Abstract
The human (h) growth hormone/chorionic somatomammotropin (GH/CS) gene locus presents a unique model to gain insight into the molecular mechanisms that have allowed a closely related family of genes to be expressed in two distinct cell lineages/tissues: pituitary somatotrophs and placental syncytiotrophoblasts. However, studies of external factors that regulate gene expression have been somewhat limited by (i) a lack of human cell lines expressing endogenous GH or CS appropriately; and (ii) the fact that the GH/CS locus is unique to primates and thus does not exist in rodents. In the current study, a transgenic (171hGH/CS-TG) mouse was generated containing the intact hGH/CS gene cluster and hGH locus control region (LCR) in a 171-kilobase DNA fragment. Pituitary and placental-specific expression of hGH/CS RNA was detected at embryonic day (E) 18.5. Immunostaining of hGH was seen in somatotrophs of the anterior pituitary beginning in late gestation. The presence of hCS protein was detected in the placental labyrinth in trophoblasts functionally analogous to the syncytiotrophoblast of the chorionic villi. This pattern of gene expression is consistent with the presence of essential components of the hGH/CS LCR. Transcript levels for hCS-A, hCS-B and placental hGH-variant increased in 171hGH/CS-TG placenta during gestation (E11.5–E18.5), as previously observed in human placental development. Throughout gestation, hCS-A RNA levels were proportionately higher, accounting for 91% of total CS RNA by E18.5, comparable to term human placenta. Finally, the previous correlation between the transcription factor AP-2α and hCS RNA expression observed in developing primary human cytotrophoblast cultures, was extended to pregnancy in the 171hGH/CS-TG mouse. The 171hGH/CS-TG mouse thus provides a model to investigate hGH/CS gene expression, including in pregnancy.
Keywords: Gene expression, Transgenic mice, Human growth hormone gene cluster, Pituitary, Placenta, Trophoblast
1. Introduction
The human growth hormone/chorionic somatomammotropin (hGH/CS) locus presents a unique model to investigate the molecular mechanisms by which a group of recently evolved, and thus closely related, genes have become specialized to express related hormones in two distinct tissues. The five hGH/CS gene family members, including pituitary growth hormone (GH-N) as well as placental GH variant (GH-V), and the placental lactogen or CS genes (CS-A, CS-B, and CS-L), are contained within a single 47-kilobase (kb) locus on chromosome 17 [1]. The five GH/CS genes are aligned in the same transcriptional orientation in the order 5′-GH-N/CS-L/CS-A/GH-V/CS-B-3′, share 90–99% sequence similarity and are believed to have evolved by gene duplication [1]. In spite of the sequence homology between the GH/CS genes, GH-N is expressed in pituitary somatotrophs, while CS and GH-V are expressed specifically in the villus syncytiotrophoblast of the placenta [1]. This distinguishes the GH/CS locus from the more extensively studied β-globin locus, where expressionis restrictedto a single tissue type (erythroid tissues) with varied expression during developmental stages [2].
The GH family plays fundamental roles in fetal as well as adult growth and development. Pituitary GH-N functions in post-natal growth, development and homeostatic regulation. In pregnancy, pituitary GH is replaced in the maternal bloodstream by its placental counterparts. The primary role of CS seems to be to provide a continuous supply of metabolic substrates, especially glucose, to the fetus in an indirect manner by influencing maternal metabolism through its insulin antagonistic activity [3,4]. Some evidence also supports a direct action on fetal growth and metabolism [3,4]. Maternal deletion of the placental GH/CS genes in a human mother resulted in the birth of a severely growth-retarded but otherwise normal newborn [5].
Distinct mechanisms of pituitary versus placental expression are reflected in the chromatin structure of the hGH/CS locus and participation of specific transcription factor complexes [6]. Understanding the molecular mechanisms underlying these dynamic relationships for the hGH/CS gene family has been somewhat limited by the absence of readily usable model systems, including human tissue, to study gene expression in a developmental context in situ. This is due, in part, to the absence of appropriate human pituitary or placental cell lines capable of efficient hGH or hCS expression. While rodent cell systems exist, the organization of the five pituitary and placental GH/CS genes in the human locus is peculiar to primates; the placental lactogen genes in mice and rats are more related structurally and functionally to prolactin than GH [7]. A technical difficulty has also been the length of regulatory DNA needed to drive appropriate GH/CS gene expression. While the expression of rat GH appears to be controlled by proximal sequences (within 310 base pairs upstream of the transcription initiation site) [8,9], a locus control region (LCR) including remote sequences located 14–32 kilobases (kb) upstream of the human GH gene [10], is required for tissue-specific expression of the pituitary GH-N gene and placental CS-A gene in vivo [11]. Furthermore, the LCR was shown in these transgenic mouse studies to confer expression of ‘appropriate’ levels of pituitary GH, reflecting combined products of human and mouse GH-N genes, such that no apparent difference in animal size was detected [10].
The GH LCR contains regulatory sequences associated with five nuclease hypersensitive sites (HSI–V) including those reported to be specific to the pituitary (HSI/II) and placenta (HSIV) as well as constitutive (HSIII and HSV) in nature [10,12]. These sites are found in the loci of the adjacent lymphocyte-specific CD79b and skeletal muscle sodium channel α-subunit SCN4A genes [13,14]. This chromosomal arrangement has been conserved, at least in mammals [15,16], which is consistent with the idea that this organization of genes is not without regulatory significance.
In the case of the hGH-N gene, binding of the pituitary-specific POU homeodomain transcription factor Pit-1 to sites within HSI/II [17] is essential for further histone modifications and generation of an active and efficient hGH-N promoter in pituitary somatotrophs in vivo [11]. Placenta-specific expression, including that of human hormone genes such as chorionic gonadotropin and corticotropin-releasing hormone, has been linked with members of the activator protein 2 (AP-2) transcription factor family [18–20]. The AP-2 family was also shown to regulate levels of both rat [21] and human placental lactogen (hCS in humans), through elements in both the proximal promoter [22,23] and remote sequences in the upstream LCR [24]. Direct involvement of AP-2 in hCS expression was indicated by downregulation of hCS RNA levels in cultured human cyto/syncytiotrophoblasts treated with a dominant negative AP-2 adenoviral vector ([25] and our unpublished observations).
Previous studies to assess hGH/CS expression and dissect the mechanism of tissue-specific expression have relied largely on human choriocarcinoma cell lines, short-term primary human placental cultures and transgenic mice containing truncated hGH or an incomplete hGH/CS gene locus. The latter includes the absence of the hCS-B gene and thus potential placental enhancer activity associated with downstream flanking sequences, as identified through reporter gene and transfection studies of human placental cell cultures. Here we describe the characterization of a transgenic mouse model (171hGH/CS-TG) containing all five hGH/CS genes as well as flanking CD79b and SCN4A genes, encompassing the HSI–V regions that comprise the hGH LCR. The pattern of pituitary hGH-N and placental hCS gene expression observed, as well as the apparent normal growth of the 171hGH/CS-TG mice, are consistent with the presence of an intact hGH LCR.
