Abstract
Tbx20 is a T-box transcription factor that plays essential roles in the development and maintenance of the heart. Although it is expressed by cardiac progenitors in all species examined, an involvement of Tbx20 in cardiac progenitor formation in vertebrates has not been previously described. Here we report the identification of a zebrafish tbx20 mutation that results in an inactive, truncated protein lacking any functional domains. The cardiac progenitor population is strongly diminished in this mutant, leading to the formation of a small, stretched-out heart. We found that overexpression of Tbx20 results in an enlarged heart with significantly more cardiomyocytes. Interestingly, this increase in cell number is caused by both enhanced cardiac progenitor cell formation and the proliferation of differentiated cardiomyocytes, and is dependent upon the activity of Tbx20’s T-box and transcription activation domains. Together, our findings highlight a previously unappreciated role for Tbx20 in promoting cardiac progenitor formation in vertebrates and reveal a novel function for its activation domain in cardiac cell proliferation during embryogenesis.
Keywords: Tbx20, heart development, cardiogenesis, cardiac progenitor, cardiomyocyte proliferation, zebrafish
Introduction
The T-box transcription factor Tbx20 is a central regulator of the gene programs that maintain cardiomyocyte structure and function. Ablation of Tbx20 activity in the adult myocardium of both flies and mice results in the downregulation of genes encoding cardiac transcription factors, myofibrillar proteins and ion channels, leading to a range of cardiac functional defects including arrhythmia [1, 2]. Similarly, dysregulation of Tbx20 (both loss- and gain-of-function) is associated with diverse cardiac pathologies including arrhythmia and cardiomyopathy in humans [1, 3–9].
The cardiac expression of Tbx20 is initiated in the heart-forming region at the earliest stages during heart development [10–13]. Nmr1 and nmr2 are two Tbx20 homologs in Drosophila whose expression is detected bilaterally in the cardiac progenitors [11, 14]. Flies deficient in both nmr1 and nmr2 lack lbe-positive cardiac progenitors whereas overexpression of Nmr/Tbx20 expands the cardiac progenitor population, indicating a critical requirement for nmr/tbx20 in the formation of cardiac progenitors [14]. In mouse embryos, tbx20 expression is present in the cardiogenic mesoderm at E7.5, and persists throughout heart development [12]. Overexpression of Tbx20 in the developing mouse heart increases cardiomyocyte proliferation and thickens the compact myocardium [15]. However, unlike what has been observed in flies, mice lacking Tbx20 activity have normal patterning of the cardiac crescent and linear heart tube, but the heart tube fails to loop and displays defects in cardiomyocyte proliferation and AV canal differentiation [16–19]. Knockdown of Tbx20 in fish and Xenopus also results in a linear heart tube with defects in looping and chamber remodeling [20, 21]. These observations have led to the thought that Tbx20 activity is dispensable for the production of cardiac progenitors in vertebrates.
In this study, we report the identification of a zebrafish tbx20 null mutation that causes a severe cardiac progenitor deficit, highlighting an early requirement of Tbx20 during vertebrate cardiogenesis. We found that Tbx20 overexpression prior to cardiogenesis potentiates the expression of cardiac transcription factors and expands the cardiac progenitor population, leading to an enlarged heart with significantly more cardiomyocytes. Remarkably, forced expression of Tbx20 in committed cardiomyocytes also causes cardiac hyperplasia, indicating that tbx20 can further promote cardiomyocyte production after the cardiomyocytes have been specified. Furthermore, our structure-function study revealed that the cardiac expansion induced by Tbx20 overexpression requires its T-box DNA binding and transcription activation domains. These results highlight the important role of Tbx20 in promoting cardiac progenitor cell formation and cardiomyocyte proliferation and suggest that Tbx20 may enhance cardiomyocyte production as a transcriptional activator.
