Abstract
Cyclin D1 binds and activates cyclin-dependent kinases 4/6 (Cdk4/6) to phosphorylate the retinoblastoma (RB) family proteins, relieving E2F/DPs from the negative restraint of RB proteins and histone deacetylases. The cyclin D-Cdk4/6 complexes activate cyclin E/Cdk2 through titration of the Cdk inhibitors p21Cip1/p27Kip1. Cyclin E/Cdk2 further phosphorylates RBs, thereby activating E2F/DPs, and cells enter the S phase of the cell cycle. Cyclin D-Cdk4/6 also phosphorylates MEP50 subunit of the protein arginine methyltransferase 5 (PRMT5), which cooperates with cyclin D1 to drive lymphomagenesis in vivo. Activated PRMPT5 causes arginine methylation of p53 to suppress expression of pro-apoptotic and anti-proliferative target genes, explaining the molecular mechanism for tumorigenesis. Cyclin D1 physically interacts with transcription factors such as estrogen receptor, androgen receptor, and Myb family proteins to regulate gene expression in Cdk-independent fashion. Dmp1 is a Myb-like protein that quenches the oncogenic signals from activated Ras or HER2 by inducing Arf/p53-dependent cell cycle arrest. Cyclin D1 binds to Dmp1α to activate both Arf and Ink4a promoters to induce cell cycle arrest or apoptosis in non-transformed cells to prevent them from neoplastic transformation. Dmp1-deficiency significantly accelerates mouse mammary tumorigenesis with reduced apoptosis and increased metastasis. Cyclin D1 interferes with ligand activation of PPARγ involved in cellular differentiation; it also physically interacts with histone deacetylases (HDACs) and p300 to repress gene expression. It has also been shown that cyclin D1 accelerates tumorigenesis through transcriptional activation of miR-17/20 and Dicer1 which, in turn, represses cyclin D1 expression. Identification of cyclin D1-binding proteins/promoters will be essential for further clarification of its biological activities.
Keywords: cyclin D1, overexpression, breast cancer, prognosis, DMP1 (DMTF1), transcription, PRMT5, microRNA, Dicer
Introduction
D-type cyclins bind and activate cyclin dependent kinases (Cdk4 and Cdk6) to phosphorylate the retinoblastoma (RB) family proteins, regulating the G1/S-phase transition. The cyclin D-Cdk4/6 complexes also titrate the Cdk inhibitors p21Cip1 and p27Kip1, thus activates cyclin E/Cdk2 (1, 2). Of note, both p21Cip1 and p27Kip1 behave as assembly factors for cyclin D-Cdk4/6 as demonstrated by the study using mouse embryonic fibroblasts (MEFs) from p21/p27 double-knockout mice (3). The cyclin E/Cdk2 complex completes the phosphorylation of pRB and histone deacetylases (HDACs), relieving E2F/DPs from their negative constraint. p27kip1 is phosphorylated by cyclin E/Cdk2 for proteasomal degradation (4), and when the levels of p27Kip1 decreases to a certain threshold level, the G1-S progression becomes irreversible, and cells enter the S phase of the cell cycle (5).
There are at least three known mechanisms that increase cyclin D levels and thus activation of CDK4/6: i) increased transcription of the cyclin D genes driven by a variety of transcription factors, primarily those that are involved in mitogenic signaling pathways, i.e., Fos/Jun, Ets-2, STATs, NF-κB, and others in cyclin D1, ii) elevated cyclin D protein translation, mediated through PI3K-Akt-mTOR-S6K1 signaling (6, 7), and iii) stabilization and nuclear localization of cyclin D proteins, regulated by C-terminal phosphorylation of the protein (Thr286 in cyclin D1) (8, 9). The signaling pathways regulate nuclear cyclin D protein levels through site-specific phosphorylation by GSK3β or other Thr286 kinases; these kinases phosphorylate D-type cyclins, thereby promoting its nuclear export, polyubiquitination, and proteasomal degradation in the cytoplasm. This phosphorylation-dependent proteolysis of D-cyclins is mediated through at least three distinct F-box proteins (FBXO5, FBXW8, and FBXO31), all of which recruit phosphorylated D-cyclin to the Skp, Cullin, and F-box (SCF) ubiquitin ligase complex (10).
Gene knockout studies in mice have shown an essential role for cyclin D1 in whole body development; those of the retina and the central nervous system, and terminal differentiation/proliferation of mammary buds (11). Knock-in mice using a kinase-deficient cyclin D1 mutant (D1K112E) established a critical role for cyclin D1/Cdks in tumorigenesis driven by HER2/neu, but not normal mammary gland development (12). Recently the same group demonstrated that cyclin D1 inhibits autophagy since immortalized cyclin D1KE/KE mouse mammary epithelial cells retained high rates of autophagy and reduced ErbB2-mediated transformation in vivo (13). Autophagy upregulation was also found in human mammary epithelial cells subjected to inhibition of Cyclin D1/CDK activity, and simultaneous pharmacologic inhibition of CDK and autophagy enhanced senescence response, suggesting that inhibition of both activities will give more efficacious treatment for breast cancer (13).
In humans, Cyclin D1 is the product of the CCND1 gene located on chromosome 11q13, and is amplified in ~15% of breast cancers (14–16). 11p13 is one of the four representative chromosomal loci where high levels of gene amplification and/or overexpression is strongly associated with short survival of breast cancer (17, 18). However, Cyclin D1 is overexpressed at the protein levels in ~50% of breast cancers in the presence or absence of gene amplification (19). The difference in the frequency of CCND1 gene amplification and protein overexpression can be explained by the Cyclin D1 promoter activation by aberrant mitogenic signaling (e.g. HER2 or EGFR amplification/overexpression) mediated by MAPK-AP1/Ets and PI3K-NF-κB pathways (20). Moreover, a recent study showing that inhibition of cyclin D1 degradation by mutations of Fbx4 (cyclin D1 E3 ligase) could explain cyclin D1 overexpression without gene amplification (21, 22). Overexpression of cyclin D1 occurs in other human malignancies than the breast including carcinomas of the esophagus, colon, and lung (23, 24). It is generally believed that overexpression of cyclin D1 contributes to tumorigenesis; however, the prognostic value of Cyclin D1 in cancer is still controversial. Cyclin D1 expression is generally a favorable prognostic factor (25); however, Cyclin D1 overexpression, when caused by CCND1 genomic amplification, is associated with shorter survival of cancer patients (16, 19, 26). Conversely, Cyclin D1b protein overexpression is independent of Cyclin D1a, and associated with poor disease outcome (27).
In addition to the established roles of cyclin D1/Cdks for controlling in G1-S progression through pRB/HDAC phosphorylation and activation of E2F/DPs, the enzyme complex causes histone methylation through activation of protein arginine methyltransferase 5 (PRMT5)/MP50 (28). PRMT5 also causes p53 inactivation through methylation and accelerates tumorigenesis (29). These novel findings will be a topic of discussion.
Many of its biological activities of cyclin D1 have been attributed to transcriptional regulation independent of Cdks. For instance, cyclin D1 regulates the growth of estrogen-responsive tissues through interaction with DNA-binding proteins, e.g., by activating the estrogen receptor α (ERα) in the absence of estrogen (30). Cyclin D1 modifies the activities of Dmp1 (cyclin D binding myb-like protein 1; Dmtf1) transcription factor independent of Cdk4/6 (31–33). Dmp1 (Dmp1α) is a physiological regulator of the Arf-p53 pathway that receives oncogenic hyperproliferative signals from mutant Ras or HER2 overexpression and induces cell Arf/p53- dependent cycle arrest or apoptosis to eliminate early stage cancer cells to avoid neoplastic transformation (reviewed in 33, 34; for Arf. see 35–37). Cyclin D1 accelerates G1-S progression when expressed at physiological levels through phosphorylation of RB and activation of E2F/DPs (1–5); overexpression of cyclin D1 was expected to activate the Arf-p53 pathway when it is overexpressed since Arf receives nearly all oncogenic hyperproliferative signals to p53 (35–37). We recently conducted a series of experiments to prove that overexpressed cyclin D1 binds to Dmp1 to activate the Arf and Ink4a promoters, and as a consequence, cyclin D1-induced mammary carcinogenesis is accelerated in mice that were deficient for Dmp1 (38). The results for these assays are discussed in this review. These Cdk-independent activities of cyclin D1 could underlie its oncogenic role in human cancers.
Recent studies show that cyclin D1 regulates gene expression through physical interaction of HDACs and p300. It has also been reported that cyclin D1 binds to the regulatory region of microRNAs/Dicer and affects their gene expression. These advances on novel functions of cyclin D1 in cell cycle regulation and tumorigenesis will be discussed.