2. Materials and methods
2.1. Placental tissue and cells
Collection of term placental tissue was carried out under the approval of the Bannatyne Campus Research Ethics Board of the University of Manitoba. Term placentas were collected from routine births at the Women’s Hospital (Winnipeg, MB) as discarded tissue with no connection to the identity or family history of mother or baby. Early placental tissue (12 weeks) was obtained as a donation of discarded tissue from the clinic of Dr. Chui Kin Yuen of the Women’s Hospital.
2.2. Generation of 171hGH/CS-TG mice
All animals were housed and treated according to standards and guidelines set by the Canadian Council for Animal Care. The investigation was conducted in accordance with the National Research Council (NRC) publication Guide for Care and Use of Laboratory Animals (copyright 1996, National Academy of Science). All protocols and procedures involving animals were reviewed and approved by the Bannatyne Campus Protocol Management and Review Committee at the University of Manitoba.
A BAC clone (RP11-1119G22, 180 kb total length including vector) containing the entire hGH/CS gene cluster and upstream LCR was obtained with kind assistance from the TCAG Genome Resource Facility (Hospital for Sick Children, Toronto, ON). Through sequence alignment with Genbank Accession numbers AC005803 and J03071, the BAC DNA fragment was found to encompass 171 kb of human genomic DNA corresponding to sequence from 110.7 kb upstream to 61.3 kb downstream of the hGH-N transcription initiation site. The clone was linearized by the rare-cutting enzyme P1-SceI, followed by purification using a Clontech tip20, then introduced into the pronuclei of single-cell zygotes from CD1 mice. Injected embryos were subsequently transferred to the oviduct of surrogate mothers and brought to term [17]. Genomic DNA was extracted from tail tissue using DNeasy Blood & Tissue Kit (Qiagen). The presence of human DNA sequences in the genomes of the resulting founder animals was detected by DNA (Southern) blotting, using BamH1 and EcoRI digestion and hybridization with radioactive-labeled hGH cDNA, HSV, HSIII, and HSI/II fragments, respectively. Two transgenic male founders were bred with female CD1 mice. Mice from the F1 generation carrying the intact hGH/CS locus were crossbred and maintained in a CD1 genetic background. Mice used for assessment of RNA by PCR were heterozygous; for immunohistochemistry, homozygotes were bred.
2.3. Detection of RNA by reverse transcription and quantitative polymerase chain reaction (PCR)
Total RNA was obtained using the RNeasy Plus Mini Kit (Qiagen), from embryonic (E11.5, E13.5, and E18.5) and adult tissues of wild type and BAC transgenic mice. Placental tissue was enriched for labyrinthine cells by careful dissection using the anatomy described by others [26]. RNA (1 μg) from different tissues was used for reverse transcription by the QuantiTect Reverse Transcription Kit (Qiagen), according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) analyses were performed in an iCycler (Bio-Rad) with specific primers (see Table 1) and 0.1 μg cDNA in each reaction. Reaction volumes of 30 μl consisted of the following components: 2.5 mM MgCl2, 0.025% DMSO, 0.6 μl (1:1000 dilution) of SYBR green (Sigma, S9430), 0.3 μl (1:1000 dilution) of fluorescein calibration dye (Bio-Rad, Cat# 170-8780) and 0.75 units platinum Taq DNA polymerase (Invitrogen). Thermal cycling was initiated with a 4-min denaturation at 95 °C, then followed by 40 cycles of 95 °C for 30 s, annealing at 60 °C for 15 s and 72 °C for 30 s. Standard curves were generated using plasmids containing the amplicon sequences for CS-A, CS-B, GH-V, AP-2α, AP-2γ and GAPDH. Minus RT controls were performed using the same PCR primers and thermal cycle conditions and no genomic DNA contamination was detected. Specific amplifications were identified by a single peak in the melting curve and a single band in the final PCR product visualized on an agarose gel. The gene expression level in each sample was calculated from the standard curve and normalized to mouse or human GAPDH expression as appropriate. Tests were run in duplicate on independent samples (multiple pups/placentas from two or more pregnant mothers). All amplicon sequences were confirmed by sequencing.
Table 1.
Oligonucleotide primers.
| Name | Sequence |
|---|---|
| GH/CS-F | 5′-CAGAAGTATTCATTCCTGCA-3′ |
| GH/CS-R | 5′-TTTGGATGCCTTCCTCTAG-3′ |
| CSA-F | 5′-GGCTTCTAGGTGCCCGAGTA-3′ |
| CSA-R | 5′-GCACTGGAGTGGCACCTTCA-3′ |
| CSB-F | 5′-CAGCAAGTTTGACACAAACTCA-3′ |
| CSB-R | 5′-AGAAGCCACAGCTACCCTCT-3′ |
| GH-V-F | 5′-GTTTGAAGAAGCCTATATCCTG-3′ |
| GH-V-R | 5′-TCACCCTGTTGGAAGGTGTT-3′ |
| h/mAP-2α-F | 5′-GTGTCCCTGTCCAAGTCCAAC-3′ |
| h/mAP-2α-R | 5′-CTGAGGAGCGAGAGGCGACC-3′ |
| h/mAP-2γ-F | 5′-GATCAGACAGTCATTCGCAAAGG-3′ |
| h/mAP-2γ-R | 5′-AGGGACCGAGCAGAAGACCTCA-3′ |
| mGAPDH-F | 5′-TCACCACCATGGAGAAGGC-3′ |
| mGAPDH-R | 5′-GCTAAGCAGTTGGTGGTGCA-3′ |
| hGAPDH-F | 5′-GAGTCAACGGATTTGGTCGT-3′ |
| hGAPDH-R | 5′-TTGATTTTGGAGGGATCTCGC-3′ |
| HSI and II-rt-F | 5′-GTGGCTGGAGAGAGACTGACCCG-3′ |
| HSI and II-rt-R | 5′-GTTAATGAAGGACTCTGTCGTTAG-3′ |
| Glucagon-F(h and m) | 5′-GGAATAACATTGCCAAACGTC-3′ |
| Glucagon-R(h and m) | 5′-CTCGCCTTCCTCGGCCTTTC-3′ |
Transgene copy number was estimated by the 2−ΔΔCt method using quantitative PCR [27]. ΔCt was calculated from the cycle threshold (Ct) of the HSI/II region minus the Ct for the endogenous glucagon gene using DNA from F-74 transgenic mice, and this was compared to the value of ΔCt in a human placental (JAR) cell line; the primers used to assess the mouse and human glucagon genes were identical (see Table 1 for primer sequences). This assessment was confirmed by DNA (Southern) blotting.
