Abstract
Genetic predisposition increases the risk of familial breast cancer. Recent studies indicate that genetic predisposition for familial breast cancer can be ethnic-specific. However, current knowledge of genetic predisposition for the disease is predominantly derived from Western populations. Using this existing information as the sole reference to judge the predisposition in non-Western populations is not adequate and can potentially lead to misdiagnosis. Efforts are required to collect genetic predisposition from non-Western populations. The Egyptian population has high genetic variations in reflecting its divergent ethnic origins, and incident rate of familial breast cancer in Egypt is also higher than the rate in many other populations. Using whole exome sequencing, we investigated genetic predisposition in five Egyptian familial breast cancer families. No pathogenic variants in BRCA1, BRCA2 and other classical breast cancer-predisposition genes were present in these five families. Comparison of the genetic variants with those in Caucasian familial breast cancer showed that variants in the Egyptian families were more variable and heterogeneous than the variants in Caucasian families. Multiple damaging variants in genes of different functional categories were identified either in a single family or shared between families. Our study demonstrates that genetic predisposition in Egyptian breast cancer families may differ from those in other disease populations, and supports a comprehensive screening of local disease families to determine the genetic predisposition in Egyptian familial breast cancer.
Introduction
Familial breast cancer is a hereditary disease and genetic predispositions play major roles in increasing the risk of the disease in the carriers. Genetic predispositions for approximately half of familial breast cancers have been determined, and studies are actively going on to determine the unknown genetic predispositions for the remaining cases [1–3]. Recent studies demonstrate that genetic predispositions for familial breast cancer can be ethnic-specific, as well exemplified by the different spectrum of germline mutation in BRCA1 and BRCA2 between different ethnic populations [4–10]. Knowledge of ethnic-specific genetic predispositions for familial breast cancer is important, as it directly affects the accuracy of clinical diagnosis and intervention in patients of different ethnicities. However, current predisposition information is predominantly derived from Western populations. Using the information as the sole reference is not adequate and can potentially lead to misdiagnosis for the patients of non-Western ethnicities, which constitute the majority of human populations.
Egypt population has high-degree of genetic diversity due to its complex and diverse ethnic origins. The population has substantial variations from other populations including its proximal Ethiopia population and distal Yoruba population within African continent [11]. Breast cancer is the most common cancer in Egyptian females with unique characters. While its incidence rate of 45.4 per 100,000 is moderate comparing to other ethnic populations [12], it has high-degree family history of breast cancer, possibly related to high rate of consanguineous marriage in the population [13], and it has high-degree of inflammatory breast cancer [14]. Efforts have been made to study genetic predisposition for Egyptian familial breast cancer, mostly focused on BRCA1 and BRCA2 [15], but comprehensive data at genomic level from local patients are lacking.
We used Egyptian familial breast cancer as a model to investigate ethnic-specific genetic predisposition in familial breast cancer. In the study, we applied exome sequencing to analyze genomic variations across all coding genes in five Egyptian breast cancer families. Our study revealed that these disease families have high genetic variability, and they do not contain currently known predispositions for the disease but carry Egyptian-specific genetic variants, some of which may represent Egyptian-specific predispositions. The study supports the concept of ethnic-specific predispositions in familial breast cancer.
Methods
Breast cancer families used in the study
The Institutional Review Board of University of Nebraska Medical Center approved the study (049-14-EP). All participants provided verbal informed consent that was read by a study nurse with another nurse or a relative witnessing the delivery of the consent. Written consent was not obtained because of the high illiteracy rate among women in the study population in Egypt. Signatures of the nurse/relative witnessing the interviews were obtained. The local IRB committee in Egypt approved this consent procedure. Five Egyptian breast cancer families from Gharbiah district, Egypt, participated in the study. The families were identified from the Gharbiah Cancer Registry, Egypt. Each participant was interviewed by local oncologists and answered the questions in a standard questionnaire. Venous blood was collected from each participant during the interview process.
Exome sequencing
DNA was extracted from blood cells using a FlexiGene DNA kit (Qiagen, Valencia, USA). Exome sequences were collected according to previously described procedures [15]. Briefly, genomic DNA was fragmented using a Covaris II system (Covaris, Woburn, MA, USA). Exon templates were isolated using the TruSeq Exome Enrichment Kit (Illumina, San Diego, CA, USA) and exome sequences were collected in a HiSeq2500 sequencer (paired-end 2×150) at 100x coverage. The total variants called from the exome data from this study have been deposited in DRYAD Digital Repository with accession ID: doi:10.5061/dryad.p236p.
Variant identification
Three controls were used in the study, including 1) the human variation databases of dbSNP, 1000 Genomes and ESP6500 were used to filter out population polymorphism; 2) the Egyptian genome variation data were used to filter out Egyptian-specific normal polymorphism; 3) the variants from 27 Caucasian familial breast cancer probands were used to compare the genomic variation in familial breast cancer families between the two ethnic populations.