The genomic structure of the CCND1 locus – alternative splicing of CCND1 in human cancer
The genomic structure for the human Cyclin D1 (CCND1) locus is shown in Fig. 1. There have been over 100 single nucleotide polymorphisms (SNPs) identified at the CCND1 locus without altering the primary amino acid sequence of the protein (39, 40). The SNP at G/A870 (Pro241) located at the splice donor site of intron 4 has been a subject of alternative splicing (Fig. 1). Although the polymorphic residue is silent for the amino acid sequence, the “A” allele reduces the efficiency of the splice donor site, and favors the production of an alternative transcript known as Cyclin D1b (40–43). The Cyclin D1b protein has new 33 amino acid stretch at the C-terminus from the intron 4 (Fig. 1B, I4). The lack of exon 5 makes Cyclin D1b a constitutively nuclear protein thought to be equivalent to mouse cyclin D1T286A (8) since Cyclin D1b lacks a PEST (Pro-Glu-Ser-Thr) motif and the Thr286 residue that are required for nuclear export and degradation of the protein (40–43). Despite the lack of this region, the half-life of cyclin D1b is only slightly greater than that of cyclin D1a (~24 min). Cyclin D1b binds to CDK4/6 for activation, but at lower efficiency in phosphorylating pRb when compared with Cyclin D1a (40). The ER regulatory function for cyclin D1a is reportedly disrupted in Cyclin D1b since it lacks the LxxLL motif which is essential for its ligand-dependent interaction with nuclear receptors (40). However, stable overexpression of Cyclin D1b in NIH 3T3 cells promotes anchorage-independent growth (43), and leads to the focus formation after 12 passages (23). It is generally considered that overexpression of Cyclin D1a alone is not sufficient for cellular transformation; indeed it takes >20 months for parous MMTV-Cyclin D1a transgenic mice to develop mammary tumors (44–46). Rather, the constitutive nuclear localization of cyclin D1 is critical to reveal its oncogenic potential since this can lead to DNA re-replication, genomic instability and loss of contact inhibition (40, 42, 47). Thus Cyclin D1b can cause cellular transformation and has been linked to human cancers, and produces tumors in vivo (23, 42, 43).
Figure 1. The genomic and domain structures for Cyclin D1a (A) and b (B).
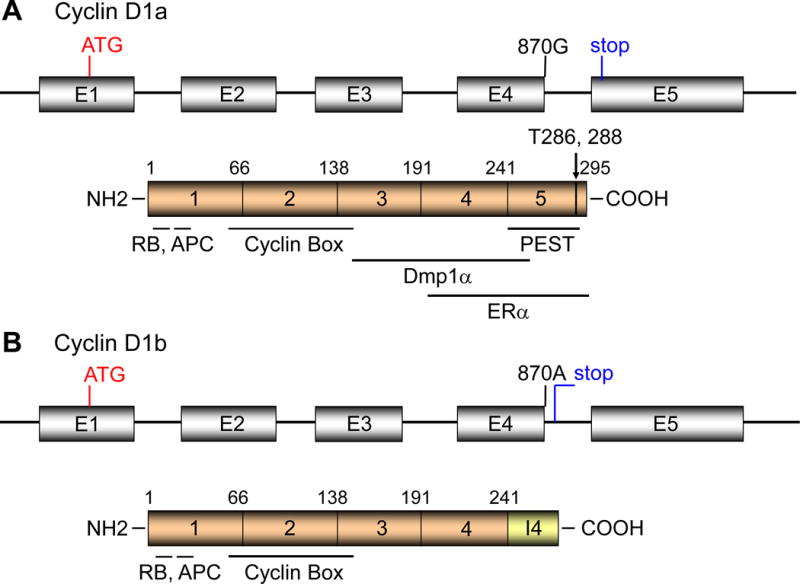
A. The human Cyclin D1 locus consists of 5 exons, encoding the full length product of 295 amino acids. The LxCxE RB-binding motif is at the N-terminus (a.a. 5–9), the RxxL motif responsible for APC interaction is at a.a. 29–32, the cyclin box responsible for Cdk interaction is at a.a. 56–145. The Dmp1α-binding and ERα-binding domains have been mapped outside the cyclin box (a.a. 142–253). The LxxLL motif that is essential for its ligand-dependent interaction with nuclear receptors is at a.a. 251–255. The PEST domain (a.a. 241–290) and Thr286 responsible for cyclin D1 degradation are located at the carboxyl terminus of the protein.
B. The SNP at G/A870 (Pro241) located at the splice donor site of intron 4 causes alternative splicing. The Cyclin D1b protein has new 33 amino acid stretch at the C-terminus as contributed by the intron 4 and the stop codon. Although Cyclin D1b lacks the C-terminal PEST, Thr286, Thr288 responsible for phosphorylation/degradation, the half-life of the protein is not significantly different from that of Cyclin D1a. It is considered that the constant nuclear localization of Cyclin D1b contributes to its oncogenic activity.
Regulation of cyclin D1
Cyclin D1 is regulated at both transcriptional and post-transcriptional levels. The human and mouse cyclin D1 promoter sequences are homologous and transcription factor-binding sites have been mapped to their promoter regions (48). Significantly, these studies have revealed enormous diversity in the transcriptional programs controlling cyclin D1 transcription, with a wide variety of signaling pathways converging on this promoter, dependent on the cell type where cyclin D1 is expressed. The signaling cascades that affect cyclin D1 transcription include MAP kinases, phosphoinositide 3-kinase/protein kinase B (PI3K/Akt) signaling, IKK/IκB/NF-κB pathway, Wnt/β-catenin signaling, STAT signaling, and nuclear hormone receptors. Consistent with these, the cyclin D1 promoter has consensus sequences for Ets, Fos/Jun, NF-κB, STAT, TCF, E2F, Sp1, EGR, and ERα (48).
In addition to transcriptional regulation, cells employ a vast array of post-transcriptional mechanisms that can rapidly modulate cyclin D1’s activity allowing cell cycle progression in response to mitogens. Translation of the cyclin D1 mRNA is regulated by PI3K-Akt-mTOR-S6K1 signaling, which determines the association of cyclin D1 with CDK4/6, nucleocytoplasmic transport, and degradation. Accumulating pieces of evidence have shown that DNA transcription, mRNA processing and translation are all closely interconnected with regulatory information flowing bidirectionally (49).
The cyclin D1 protein is inherently unstable (half-life: 24 minutes; 8, 9), providing an additional level of regulation. Instability of the protein has been attributed to the PEST sequence (41, 50) and the presence of a specific GSK-3β phosphorylation site (Thr 286) at the C-terminus (8, 9). Phosphorylation of the Thr 286 has also been shown to target the cyclin D1 protein for nuclear export and proteasomal degradation (51). Thus phosphorylation-dependent destruction of cyclin D1 provides a critical indicator to prevent overexpression of cyclin D1 resulting from continuous mitogen-dependent cyclin D1 transcription.
Thr286 is phosphorylated by other kinases than GSK3β for proteasomal degradation. p38SAPK2 phosphorylates cyclin D1 in vitro at Thr286 to trigger the ubiquitination and degradation (52), which explains the molecular mechanisms by which stress transduction pathways regulate the cell cycle machinery and take control of cell proliferation. Although cyclin D1 is phosphorylated by GSK3β at Thr286 in non-transformed cells (9), it is phosphorylated by MAPK and the F-box protein FBXW8 for degradation in cancer cells (53). Hence FBXW8 plays an essential role in cancer cell proliferation through proteolysis of cyclin D1, raising a possibility to develop cancer therapy by targeting Cyclin D1.
Other kinases also phosphorylate cyclin D1 for degradation. Zou et al. (54) reported an arginine-directed protein kinase Mirk/Dyrk1b in regulation of cyclin D1 stability. Mirk/Dyrk1b is active at early G1 phase of the cell cycle and phosphorylates cyclin D1 at Thr288 rather than Thr286. Mirk even phosphorylated cyclin D1 mutated at the GSK3β phosphorylation site and in the presence of the GSK3β inhibitor LiCl. Mirk will thus function together with GSK3β to assist cell arrest in G0/G1 by destabilizing cyclin D1 (54).
Cyclin D1 is marked for degradation after genomic insult (55), thus preventing cell cycle progression in the presence of DNA damage. This degradation differs from cell cycle-related proteolysis since it requires the anaphase-promoting complex and is independent of GSK3β (55). These mechanisms contribute to the accumulation of cyclin D1 protein and are considered to be critical for precise control of G1-S phase of the cell cycle in response to mitogenic stimuli and DNA damage.
Cyclin D1 and histone arginine methyltransferases
Cyclin D1/CDK4 phosphorylates a co-regulatory factor MEP50 for PRMT5, an enzyme associated with histone methylation and transcriptional repression. Aggarval et al. revealed a molecular relationship between cyclin D1/CDK4 and PRMT5 in vivo by establishing a novel B cell lymphoma model Eμ-D1T286A and analysis of human cancer specimens (28; Fig. 2A). They showed that the tumor cells had increased histone arginine methylation (56), CDT1 overexpression, CUL4 repression, and DNA re-replication (57) reflecting increased PRMT5 activity. Human esophageal cancers having Fbx4 mutation (cyclin D1 E3 ligase) expressed cyclin D1 in the nucleus with increased PRMT5 activity, suggesting that signaling through PRMT5/MEP50 is not limited to murine lymphoid tumors (28).
Figure 2. Novel signaling pathways mediated by cyclin D1 involved in tumor initiation and progression.