Results
Identification of a zebrafish mutant with severe defects in cardiogenesis
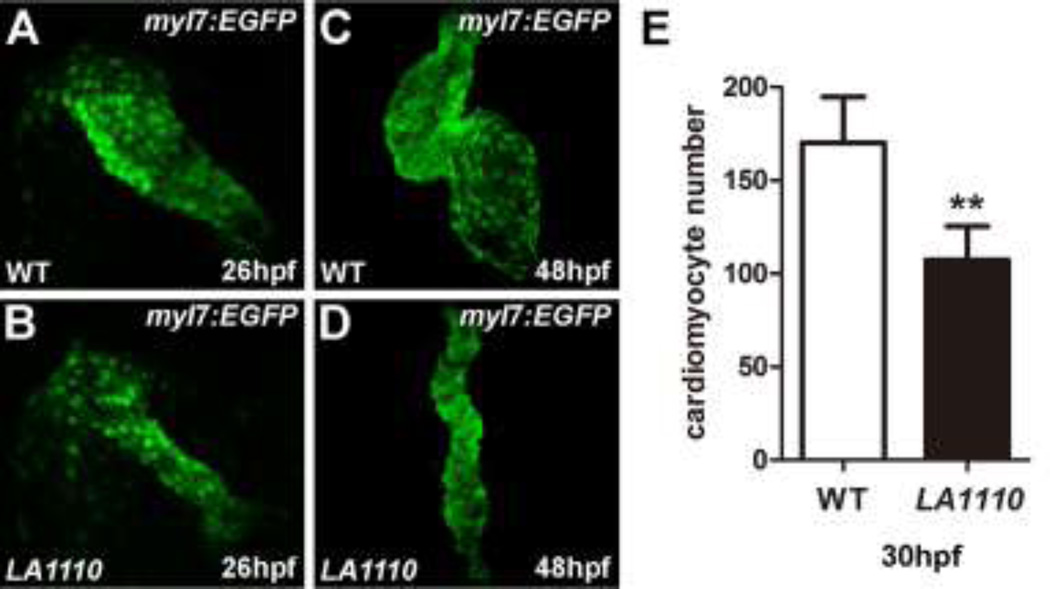
In search of genes critical for early cardiogenesis, we performed an N-ethyl-N-nitrosourea (ENU) mutagenesis screen [22, 23]. Zebrafish mutant LA1110 was identified in this screen based on its severe cardiac defects. In zebrafish, a primitive heart tube capable of propelling circulation through the body has formed by 26 hours post fertilization (hpf), but LA1110 mutant hearts are small and can barely support circulation (Fig.1A,B, and data not shown). By 48 hpf, LA1110 mutant hearts are stretched into a string-like structure (Fig.1C,D), resulting in severe cardiac edema (Fig.S1) and embryonic lethality. By directly counting cardiomyocytes in an myl7:EGFP transgenic background, we found that LA1110 mutants exhibit a 40% reduction in the number of cardiomyocytes at 30 hpf compared to their wild type siblings (170.2±24.5 in wild type hearts v.s. 107.2±18.14 in LA1110 hearts, n=5, p<0.01) (Fig.1E), indicating a critical role for LA1110 in cardiomyocyte production. This hypoplastic cardiac phenotype is fully penetrant as ~25% of embryos from all clutches examined thus far displayed the same phenotype.
Fig.1.
LA1110 mutants exhibit a reduction in cardiomyocyte cell number. (A and B) Tbx20 mutant embryos (B) form a thin and small heart tube at 26hpf compared with that of wild-type siblings (A), as indicated by the fluorescent labeling of differentiated cardiomyocytes by the myl7:EGFP transgene. (C and D) Wild type embryos display a looped two-chambered heart at 48hpf (C), whereas LA1110 mutant embryos (D) exhibit a thin, stretched and string-like heart. (E) Graph of the average number of GFP-positive cardiomyocytes in wild type and LA1110 mutant embryos at 30hpf (n=5). Error bars indicate standard deviations. Asterisks indicate a significant difference (**p<0.01).