2.4. Immunohistochemistry
Adult mouse pituitary, E18.5 mouse placenta and human term placenta were fixed in 10% neutral buffered formalin for 24 h at room temperature. Paraffin sections (5 μm) were deparaffinized and rehydrated, followed by microwave antigen retrieval [28]. The sections were pretreated with 3% H2O2 to deactivate endogenous peroxidase and incubated with 5% normal serum from the same species as the secondary antibody for 30 min at room temperature or 4 °C overnight for blocking the non-specific binding of IgG. For immunoperoxidase staining, rabbit anti-rat GH antibody cross-reacting with mGH (1:20,000 dilution, National Hormone and Pituitary Program (NIDDK), Torrance, CA) or rabbit anti-human placental lactogen (1:10,000 dilution, DakoCytomatin, Denmark) was applied overnight at 4 °C after removing normal serum. This was followed by incubation (30 min) with biotinylated anti-rabbit IgG (1:500 dilution, Jackson ImmunoResearch Laboratories). A mouse-on-mouse kit (BioGenex, San Ramon, CA) was applied according to the manufacturer’s instructions for mouse anti-hGH (1:400 dilution, AbCam, Cambridge, MA) on mouse tissue for optimum staining. Positive signals were visualized with streptavidin–biotin–peroxidase and revealed with chromogen 3,3′-diaminobenzidine (DAB) and counter stained with haematoxylin. For immunofluorescent staining, sections were incubated with rabbit anti-rat GH overnight at 4 °C after quenching endogenous peroxidase and blocking non-specific binding. The next day, mouse anti-human GH (1:400 dilution) was applied to sections after biotin labeling using a mouse-on-mouse kit, and were picked up by streptavidin–horse-radish peroxidase that catalyzes the deposition of fluorescein-labeled tyramide amplification reagent (Tyramide Fluorescence systems, NEN Life Science Products Technical). Mouse GH was visualized by anti-rabbit Cy3 (1:400 dilution, Invitrogen). For both peroxidase and fluorescence, specificity of staining was checked routinely by omitting primary antibody and confirming a loss of signal.
2.5. Statistical analysis
Data were analyzed using Instat (GraphPad) software by one-way ANOVA with Tukey–Kramer post-tests for multiple comparisons. Differences with a p value <0.05 were considered statistically significant.
3. Results
3.1. Detection of the human GH/CS gene locus in transgenic mice
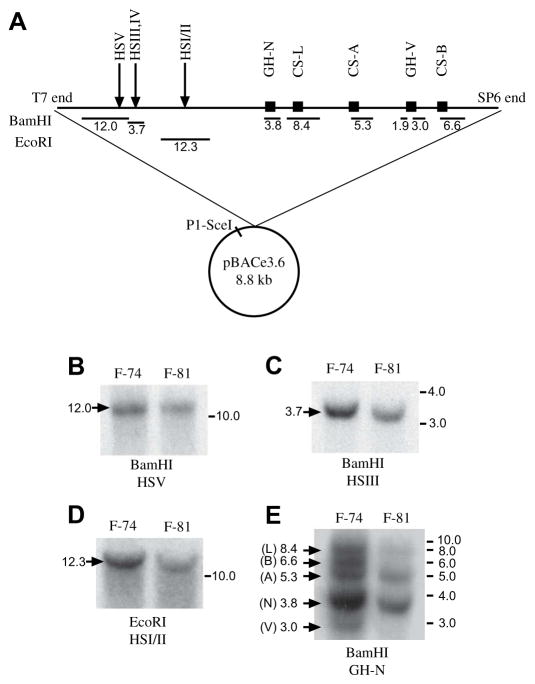
A 171-kb stretch of human genomic DNA was identified from a BAC library to contain the intact hGH/CS locus (described in Section 2), purified and introduced into mouse zygotes by pronuclear injection. Out of 45 potential transgenic founder mice, two (F-74 and F-81) containing the hGH LCR were identified by PCR detection of the HSI/II region. Descendents of each of these founders appeared indistinguishable in size from non-transgenic mice, bred normally, and gave rise to normal litters (8–12 mice). Long-range integrity of the hGH/CS gene locus was assessed in these animals by restriction endonuclease digestion and DNA blotting (Fig. 1). Based on the expected fragment sizes shown in Fig. 1A, digestion with either BamHI or EcoRI revealed that both F-74 and F-81 contained all five hypersensitive sites, and thus an intact hGH LCR (Fig. 1B–D). However, when BamHI-digested DNA was probed with hGH-N cDNA, samples from only one of the mice (F-74) yielded fragments corresponding to each of the five members of the hGH/CS family [29]: specifically 8.4 kb for hCS-L, 6.6 kb for hCS-B, 5.3 kb for hCS-A, 3.8 kb for hGH-N, and 3.0 kb for hGH-V (Fig. 1E). Fragments corresponding to hGH-V and hCS-B were not detected in F-81 DNA, indicating a truncated hGH/CS locus. Transgene copy number was estimated in a heterozygous F-74 mouse using quantitative PCR [27]. Calculation of 2−ΔΔCt, revealed that the quantity of HSI/II DNA in transgenic mice was half that found in JAR cells, representing the homozygous situation (0.580 ± 0.004 versus 1.00 ± 0.24 respectively). These data are consistent with the presence of a single intact hGH LCR extending 43 kb upstream of the hGH-N promoter, and thus, expected to contain the intact human CD79b and SCN4A genes in addition to those for GH-N, CS-L, CS-A, GH-V and CS-B in the F-74 mice.
Fig. 1.
Genomic integration of the 180-kb BAC clone from human chromosome 17. (A) Schematic (not to scale) of the vector and insert used in the generation of the 171hGH/CS-TG mice. A 171-kb contiguous region of human chromosome 17 was inserted into the vector pBACe3.6 (8.8 kb). The BAC clone was then linearized with the rare cutter P1-SceI for microinjection. The locations of the five members of the hGH/CS gene family, as well as the hypersensitive sites (HSI–V) are shown. Below the schematic, fragments sizes (kb) generated with digestion of BAC DNA with either BamHI or EcoRI are shown. (B–E) Genomic DNA from two founder mice (F-74 and F-81) was digested with BamHI or EcoRI and assessed by DNA blotting and radiolabeled probes corresponding to the regions listed beneath the restriction enzyme (HSV, HSIII, HSI/II, and GH-N coding sequences, respectively). The sizes of the fragments according to the schematic in part (A) are given at the left of each scan, and the migration positions of size markers (kb) is given at the right. In (E), the probe is a radiolabeled hGH cDNA, which hybridizes to all five members of the hGH/CS family [29].
3.2. Pituitary and placental expression of the human GH/CS transgene
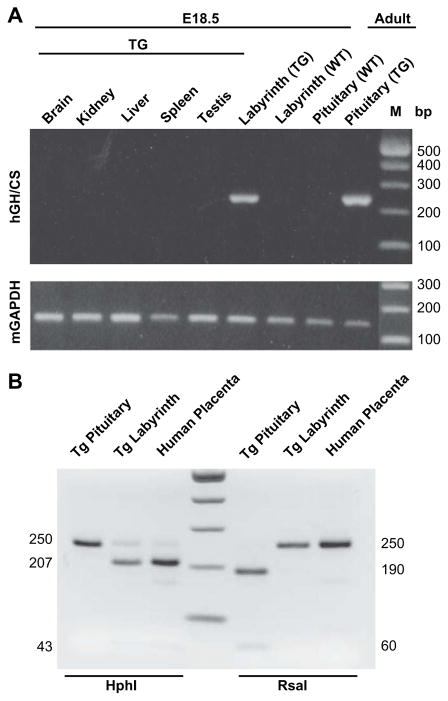
To assess for tissue-specific expression of hGH/CS RNA, gestation was terminated at embryonic day (E) 18.5 and RNA was isolated from embryonic tissues (brain, kidney, liver, spleen, testis and placental labyrinth) as well as adult pituitary of F-74 (now referred to as 171hGH/CS-TG) and non-transgenic (non-TG) mice. RNA was screened by RT-PCR with primers used previously to detect transcripts from any of the hGH/CS genes [30], and mouse (m) GAPDH RNA from each tissue was also amplified to correct for variation (Fig. 2A). The presence of hGH/CS transcripts was readily detected in pituitary and placenta tissue, but not in other tissues as analyzed (Fig. 2A). In addition, restriction enzyme analysis of the hGH/CS RT-PCR product confirmed the absence of hGH in transgenic labyrinth, and the absence of hCS in transgenic pituitary (Fig. 2B).