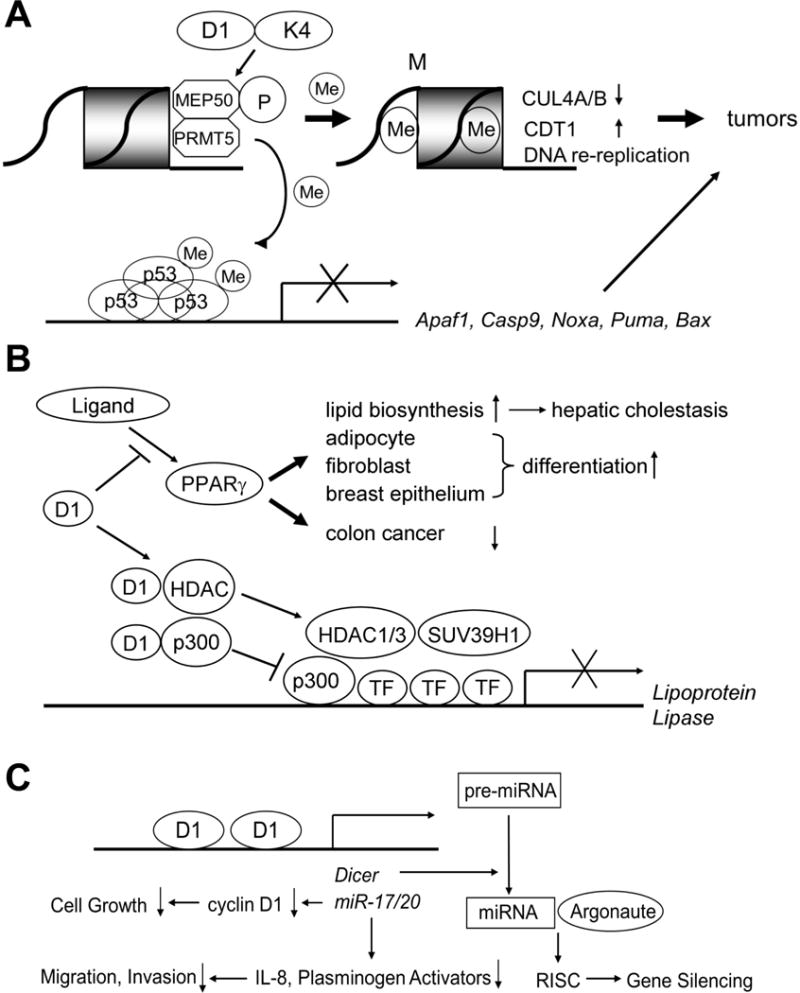
A: (Upper) Cyclin D1/CDK4 phosphorylates the MEP50 co-regulatory factor for PRMT5 to increase its methyltransferase activity. This mediates key events associated with cyclin D1-dependent neoplastic growth, including CUL4 repression, CDT1 overexpression, and DNA re-replication.
(Lower) PRMT5 also causes methylation of p53, which selectively suppresses the expression of pro-apoptotic target genes such as Apaf1, Caspase 9, Noxa, Puma, and Bax.
B: Cyclin D1 inhibits ligand-mediated PPARγ activation in Cdk-independent fashion. PPARs are ligand-activated nuclear proteins that stimulate lipid biosynthesis and accelerate differentiation of adipocytes, fibroblasts, and breast epithelial cells. Increased lipid biosynthesis leads to hepatic steatosis in mice. PPARγ also inhibits the growth of colon cancer cells. Cyclin D1 physically interacts with both HDACs and p300 to inhibit transactivation of the lipoprotein lipase gene, which occurs in Cdk-independent fashion.
C: Cyclin D1 regulates the expression of microRNAs. Cyclin D1 binds to the promoter of miR17/20 gene clusters and increases their transactivation. These microRNAs, in turn, inhibit the transcription of cyclin D1, which results in inhibition of cell growth. MiR17/20 genes also repress the transcription of IL-8 and Plasminogen Activators, resulting in decreased cellular migration and invasion, i.e. inhibition of cancer metastasis. Cyclin D1 transactivates the Dicer1 gene, the product of which accelerates the conversion of pre-mRNAs to miRNAs. Mature miRNAs then associate with argonaute proteins and conduct gene silencing through RNA-induced silencing complex (RISC).
Very recently Li et al. reported that inactivation of PRMT5 inhibits colony-forming activity driven by oncogene activation by cyclin D1, c-Myc, Notch1, and MLL-AF9 (29). PRMT5 overexpression specifically cooperated with cyclin D1 to drive lymphomagenesis in vivo, revealing inherent neoplastic activity. Arginine methylation of p53 selectively suppressed expression of pro-apoptotic and anti-proliferative target genes in lymphoma cells, thereby sustaining self-renewal and proliferation and with wild-type p53 (Fig. 2A). Analysis of human mantle cell lymphoma specimen revealed a strong correlation between cyclin D1 overexpression and p53 methylation, supporting the physiological relevance of the cyclin D1-PRMT5 pathway (29). Thus a molecular basis for therapeutic intervention of cancer through PRMT5 inhibition has been established. Indeed, PRMT5 inhibition induced lymphoma cell death through cyclin D1 transcriptional repression, reactivation of the RB pathway, and polycomb repressor complex 2 silencing (58).
Cyclin D1 binds to transcription factors and controls their activity in CDK-independent fashion
Cyclin D1 interacts with ER and Myb family proteins
Direct physical interaction of cyclin D1 with transcription factors (estrogen receptor α [ERα], androgen receptor [AR], thyroid receptor [TR], Dmp1, v-Myb, B-Myb) have been reported (30, 59–63). Cyclin D1 directly binds to the hormone-binding domain of the ERα, which results in an increased binding of the receptor to estrogen-response element sequences and upregulates ER-mediated transcription (30). Thus Cyclin D1 activates ER-mediated transcription in the absence of estrogen, which is not inhibited by anti-estrogens. Cyclin D1 also interacts with steroid receptor co-activators (SRCs) and recruits them to ER in the absence of ligand by acting as a bridging factor between ER and SRCs (64). These data explain the molecular mechanisms how cyclin D1 contributes to ER activation in breast cancers when overexpressed. Conversely, cyclin D1 forms a specific complex with the AR, and inhibits AR-mediated transactivation (65a) of the Prostate-Specific Antigen promoter (65b). Cyclin D1 caused thyroid receptor repression by serving as a ligand-independent bridging factor to selectively recruit HDAC3 to form ternary complexes (60). Of note, all of these regulatory functions of hormone receptors by cyclin D1 occurred in a Cdk-independent fashion indicating that significant parts of cyclin D1 activities take place independent of Rb phosphorylation or E2F activation.
D-type cyclins also physically interact with Myb proteins, Dmp1, v-Myb, and B-Myb (61–63). We recently conducted studies on cyclin D1-Dmp1 interaction, and found that Dmp1α binds to cyclin D1 and acts as a novel transcription factor for Arf and Ink4a (38). In v-Myb, cyclin D1 and D2 bound to the DNA-binding domain of v-Myb, resulting in a stabilization of the Myb proteins and decrease of Myb-driven transactivation of target genes in a Cdk-independent fashion (62). The activity of B-Myb is stimulated by cyclin A/Cdk2-dependent phosphorylation of the carboxyl-terminus of B-Myb (66). It was also reported that B-Myb is activated by cyclins A/E (67). Cyclin D1 strongly inhibits the activity of B-Myb-dependent transcription by forming a specific complex of B-Myb and cyclin D1 (63). Hence the activity of B-Myb during the cell cycle is controlled by the antagonistic effects of cyclins D1 and A/E.
In addition to signaling cascades mediated by Myb proteins, cyclin D1 represses cytokine signaling by repressing STAT3 activation, which also occurs in CDK-independent fashion (68). The significance and impact of cyclin D1 (or D-type cyclins in general) in transcription factor functions remain to be determined in vivo for these cyclin D-interacting proteins.
Regulation of gene transcription by cyclin D1: binding of cyclin D1 to HDACs and p300
The peroxisome proliferator activator (PPA) receptors: PPARα, PPARγ, and PPARδ are ligand-activated nuclear proteins (69). Each molecule consists of an N-terminal domain, a DNA-binding domain, and a carboxyl terminal ligand-binding domain. Among these, PPARγ was cloned as a transcription factor involved in adipocyte differentiation (70–72; Fig. 2B). Forced expression of PPARγ in the liver induces hepatic steatosis in mice, suggesting a critical role for PPARγ in hepatocellular lipid biosynthesis (72). Importantly, PPARγ agonists inhibit the growth of human colorectal cancer cells (73) and promote fibroblast and breast epithelial cell differentiation (74, 75; Fig. 2B). Thus inhibition of cell proliferation using PPARγ ligands is a reasonable approach in cancer therapy.
Recent studies show the regulation of PPAR signaling by cyclin D1. PPARγ transactivation induced by the ligand BRL49653 was inhibited by cyclin D1 through a Cdk-independent mechanism (76; Fig. 2B). Adipocyte differentiation by PPARγ-specific ligands was enhanced in fibroblasts that lack cyclin D1 and caused hepatic cholestasis in vivo. Thus inhibition of PPARγ is a novel signal pathway for cyclin D1 to control cellular differentiation (76).
To elucidate the molecular mechanisms for cell proliferation by cyclin D1 through PPARγ, Fu et al. studied the molecular interaction between cyclin D1 and histone deacetylases (HDACs) for transcriptional regulation (77; Fig. 2B). Cyclin D1 physically associated with histone HDACs in vivo, and cyclin D1 enhanced recruitment of HDAC1/3 and histone methyltransferase SUV39H1 to the PPAR-response element of the lipoprotein lipase promoter for transcriptional repression and decreased acetylation of histone H3. Taken together, their studies suggest an important role of cyclin D1 in regulation of PPARγ-mediated adipocyte differentiation through recruitment of HDACs to regulate PPAR-response elements and PPARγ function (77). The same group also showed that cyclin D1 physically interacted with p300 and represses p300-mediated transactivation on the lipoprotein lipase promoter (78; Fig. 2B). Cyclin D1 inhibited the histone acetyltransferase activity of p300 independent of Cdks. Thus cyclin D1 plays an important role in cellular proliferation and differentiation through direct regulation of p300 independent of Cdks (78).
To analyze the transcriptional regulatory mechanisms by cyclin D1 in vivo, Bienvenu et al. developed FLAG- and HA-tagged cyclin D1 knock-in mouse strains that allowed high-throughput mass spectrometric approach to search for cyclin D1-binding proteins in vivo, and identified STAT, NF-Y, STAT, CREB2, ELK1, ZNF423 and CUX1 (79). Numerous transcription factors were controlled by cyclin D1 expression for their activities without direct binding to cyclin D1. The mechanism appears to involve the recruitment to genomic DNA of transcription factors and associated chromatin-modifying enzymes (79).