Exome sequences were mapped to the human reference genome sequences hg19 [16] using the Burrows-Wheeler Aligner [17] and pre-processed with Picard Toolkit [18]. Variants were called using Freebayes [19], and filtered with a minimum read depth of 10, a minimum of four reads mapped to the location and a minimum of four reads on opposite strands, and a minimum base quality score of 30. Qualified variants were annotated with ANNOVAR [20] against the following reference databases: RefSeq (February 4, 2016), 1000 Genomes (August 2015), NHLBI Exome Sequencing Project (ESP6500) version 2, dbSNP Build 144, and ClinVar (May 5, 2016). Variants causing codon changes were identified, and further filtered by 1000 Genomes with a minor allele frequency (MAF) < = 1%. The remaining variants were further filtered through Egypt population polymorphism data. The Egyptian variant dataset containing 1,422 Egyptian-specific variants was derived from whole genome sequences of 25 Egyptian individuals. Each was sequenced by Ion Torrent technology with base quality score (50+) at average depth of 20X. Variants were called by using the Torrent Suite software following manufacturer’s instruction and the variants present in other ethnic populations at the frequency > 0.01 were eliminated [21]. Annotation was made by using ANNOVAR and in-house programs. Damaging variants were predicted using SIFT [22, score < 0.05] and PolyPhen2 [23, score > 0.909]. Only variants shared by at least two breast cancer-affected members in the same family were included in the final list of damaging variants. Pathways affected by variant-affected genes were identified by searching in the Reactome pathway database (version 57) [24].
Results
Breast cancer families used in the study
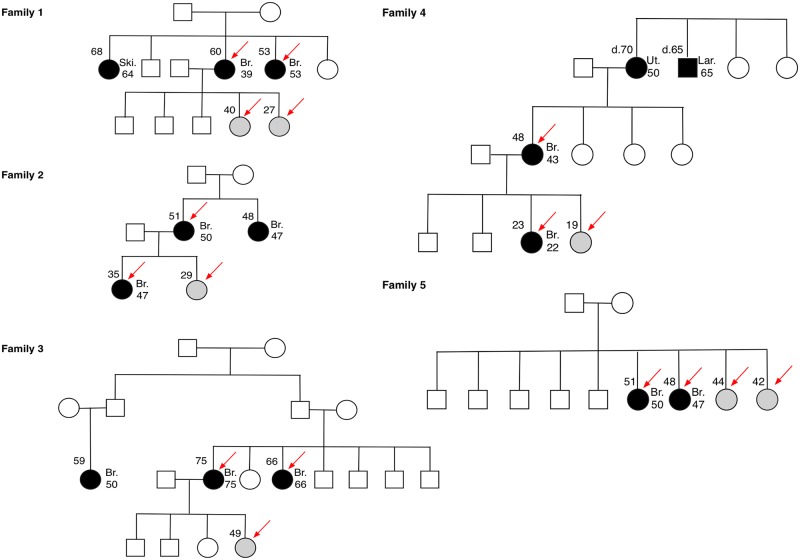
We analyzed five Egyptian breast cancer families (Fig 1, Table 1). Familial breast cancer was diagnosed using the inclusion criteria of at least one first-degree relative with breast cancer irrespective of age. In Family 1, three of the four sisters were affected by cancer, of whom two were breast cancer; in Family 2, both sisters and one daughter were affected by breast cancer; in Family 3, two sisters and one cousin were affected by breast cancer; in Family 4, grandmother, grandmother’s brother, mother and a daughter were affected by cancer, of whom mother and daughter had breast cancer; in Family 5, two sisters were affected by breast cancer. Of the 12 breast cancer-affected cases in the five families, eight were diagnosed at an age of younger than or at 50 years old. Based on the availability of DNA samples, 10 breast cancer-affected and seven breast cancer-unaffected family members were included for exome sequencing.
Fig 1. Pedigrees of the five Egyptian familial breast cancer families used in the study.
Dark circle: cancer-affected family member; gray circle: cancer-unaffected family member: red arrow: cased used for exome sequencing. Br: breast cancer; Ski: skin cancer; Ut: uterus cancer; Lar: laryngcarcinoma; d: age of death.
Table 1. Clinical data of the breast cancer-affected cases used in exome sequencing.