LA1110 leads to cardiac progenitor defects
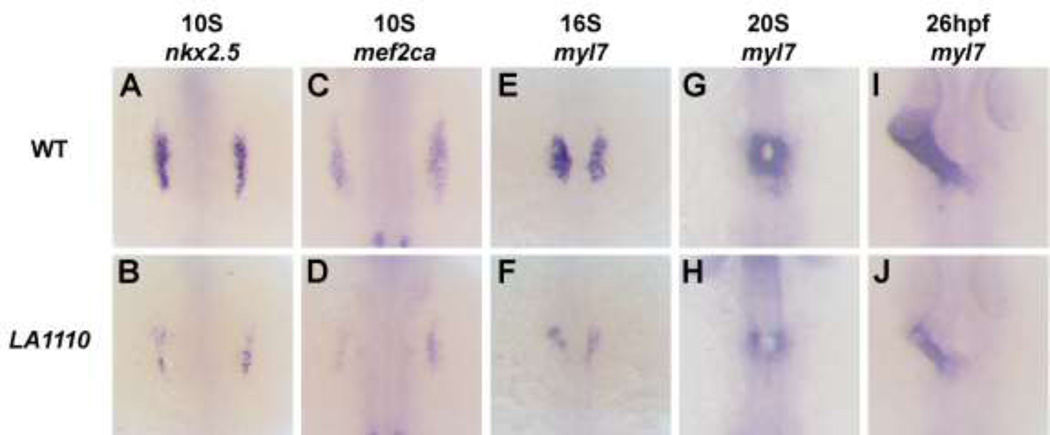
We next investigated the potential causes of the myocardial deficit observed in LA1110. We performed in situ hybridization using the early cardiac markers nkx2.5, mef2ca, tbx5a and bmp4. These genes are expressed as bilateral stripes in the heart-forming region at the time when cardiac progenitors emerge from the anterior lateral plate mesoderm (ALPM). We found that the expression of these genes was strongly diminished in LA1110 mutants (Fig. 2A–D and Fig. S2), indicating a loss of most of the cardiac progenitor population in LA1110.
Fig.2.
The LA1110 gene is required for cardiac progenitor formation. (A–J) Dorsal views of nkx2.5 (A and B), mef2ca (C and D) and myl7 (E–J) expression in wild type and LA1110 mutant embryos. In wild type embryos, expression of nkx2.5 (A) and mef2ca (C) are detected in the cardiac progenitors within the ALPM at the 10-somite stage. These cells then differentiate into myl7-expressing cardiomyocytes (E) by the 16-somite stage and migrate toward the midline and fuse to form a cardiac cone (G) which then grows into a linear heart tube by 26hpf (I). In LA1110 mutant embryos, expression of nkx2.5 (B) and mef2ca (D) are strongly reduced. Even though the expression of myl7 is far lower (F), the myl7-expressing cardiomyocytes in LA1110 mutant embryos migrate to the midline and form the cardiac cone at the 20-somite stage (H) and the linear heart tube at 26hpf (J).
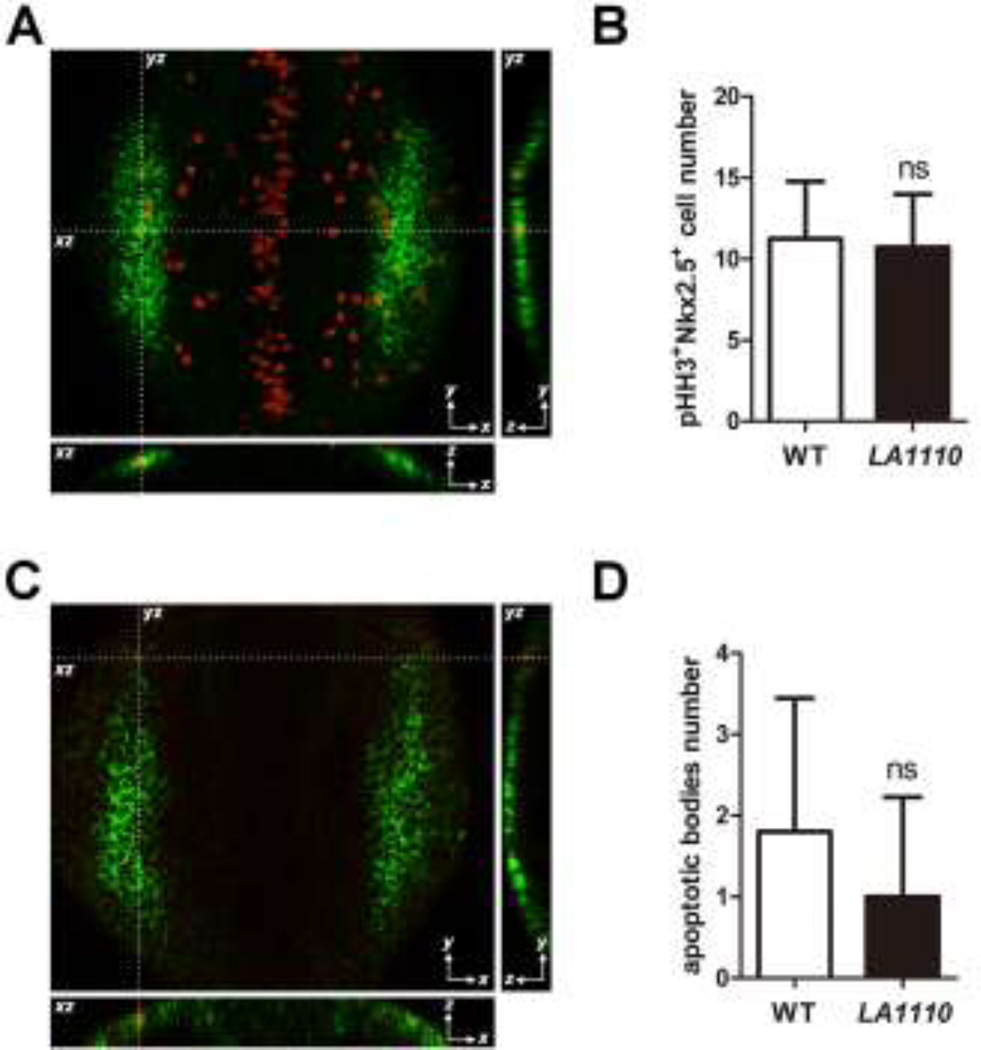
How might LA1110 regulate the production of cardiac progenitor cells? We explored the possibilities that LA1110 might influence the proliferation and/or the survival of cardiac progenitors. We assessed cardiac progenitor cell proliferation by counting cells in the heart-forming region that are double positive for nkx2.5 and Phospho-Histone H3 (pHH3), a marker for cells undergoing mitosis, at the 10-somite stage (when cardiac progenitor cells are emerging from ALPM), but did not detect a significant difference between LA1110 and their wild type siblings (Fig. 3A,B). Apoptotic bodies were rare in the heart-forming region, and no significant difference in the number of apoptotic nkx2.5 positive cells was noted between wild type and LA1110 mutant embryos (Fig. 3C,D), suggesting that changes in cell proliferation or cell death may not be the primary causes of LA1110’s cardiac progenitor deficiency.