Cyclin D1 induces Dicer and controls microRNA processing
MicroRNAs (miRNAs) are a class of non-coding small RNAs, functional RNA molecules that is not translated into a protein. They are initially transcribed to primary-miRNAs, which are then cleaved by the endonuclease Drosha and its partner DiGeorge Syndrome Critical Region 8 (DGCR8) to generate hairpin-folded pre-miRNAs of 60–70 nucleotides in length (80). Following transport to the cytoplasm, pre-miRNAs are processed by Dicer with its partner Human Immunodeficiency Virus Transactivating Response RNA-Binding Protein (TRBP) to generate the 20–22 nucleotide mature miRNAs (80–83). Mature miRNAs then associate with Argonaute proteins to regulate target mRNA expression through the RNA-Induced Silencing Complex (RISC; Fig. 2C). miRNAs regulate gene expression through controlling mRNA stability by base-pair binding to the complementary sequence in the 3′ untranslated region of mRNAs. Hence miRNAs contribute to tissue development, cell cycle progression, apoptosis, stem cell self-renewal, and finally cancer initiation/metastasis (84–87). The expression of miRNA is regulated during cell-cycle transition and cellular contact at least in part through active degradation (88).
Decreased expression of specific miRNAs occurs in human tumors, which suggests their roles in tumor suppression. Levels of the miR-17-5p/miR-20a miRNA cluster were inversely correlated to cyclin D1 abundance in human breast tumors and cell lines (89; Fig. 2C). miR-17/20 suppressed breast cancer cell proliferation by negatively regulating cyclin D1 translation via a conserved 3′ untranslated region miRNA-binding site, thereby inhibiting serum-stimulated S-phase entry. This effect of miR-17/20 was abrogated by cyclin D1 depletion, thus is cyclin D1-dependent. Cyclin D1 expression induced miR-17-5p and miR-20a expression in mammary epithelial cells in vivo, and chromatin immunoprecipitation (ChIP) showed that cyclin D1 bound the miR-17/20 cluster promoter regulatory region (Fig. 2C). This cyclin D1/miR-17/20 regulatory feedback loop links a specific miRNA cluster to the regulation of oncogenesis.
The RNase III endoribonuclease Dicer cleaves long double-stranded RNA (dsRNA) or stem-loop-stem structured pre-miRNA to form mature miRNAs. Depletion of Dicer in human cells led to defects in both miRNA production and shRNA-mediated RNAi (90, 91). Of note, Dicer is also transcriptionally activated by cyclin D1 (92), and cyclin D1 and Dicer maintain heterochromatic histone modification (Tri-m-H3K9). Cellular proliferation and migration mediated by cyclin D1 was Dicer-dependent. Elevated Dicer expression and high Cyclin D1 expression significantly correlated with the luminal A subtype of breast cancer whereas reduced Dicer expression and low levels of Cyclin D1a were significantly associated with the basal-like subtype (92), suggesting the translational value of the study.
Cyclin D1-Dmp1 interaction and breast carcinogenesis
The Myb-like transcription factor Dmp1 has been isolated in a yeast two-hybrid screen using cyclin D2 as bait (61). Dmp1 (Dmp1α) binds to the DNA consensus sequences CCCG(G/T)ATG(T/C), a subset of which are also recognized by Ets proteins (61). Dmp1 physically interacts with any of the three D-type cyclins, and cyclin D/Cdk4 phosphorylates Dmp1 in Sf9 cells (61). The human DMP1 locus encodes three independent transcripts, namely DMP1α, DMP1β, and DMP1γ (93). Dmp1α (and also DMP1α) directly binds to the Arf promoter for transactivation, and thereby induces Arf-, p53-dependent cell cycle arrest (94). Although D-type cyclins generally inhibit the activity of Dmp1α independent of Cdks (31, 32), Dmp1α and D-type cyclins collaborate on the Arf promoter in a Cdk-dependent fashion (95). Thus, the mode of Dmp1-cyclin D interaction depends on the context of the target promoter.
The Dmp1 promoter is activated by oncogenic Ras, HER2/neu (95, 96), but repressed by E2F and NF-κB signaling (97, 98). When crossed onto a Dmp1+/− or Dmp1−/− background, tumors induced by c-Myc, K-RasVal12, or HER2/neu transgene were greatly accelerated, with no differences between groups lacking one or two Dmp1 alleles (96, 100, 101). Thus, Dmp1 is haplo-insufficient for tumor suppression in mice (33, 96, 99–105; see 106, 107 for other Dmp1-related references). In all three transgenic mouse models, the combined frequencies of p53 mutation and Arf deletion in the Dmp1−/− and Dmp1+/− cohorts were significantly less than those in Dmp1+/+ littermates, indicating that Dmp1 is a physiologic regulator of the Arf-p53 pathway in vivo (33, 96, 99–105). Consistent with tumor-prone phenotypes of Dmp1-deficient mice, the human DMP1 (hDMP1; hDMTF1) gene is located on human chromosome 7q21.12, a locus often deleted in human breast cancers and myeloid leukemias (108–110). Our studies show that loss of heterozygosity (LOH) of hDMP1 occurs in nearly ~45% of breast cancers (111) and ~35% of non-small cell lung carcinomas (101), suggesting primary involvement of hDMP1 in human malignancies. Of note, LOH of hDMP1 was found in mutually exclusive fashion with those of p14ARF and/or TP53, suggesting that DMP1 physiologically regulates the ARF-p53 pathway in humans as well as in mice (101, 111).
Dmp1 is essential to cyclin D1-mediated activation of the Arf and Ink4a promoters
Dmp1α directly binds and activates the Arf promoter (94). To determine whether Dmp1 also regulates Ink4a gene expression, we analyzed the sequence for the murine Ink4a promoter and found a candidate Dmp1/Ets-consensus sequence (TCCGGATGG) (38). Recombinant Dmp1α directly bound to this sequence at lower affinity than its binding to the Arf promoter (38). Dmp1α activated the Ink4a promoter by 6–8 folds, which was not found in constructs with the Dmp1/Ets site mutation or deletion (38). We also found two consensus sequences for Dmp1 in the human INK4a promoter, which were bound and activated by Dmp1α (39). In summary, Dmp1 activates INK4a expression in both humans and mice.
Cyclin D1 induces cell cycle arrest, senescence, or apoptosis when overexpressed (Fig. 3A). Both wild-type cyclin D1 and its constitutively active mutant D1T286A (8) activated the Arf and Ink4a promoters, although the former showed better response than the latter (7- versus 4-fold; Fig. 3B). A cyclin D1 mutant, D1Δ142–253 that is deficient in Dmp1-binding (31), failed in activating either the Arf or Ink4a promoter, suggesting that the effect of cyclin D1 on these promoters depended on cyclin D1-Dmp1α interaction (Fig. 3B, D1Δ). Moreover, the transcription of endogenous p19Arf and p16Ink4a was induced by cyclin D1 or D1T286A in wild-type MEFs, but this effect was not found in Dmp1-null MEFs (Fig. 3C). Consistently, we observed binding of cyclin D1 on the Arf and Ink4a promoters in ChIP assays in wild-type MEFs expressing HA-tagged cyclin D1, but this binding was not detected in Dmp1-null MEFs (Fig. 3D). Collectively, these data suggest that cyclin D1 binds and activates both the Arf and Ink4a promoters, and this regulation depends on the presence of Dmp1.
Figure 3. Activation of p19Arf and p16Ink4a promoters by cyclin D1 and cyclin D1T286A.
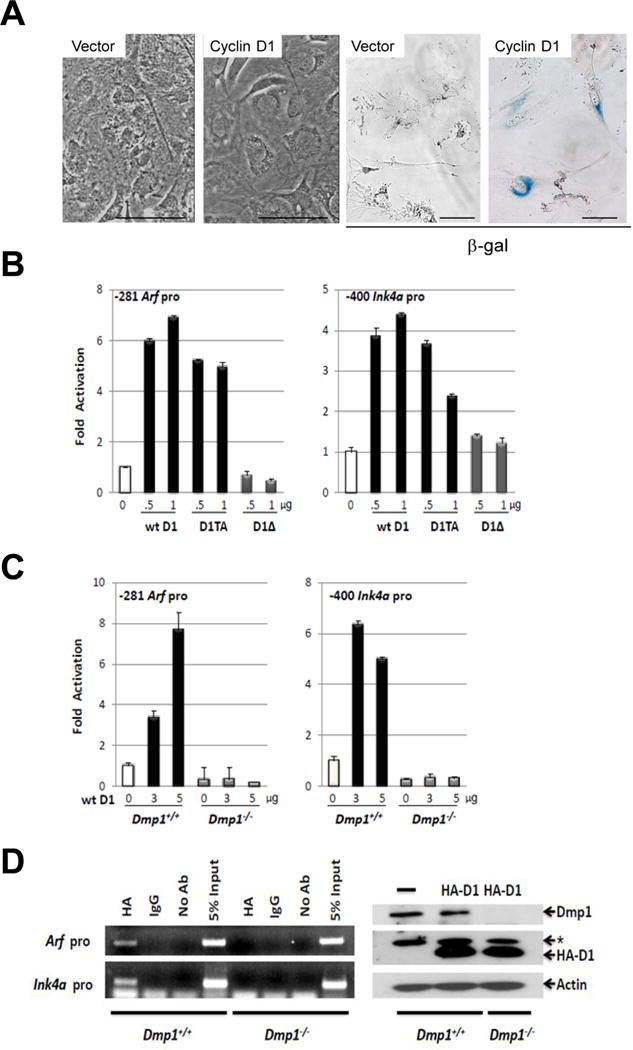
A: Sustained overexpression of wild-type cyclin D1 causes senescence in primary cells.