Fig. 2.
Pituitary and placental-specific expression of hGH/CS in 171hGH/CS-TG mice. (A) Total RNA from E18.5 transgenic (TG) and wild type (WT) embryos and placentas as well as WT and TG adult pituitary was isolated from the tissues indicated and used as a template for RT-PCR, using a primer pair that will amplify any member of the hGH/CS family (product size 250 bp). Murine GAPDH (mGAPDH) was assessed by RT-PCR for each tissue and used as a control for RNA loading (product size 168 bp). M, size markers, indicated in base pairs (bp). (B) hGH RNA can be distinguished from hCS by enzyme digestion [30]. With HphI digestion, the hGH is left uncut at 250 bp while the hCS products are cleaved to fragments of 207 and 43 bp (some residual uncut product remains). With RsaI digestion, the hCS product is uncut (250 bp) while hGH is cleaved to fragments of 190 and 60 bp.
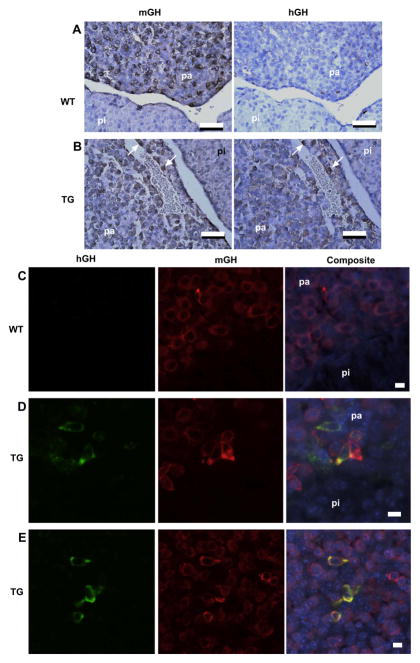
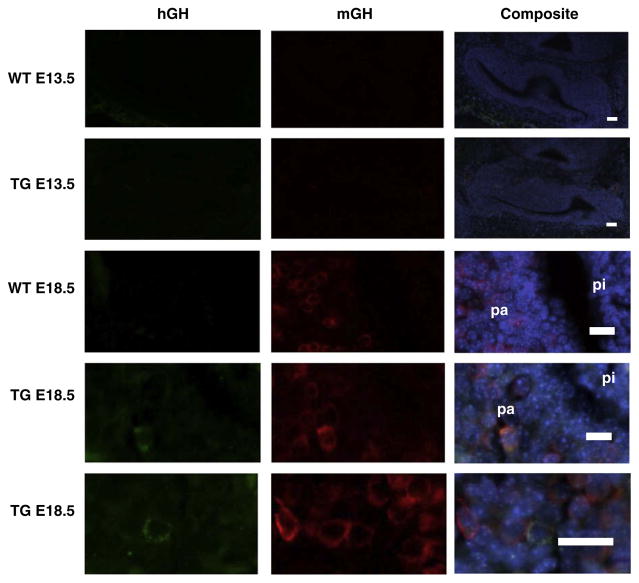
The distribution of hGH/CS in transgenic pituitary and placenta tissue was examined by immunohistochemistry. Histological sections of anterior pituitary tissue from 171hGH/CS-TG and non-TG mice were detected using primary antibodies against either mGH or hGH (Fig. 3). Evidence of cells staining for mGH, and thus detection of somatotrophs, were apparent in the anterior pituitary of both 171hGH/CS-TG and non-TG mice. In contrast, detection of hGH was specific to 171hGH/CS-TG mice (Fig. 3). Expression of both mGH and hGH in the same cell (as determined from colocalization of immunofluorescence, Fig. 3C–E) indicated that hGH was expressed in the somatotrophs of the anterior pituitary of 171hGH/CS-TG mice. Analysis of embryonic pituitaries by similar means showed the appearance of hGH in the anterior pituitary between E13.5 and E18.5 (Fig. 4).
Fig. 3.
Immunohistochemical analysis demonstrates specific expression of hGH by somatotrophs in 171hGH/CS-TG anterior pituitary. (A and B) Sequential serial sections of paraffin-embedded pituitaries from wild type (WT, A) and 171hGH/CS-TG (TG, B) mice were incubated with antibodies against murine (mGH) and human (hGH) growth hormone. pa, pars anterior; pi, pars intermedia. White arrows indicate cells in the pars anterior of transgenic animals that express both mGH and hGH, thus identifying hGH expression in somatotrophs. Bars are 50 μm. (C–E) Immunofluorescence confirms presence of hGH in somatotrophs of the anterior pituitary (pa) through colocalization with mGH.
Fig. 4.
Human growth hormone expression appears in anterior pituitary in late gestation. Pituitaries from wild type (WT) and 171hGH/CS transgenic (TG) mice at E13.5 and E18.5 were sectioned and stained with antibody against human (hGH, green) and murine (mGH, red) growth hormone. Composite images with DAPI staining of nuclei (blue) are shown on the right. For E13.5, bars are 200 μm; for E18.5, bars are 50 μm, pa, pars anterior; pi, pars intermedia. The appearance of hGH in some mGH-expressing cells in the pa is evident at E18.5.
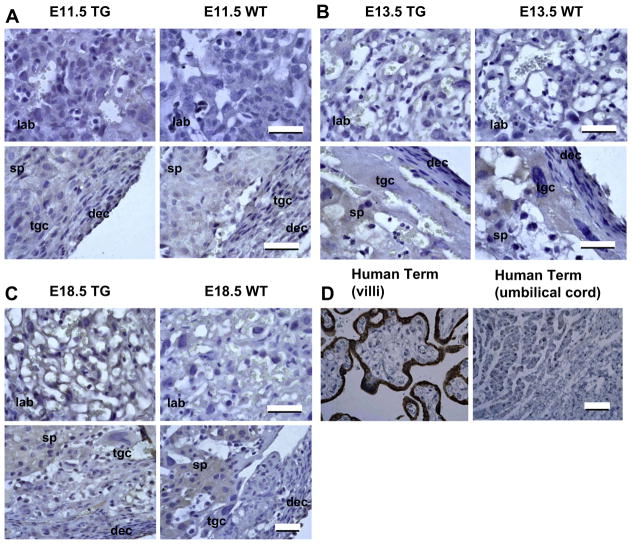
Regions of wild type and 171hGH/CS-TG mouse placenta were screened with primary antibodies to hCS (human placental lactogen) (Fig. 5). A specific signal above background was evident in the placental labyrinth of TG compared to non-TG mice at E18.5 (Fig. 5C). Cells in the labyrinth that gave specific staining for hCS were positioned adjacent to the maternal–fetal interface. Although some staining was observed in trophoblast giant cells and spongiotrophoblasts, this was considered ‘background’ staining, as levels were the same as those seen in non-TG mouse placenta (Fig. 5A–C). Sections through human term placenta and umbilical cord were also assessed as positive and negative controls, respectively (Fig. 5D). As expected, the outer syncytiotrophoblast layer of placental villi stained intensely for hCS, and no significant signal was detected in the umbilical cord sections.