Fig.3.
Quantification of cardiac progenitor proliferation and apoptosis in LA1110 mutants and wild type siblings. (A) Dorsal view of pHH3 immunostaining (red; nuclei) on a 10-somite stage wild type embryo. The cardiac progenitor cells are visualized by nkx2.5 fluorescent in situ hybridization (green; cytoplasmic). Orthogonal reconstructions are shown on the bottom and the right side of the image. (B) Graph of the average number of pHH3-positive cardiac progenitor cells in wild type (n=9) and LA1110 mutant embryos (n=7). (C) Dorsal view of apoptotic bodies (red) detected by TUNEL assay in a 10-somite stage wild type embryo. Cardiac progenitor cells are marked by nkx2.5 fluorescent in situ hybridization (green; cytoplasmic). Orthogonal reconstructions are shown on the bottom and the right side of the image. (D) Graph of the average number of apoptotic bodies in the cardiac progenitor region in wild type (n=5) and LA1110 mutant embryos (n=5). Error bars indicate standard deviations. ns, not significant.
LA1110 is not required for early cardiac morphogenesis
We wondered whether the cardiac progenitor deficit in LA1110 would have an impact on cardiomyocyte production and/or cardiac morphogenesis. LA1110 embryos had smaller patches of myl7-positive cardiomyocyte precursors at the 16-somite stage (Fig. 2E,F), a result consistent with the reduction in cardiac progenitors (Fig.2A–D and Fig.S2) and the decreased myocardial cells in the mutant hearts (Fig.1E). However, to our surprise and despite their greatly reduced number, LA1110 cardiomyocytes migrate to and fuse at the midline (Fig. 2G and H) and form a linear heart tube on time (Fig. 2I,J), indicating that LA1110 is dispensable for early cardiac morphogenetic processes like migration, fusion and the formation of a tubular structure.