| Case | Diagnosis age | ER | PR | HER2/neu | Lymphnode | Ki-67 | Diagnosis |
|---|---|---|---|---|---|---|---|
| Family 1 | |||||||
| 1 | 39 | + | + | - | 6/15 | 5% | Ductual carcinoma, invasive & In situ |
| 2 | 53 | + | + | - | 6/19 | 40% | Ductual carcinoma, invasive & In situ |
| Family 2 | |||||||
| 1 | 50 | + | + | - | 0/18 | 80% | Ductual carcinoma, invasive |
| 2 | 47 | - | - | + | 2/24 | NA | Ductual carcinoma, invasive & In situ |
| Family 3 | |||||||
| 1 | 75 | + | + | - | 0/18 | 80% | Ductual carcinoma, invasive |
| 2 | 66 | + | + | - | 13/14 | NA | Ductual carcinoma, invasive & In situ |
| Family 4 | |||||||
| 1 | 43 | + | + | unknown | NA | NA | Ductual carcinoma, invasive |
| 2 | 22 | - | - | - | 0/20 | 5% | Ductual carcinoma, invasive |
| Family 5 | |||||||
| 1 | 50 | + | + | - | 1/19 | 30% | Ductual carcinoma, invasive & In situ |
| 2 | 47 | - | - | - | 0/20 | 5% | Ductual carcinoma, invasive |
Variants in BRCA1, BRCA2 and other known breast cancer predisposition genes
A total of 938,606 unique variants were called from exome sequences of all cases through bioinformatics analysis. To determine if any of these five families carried BRCA mutations, we searched the entire variants and identified 18 variants in BRCA1 and 20 variants in BRCA2. Based on Breast Cancer Information Core (BIC) and ClinVar databases, none of the variants was classified as pathogenic (Table 2). We further identified 340 variants in other known predisposition genes of ATM, BARD1, BRIP1, CDH1, CHEK2, MRE11A, MUTYH, NBN, NF1, PALB2, PTEN, RAD50, RAD51C, RAD51D, STK11, and TP53. Six variants were identified in BRIP1, MRE11A, NBN, PTEN, TP53, of which only one in NBN (chr8:90990521T>C, NM_002485, c.A511G, p.I171V) was predicted as deleterious by both SIFT and Polyphen2 programs and classified as pathogenic by ClinVar database but this variant was present only in one breast cancer-affected case (member 2 in Family 3). All other variants were predicted as possibly damaging or deleterious by a single program and classified as unknown, untested, non-pathogenic by ClinVar (S1 Table). We also searched the variants affecting 160 cancer-related genes (https://dnapittcrew.upmc.com/db/hsa.php), and identified three coding-change variants affected RECQL4, a DNA helicase involved in DNA replication and repair and known to relate with breast cancer [25]. However, the homozygote G-del variant was present in all ten cancer-affected and seven cancer-unaffected cases, the C to T variant was present in an affected and an unaffected members in family 4, and the G to A variant was present in one affected and two unaffected members in family 1. A C to T variant was also identified in RRM2B, a gene involved in a TP53-dependent DNA repair process. This variant was present in one affected and two unaffected members in family 5. None of the variants were predicted to damage the function of RECQL4 and RRM2B. Therefore, these variants were unlikely the potential predisposition but the normal variation in these families (S2 Table). The lack of pathogenic variants in BRCA1, BRCA2 and other predisposition genes indicates that these five families are all BRCAx breast cancer family [15].
Table 2. Variants in BRCA1 and BRCA2.
| Position | Exon | Variation type | Variant (HGVS) | Frequency | dbSNP144 | Zygocity | Total cases | Classification | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| cDNA | Protein | Affected | Unaffected | BIC | ClinVar | ||||||
| BRCA1 | |||||||||||
| Exonic | 11B | nonsynonymous | c.2077G>A | p.Asp693Asn | 0.0335 | rs4986850 | het | 1 | 1 | Pending | Benign |
| Exonic | 11B | synonymous | c.2082C>T | p.Ser694= | 0.3365 | rs1799949 | het | 7 | 5 | Class 1 | Benign |
| Exonic | 11B | synonymous | c.2311T>C | p.Leu771= | 0.3353 | rs16940 | het | 7 | 5 | Class 1 | Benign |
| Exonic | 11C | nonsynonymous | c.2612C>T | p.Pro871Leu | 0.5439 | rs799917 | het | 8 | 7 | Class 1 | Benign |
| Exonic | 11C | nonsynonymous | c.3113A>G | p.Glu1038Gly | 0.3357 | rs16941 | het | 7 | 5 | Pending | Benign |
| Exonic | 11D | nonsynonymous | c.3548A>G | p.Lys1183Arg | 0.3526 | rs16942 | het | 7 | 5 | Pending | Benign |
| Exonic | 13 | synonymous | c.4308T>C | p.Ser1436= | 0.3363 | rs1060915 | het | 7 | 5 | Class 1 | Benign |