Fig. 5.
Immunohistochemical analysis of human CS in 171hGH/CS-TG (TG) and wild type mouse (WT) placenta. Placenta from embryonic stages E11.5 (A), E13.5 (B), and E18.5 (C) were sectioned and stained with primary antibodies to hCS. The visible regions of the placenta are the decidua (dec), trophoblast giant cells (tgc), spongiotrophoblasts (sp), and labyrinth (lab). Cells in the 171hGH/CS-TG labyrinth that show specific staining for hCS are adjacent to the maternal–fetal interface, consistent with their functional equivalence to the hCS-expressing syncytiotrophoblasts of the human chorionic villi (D). Bars are 50 μm.
3.3. Expression profiles for placental GH/CS genes during gestation in 171hGH/CS-TG mice
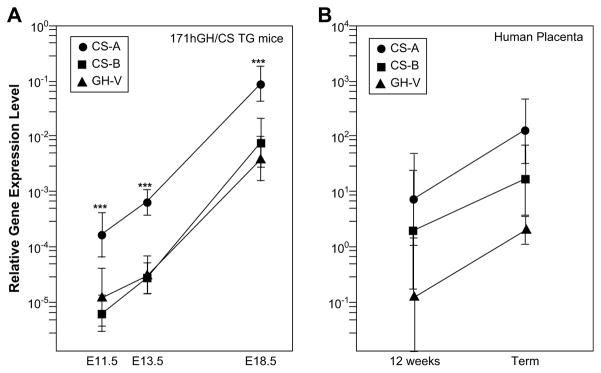
The relative expression of hCS-A, hCS-B and hGH-V RNA in the placental labyrinth region of 171hGH/CS-TG mice was determined by qRT-PCR at different stages of pregnancy/embryonic development (Fig. 6A). Levels of all three hCS/GH-V RNAs (relative to GAPDH) increase as gestation progresses; however, the amount of hCS-A transcripts detected exceeds that of hCS-B and hGH-V at all three time points by at least an order of magnitude. For further comparison, the expression of hCS-A and hCS-B was assessed by qRT-PCR in human term placenta (Fig. 6B). The low level of RNA expression detected in TG mice compared to human term placenta is consistent with the intensity of hCS staining detected by immunohistochemistry (Fig. 5). However, the proportion of hCS-A in human term and E18.5 transgenic mouse placentas is comparable, with values of 88.1 ± 5.3 (n = 4) and 91.3 ± 4.0% (n = 10) of total hCS RNA, respectively.
Fig. 6.
Expression of placental members of the GH/CS gene family in 171hGH/CS-TG mice. Quantitative RT-PCR was used to analyze total RNA from transgenic mouse placentas from mid to late gestation (E11.5, E13.5, and E18.5 as indicated). The expression level of each gene is expressed relative to GAPDH at each time point and plotted on a logarithmic scale. ***, p < 0.001 for CS-A relative to CS-B or GH-V at a each time point.
3.4. Expression of AP-2α but not AP-2γ correlates with placental GH/CS expression
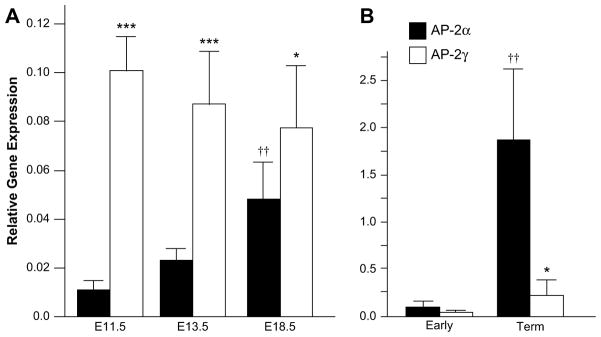
The transcription factor AP-2 has been linked with hCS expression in human syncytiotrophoblast cells [22–24], and both AP-2α and AP-2γ were found to increase hCS promoter activity in vitro. Here, we used 171hGH/CS-TG mice to further explore a link between AP-2α versus AP-2γ and hCS gene expression during pregnancy. The relative AP-2α and AP-2γ RNA levels were assessed in the placental labyrinth region from at least three independent 171hGH/CS-TG mice at different stages of pregnancy (E11.5, E13.5 and E18.5) by qRT-PCR (Fig. 7A). AP-2γ transcripts exceed those of AP-2α at all time points assessed including (close to term) at E18.5. Although there appears to be a decrease in AP-2γ RNA levels as gestation progresses, there was, however, no significant difference between the levels of AP-2γ transcripts detected at E11.5, E13.5 and E18.5. In contrast, the amount of AP-2α transcripts appears to increase significantly from E11.5 to E18.5, which correlates with the increase in hCS RNA levels. This is reflected in the increasing proportion of AP-2α transcripts: AP-2α RNA represents 9.8 ± 2.6% of total AP-2 at E11.5, 21.2 ± 4.6% at E13.5 and 38.4 ± 5.3% at E18.5 (n = 6 for each). In spite of this, and an apparent increase in total AP-2 (α plus γ) at E18.5, there was no significant difference between values for total AP-2 at E11.5, E13.5 and E18.5 (n = 6). For comparison, AP-2α versus AP-2γ RNA relative to GAPDH was assessed in samples of human placenta at 12 and 40 weeks (term) by qRT-PCR (Fig. 7B). Although AP-2α transcripts were far more abundant than those of AP-2γ, like mouse, the proportion of AP-2α increased from 61.11 ± 23.51% at 12 weeks (n = 6) to 88.77 ± 7.88% of total AP-2 RNA at term (n = 4).
Fig. 7.
AP-2α and AP-2γ are expressed at distinct relative levels in mouse and human placenta. (A) Total RNA from the placental labyrinths of mice at mid to late gestation (E11.5, E13.5 and E18.5 as shown) were analyzed by qRT-PCR for the expression of AP-2α and AP-2γ, corrected to the expression of GAPDH. ***, p < 0.001 or *, p < 0.05 for AP-2γ relative to AP-2α at the same time point. ††, p < 0.01 for AP-2α relative to E11.5 or E13.5. (B) Total RNA from human placenta (Early, 12 weeks, n = 6; Term, 39–40 weeks, n = 4) was similarly analyzed by qRT-PCR for AP-2 expression. *, p < 0.05 for AP-2γ relative to AP-2α at Term. ††, p < 0.01 for AP-2α at Term relative to Early placenta.