Left panels. Wild-type mouse embryonic fibroblasts (MEFs) (passage 5) were infected with mock or retrovirus encoding cyclin D1, selected with G418, and cultured for 7 days. Cyclin D1-virus infected cells stopped growing showing enlarged nuclei.
Right panels. Cyclin D1-virus infected cells were positive for β-galactosidase indicating irreversible cell cycle arrest. Scale bar shows 50 μm.
B: Left: The Arf promoter (−281) or Ink4a (-400) luciferase constructs were co-transfected with indicated amounts of cyclin D1, D1T286A, or D1Δ142–253 expression vectors in NIH 3T3 cells.
C: Arf (-281) or Ink4a (-400) promoter luciferase construct was co-transfected with increasing amounts of cyclin D1 expression vector in Dmp1+/+ or Dmp1−/− MEFs.
D: Left: ChIP analysis of cyclin D1 binding to the Arf and Ink4a promoters in Dmp1+/+ and Dmp1−/− MEFs infected with lentivirus expressing HA-cyclin D1. Right: Lysates derived from Dmp1+/+ and Dmp1−/− MEFs were immunoblotted for Dmp1 and HA. β-Actin was used as a loading control. *: non-specific band.
Both p14ARF and p16INK4a mRNA levels increased in human mammary epithelial cells when cyclin D1 was highly expressed; however, ectopic cyclin D1 or D1T286A failed to increase p14ARF or p16INK4a when endogenous DMP1 was depleted by shRNA (38). Similar findings were obtained in mouse studies using wild-type and Dmp1-deficient MEFs (38). Thus our data indicates that elevated cyclin D1 or D1T286A expression activates the ARF/INK4a genes, and induces G2/M delay or apoptosis in a DMP1-dependent fashion.
Cyclin D1 and D1T286A-induced mammary carcinogenesis is accelerated in Dmp1+/− mice
We saw highly dysplastic mammary glands with abnormal cell proliferation with little milk production in pre-cancerous mammary glands from MMTV-cyclin D1 (T286A); Dmp1+/− and Dmp1−/− mice (Fig. 4A, upper panels). We also found significant reduction of both Arf and Ink4a in Dmp1-deficient mammary glands (38). We, therefore, established long-term murine cohorts to assess the effect of cyclin D1 overexpression and Dmp1-loss on tumor development. Observation of mammary tumor development in Dmp1+/− mice was clinically relevant since human breast cancers showed hemizygous deletion of hDMP1 in nearly half of the cases while biallelic deletion of hDMP1 was rare (111). The cyclin D1 or D1T286A-induced mammary tumor development was significantly accelerated in Dmp1+/− mice with median disease-free survival of 810 to 600 days (p = 0.0238) for wild type cyclin D1, and from 730 to 645 days in D1T286A (p = 0.0284; Fig. 4B). Mammary tumors of Dmp1+/− mice retained a wild-type Dmp1 allele in all the seven tumors examined (38), confirming the haploid insufficiency of Dmp1 in suppressing cyclin D1-driven tumors. Mammary tumors from Dmp1+/+; MMTV-cyclin D1 and D1T286A mice showed squamous metaplasia and microinvasion as indicated by blurred epithelial-stromal interface in some glands and a corresponding tissue desmoplasia (38), while tumors from Dmp1+/−; MMTV-cyclin D1 or D1T286A mice showed a more aggressive phenotype as measured by an increased nuclear/cytoplasmic ratio, nuclear pleomorphism, and layers of disorganized epithelium (38). Published studies have shown that breast cancers that overexpress Ki67 in more than 20–50% of the cells are at high risk of developing recurrent disease, showing correlation with clinical outcomes, such as short disease-free survival or overall survival (112, 113). More Ki67-positive cells were found in the mammary glands of Dmp1+/−; MMTV-cyclin D1 and D1T286A mice than those in Dmp1+/+; MMTV-cyclin D1 and D1T286A mice between the ages of 8 and 10 months (38). Moreover, mammary tumors from Dmp1+/+; MMTV-cyclin D1 and D1T286A mice displayed a significantly higher rate of apoptosis compared to those from Dmp1+/−; MMTV-cyclin D1 and D1T286A mice, as indicated by cleaved caspase-3 staining (38).
Figure 4. Cyclin D1 and D1T286A-induced mammary carcinogenesis is accelerated in Dmp1+/− mice.
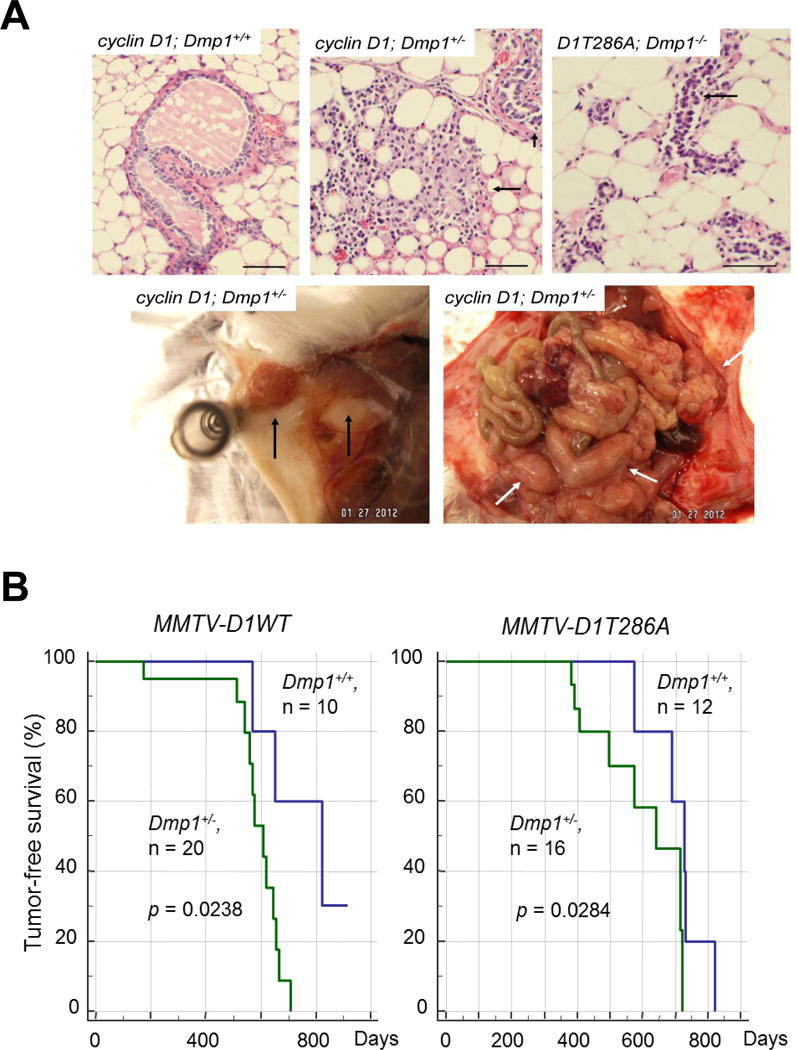
A: Mammary gland dysplasia and tumors from MMTV-cyclin D1(D1T286A) transgenic mice. Upper Panels. Left: Hyperplastic mammary gland (arrow) from a wild-type MMTV-cyclin D1 virgin female (12 months old). Middle: Highly dysplastic mammary glands from a Dmp1+/−; MMTV-cyclin D1 virgin female (5 months old). The mammary gland has lost its normal architecture. Abnormal cell proliferation is found with little milk production (arrows). Right: Dysplastic mammary glands from a Dmp1−/−; MMTV-cyclin D1T286A virgin female (4 months old). Abnormal epithelial cell proliferation, but no milk production was found in this mammary gland (arrow). Lower panels. Metastatic mammary adenocarcinoma found in 8-month-old Dmp1+/−; MMTV-cyclin D1 female. Tumor cells disseminated throughout the body in this animal, including the uterus and ovaries. Left: primary mammary tumor (arrows); Right: Metastasis of tumor cells throughout the abdomen (white arrows).
B: Left: tumor-free survival of Dmp1+/+ (blue), Dmp1+/− (green); MMTV-cyclin D1 (left) and DT286A (right) compound transgenic mice. Cyclin D1-mediated-tumor development was significantly accelerated in Dmp1+/− than in Dmp1+/+ genetic background.
Of note, mammary tumors from Dmp1+/−; MMTV-cyclin D1 (multiparous) and from Dmp1+/−; MMTV-D1T286A (nulliparous) frequently metastasized to other organs including liver, ovary and uterus (38; Fig. 4A, lower panels). Intense cyclin D1 staining was detected in the metastatic tumors, but not in adjacent normal tissues indicating that loss of one allele for Dmp1 reveals the metastatic potential of cyclin D1 (or D1T286A)-initiated mammary tumors.
The working hypotheses for cyclin D1-Dmp1 interaction are shown in Fig. 5. The primary role of cyclin D1 is to accelerate G1-S progression through activation of Cdk4/6 and phosphorylation of RB proteins, and activating E2Fs (red arrows). Dmp1α regulates the Arf-p53 pathway by activating Arf (94) and physically interacts with p53 to neutralize its antagonism by Mdm2 (107). Physical interaction between cyclin D1 and Dmp1α is essential in the activation of both Ink4a, Arf promoters to quench oncogenic signaling since the cyclin D1 mutant that the mutant D1Δ142–253, deficient in interacting with Dmp1, did not activate these two promoters, and cyclin D1 did not bind to these promoters in Dmp1 deficient cells. This is a self-autonomous safeguard mechanism to eliminate incipient cancer cells to prevent carcinogenesis in a living animal.
Figure 5. Signaling pathways involving cyclin D1 and Dmp1.