| Exonic | 16 | nonsynonymous | c.4837A>G | p.Ser1613Gly | 0.3558 | rs1799966 | het | 7 | 5 | Pending | Benign |
| Intronic | 6 | - | c.213-161A>G | 0.5485 | rs799912 | het | 2 | 3 | Class 1 | Benign | |
| Intronic | 8 | - | c.442-34C>T | 0.0986 | rs799923 | het | 4 | 4 | Class 1 | Benign | |
| Intronic | 8 | - | c.547+146A>T | 0.3526 | rs8176140 | hom | 1 | 0 | Class 1 | Benign | |
| Intronic | 9 | - | c.548-58delT | 0.3349 | rs273902772 | het | 7 | 5 | Pending | Benign | |
| Intronic | 13 | - | c.4357+117G>A | 0.0643 | rs3737559 | het | 5 | 2 | Class 1 | Benign | |
| Intronic | 15 | - | c.4485-63A>G | 0.3534 | rs273900734 | het | 7 | 5 | Pending | Benign | |
| Intronic | 17 | - | c.5774+6C>G | . | rs80358032 | het | 1 | 0 | Pending | Benign / Uncertain | |
| Intronic | 17 | - | c.4987-68G>A | 0.3546 | rs8176234 | het | 7 | 5 | Pending | Benign | |
| Intronic | 17 | - | c.4987-92A>G | 0.3546 | rs8176233 | het | 7 | 5 | Pending | Benign | |
| Intronic | 18 | - | c.5152+66G>A | 0.3425 | rs3092994 | het | 7 | 5 | Class 1 | Benign | |
| BRCA2 | |||||||||||
| Exonic | 10 | nonsynonymous | c.865A>C | p.Asn289His | 0.0737 | rs766173 | het | 1 | 1 | Pending | Benign |
| Exonic | 10 | nonsynonymous | c.1114A>C | p.His372Asn | 0.2494 | rs144848 | het | 7 | 4 | Class 1 | Benign |
| Exonic | 10 | synonymous | c.1365A>G | p.Ser455= | 0.0737 | rs1801439 | het | 1 | 1 | Class 1 | Benign |
| Exonic | 11A | synonymous | c.2229T>C | p.His743= | 0.0735 | rs1801499 | het | 1 | 1 | Class 1 | Benign |
| Exonic | 11B | nonsynonymous | c.2971A>G | p.Asn991Asp | 0.0801 | rs1799944 | het | 1 | 1 | Class 1 | Benign |
| Exonic | 11B | synonymous | c.3396A>G | p.Lys1132= | 0.2668 | rs1801406 | het | 4 | 3 | Class 1 | Benign |
| Exonic | 11C | synonymous | c.3807T>C | p.Val1269= | 0.1681 | rs543304 | het | 1 | 2 | Class 1 | Benign |
| Exonic | 11D | synonymous | c.4563A>G | p.Leu1521= | 0.9740 | rs206075 | hom | 10 | 7 | Class 1 | Benign |
| Exonic | 11F | synonymous | c.6513C>G | p.Val2171= | 0.9736 | rs206076 | hom | 10 | 7 | Pending | Benign |
| Exonic | 14 | synonymous | c.7242A>G | p.Ser2414= | 0.2326 | rs1799955 | het | 4 | 3 | Class 1 | Benign |
| Exonic | 14 | nonsynonymous | c.7397C>T | p.Ala2466Val | 0.9758 | rs169547 | hom | 10 | 7 | Class 1 | Benign |
| Intronic | 2 | - | c.67+82C>G | 0.0010 | rs189026060 | het | 1 | 0 | Pending | Benign | |
| Intronic | 4 | - | c.425+67A>C | 0.0743 | rs11571610 | het | 1 | 1 | Class 1 | Benign | |
| Intronic | 5 | - | c.426-89T>C | 0.0743 | rs3783265 | het | 1 | 1 | Class 1 | Benign | |
| Intronic | 8 | - | c.681+56C>G | 0.1859 | rs2126042 | het | 2 | 0 | Pending | Benign | |
| Intronic | 14 | - | c.7435+53C>T | 0.0725 | rs11147489 | het | 1 | 1 | Pending | Benign | |
| Intronic | 17 | - | c.7806-14T>C | 0.5316 | rs9534262 | hom | 9 | 5 | Pending | Benign | |
| Intronic | 22 | - | c.8755-66T>C | 0.5116 | rs4942486 | hom | 7 | 3 | Class 1 | Benign | |
| UTR5 | 2 | - | c.-26G>A | 0.2093 | rs1799943 | het | 3 | 3 | Class 1 | Benign | |
| UTR3 | 27 | - | c.10362A>C | 0.1607 | rs15869 | het | 2 | 2 | Class 1 | Benign | |
Removal of Egyptian-specific normal polymorphism
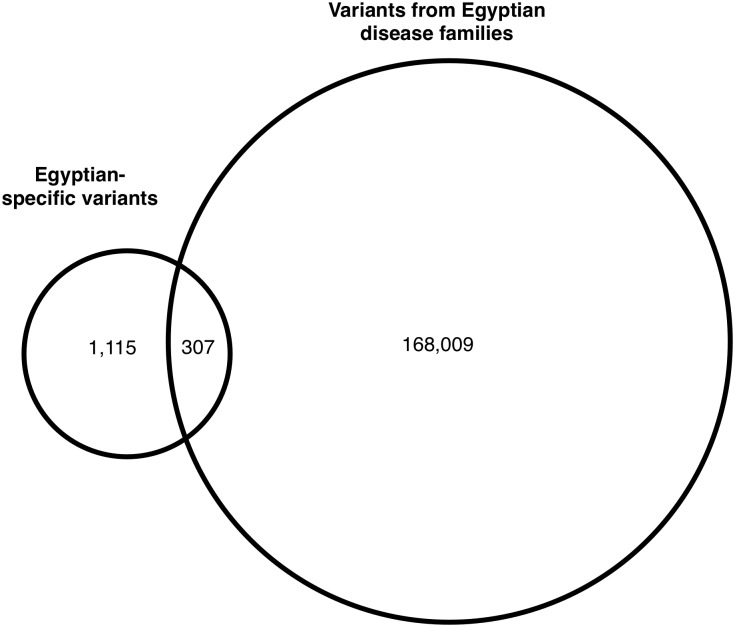
The total variants called from the exome sequences were filtered from the normal population polymorphisms from 1000 Genomes and NHLBI Exome Sequencing Project (ESP6500). As the Egypt genomic variation data were not well represented in public databases, the remaining 168,009 variants were further filtered against the 1,422 Egyptian-specific normal variant data derived from Egyptian genome study [21, S3 Table]. This step eliminated 307 Egyptian-specific normal variants, of which 13 were coding-change variants (Fig 2). From the remaining variants, we identified 421 rare, coding-change variants in the five families (S4 Table).
Fig 2. Removal of Egyptian-specific polymorphism.
The variants called from exome data and filtered from 1000 Genomes and ESP6500 databases were further filtered through the Egyptian-specific normal variants from Egyptian population. This step eliminated 307 Egyptian-specific normal variants, of which 13 were coding-change variants, from the variants called from the disease families.