4. Discussion
Transgenic mice containing a 171-kb region of the long arm of chromosome 17 from a BAC clone have been generated. This region encompasses the entire human growth hormone/chorionic somatomammotropin gene cluster (containing the pituitary GH-N as well as the placental CS-A, CS-B, CS-L and GH-V genes) and the upstream LCR including the five previously described hypersensitive sites (HSI–HSV). The LCR was reported previously to confer appropriate expression of pituitary GH and, consistent with this, 171hGH/CS-TG mice appeared normal in size. The equivalent designation for hCS and the placental hGH/CS LCR cannot be determined as, strictly, there is no GH-related mouse CS for comparison; mouse placental lactogen (PL) is more structurally related to prolactin [7]. However, essential components of the hGH/CS LCR were detected. The 171hGH/CS-TG mice are fertile, deliver normal litter sizes and exhibited preferential expression of hCS/placental lactogen in the placental labyrinth as well as GH-N in the anterior pituitary. Detection of hCS-B and hGH-V transcripts in addition to previously described hCS-A RNA is consistent with inclusion of essential components of LCRs for both pituitary and placental members in the fragment of DNA used. Also, levels of hGH-V and hCS, predominantly hCS-A, increase from mid to late gestation in the transgenic placenta, as with human placenta.
Cooke and colleagues showed previously, using a truncated hGH locus, that 40 kb of upstream flanking hGH-N DNA is necessary and sufficient for appropriate levels of pituitary-specific expression of the hGH-N gene [31]. This helped to define the hGH LCR to the region 14.5–32 kb upstream of the hGH-N transcription initiation site. Subsequent elegant studies using transgenic mice expressing a truncated hGH/CS locus (minus the hCS-B gene), and often crossed with an overexpressing GH releasing hormone mouse, helped to define the role of Pit-1 and chromatin remodeling through posttranslational modifications of histones and looping, in pituitary-specific expression [32–34]. While transgenic mice containing the intact hGH/CS locus have been generated, to our knowledge, detection of hGH and hCS protein by immunohistochemistry in these mice has not been reported [12]. Thus, the immunohistochemical data presented here are, to our knowledge, the first demonstrating the appearance of hGH protein by E18.5 as well as specifically in the pituitary of adult transgenic mice containing all five hGH/CS genes and the hGH LCR (Figs. 3 and 4).
While there are definite differences between human and rodent placental structure, the various cell types can be grouped according to function [35]. The syncytiotrophoblast cells that line the chorionic villi of the human placenta serve as the interface between maternal and fetal blood circulation. An analogous role is played by the labyrinthine trophoblast layer in the mouse placenta [36]. The trophoblast portion of the maternal–fetal interface in the mouse is made up of three layers, the first of which lines the interface and is made up of mononuclear trophoblasts. Layers II and III are syncytiotrophoblastic and function as a single layer, although there is evidence that layer II shares direct contact with maternal blood, along with layer I. The layers are distinguished with high-power electron microscopy [36]. We present, for the first time to our knowledge, chromogenic staining and detection of hCS by light microscopy in mice containing the intact hGH/CS locus (Fig. 5). Furthermore, staining in the 171hGH/CS-TG mice is restricted to labyrinthine cells that line the maternal–fetal interface, the closest murine approximation to the human villus syncytiotrophoblast. The existence of sufficient transcription factors in the mouse for appropriate expression of a human gene has also been demonstrated for the aromatase gene hCYP19 [37]. In the mouse, aromatase expression is confined to the gonads and brain; in humans its expression pattern includes the syncytiotrophoblast layer of the placenta. Transfer of the hCYP19 gene to the mouse genome resulted in aromatase expression in the trophoblast layer of the murine labyrinth, which is functionally analogous to human syncytial trophoblast. The observation was made that the placental transcription factors that direct hCYP19 expression to the placenta are evidently conserved between mouse and human, even though the genetic elements that correspond to those factors are not. It also appears then that the factors required for expression of hGH/CS genes in the placenta also exist in the mouse, in spite of the lack of a GH/CS locus in the murine genome.
The importance of considering the greater chromatin configuration in placental gene expression with the enhancer and/or repressor function of individual or multiple DNA elements associated with HSI–V, 3′-enhancer regions and P sequences identified in cell culture, is becoming increasingly apparent. Inclusion of HSIII–V juxtaposed to the CS-A gene with all known regulatory sequences intact did not result in appropriate tissue expression of CS-A in transgenic placenta [12]. This emphasizes the need for a readily available model in which to study the entire intact locus, if we are to understand the mechanisms, and the possible importance, of distinct transcriptional control of GH and CS genes. The inclusion of the intact hGH/CS locus and specifically the hCS-B gene locus in the 171hGH/CS-TG mice might have been expected to affect placental gene expression. There is considerable data from the transient transfection of human choriocarcinoma cells, including from our laboratory, to support the presence of a strong placental enhancer located in the 3′-flanking DNA of the hCS-B gene [38–41]. Further support for the possible importance of the downstream CS gene enhancer sequences in vivo, was the detection of histone hyperacetylation and methylation of this region in human placenta chromatin [42]. Historically, the comparative study of hCS-A and hCS-B expression has been puzzling. RT-PCR suggested equivalent expression at eight weeks pregnancy, but by term CS-A levels are significantly higher [43]. Our data confirm this result in human placenta (Fig. 6). More importantly, hCS-A continued to be the predominant CS RNA species in 171hGH/CS-TG mouse placenta (Fig. 6), regardless of the presence of the intact hGH/CS locus and specifically the hCS-B gene and putative downstream enhancer sequences.
It has been reported previously that expression of the placental hGH/CS genes increases during gestation [43]. In the 171hGH/CS-TG placenta, the expression of hCS-A, hCS-B and hGH-V all increase during gestation, but the ratios do not change significantly, with hCS-A representing about 90% of total hCS RNA expression from E11.5 to E18.5. The proportionate expression of hCS-A RNA in 171hGH/CS-TG placentas in late gestation is comparable to the proportionate expression of hCS-A transcripts observed in human term placenta. Although the expression patterns for hCS-A and -B in the transgenic mouse placenta closely parallel that seen in the human placenta in functional location, timing and relative levels, the absolute levels of mRNA and protein are lower than those observed in the human. The low level of expression is consistent with other models of hCS expression, including human cytotrophoblast cell cultures [12,29]. It is possible that while the murine genome contains the factors necessary for placental-specific expression of hCS, the appropriate levels or combinations of these factors may not be present to drive hCS expression to the level seen in human syncytiotrophoblast. The mouse placenta expresses a large family of placental-specific, prolactin-related genes, including placental lactogen 2 (Prl3b1). Prl3b1 is hypothesized to carry out similar functions to the hCS and hGH-V during pregnancy and is expressed in the labyrinth region [44]. Little is yet known about the regulation of specific genes in gene family, but there may be mechanisms that modulate expression levels and which may have similar effects on hCS expression. Mating our mouse model with a Prl3b1 null mouse, when available, could address the question of expression levels and provide further functional information on this member of the mouse gene family.
While both AP-2α and AP-2γ have been linked to hCS gene promoter activity in gene transfer studies in vitro, an increase in AP-2α levels alone was shown to correlate with the appearance and increase in hCS RNA levels as human cytotrophoblasts develop into syncytiotrophoblasts in primary placental culture [22,23]. Using the 171hGH/CS-TG mouse model we have been able to extend this observation of AP-2α and human GH/CS RNA expression beyond cytotrophoblast differentiation in primary culture to pregnancy. Our results showed that AP-2α transcripts were far more abundant than AP-2γ in term human placenta, and the levels of both isoforms increase from mid to late gestation. In the mouse, by contrast, AP-2γ is consistently expressed at a higher level than AP-2α at all time points. However, only AP-2α increases from mid to late gestation while there is no significant change in AP-2γ expression. These data implicate an increase in AP-2α but not AP-2γ in stimulating placental GH/CS RNA levels during pregnancy, since hGH/CS expression also increased in transgenic placenta from mid to late gestation. This does not, however, rule out a role for AP-2γ in placental hGH/CS expression in normal human development, but suggests that an increase in AP-2γ may not be necessary for augmentation of hGH/CS expression in the placenta during pregnancy (Fig. 7).
