspot mutations in non-epithelial gonadal tumours. Br J Cancer. 2013;109:2744–50. doi: 10.1038/bjc.2013.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU C, SO J, DAVIS-DUSENBERY BN, QI HH, BLOCH DB, SHI Y, LAGNA G, HATA A. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol Cell Biol. 2011a;31:4760–74. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU D, TAO J, XU B, LI P, LU Q, ZHANG W. Downregulation of Dicer, a component of the microRNA machinery, in bladder cancer. Mol Med Rep. 2012;5:695–9. doi: 10.3892/mmr.2011.711. [DOI] [PubMed] [Google Scholar]
- WU JF, SHEN W, LIU NZ, ZENG GL, YANG M, ZUO GQ, GAN XN, REN H, TANG KF. Down-regulation of Dicer in hepatocellular carcinoma. Med Oncol. 2011b;28:804–9. doi: 10.1007/s12032-010-9520-5. [DOI] [PubMed] [Google Scholar]
- WU MK, SABBAGHIAN N, XU B, ADDIDOU-KALUCKI S, BERNARD C, ZOU D, REEVE AE, ECCLES MR, COLE C, CHOONG CS, CHARLES A, TAN TY, IGLESIAS DM, GOODYER PR, FOULKES WD. Biallelic DICER1 mutations occur in Wilms tumours. J Pathol. 2013;230:154–64. doi: 10.1002/path.4196. [DOI] [PubMed] [Google Scholar]
- YAN M, HUANG HY, WANG T, WAN Y, CUI SD, LIU ZZ, FAN QX. Dysregulated expression of dicer and drosha in breast cancer. Pathol Oncol Res. 2012;18:343–8. doi: 10.1007/s12253-011-9450-3. [DOI] [PubMed] [Google Scholar]
- YANG JS, LAI EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle. 2010;9:4455–60. doi: 10.4161/cc.9.22.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG JS, LAI EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG JS, MAURIN T, ROBINE N, RASMUSSEN KD, JEFFREY KL, CHANDWANI R, PAPAPETROU EP, SADELAIN M, O’CARROLL D, LAI EC. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci U S A. 2010;107:15163–8. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZENG Y, SANKALA H, ZHANG X, GRAVES PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413:429–36. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- ZHANG X, WAN G, BERGER FG, HE X, LU X. The ATM kinase induces microRNA biogenesis in the DNA damage response. Mol Cell. 2011;41:371–83. doi: 10.1016/j.molcel.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHU DX, FAN L, LU RN, FANG C, SHEN WY, ZOU ZJ, WANG YH, ZHU HY, MIAO KR, LIU P, XU W, LI JY. Downregulated Dicer expression predicts poor prognosis in chronic lymphocytic leukemia. Cancer Sci. 2012;103:875–81. doi: 10.1111/j.1349-7006.2012.02234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]