Comparison of variants between Egypt and Caucasian BRCAx familial breast cancer cases
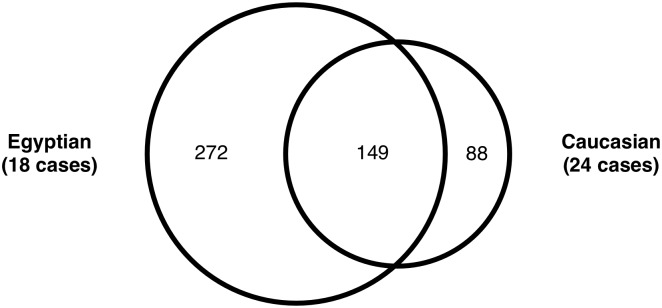
We compared the 421 coding-change variants with these from 24 Caucasian BRCAx cases we identified previously by exome sequencing [17, S5 Table]. Despite the fact that the 18 cases were from five families and the 24 cases were the probands representing 24 families, the number of variants in Egyptian cases (421 variants) was much larger than these in the Caucasian cases (237 variants). There were 149 variants shared between the two groups, but these shared variants accounted for only 35.4% of the total variants in Egyptian group comparing to 62.8% in the Caucasian group. The information indicated that the coding-change variants in Egyptian BRCAx familial breast cancer families were more heterogeneous than in the Caucasian BRCAx familial breast cancer families (Fig 3).
Fig 3. Comparison between coding-damage variants identified in Egyptian and Caucasian familial breast cancer groups.
More variants were present in Egyptian group than in Caucasian group, despite the smaller size of Egyptian group than Caucasian group. Nearly two-thirds of Caucasian variants were shared in Egyptian group, but these shared ones accounted for only about a third in Egyptian group.
Identification of damaging variants in each family
Damaging variants were predicted from the coding-change variants using SIFT and Polyphen2 programs. Those only present in a single case in each family were removed to avoid individual differences, and the remaining ones were present in at least two breast cancer-affected members in each family. The inclusion of unaffected family members aimed to know the status of the damaging variants identified in the cancer-affected members in the family: negative implies they did not carry the potential risk imposed by these damaging variants, positive implies they carried the potential risk considering that they were all at the age of younger than 50 year old. Specific conditions used in each family were:
-
Family 1:
a variant must be shared between both affected sisters, but is not required in either unaffected daughters;
-
Family 2:
a variant must be shared in the affected mother and daughter 1, but is not required in the unaffected daughter 2;
-
Family 3:
a variant must be shared between the two affected sisters, but is not required in the unaffected daughter;
-
Family 4:
a variant must be shared between the affected mother and daughter, but is not required in the unaffected daughter;
-
Family 5:
a variant must be shared between the affected sister 1 and sister 2, but is not required in the unaffected sisters 3 and 4.
A total of 26 distinct damaging variants were identified from the five disease families, of which 19 (73.1%) were known variants in the dbSNP database, 22 (84.6%) were nonsynonymous single nucleotide variants, 25 (96.2%) were heterozygous (Table 3). These variants were distributed at the frequencies of 4 to 9 per family. None of these variants was listed in the ClinVar database (Tables 3 and 4).
Table 3. Summary of damaging variants and the affected gene.
| Item | Number (%) |
|---|---|
| Total unique variants | 26 (100) |
| Known | 19 (73.1) |
| Novel | 7 (26.9) |
| Nonsynonyous SNV | 22 (84.6) |
| Frameshift deletion | 3 (11.5) |
| Frameshift substitution | 1 (3.8) |
| Heterozygote | 24 (92.3) |
| Homozygote | 2 (7.7) |
| Gene affected | 23 |
Table 4. Damaging mutations identified in five Egyptian familial breast cancer families*.
| Gene | Affected | Member | Chr. | Position | Ref. | Variant | Genotype | Var type | Coding change | dbSNP144 | Damage | Frequency | ClinVar | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIFT | PolyPhen2 | All | African | Egypt | |||||||||||||||
| Family 1 | No. | 1 | 2 | 3 | 4 | ||||||||||||||
| Status | A. | A. | U. | U. | |||||||||||||||
| ABCA10 | 2 | + | + | - | + | 17 | 67145196 | AG | - | Het. | Frameshift deletion | c.4510_4511del:p.L1504fs | - | - | - | - | - | - | - |
| CHST15 | 2 | + | + | - | - | 10 | 125780753 | GG | - | Hom. | Frameshift deletion | c.1365_1366del:p.P455fs | rs746518074 | - | - | - | - | - | - |
| GRIP1 | 2 | + | + | + | + | 12 | 66935707 | C | T | Het. | Nonsynonymous SNV | c.G160A:p.V54I | rs199768740 | - | D | 0.0004 | - | - | - |
| LOC100129697 | 2 | + | + | - | - | 16 | 89016677 | G | C | Het. | Nonsynonymous SNV | c.G151C:p.V51L | rs71395347 | D | - | - | - | - | - |
| LOC100129697 | 2 | + | + | - | - | 16 | 89017569 | G | C | Het. | Nonsynonymous SNV | c.G1043C:p.G348A | rs34847212 | D | - | - | - | - | - |
| LOC100129697 | 2 | + | + | + | - | 16 | 89017602 | C | T | Het. | Nonsynonymous SNV | c.C1076T:p.P359L | rs188085712 | D | - | - | - | - | - |