LA1110 carries a nonsense mutation in Tbx20
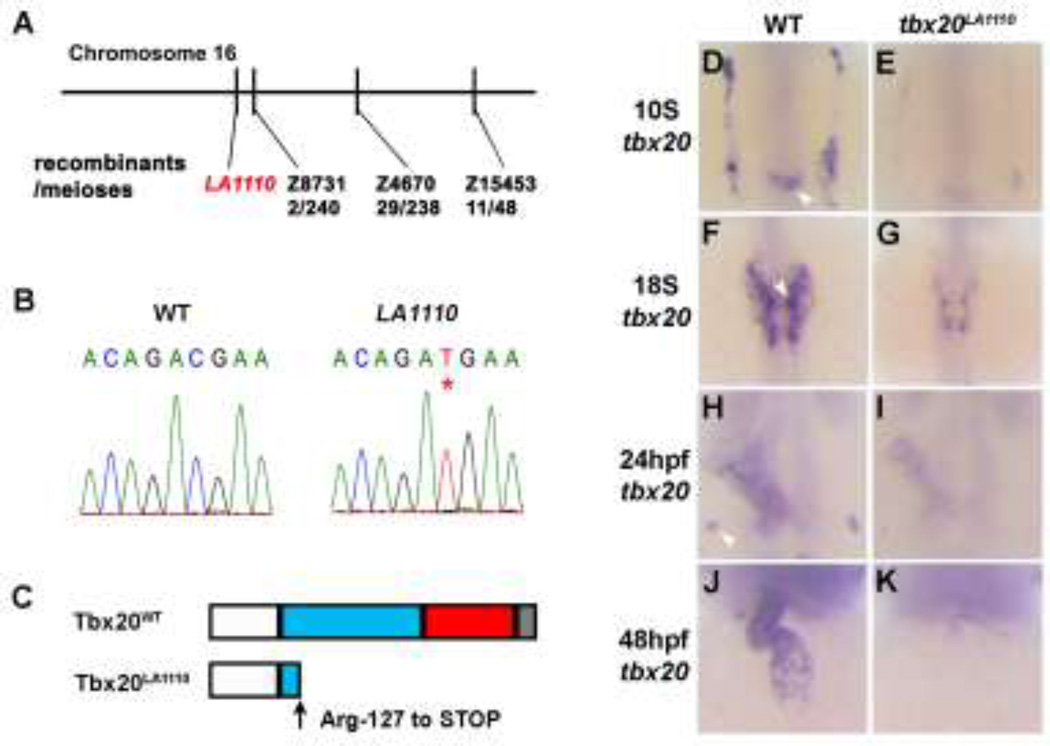
To identify the molecular lesion of LA1110, we performed linkage analysis and placed the LA1110 mutation on Chromosome 16 approximately 0.8 cM from marker Z8731 (Fig. 4A). This marker is in close proximity to tbx20, one of the earliest genes expressed in the heart-forming region. In 10-somite stage zebrafish embryos, the expression of tbx20 is noted as two bilateral stripes within the ALPM, which encompasses the heart primordia (Fig. 4D). The expression of tbx20 persists in the heart as cardiomyocytes migrate toward the midline, form the primitive heart tube and further differentiate into two distinct chambers [10, 11] (Fig. 4F,H and J), making it a good candidate gene for the LA1110 lesion. Supporting this hypothesis, we found a C to T transition at nucleotide 379 of tbx20 in LA1110, producing a premature stop codon and a truncated peptide lacking the C-terminal transcription activation and repression domains and the majority of the T-box domain (Fig. 4B,C). In addition, tbx20 transcripts were markedly reduced in LA1110 embryos at all stages examined (Fig. 4E,G, I and K), likely due to RNA degradation via the nonsense-mediated decay mechanism.
Fig.4.
Positional cloning of the tbx20LA1110 mutant. (A) Diagram showing the location of the LA1110 locus with respect to markers on Chromosome 16. (B) Sequencing of the tbx20 locus in wild type and LA1110 mutant embryos identified a C to T transition, resulting in a premature stop codon (*). (C) Schematic diagram of the Tbx20WT and Tbx20LA1110 proteins. Zebrafish Tbx20WT contains an N-terminal domain (white), a T-box domain (blue), and predicted transactivation (red) and transrepression (grey) domains in the C-terminal portion of the protein. The Tbx20LA1110 protein is truncated within the T-box DNA binding domain. (D–K) Expression of tbx20 in wild type and LA1110 embryos showing the anterior dorsal view (D–I) and frontal view (J,K). In wild type embryos, expression of tbx20 is visible as bilateral strips in the cardiac primordia within the ALPM at the 10-somite stage (D). These strips of cells migrate toward and fuse at the midline (F). By 24 hpf, the entire heart tube displays expression of tbx20 (H,J). Additional tbx20 expression is present within the hindbrain at the 10-somite stage (arrowhead in D), hindbrain branchiomotor neurons at the 18-somite (F) and the primordia of the pituitary gland at 24 hpf (arrowhead in H). (E, G, I and K) In tbx20LA1110 mutant embryos, tbx20 transcripts are dramatically reduced.