The availability and use of multiple human placental cell systems, including the BeWo, JAR and JEG-3 lines as well as primary cultures, continue to play an important role in understanding placental gene expression and, perhaps to a lesser extent, function. The availability of transgenic mouse models can complement these cell systems and provide the capacity to expand studies in the context of pregnancy. Thus, the 171hGH/CS-TG mouse has the potential to provide a valuable model to explore pituitary as well as placental hGH/CS gene expression in vivo given limitations on the availability of human tissues.
Acknowledgments
This work was supported by a grant from the Canadian Institutes of Health Research (MT-10853). SYL is the recipient of a postdoctoral fellowship from the Manitoba Institute of Child Health. The sponsors of this study had no role or involvement in the collection, analysis or interpretation of data.
References
- 1.Chen EY, Liao YC, Smith DH, Barrera-Saldana HA, Gelinas RE, Seeburg PH. The human growth hormone locus: nucleotide sequence, biology, and evolution. Genomics. 1989;4:479–97. doi: 10.1016/0888-7543(89)90271-1. [DOI] [PubMed] [Google Scholar]
- 2.Bulger M, van Doorninck JH, Saitoh N, Telling A, Farrell C, Bender MA, et al. Conservation of sequence and structure flanking the mouse and human beta-globin loci: the beta-globin genes are embedded within an array of odorant receptor genes. Proc Natl Acad Sci U S A. 1999;96:5129–34. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker WH, Fitzpatrick SL, Barrera-Saldana HA, Resendez-Perez D, Saunders GF. The human placental lactogen genes: structure, function, evolution and transcriptional regulation. Endocr Rev. 1991;12:316–28. doi: 10.1210/edrv-12-4-316. [DOI] [PubMed] [Google Scholar]
- 4.Handwerger S, Freemark M. The roles of placental growth hormone and placental lactogen in the regulation of human fetal growth and development. J Pediatr Endocrinol Metab. 2000;13:343–56. doi: 10.1515/jpem.2000.13.4.343. [DOI] [PubMed] [Google Scholar]
- 5.Rygaard K, Revol A, Esquivel-Escobedo D, Beck BL, Barrera-Saldana HA. Absence of human placental lactogen and placental growth hormone (HGH-V) during pregnancy: PCR analysis of the deletion. Hum Genet. 1998;102:87–92. doi: 10.1007/s004390050658. [DOI] [PubMed] [Google Scholar]
- 6.Cattini PA, Yang X, Jin Y, Detillieux KA. Regulation of the human growth hormone gene family: possible role for Pit-1 in early stages of pituitary-specific expression and repression. Neuroendocrinology. 2006;83:145–53. doi: 10.1159/000095522. [DOI] [PubMed] [Google Scholar]
- 7.Goffin V, Shiverick KT, Kelly PA, Martial JA. Sequence–function relationships within the expanding family of prolactin, growth hormone, placental lactogen, and related proteins in mammals. Endocr Rev. 1996;17:385–410. doi: 10.1210/edrv-17-4-385. [DOI] [PubMed] [Google Scholar]
- 8.West BL, Catanzaro DF, Mellon SH, Cattini PA, Baxter JD, Reudelhuber TL. Interaction of a tissue-specific factor with an essential rat growth hormone gene promoter element. Mol Cell Biol. 1987;7:1193–7. doi: 10.1128/mcb.7.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behringer RR, Mathews LS, Palmiter RD, Brinster RL. Dwarf mice produced by genetic ablation of growth hormone-expressing cells. Genes Dev. 1988;2:453–61. doi: 10.1101/gad.2.4.453. [DOI] [PubMed] [Google Scholar]
- 10.Jones BK, Monks BR, Liebhaber SA, Cooke NE. The human growth hormone gene is regulated by a multicomponent locus control region. Mol Cell Biol. 1995;15:7010–21. doi: 10.1128/mcb.15.12.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su Y, Liebhaber SA, Cooke NE. The human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in the transgenic mouse. J Biol Chem. 2000;275:7902–9. doi: 10.1074/jbc.275.11.7902. [DOI] [PubMed] [Google Scholar]
- 12.Kimura AP, Sizova D, Handwerger S, Cooke NE, Liebhaber SA. Epigenetic activation of the human growth hormone gene cluster during placental cytotrophoblast differentiation. Mol Cell Biol. 2007;27:6555–68. doi: 10.1128/MCB.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennani-Baiti IM, Jones BK, Liebhaber SA, Cooke NE. Physical linkage of the human growth hormone gene cluster and the skeletal muscle sodium channel alpha-subunit gene (SCN4A) on chromosome 17. Genomics. 1995;29:647–52. doi: 10.1006/geno.1995.9954. [DOI] [PubMed] [Google Scholar]
- 14.Bennani-Baiti IM, Cooke NE, Liebhaber SA. Physical linkage of the human growth hormone gene cluster and the CD79b (Ig beta/B29) gene. Genomics. 1998;48:258–64. doi: 10.1006/geno.1997.5171. [DOI] [PubMed] [Google Scholar]
- 15.Surabhi RM, Bose S, Kuschak BC, Cattini PA. Physical linkage of the human growth hormone gene family and the thyroid hormone receptor interacting protein-1 gene on chromosome 17. Gene. 1998;212:67–75. doi: 10.1016/s0378-1119(98)00149-8. [DOI] [PubMed] [Google Scholar]
- 16.Surabhi RM, Daly LD, Cattini PA. Evidence for evolutionary conservation of a physical linkage between the human BAF60b, a subunit of SWI/SNF complex, and thyroid hormone receptor interacting protein-1 genes on chromosome 17. Genome. 1999;42:545–9. [PubMed] [Google Scholar]
- 17.Jin Y, Surabhi RM, Fresnoza A, Lytras A, Cattini PA. A role for A/T-rich sequences and Pit-1/GHF-1 in a distal enhancer located in the human growth hormone locus control region with preferential pituitary activity in culture and transgenic mice. Mol Endocrinol. 1999;13:1249–66. doi: 10.1210/mend.13.8.0332. [DOI] [PubMed] [Google Scholar]
- 18.Johnson W, Albanese C, Handwerger S, Williams T, Pestell RG, Jameson JL. Regulation of the human chorionic gonadotropin alpha- and beta-subunit promoters by AP-2. J Biol Chem. 1997;272:15405–12. doi: 10.1074/jbc.272.24.15405. [DOI] [PubMed] [Google Scholar]
- 19.LiCalsi C, Christophe S, Steger DJ, Buescher M, Fischer W, Mellon PL. AP-2 family members regulate basal and cAMP-induced expression of human chorionic gonadotropin. Nucleic Acids Res. 2000;28:1036–43. doi: 10.1093/nar/28.4.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng YH, Handwerger S. AP-2alpha modulates human corticotropin-releasing hormone gene expression in the placenta by direct protein–protein interaction. Mol Cell Endocrinol. 2002;191:127–36. doi: 10.1016/s0303-7207(02)00081-3. [DOI] [PubMed] [Google Scholar]