| LOC388813 | 2 | + | + | + | - | 21 | 15974379 | A | C | Het. | Nonsynonymous SNV | c.T280G:p.Y94D | rs370454293 | D | - | 0.0020 | 0.0008 | - | - |
| NBPF10 | 2 | + | + | + | - | 1 | 145296478 | G | T | Het. | Nonsynonymous SNV | c.G400T:p.D134Y | rs6663523 | D | - | - | - | - | - |
| PABPC3 | 2 | + | + | - | - | 13 | 25671271 | AAGC | GAT | Het. | Frameshift substitution | c.935_938GAT | - | - | - | - | - | - | - |
| Family 2 | No. | 1 | 2 | 3 | |||||||||||||||
| Status | A. | A. | U. | ||||||||||||||||
| C16orf62 | 2 | + | + | - | 16 | 19566981 | C | G | Het. | Nonsynonymous SNV | c.C197G:p.A66G | rs564734737 | D | - | 0.0002 | - | - | - | |
| KRTAP21-3 | 2 | + | + | - | 21 | 32090989 | C | T | Het. | Nonsynonymous SNV | c.G89A:p.C30Y | rs760518532 | D | - | - | - | - | - | |
| LOC100129697 | 2 | + | + | - | 16 | 89016677 | G | C | Het. | Nonsynonymous SNV | c.G151C:p.V51L | rs71395347 | D | - | - | - | - | - | |
| LOC100129697 | 2 | + | + | - | 16 | 89017569 | G | C | Het. | Nonsynonymous SNV | c.G1043C:p.G348A | rs34847212 | D | - | - | - | - | - | |
| NBPF10 | 2 | + | + | + | 1 | 145296478 | G | T | Het. | Nonsynonymous SNV | c.G400T:p.D134Y | rs6663523 | D | - | - | - | - | - | |
| NPIPB11 | 2 | + | + | + | 16 | 29394758 | G | T | Hom. | Nonsynonymous SNV | c.C1495A:p.P499T | - | D | - | - | - | - | - | |
| PABPC3 | 2 | + | + | - | 13 | 25671271 | AAGC | GAT | Het. | Frameshift substitution | c.935_938GAT | - | - | - | - | - | - | - | |
| PDE4DIP | 2 | + | + | + | 1 | 144852379 | G | T | Het. | Nonsynonymous SNV | c.C7064A:p.P2355H | - | D | - | - | - | - | - | |
| Family 3 | No. | 1 | 2 | 3 | |||||||||||||||
| Status | A. | U. | U. | ||||||||||||||||
| CCDC7 | 2 | + | + | + | 10 | 33136820 | AA | - | Het. | Frameshift deletion | c.1477_1478del:p.K493fs | - | - | - | - | - | - | - | |
| CFAP46 | 2 | + | + | + | 10 | 134736204 | G | A | Het. | Nonsynonymous SNV | c.C1265T:p.T422M | rs140185143 | D | - | 0.0022 | - | - | - | |
| CXorf23 | 2 | + | - | + | X | 19984574 | G | T | Het. | Nonsynonymous SNV | c.C235A:p.P79T | rs143234295 | - | D | 0.0082 | 0.0299 | - | - | |
| LOC100129697 | 2 | + | + | + | 16 | 89017334 | C | T | Het. | Nonsynonymous SNV | c.C808T:p.R270W | rs28617399 | D | - | - | - | - | - | |
| NBPF10 | 2 | + | + | + | 1 | 145296478 | G | T | Het. | Nonsynonymous SNV | c.G400T:p.D134Y | rs6663523 | D | - | - | - | - | - | |
| NPIPB11 | 2 | + | + | + | 16 | 29394758 | G | T | Hom. | Nonsynonymous SNV | c.C1495A:p.P499T | - | D | - | - | - | - | - | |
| PABPC3 | 2 | + | + | + | 13 | 25671271 | AAGC | GAT | Het. | Frameshift substitution | c.935_938GAT | - | - | - | - | - | - | - | |
| SMIM13 | 2 | + | + | + | 6 | 11134683 | C | T | Het. | Nonsynonymous SNV | c.C124T:p.R42W | - | - | D | - | - | - | - | |
| Family 4 | No. | 1 | 2 | 3 | |||||||||||||||
| Status | A. | A. | U. | ||||||||||||||||
| C16orf62 | 2 | + | + | - | 16 | 19566981 | C | G | Het. | Nonsynonymous SNV | c.C197G:p.A66G | rs564734737 | D | - | 0.0002 | - | - | - | |
| GAGE2A | 2 | + | + | + | X | 49237474 | A | T | Het. | Nonsynonymous SNV | c.A179T:p.D60V | rs782582454 | D | - | - | - | - | - | |
| NPIPB11 | 2 | + | + | + | 16 | 29394758 | G | T | Hom. | Nonsynonymous SNV | c.C1495A:p.P499T | - | D | - | - | - | - | - | |
| PHIP | 2 | + | + | + | 6 | 79671522 | C | A | Het. | Nonsynonymous SNV | c.G3541T:p.A1181S | rs147526156 | - | D | 0.0002 | - | - | - | |
| SLC15A5 | 2 | + | + | - | 12 | 16430451 | A | T | Het. | Nonsynonymous SNV | c.T169A:p.F57I | rs79942763 | D | - | 0.0066 | 0.0227 | - | - | |
| ZNF750 | 2 | + | + | - | 17 | 80789339 | T | C | Het. | Nonsynonymous SNV | c.A992G:p.Y331C | - | - | D | - | - | - | - | |
| Family 5 | No. | 1 | 2 | 3 | 4 | ||||||||||||||
| Status | A. | U. | A. | U. | - | ||||||||||||||
| ATP10B | 2 | + | + | + | + | 5 | 160016684 | A | G | Het. | Nonsynonymous SNV | c.T3665C:p.I1222T | rs144497343 | - | D | 0.0094 | 0.0061 | - | - |
| NPIPB11 | 2 | + | + | + | + | 16 | 29394758 | G | T | Hom. | Nonsynonymous SNV | c.C1495A:p.P499T | - | D | - | - | - | - | - |
| PIGN | 2 | + | + | + | - | 18 | 59828420 | G | A | Het. | Nonsynonymous SNV | c.C167T:p.A56V | rs61755362 | - | D | 0.0044 | - | - | - |
| PRR14L | 2 | + | - | + | + | 22 | 32108801 | G | T | Het. | Nonsynonymous SNV | c.C5024A:p.A1675E | rs750572033 | - | D | - | - | - | - |
*A.: affected; U.: unaffected; Chr.: Chromosome; Ref.: Reference genome hg19; Het.: Heterozygote; Hom.: homozygote; D: Damaging variant