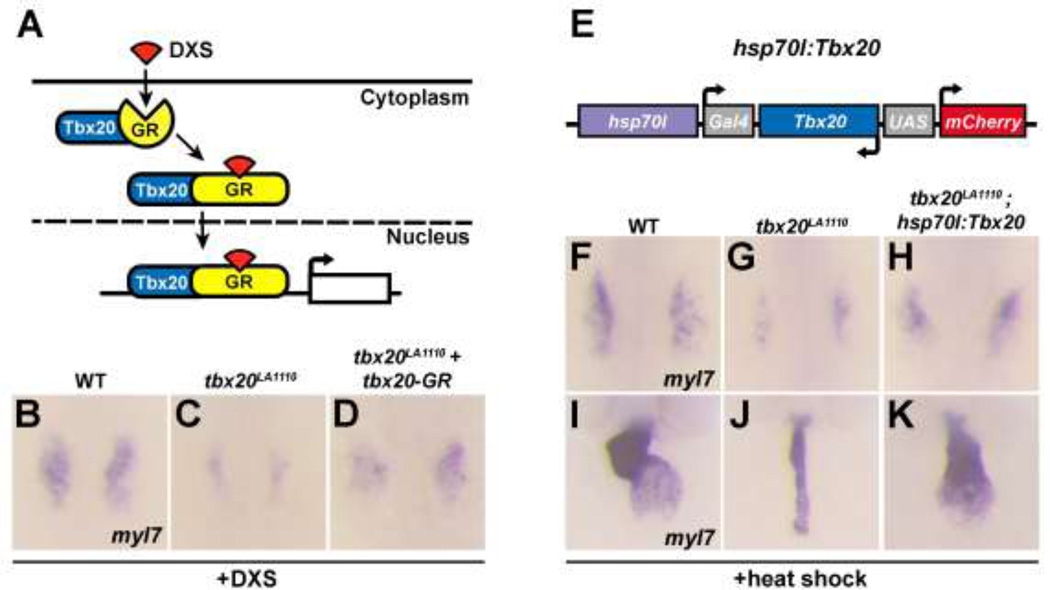
Overexpression of tbx20 restores cardiac progenitor cells in tbx20LA1110 embryos
To determine the causative relationship between LA1110’s cardiac defects and tbx20 deficiency, we examined whether replenishing Tbx20 expression could restore the cardiac progenitor cell population in LA1110 embryos. Surprisingly, zebrafish embryos injected with tbx20 RNA displayed gastrulation defects leading to early lethality, which prevented further analysis (Fig. S3A,B). To circumvent this problem, we fused the coding region of tbx20 with the ligand-binding domain of glucocorticoid receptor (GR) [20]. The Tbx20-GR fusion protein is sequestered in the cytoplasm, but is translocated into the nucleus upon dexamethasone (DXS) induction (Fig. 5A) [24, 25]. We injected tbx20-GR RNA into embryos collected from tbx20LA1110 heterozygous crosses. As expected, in the absence of DXS, a quarter of the injected embryos showed a characteristic tbx20LA1110 cardiac phenotype (Fig. S3D) whereas the other three quarters of embryos developed normally and did not exhibit obvious morphological defects (Fig. S3C). We next administered DXS at the 90% epiboly stage (9hpf) to induce Tbx20 nuclear translocation and observed an expansion of myl7 expression in tbx20LA1110 homozygous embryos at the 16-somite stage (confirmed by genotyping, n=9) (Fig. 5B–D), demonstrating the ability of Tbx20 overexpression to partially rescue the myocardial deficit in LA1110.
Fig.5.