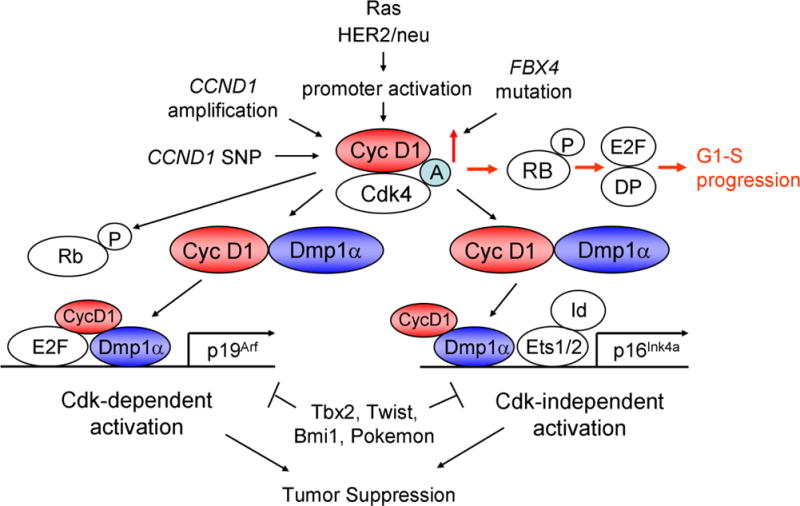
Human cyclin D1 is activated by Cyclin1 gene amplification, SNP-G/A870, promoter activation by mutant Ras or HER2/neu overexpression (95, 96), or by inhibition of protein degradation by FBX4 mutations (21). Cyclin D1 binds and activates Cdk4/6 to phosphorylate the RB family proteins, relieving E2F/DPs from the negative restraint of RB proteins and histone deacetylases, thus accelerates G1-S progression (red arrows). Cyclin D1/Cdk4 phosphorylates Dmp1α and activated Dmp1 will synergize with E2Fs to activate the Arf promoter (95). Overexpressed cyclin D1 protein in pre-neoplastic cells will interact with Dmp1 and other nuclear proteins to activate the Ink4a promoter, independent of Cdk4 activation (38). Dmp1α plays critical roles in both Arf and Ink4a promoter activation by wild type or mutant cyclin D1, and thus, cyclin D1- or D1T286A-mediated mammary tumorigenesis was accelerated in Dmp1-deficient mice (38). This process was significantly delayed in MMTV-FlagDmp1α mice (115). A: assembly factor
How to control metastasis is a major goal in cancer therapy for solid tumors. Our study shows the significantly increased metastasis of keratin-positive, cyclin D1-induced mammary tumors in Dmp1+/− mice in comparison to Dmp1+/+ mice. Most cyclin D1 tumors were adenosquamous ductal carcinomas which metastasized to the liver, ovary, uterus and intestines, although we did not see any brain or bone metastasis which is commonly found in human breast cancer patients. Nevertheless, our results imply an essential role for Dmp1 in preventing mammary tumor metastasis induced by cyclin D1 and provide a potential mechanism of such progress.
Concluding remarks and future directions
Accumulating pieces of information stress the importance of regulation of gene transcription by cyclin D1, which occurs in both Cdk-dependent and -independent fashion. The RB-E2F/DP pathway is a classical target for cyclin D/Cdks; however recent studies have demonstrated the importance of histone arginine methylation by MEP50/PRIMT5 that is a novel target contributing to tumorigenesis. PRTMT5 also inactivates p53 by methylation; these data justify the therapeutic intervention of PRTM5 for cancer therapy. Cyclin D1 binds to both p300 and HDACs, playing roles in cell proliferation and differentiation in a Cdk-independent fashion.
Recent studies indicate that cyclin D1 induces Dicer and controls miRNA processing; cyclin D1 binds to the promoter of miR-17/20 cluster, induces their production, which, in turn, repressed cyclin D1 expression by binding to 3’ untranslated region. Further studies are expected in the near future on the roles of cyclin D1 in the regulation of non-coding genomic DNA sequences.
Overexpression of cyclin D1 causes cell cycle arrest or apoptosis, but the mechanism of which remained poorly understood. Although published studies show the inhibitory roles D-cyclins in Myb protein-mediated transcriptions, our recent study shows that cyclin D1 interacts with Dmp1α to activate the Ink4a and Arf promoters. The binding of cyclin D1 to the Ink4a/Arf promoters are Dmp1-dependent, indicating that cyclin D1 can act as a co-factor for Dmp1α in this biological setting. Mapping the cyclin D1-binding site on Dmp1α on DNA will reveal the novel function of cyclin D1 in gene regulation.
The DMP1 locus produces three splice variants, namely DMP1α, β, and γ (93). We have recently created MMTV-DMP1β transgenic mice and found that the DMP1β splice variant has oncogenic activity in mammary epithelial cells rather than acting as a tumor suppressor (114). Thus it is possible that DMP1β interacts with some of the transcription factors that regulate the cyclin D1 promoter. Characterization of the signaling pathways between DMP1β and cyclin D1 will thus be important for elucidation of the mechanism of DMP1β action.
Acknowledgments
We thank all other members of Dr. Inoue’s laboratory for sharing unpublished research findings.
Financial Support
K. Inoue was supported by NIH/NCI 2R01CA106314, ACS RSG-07-207-01-MGO, and KG080179.
Abbreviations
- STAT
signal transduction and activator of transcription
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- PI3K
phosphoinositide 3 kinase
- Akt
the cellular counterpart of a retroviral oncogene v-Akt; protein kinase B
- mTOR
mammalian target of rapamycin
- S6K1
ribosomal S6 kinase 1
- GSK3β
glycogen synthase kinase-3beta
- FBXO5
F box protein 5
- FBXW8
F-Box and WD repeat domain containing 8
- MAPK
mitogen-activated protein kinase
- MP50
methylosome protein 50
- MMTV
mouse mammary tumor virus
- IKK
IκB kinase
- IκB
inhibitor of NF-κB
- TCF
T cell factor
- EGR
early growth response
- PEST
proline-glutamic acid-serine-threonine
- SAPK2
stress-activated protein kinase 2
- Mirk/Dyrk1b
minibrain-related kinase/dual-specificity tyrosine-regulated kinase 1b
- LiCl
lithum chloride
- MLL-AF9
mixed lineage leukemia – ALL1 fused gene from chromosome 9
- SUV39H1
suppressor of variegation 3-9 homolog 1
- NF-Y
nuclear factor Y
- CREB2
cAMP response element-binding protein 2
- ELK1
Ets-like kinase 1
- ZNF423
zinc finger protein 423
- CUX1
cut-like homeobox 1
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest related to this work.
Author Contributions
Both authors contributed to the conceptual design, writing the text, creating figures, and collecting literature for this review.
Literature Cited
- 1.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18(22):2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 2.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 3.Cheng M, Olivier P, Diehl JA, et al. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18(6):1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11(11):1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 5.Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60(14):3689–3695. [PubMed] [Google Scholar]
- 6.Witzel I-I, Koh LF, Perkins ND. Regulation of cyclin D1 gene expression. Biochem Soc Trans. 2010;38(Pt 1):217–222. doi: 10.1042/BST0380217. [DOI] [PubMed] [Google Scholar]
- 7.Choi YJ, Anders L. Signaling through cyclin D-dependent kinases. Oncogene. 2014;33(15):1890–1903. doi: 10.1038/onc.2013.137. [DOI] [PubMed] [Google Scholar]
- 8.Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11(8):957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 9.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12(22):3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin DI, Barbash O, Kumar KG, et al. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCF (FBX4-alphaB crystallin) complex. Mol Cell. 2006;24(3):355–366. doi: 10.1016/j.molcel.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sicinski P, Donaher JL, Parker SB, et al. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82(4):621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 12.Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer Cell. 2006;9(1):13–22. doi: 10.1016/j.ccr.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 13.Brown NE, Jeselsohn R, Bihani T, et al. Cyclin D1 activity regulates autophagy and senescence in the mammary epithelium. Cancer Res. 2012;72(24):6477–6489. doi: 10.1158/0008-5472.CAN-11-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillett C, Fantl V, Smith R, et al. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res. 1994;54(7):1812–1817. [PubMed] [Google Scholar]
- 15.Ormandy CJ, Musgrove EA, Hui R, Daly RJ, Sutherland RL. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res Treat. 2003;78(3):323–335. doi: 10.1023/a:1023033708204. [DOI] [PubMed] [Google Scholar]
- 16.Roy PG, Thompson AM. Cyclin D1 and breast cancer. Breast. 2006;15(6):718–727. doi: 10.1016/j.breast.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10(6):529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Holm K, Staaf J, Jönsson G, et al. Characterization of amplification patterns and target genes at chromosome 11q13 in CCND1-amplified sporadic and familial breast tumours. Breast Cancer Res Treat. 2012;133(2):583–594. doi: 10.1007/s10549-011-1817-3. [DOI] [PubMed] [Google Scholar]
- 19.Arnold A, Papanikolau A. Cyclin D1 in breast pathogenesis. J Clin Oncol. 2006;23:4215–4224. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 20.Citri A, Skaria KB, Yarden Y. The deaf and dumb: the biology of ErbB-2 and ErbB-3. Exp Cell Res. 2003;284:54–65. doi: 10.1016/s0014-4827(02)00101-5. [DOI] [PubMed] [Google Scholar]
- 21.Barbash O, Zamfirova P, Lin DI, et al. Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell. 14(1):68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbash O, Diehl JA. SCF (Fbx4/alphaB-crystallin) E3 ligase: when one is not enough. Cell Cycle. 2008;7(19):2983–2986. doi: 10.4161/cc.7.19.6775. [DOI] [PubMed] [Google Scholar]
- 23.Kim JK, Diehl JA. Nuclear cyclin D1: an oncogenic driver in human cancer. J Cell Physiol. 2009;220(2):292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casimiro MC, Velasco-Velázquez M, Aguirre-Alvarado C, Pestell RG. Overview of cyclins D1 function in cancer and the CDK inhibitor landscape: past and present. Expert Opin Investig Drugs. 2014;23(3):295–304. doi: 10.1517/13543784.2014.867017. [DOI] [PubMed] [Google Scholar]