The 26 damaging variants affected 23 genes. The variants-affected genes are distributed in various functional categories, including RNA binding (NBPF10, PABPC3), transcriptional regulation (ZNF750), extracellular matrix (CHST15), structural protein (NPIPB11, GRIP1, CFAP46), and signal transduction (PDE4DIP, PHIP). As the examples, copy number change in NBPF10 is associated with multiple developmental and neurogenetic diseases, PABPC3 is involved in regulation of mRNA stability and translation initiation, and NPIPB11 is involved in forming nuclear pore complex. None of these genes are involved in DNA damaging repair pathways, in which the predisposition genes are traditionally considered to be located. Several variant-affected genes affected a few pathways mostly involved in housekeeping function. Whether any of these variant-affected genes can be predisposition gene candidates remains to be determined (Table 4, S4 Table).
Two homozygous damaging variants were present in CHST15 and NPIPB11. The variant rs746518074 in CHST15 was present in two affected members in Family 1, and the novel variant in NPIPB11 was present in both affected and unaffected members in families 2, 3, 4 and 5. CHST15 is an extracellular matrix component [26], and NPIPB11 has unknown function. The high frequency of the novel variant in NPIPB11 suggests that this variant is likely to be a normal homozygous polymorphism in Egyptian population. Little evidence exists for the roles of CHST15 and NPIPB11 in genetic predisposition in familial breast cancer.
We also compared the variant-affected genes with the mutation data from The Cancer Genome Atlas (TCGA) study [27]. Although none of the 825 breast cancer cases were marked as familial breast cancer cases, 49 germline mutations in the classical predisposition genes of ATM, BRCA1, BRCA2, BRIPi, CHEK2, NBN, PTEN, RAD51C and TP53 were identified in 47 of 507 blood samples paired with breast cancer cases. None of the same variants were present in the Egyptian families we analyzed.
Discussion
Decades’ study has well concluded that genetic predisposition plays the major roles in the development of familial breast cancer. As demonstrated by the extensive BRCA study, identification of the predisposition is essential for early diagnosis and prevention of breast cancer as it allows frequent monitoring the carrier health for early sign of the disease, blocking the tumorigenesis process by using chemo-prevention including tamoxifen and poly (ADP-ribose) polymerase (PARP) inhibitors, and applying preventive surgery to remove cancer susceptible tissues. Due largely to the scientific and economic advantages, current knowledge of genetic predisposition are largely derived from the developed countries of European and North American populations. Increased data from recent studies in Latino, Africa, and Asia populations demonstrate that genetic predisposition for familial breast cancer can be ethnic-specific in reflecting human evolution and geographic differences [4–10]. Without the information from different ethnic populations, our understanding of genetic predisposition for familial breast cancer will remain incomplete; and relying on the existing information as the solely references is not adequate to identify the patients from other ethnicities.
The Egyptian population has many unique genetic features developed during its evolution history and specific geographic location across Asian and African continents. Our study selected Egypt breast cancer families as a model to test if the genetic predisposition in populations of this area is the same as, or similar to, or very different from existing data of other ethnic populations. Our study showed the absence of mutations in BRCA1, BRCA2, and other classical predisposition genes, and the presence of the damaging variants in genes not involved in DNA damage repair in Egyptian patient families. We consider that the genetic predisposition in Egyptian familial breast cancer can be substantially different from the ones currently known from other ethnic populations.
Our study analyzed only five disease families. It is known that many predispositions are rare in the disease population. A possibility exists that certain known predispositions in the classical genes could be present in Egyptian familial breast cancer population but not detected due to the size limitation. Other possibility could be that the predisposition is located in non-coding region of the genome, which cannot be detected by exome sequencing method.
Conclusions
Our study provides proof-of-principal evidence for the presence of specific genetic predisposition for familial breast cancer in Egyptian patients, and supports a scale-up study to characterize substantial numbers of disease families from local population in order to determine the nature of Egypt-specific predispositions in this population.