- 21.Ozturk A, Donald LJ, Li L, Duckworth HW, Duckworth ML. Proteomic identification of AP2 gamma as a rat placental lactogen II trophoblast cell-specific enhancer binding protein. Endocrinology. 2006;147:4319–29. doi: 10.1210/en.2006-0492. [DOI] [PubMed] [Google Scholar]
- 22.Richardson BD, Langland RA, Bachurski CJ, Richards RG, Kessler CA, Cheng YH, et al. Activator protein-2 regulates human placental lactogen gene expression. Mol Cell Endocrinol. 2000;160:183–92. doi: 10.1016/s0303-7207(99)00209-9. [DOI] [PubMed] [Google Scholar]
- 23.Richardson BD, Cheng YH, Langland RA, Handwerger S. Differential expression of AP-2gamma and AP-2alpha during human trophoblast differentiation. Life Sci. 2001;69:2157–65. doi: 10.1016/s0024-3205(01)01299-1. [DOI] [PubMed] [Google Scholar]
- 24.Jin Y, Norquay LD, Yang X, Gregoire S, Cattini PA. Binding of AP-2 and ETS-domain family members is associated with enhancer activity in the hypersensitive site III region of the human growth hormone/chorionic somatomammotropin locus. Mol Endocrinol. 2004;18:574–87. doi: 10.1210/me.2003-0405. [DOI] [PubMed] [Google Scholar]
- 25.Cheng YH, Aronow BJ, Hossain S, Trapnell B, Kong S, Handwerger S. Critical role for transcription factor AP-2alpha in human trophoblast differentiation. Physiol Genomics. 2004;18:99–107. doi: 10.1152/physiolgenomics.00181.2003. [DOI] [PubMed] [Google Scholar]
- 26.Matt DW, Macdonald GJ. Placental steroid production by the basal and labyrinth zones during the latter third of gestation in the rat. Biol Reprod. 1985;32:969–77. doi: 10.1095/biolreprod32.4.969. [DOI] [PubMed] [Google Scholar]
- 27.Ballester M, Castello A, Ibanez E, Sanchez A, Folch JM. Real-time quantitative PCR-based system for determining transgene copy number in transgenic animals. Biotechniques. 2004;37:610–3. doi: 10.2144/04374ST06. [DOI] [PubMed] [Google Scholar]
- 28.Brown RW, Chirala R. Utility of microwave-citrate antigen retrieval in diagnostic immunohistochemistry. Mod Pathol. 1995;8:515–20. [PubMed] [Google Scholar]
- 29.Nickel BE, Cattini PA. Nuclease sensitivity of the human growth hormone-chorionic somatomammotropin locus in pituitary and placenta suggest different mechanisms for tissue-specific regulation. Mol Cell Endocrinol. 1996;118:155–62. doi: 10.1016/0303-7207(96)03778-1. [DOI] [PubMed] [Google Scholar]
- 30.Lytras A, Bock ME, Dodd JG, Cattini PA. Detection of placental growth hormone variant and chorionic somatomammotropin ribonucleic acid expression in human trophoblastic neoplasms by reverse transcriptase–polymerase chain reaction. Endocrinology. 1994;134:2461–7. doi: 10.1210/endo.134.6.7515000. [DOI] [PubMed] [Google Scholar]
- 31.Bennani-Baiti IM, Asa SL, Song D, Iratni R, Liebhaber SA, Cooke NE. DNase I-hypersensitive sites I and II of the human growth hormone locus control region are a major developmental activator of somatotrope gene expression. Proc Natl Acad Sci U S A. 1998;95:10655–60. doi: 10.1073/pnas.95.18.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho Y, Elefant F, Cooke N, Liebhaber S. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol Cell. 2002;9:291–302. doi: 10.1016/s1097-2765(02)00447-1. [DOI] [PubMed] [Google Scholar]
- 33.Ho Y, Elefant F, Liebhaber SA, Cooke NE. Locus control region transcription plays an active role in long-range gene activation. Mol Cell. 2006;23:365–75. doi: 10.1016/j.molcel.2006.05.041. [DOI] [PubMed] [Google Scholar]
- 34.Ho Y, Tadevosyan A, Liebhaber SA, Cooke NE. The juxtaposition of a promoter with a locus control region transcriptional domain activates gene expression. EMBO Rep. 2008;9:891–8. doi: 10.1038/embor.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, et al. Genes, development and evolution of the placenta. Placenta. 2003;24:123–30. doi: 10.1053/plac.2002.0887. [DOI] [PubMed] [Google Scholar]
- 36.Georgiades P, Ferguson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- 37.Kamat A, Smith ME, Shelton JM, Richardson JA, Mendelson CR. Genomic regions that mediate placental cell-specific and developmental regulation of human Cyp19 (aromatase) gene expression in transgenic mice. Endocrinology. 2005;146:2481–8. doi: 10.1210/en.2004-1606. [DOI] [PubMed] [Google Scholar]
- 38.Walker WH, Fitzpatrick SL, Saunders GF. Human placental lactogen transcriptional enhancer. Tissue specificity and binding with specific proteins. J Biol Chem. 1990;265:12940–8. [PubMed] [Google Scholar]
- 39.Jacquemin P, Oury C, Peers B, Morin A, Belayew A, Martial JA. Characterization of a single strong tissue-specific enhancer downstream from the three human genes encoding placental lactogen. Mol Cell Biol. 1994;14:93–103. doi: 10.1128/mcb.14.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang SW, Eberhardt NL. The human chorionic somatomammotropin gene enhancer is composed of multiple DNA elements that are homologous to several SV40 enhansons. J Biol Chem. 1994;269:10384–92. [PubMed] [Google Scholar]
- 41.Lytras A, Cattini PA. Human chorionic somatomammotropin gene enhancer activity is dependent on the blockade of a repressor mechanism. Mol Endocrinol. 1994;8:478–89. doi: 10.1210/mend.8.4.8052268. [DOI] [PubMed] [Google Scholar]
- 42.Kimura AP, Liebhaber SA, Cooke NE. Epigenetic modifications at the human growth hormone locus predict distinct roles for histone acetylation and methylation in placental gene activation. Mol Endocrinol. 2004;18:1018–32. doi: 10.1210/me.2003-0468. [DOI] [PubMed] [Google Scholar]
- 43.MacLeod JN, Lee AK, Liebhaber SA, Cooke NE. Developmental control and alternative splicing of the placentally expressed transcripts from the human growth hormone gene cluster. J Biol Chem. 1992;267:14219–26. [PubMed] [Google Scholar]
- 44.Simmons DG, Rawn S, Davies A, Hughes M, Cross JC. Spatial and temporal expression of the 23 murine Prolactin/Placental Lactogen-related genes is not associated with their position in the locus. BMC Genomics. 2008;9:352. doi: 10.1186/1471-2164-9-352. [DOI] [PMC free article] [PubMed] [Google Scholar]