Tbx20 overexpression rescues the cardiac defects present in tbx20 mutant embryos. (A) Diagram of the inducible activity of the tbx20-GR (glucocorticoid receptor) fusion protein. The addition of DXS (dexamethasone) is required for the translocation of the tbx20-GR protein into the nucleus. (B–D) Dorsal views of myl7 expression at the 16-somite stage in wild type embryos (B), tbx20 mutants (C) and tbx20 mutants injected with tbx20-GR mRNA (D). The cardiomyocyte production defects in tbx20 mutant embryos are substantially rescued by the injection of tbx20-GR mRNA. (E) Schematic representation of the hsp70l:Tbx20 transgene used for tbx20 overexpression. (F–H) Dorsal views of myl7 expression at the 16-somite stage in wild type (F), tbx20 mutant (G) and Tbx20−/−; Tg(hsp70l:Tbx20) embryos (H). (I–K) Frontal views of myl7 expression at 48hpf in wild type (I), tbx20 mutant (J) and Tbx20−/−;Tg(hsp70l:Tbx20) embryos (K). Overexpression of Tbx20 successfully restored cardiomyocyte production in tbx20 mutant embryos.
To ensure stable and persistent Tbx20 expression, we generated the hsp70l:Gal4-Tbx20-UAS-mCherry transgene (hereafter referred to as hsp70l:Tbx20) in which Tbx20 and mCherry expression is controlled by the hsp70l heat shock promoter-driven expression of Gal4 (Fig.5E). We crossed tbx20LA1110 into the hsp70l:Tbx20 background and induced the production of Gal4 at the dome stage (4.3hpf) by heat shock. Significant mCherry fluorescence was noted in tbx20LA1110; hsp70l:Tbx20 embryos four hours after heat shock and tbx20 mRNA level increased approximately 40 fold (Fig.S4). Excitingly, similar to what was observed in tbx20-GR injected tbx20LA1110 embryos, a larger domain of myl7 expression was apparent in tbx20LA1110; hsp70l:Tbx20 embryos at the 16-somite stage (confirmed by genotyping, n=6) (Fig. 5F–H). More intriguingly, heat-treatment of tbx20LA1110; hsp70l:Tbx20 partially rescued the heart phenotype that would normally be present in tbx20LA1110 mutants after two days of development (confirmed by genotyping, n=10) (Fig. 5I–K).
Tbx20 morpholino knockdown in zebrafish results in defective cardiac looping but does not have an impact on cardiac progenitor cells [20]. The morphant phenotype is significantly weaker than that observed in tbx20LA1110 mutant embryos, raising a concern about whether the truncated Tbx20LA1110 protein might function as a dominant negative form of Tbx20. To investigate this possibility, we injected tbx20LA1110-GR RNA into wild type and tbx20LA1110 embryos and induced its nuclear translocation by DXS. We did not observe signs of altered myl7 expression or gross morphological defects in Tbx20LA1110-GR-overexpressing wild type embryos (Fig. S3E and data not shown), nor did we observe a rescued cardiac phenotype in Tbx20LA1110-GR-overexpressing tbx20LA1110 mutants (Fig. S3F). These findings suggest that the truncated Tbx20LA1110 protein is not functional and does not harbor a dominant-negative effect.