Supporting Information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Abbreviations
- BIC
Breast Cancer Information Core
- BRCAx
familial breast cancer without BRCA1 and BRCA2 mutation
- ClinVar
public archive of interpretations of clinically relevant variants
- MAF
minor allele frequency
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by a pilot grant from the Fred & Pamela Buffett Cancer Center to Dr. San Ming Wang and Dr. Amr S. Soliman (http://www.unmc.edu/eppley/), a grant from the Eplley Cancer institute to Dr. San Ming Wang (http://www.unmc.edu/eppley/), and a grant from Science & Technology Development Fund in Egypt to Abdel-Rahman Nabawy Zekri (Grant number 5193). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40: 17–22. 10.1038/ng.2007.53 [DOI] [PubMed] [Google Scholar]
- 2.Madorsky-Feldman D, Sklair-Levy M, Perri T, Laitman Y, Paluch-Shimon S, Schmutzler R, et al. An international survey of surveillance schemes for unaffected BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat. 2016; Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.COMPLEXO, Southey MC, Park DJ, Nguyen-Dumont T, Campbell I, Thompson E, et al. COMPLEXO: identifying the missing heritability of breast cancer via next generation collaboration. Breast Cancer Res. 2013; 15:402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janavičius R. Founder BRCA1/2 mutations in the Europe: implications for hereditary breast-ovarian cancer prevention and control. EPMA J. 2010;1: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336: 1401–1408. 10.1056/NEJM199705153362001 [DOI] [PubMed] [Google Scholar]
- 6.Górski B, Jakubowska A, Huzarski T, Byrski T, Gronwald J, Grzybowska E, et al. A high proportion of founder BRCA1 mutations in Polish breast cancer families. Int J Cancer. 2004;110: 683–686. 10.1002/ijc.20162 [DOI] [PubMed] [Google Scholar]
- 7.Zhang J, Fackenthal JD, Zheng Y, Huo D, Hou N, Niu Q, et al. Recurrent BRCA1 and BRCA2 mutations in breast cancer patients of African ancestry. Breast Cancer Research and Treatment. 2012;134: 889–894. 10.1007/s10549-012-2136-z [DOI] [PubMed] [Google Scholar]
- 8.Villarreal-Garza C, Alvarez-Gómez RM, Pérez-Plasencia C, Herrera LA, Herzog J, Castillo D, et al. Significant clinical impact of recurrent BRCA1 and BRCA2 mutations in Mexico. Cancer. 2015; 121:372–378. 10.1002/cncr.29058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang E, Seong MW, Park SK, Lee JW, Lee J, Kim LS, et al. The prevalence and spectrum of BRCA1 and BRCA2 mutations in Korean population: recent update of the Korean Hereditary Breast Cancer [KOHBRA] study. Breast Cancer Res Treat. 2015;151: 157–168. 10.1007/s10549-015-3377-4 [DOI] [PubMed] [Google Scholar]
- 10.Kim YC, Zhao L, Zhang H, Huang Y, Cui J, Xiao F, et al. Prevalence and spectrum of BRCA germline variants in mainland Chinese familial breast and ovarian cancer patients. Oncotarget. 2016;7: 9600–9612. 10.18632/oncotarget.7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagani L, Schiffels S, Gurdasani D, Danecek P, Scally A, Chen Y, et al. Tracing the route of modern humans out of Africa by using 225 human genome sequences from Ethiopians and Egyptians. Am J Hum Genet. 2015;96: 986–91. 10.1016/j.ajhg.2015.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.http://www.iarc.fr/en/publications/pdfs-online/epi/sp164/CI5volX_Full.pdf
- 13.Bedwani R, Abdel-Fattah M, El-Shazly M, Bassili A, Zaki A, Seif HA, et al. Profile of familial breast cancer in Alexandria, Egypt. Anticancer Res. 2001;21: 3011–3014. [PubMed] [Google Scholar]
- 14.Soliman AS, Banerjee M, Lo AC, Ismail K, Hablas A, Seifeldin IA, et al. High proportion of inflammatory breast cancer in the Population-based Cancer Registry of Gharbiah, Egypt. Breast J. 2009;15: 432–434. 10.1111/j.1524-4741.2009.00755.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen H, Kim YC, Snyder C, Xiao F, Fleissner EA, Becirovic D, et al. Family-specific, novel, deleterious germline variants provide a rich resource to identify genetic predispositions for BRCAx familial breast cancer. BMC Cancer. 2014;14: 470 10.1186/1471-2407-14-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.UCSC human genome 19: http://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/
- 17.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picard Tools: http://broadinstitute.github.io/picard/, Access April 20, 2016
- 19.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. 2012; Preprint. arXiv:1207.3907.
- 20.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nature Protocols 2015;10: 1556–1566. 10.1038/nprot.2015.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egyptian Human Genome Sequencing Project: http://genomicsegypt.org
- 22.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4: 1073–1081. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 23.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7: 248–9. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, et al. The Reactome pathway Knowledgebase. Nucleic Acids Res. 2016;44(D1):D481–7. Available: www.reactome.org 10.1093/nar/gkv1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arora A, Agarwal D, Abdel-Fatah TM, Lu H, Croteau DL, Moseley P, et al. RECQL4 helicase has oncogenic potential in sporadic breast cancers. J Pathol. 2016;238: 495–501. 10.1002/path.4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salgueiro AM, Filipe M, Belo JA. N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase expression during early mouse embryonic development. Int J Dev Biol. 2006;50: 705–708. 10.1387/ijdb.062168as [DOI] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490: 61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.