- 25.Peurala E, Koivunen P, Haapasaari KM, Bloigu R, Jukkola-Vuorinen A. The prognostic significance and value of cyclin D1, CDK4 and p16 in human breast cancer. Breast Cancer Res. 2013;15(1):R5. doi: 10.1186/bcr3376.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabarestani S, Ghaderian SM, Rezvani H, et al. Prognostic and predictive value of copy number alterations in invasive breast cancer as determined by multiplex ligation-dependent probe amplification. Cell Oncol (Dordr) 2014;37(2):107–118. doi: 10.1007/s13402-013-0165-1. [DOI] [PubMed] [Google Scholar]
- 27.Millar EK, Dean JL, McNeil CM, et al. Cyclin D1b protein expression in breast cancer is independent of cyclin D1a and associated with poor disease outcome. Oncogene. 2009;28(15):1812–1820. doi: 10.1038/onc.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aggarwal P, Vaites LP, Kim JK, et al. Cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18(4):329–340. doi: 10.1016/j.ccr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Chitnis N, Nakagawa H, et al. PRMT5 is required for lymphomagenesis triggered by multiple oncogenic drivers. Cancer Discov. 2015;5(3):288–303. doi: 10.1158/2159-8290.CD-14-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zwijsen RM, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJ. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88(3):405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- 31.Inoue K, Sherr CJ. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol Cell Biol. 1998;18(3):1590–1600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue K, Sherr CJ, Shapiro LH. Regulation of the CD13/aminopeptidase N gene by DMP1, a transcription factor antagonized by D-type cyclins. J Biol Chem. 1998;273(44):29188–29194. doi: 10.1074/jbc.273.44.29188. [DOI] [PubMed] [Google Scholar]
- 33.Inoue K, Mallakin A, Frazier DP. Dmp1 and tumor suppression. Oncogene. 2007;26(30):4329–4335. doi: 10.1038/sj.onc.1210226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue K, Fry EA, Frazier DP. Transcription factors that interact with p53 and Mdm2. Int J Cancer. 2015 Jul 1; doi: 10.1002/ijc.29663. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozenne P, Eymin B, Brambilla E, Gazzeri S. The ARF tumor suppressor: structure, functions and status in cancer. Int J Cancer. 2010;127(10):2239–2247. doi: 10.1002/ijc.25511. Review. [DOI] [PubMed] [Google Scholar]
- 36.Larsson LG. Oncogene- and tumor suppressor gene-mediated suppression of cellular senescence. Semin Cancer Biol. 2011;21(6):367–376. doi: 10.1016/j.semcancer.2011.10.005. Review. [DOI] [PubMed] [Google Scholar]
- 37.Maggi LB, Jr, Winkeler CL, Miceli AP, et al. ARF tumor suppression in the nucleolus. Biochim Biophys Acta. 2014;1842(6):831–839. doi: 10.1016/j.bbadis.2014.01.016. Review. [DOI] [PubMed] [Google Scholar]
- 38.Zhu S, Mott RT, Fry EA, et al. Cooperation between cyclin D1 expression and Dmp1-loss in breast cancer. Am J Pathol. 2013;183(4):1339–1350. doi: 10.1016/j.ajpath.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Betticher DC, Thatcher N, Altermatt HJ, Hoban P, Ryder WD, Heighway J. Alternate splicing produces a novel cyclin D1 transcript. Oncogene. 1995;11(5):1005–1011. [PubMed] [Google Scholar]
- 40.Knudsen KE. The cyclin D1b splice variant: an old oncogene learns new tricks. Cell Div. 2006;1:15. doi: 10.1186/1747-1028-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hosokawa Y, Gadd M, Smith AP, Koerner FC, Schmidt EV, Arnold A. Cyclin D1 (PRAD1) alternative transcript b: full-length cDNA cloning and expression in breast cancers. Cancer Lett. 1997;113(1–2):123–130. doi: 10.1016/s0304-3835(97)04605-3. [DOI] [PubMed] [Google Scholar]
- 42.Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003;63(21):7056–7061. [PubMed] [Google Scholar]
- 43.Solomon DA, Wang Y, Fox SR, et al. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J Biol Chem. 2003;278(32):30339–30347. doi: 10.1074/jbc.M303969200. [DOI] [PubMed] [Google Scholar]
- 44.Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369(6482):669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- 45.Lin DI, Lessie MD, Gladden AB, Bassing CH, Wagner KU, Diehl JA. Disruption of cyclin D1 nuclear export and proteolysis accelerates mammary carcinogenesis. Oncogene. 2008;27(9):1231–1242. doi: 10.1038/sj.onc.1210738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taneja P, Frazier DP, Kendig RD, et al. MMTV mouse models and the diagnostic values of MMTV-like sequences in human breast cancer. Expert Rev Mol Diagn. 2009;9(5):423–440. doi: 10.1586/ERM.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alt JR, Cleveland JL, Hannink M, Diehl JA. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14(24):3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klein EA, Assoian RK. Transcriptional regulation of the cyclin D1 gene at a glance. J Cell Sci. 2008;121(Pt 23):3853–3857. doi: 10.1242/jcs.039131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136(4):688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 50.Dragnev KH, Freemantle SJ, Spinella MJ, Dmitrovsky E. Cyclin proteolysis as a retinoid cancer prevention mechanism. Ann N Y Acad Sci. 2001;952:13–22. doi: 10.1111/j.1749-6632.2001.tb02724.x. [DOI] [PubMed] [Google Scholar]
- 51.Alt JR, Gladden AB, Diehl JA. p21(Cip1) promotes cyclin D1nuclear accumulation via direct inhibition of nuclear export. J Biol Chem. 2002;277(10):8517–8523. doi: 10.1074/jbc.M108867200. [DOI] [PubMed] [Google Scholar]
- 52.Casanovas O, Miró F, Estanyol JM, Itarte E, Agell N, Bachs O. Osmotic stress regulates the stability of cyclin D1 in a p38SAPK2-dependent manner. J Biol Chem. 2000;275(45):35091–3507. doi: 10.1074/jbc.M006324200. [DOI] [PubMed] [Google Scholar]
- 53.Okabe H, Lee SH, Phuchareon J, Albertson DG, McCormick F, Tetsu O. A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS One. 2006;1:e128. doi: 10.1371/journal.pone.0000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou Y, Ewton DZ, Deng X, Mercer SE, Friedman E. Mirk/dyrk1B kinase destabilizes cyclin D1 by phosphorylation at threonine 288. J Biol Chem. 2004;279(26):27790–27998. doi: 10.1074/jbc.M403042200. [DOI] [PubMed] [Google Scholar]
- 55.Agami R, Bernards R. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell. 2000;102(1):55–66. doi: 10.1016/s0092-8674(00)00010-6. [DOI] [PubMed] [Google Scholar]
- 56.Gladden AB, Woolery R, Aggarwal P, Wasik MA, Diehl JA. Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene. 2006;25(7):998–1007. doi: 10.1038/sj.onc.1209147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aggarwal P, Lessie MD, Lin DI, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21(22):2908–2922. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chung J, Karkhanis V, Tae S, et al. Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) silencing. J Biol Chem. 2013;288(49):35534–35547. doi: 10.1074/jbc.M113.510669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knudsen KE, Cavenee WK, Arden KC. D-type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res. 1999;59(10):2297–2301. [PubMed] [Google Scholar]
- 60.Lin HM, Zhao L, Cheng SY. Cyclin D1 is ligand-independent co-repressor for thyroid hormone receptors. J Biol Chem. 2002;277(32):28733–28741. doi: 10.1074/jbc.M203380200. [DOI] [PubMed] [Google Scholar]
- 61.Hirai H, Sherr CJ. Interaction of D-type cyclins with a novel myb-like transcription factor, DMP1. Mol Cell Biol. 1996;16(11):6457–6467. doi: 10.1128/mcb.16.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ganter B, Fu Sl, Lipsick JS. D-type cyclins repress transcriptional activation by the v-Myb but not the c-Myb DNA-binding domain. EMBO J. 1998;17(1):255–268. doi: 10.1093/emboj/17.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horstmann S, Ferrari S, Klempnauer KH. Regulation of B-Myb activity by cyclin D1. Oncogene. 2000;19(2):298–306. doi: 10.1038/sj.onc.1203302. [DOI] [PubMed] [Google Scholar]
- 64.Zwijsen RM, Buckle RS, Hijmans EM, Loomans CJ, Bernards R. Ligand-independent recruitment of steroid receptor coactivators to estrogen receptor by cyclin D1. Genes Dev. 1998;12(22):3488–3498. doi: 10.1101/gad.12.22.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65a.Reutens AT, Fu M, Wang C, Albanese C, et al. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol. 2001;15(5):797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 65b.Tang S, Mishra M, Frazier DP, et al. Positive and negative regulation of prostate stem cell antigen expression by yin yang 1 in prostate epithelial cell lines. PLoS One. 2012;7(4):e35570. doi: 10.1371/journal.pone.0035570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson C, Light Y, Groves R, Mann D, Marias R, Watson R. Cell-cycle regulation of B-Myb protein expression: specific phosphorylation during the S phase of the cell cycle. Oncogene. 1996;12(9):1855–1864. [PubMed] [Google Scholar]
- 67.Sala A, Kundu M, Casella I, et al. Activation of human B-MYB by cyclins. Proc Natl Acad Sci USA. 1997;94(2):532–536. doi: 10.1073/pnas.94.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bienvenu F, Gascan H, Coqueret O. Cyclin D1 represses STAT3 activation through a Cdk4-independent mechanism. J Biol Chem. 2001;276(20):16840–1684. doi: 10.1074/jbc.M100795200. [DOI] [PubMed] [Google Scholar]
- 69.Rosen ED, Spiegelman BM. PPARγ: a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem. 2001;276(41):37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 70.Rosen ED, Sarraf P, Troy AE, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4(4):611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 71.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-x. 1994. [DOI] [PubMed] [Google Scholar]
- 72.Yu S, Matsusue K, Kashireddy P, et al. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor γ1 (PPARγ1) overexpression. J Biol Chem. 2003;278(1):498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- 73.Sarraf P, Mueller E, Smith WM, et al. Loss-of-function mutations in PPAR gamma associated with human colon cancer. Mol Cell. 1999;3(6):799–804. doi: 10.1016/s1097-2765(01)80012-5. [DOI] [PubMed] [Google Scholar]
- 74.Elstner E, Muller C, Koshizuka K, et al. Ligands for peroxisome proliferator-activated receptorγ and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proc Natl Acad Sci USA. 1998;95:8806–8811. doi: 10.1073/pnas.95.15.8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mueller E, Sarraf P, Tontonoz P, et al. Terminal differentiation of human breast cancer through PPARγ. Mol Cell. 1998;1(3):465–470. doi: 10.1016/s1097-2765(00)80047-7. [DOI] [PubMed] [Google Scholar]
- 76.Wang C, Fu M, D’Amico M, et al. Inhibition of cellular proliferation through IkappaB kinase-independent and peroxisome proliferator-activated receptor gamma-dependent repression of cyclin D1. Mol Cell Biol. 2001;21(9):3057–3070. doi: 10.1128/MCB.21.9.3057-3070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fu M, Rao M, Bouras T, et al. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem. 2005;280(17):16934–16941. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 78.Fu M, Wang C, Rao M, et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem. 2005;280(33):29728–29742. doi: 10.1074/jbc.M503188200. [DOI] [PubMed] [Google Scholar]
- 79.Bienvenu F, Jirawatnotai S, Elias JE, Meyer CA, Mizeracka K, Marson A, Frampton GM, Cole MF, Odom DT, Odajima J, Geng Y, Zagozdzon A, Jecrois M, Young RA, Liu XS, Cepko CL, Gygi SP, Sicinski P. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010 Jan 21;463(7279):374–348. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 81.Lund E, Sheets MD, Imboden SB, Dahlberg JE. Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev. 2011;25(11):1121–1131. doi: 10.1101/gad.2038811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Melo SA, Sugimoto H, O’Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilson RC, Tambe A, Kidwell MA, Noland CL, Schneider CP, Doudna JA. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell. 2015;57(3):397–407. doi: 10.1016/j.molcel.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10(2):116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Romero-Cordoba SL, Salido-Guadarrama I, Rodriguez-Dorantes M, Hidalgo-Miranda A. miRNA biogenesis: biological impact in the development of cancer. Cancer Biol Ther. 2014;15(11):1444–1455. doi: 10.4161/15384047.2014.955442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2015 doi: 10.1016/j.bbagrm.2015.06.015. pii: S1874-9399(15)00138-8. [DOI] [PubMed] [Google Scholar]
- 87.Zhou L, Liu F, Wang X, Ouyang G. The roles of microRNAs in the regulation of tumor metastasis. Cell Biosci. 2015;5:32. doi: 10.1186/s13578-015-0028-8. eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rissland OS, Hong SJ, Bartel DP. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol Cell. 2011;43(6):993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu Z, Wang C, Wang M, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182(3):509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 91.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16(8):948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu Z, Wang L, Wang C, et al. Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat Commun. 2013;4:2812. doi: 10.1038/ncomms3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tschan MP, Fischer KM, Fung VS, et al. Alternative splicing of the human cyclin D-binding Myb-like protein (hDMP1) yields a truncated protein isoform that alters macrophage differentiation patterns. J Biol Chem. 2003;278(44):42750–42760. doi: 10.1074/jbc.M307067200. [DOI] [PubMed] [Google Scholar]
- 94.Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci USA. 1999;96(7):3993–3998. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sreeramaneni R, Chaudhry A, McMahon M, Sherr CJ, Inoue K. Ras-Raf-Arf signaling critically depends on the Dmp1 transcription factor. Mol Cell Biol. 2005;25(1):220–232. doi: 10.1128/MCB.25.1.220-232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taneja P, Maglic D, Kai F, et al. Critical role of Dmp1 in HER2/neu-p53 signaling and breast carcinogenesis. Cancer Res. 2010;70(22):9084–9094. doi: 10.1158/0008-5472.CAN-10-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mallakin A, Taneja P, Matise LA, Willingham MC, Inoue K. Expression of Dmp1 in specific differentiated, nonproliferating cells and its repression by E2Fs. Oncogene. 2006;25(59):7703–7713. doi: 10.1038/sj.onc.1209750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taneja P, Mallakin A, Matise LA, Frazier DP, Choudhary M, Inoue K. Repression of Dmp1 and Arf transcription by anthracyclins: critical roles of the NF-κB subunit p65. Oncogene. 2007;26(53):7457–7466. doi: 10.1038/sj.onc.1210568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Inoue K, Wen R, Rehg JE, et al. Disruption of the ARF transcriptional activator DMP1 facilitates cell immortalization, Ras transformation, and tumorigenesis. Genes Dev. 2000;14(14):1797–1809. [PMC free article] [PubMed] [Google Scholar]
- 100.Inoue K, Zindy F, Randle DH, Rehg JE, Sherr CJ. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev. 2001;15(22):2934–2939. doi: 10.1101/gad.929901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mallakin A, Sugiyama T, Taneja P, et al. Mutually exclusive inactivation of DMP1 and ARF/p53 in lung cancer. Cancer Cell. 2007;12(4):381–394. doi: 10.1016/j.ccr.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sugiyama T, Frazier DP, Taneja P, et al. Signal transduction involving the Dmp1 transcription factor and its alteration in human cancer. Clinical Medicine Insights: Oncology. 2008;2:209–219. doi: 10.4137/cmo.s548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Inoue K, Sugiyama T, Taneja P, Morgan RL, Frazier DP. Emerging roles of DMP1 in lung cancer. Cancer Res. 2008;68(12):4487–4490. doi: 10.1158/0008-5472.CAN-07-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sugiyama T, Frazier DP, Taneja P, Morgan RL, Willingham MC, Inoue K. The role of Dmp1 and its future in lung cancer diagnostics. Expert Rev Mol Diagn. 2008;8(4):435–448. doi: 10.1586/14737159.8.4.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Quon KC, Berns A. Haplo-insufficiency? Let me count the ways. Genes Dev. 2001;15(22):2917–2921. doi: 10.1101/gad.949001. [DOI] [PubMed] [Google Scholar]
- 106.Mallakin A, Sugiyama T, Kai F, et al. The Arf-inducing transcription factor Dmp1 encodes transcriptional activator of amphiregulin, thrombospondin-1, JunB and Egr1. Int J Cancer. 2010;126(6):1403–1416. doi: 10.1002/ijc.24938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Frazier DP, Kendig RD, Kai F, et al. Dmp1 physically interacts with p53 and positively regulates p53’s stabilization, nuclear localization, and function. Cancer Res. 2012;72(7):1740–1750. doi: 10.1158/0008-5472.CAN-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bodner SM, Naeve CW, Rakestraw KM, et al. Cloning and chromosomal localization of the gene encoding human cyclin D-binding Myb-like protein (hDMP1) Gene. 1999;229(1–2):223–228. doi: 10.1016/s0378-1119(98)00591-5. [DOI] [PubMed] [Google Scholar]
- 109.Bieche I, Champeme MH, Matifas F, Hacene K, Callahan R, Lidereau R. Loss of heterozygosity on chromosome 7q and aggressive primary breast cancer. Lancet. 1992;339(8786):139–143. doi: 10.1016/0140-6736(92)90208-k. [DOI] [PubMed] [Google Scholar]
- 110.Kristjansson AK, Eiriksdottir G, Ragnarsson G, et al. Loss of heterozygosity at chromosome 7q in human breast cancer: association with clinical variables. Anticancer Res. 1997;17(1A):93–98. [PubMed] [Google Scholar]
- 111.Maglic D, Zhu S, Fry EA, et al. Prognostic value of the hDMP1-ARF-Hdm2-p53 pathway in breast cancer. Oncogene. 2013;32(35):4120–4129. doi: 10.1038/onc.2012.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Taneja P, Maglic D, Kai F, et al. Classical and novel molecular prognostic markers for human breast cancer and their clinical significance. Clinical Medicine Insights: Oncology. 2010;4:15–34. doi: 10.4137/cmo.s4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yerushalmi R, Woods R, Ravdin PM, Hayes MM, Gelmon KA. Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 2010;11(2):174–183. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- 114.Maglic D, Stovall DB, Cline JM, et al. DMP1β, a splice isoform of the tumor suppressor DMP1 locus, induces proliferation and progression of breast cancer. J Pathol. 2015;236(1):90–102. doi: 10.1002/path.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fry EA, Taneja P, Maglic D, Zhu S, Sui G, Inoue K. Dmp1α inhibits HER2/neu-induced mammary tumorigenesis. PLoS One. 2013;8(10):e77870. doi: 10.1371/journal.pone.0077870. [DOI] [PMC free article] [PubMed] [Google Scholar]