Tbx20 expands the cardiac progenitor population and promotes cardiomyocyte proliferation in wild type embryos
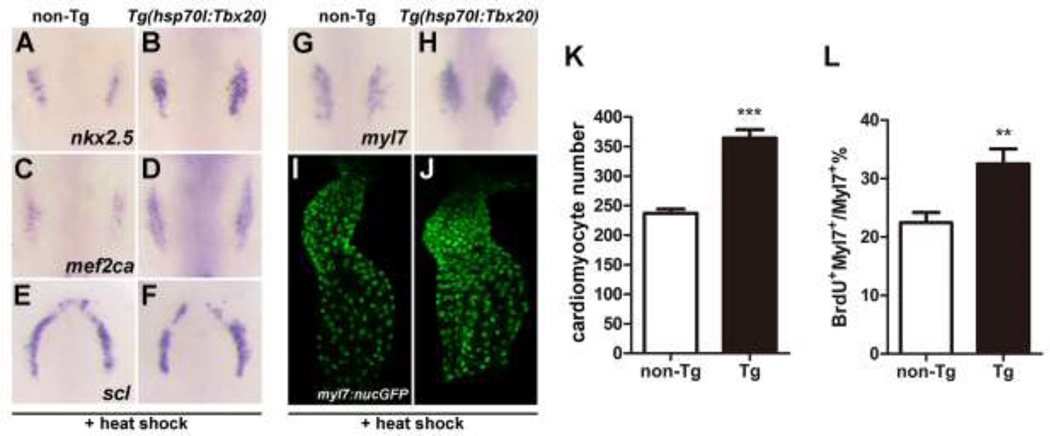
Given the ability of Tbx20 overexpression to partially rescue cardiac progenitor formation in tbx20LA1110 mutants, we wondered if overexpression of Tbx20 in wild type embryos could similarly promote the production of cardiomyocytes. We examined heat-treated hsp70l:Tbx20 embryos using in situ hybridization and found that the signals of nkx2.5, mef2ca and myl7 transcripts were elevated and expanded (Fig. 6A–D and G,H), indicating an increase of cardiac progenitor population. Since expansion of the cardiac domain can occur at the expense of neighboring lineages within the ALPM, we examined the expression of scl, a marker of the hematopoietic lineage that define the rostral boundary of the cardiac field [26]. The expression domain and level of scl (Fig. 6E,F) were similar between wild type and Tbx20-overexpressing embryos, suggesting that Tbx20 overexpression in zebrafish is sufficient to promote cardiac progenitor formation without affecting other ALPM lineages.
Fig.6.
Tbx20 overexpression significantly increases cardiomyocyte number. (A–H) Dorsal views of nkx2.5, mef2ca, scl and myl7 expression in non-transgenic (A, C, E and G) and Tg(hsp70l:Tbx20) embryos (B, D, F and H). The expression of nkx2.5 and mef2ca is expanded in Tbx20-overexpressing embryos at the 10-somite stage (A–D), whereas the expression of scl is similar to that of control embryos (E and F). Similarly, myl7 expression is enhanced in Tg(hsp70l:Tbx20) embryos at the 16-somite stage (H). (I and J) Representative confocal projections of myl7:nucGFP hearts from heat-treated non-transgenic (I) and Tg(hsp70l:Tbx20) embryos (J) at 40hpf. (K) Graph of the average number of nucGFP-positive cardiomyocytes in heat-treated non-transgenic (n=8) and Tg(hsp70l:Tbx20) embryos (n=10). Tbx20 overexpression enhanced cardiomyocyte production. (L) Graph of the ratio of BrdU-positive cardiomyocytes to the total number of cardiomyocytes in heat-treated non-transgenic and Tg(hsp70l:Tbx20) embryos (n=7). Overexpression of Tbx20 in early embryos promoted cardiomyocyte proliferation. Error bars indicate standard deviations. Asterisks indicate a significant difference (**p<0.01, ***p<0.001).
We next examined whether the increase in cardiac gene expression translated into an increased production of cardiomyocytes. We noted that heat-induced hsp70l:Tbx20 embryos have a 54% increase in the number of myl7-positive cardiomyocytes at 40 hpf (237.0±15.7 in non-transgenic embryos, n=8 vs. 364.3±45.6 in transgenic embryos, n=10, p<0.001) (Fig. 6I–K). This increase in myocardial cell number also coincided with increased proliferation of cardiomyocytes in heat-induced hsp70l:Tbx20 embryos. A 44% increase in BrdU-positive cardiomyocytes (32.5±6.2% of cardiomyocytes are BrdU-positive in hsp70l:Tbx20 embryos, n=6 vs. 22.5±4.5% in non-transgenic embryos, n=7, p<0.01) (Fig. 6L) and a 25% increase in PCNA-positive cardiomyocytes (42.0±3.4% of cardiomyocytes are PCNA-positive in hsp70l:Tbx20 embryos vs. 33.6±3.8% in non-transgenic embryos, n=6, p<0.01) (Fig. S5) were observed in 40hpf heat-treated hsp70l:Tbx20 embryos compared to control embryos. Together, these findings indicate that global Tbx20 overexpression, beginning at a time prior to when mesodermal cells have committed to the cardiac fate, promotes cardiac progenitor formation and enhances cardiomyocyte proliferation.
Cardiomyocyte-specific overexpression of Tbx20 enhances cardiomyocyte production
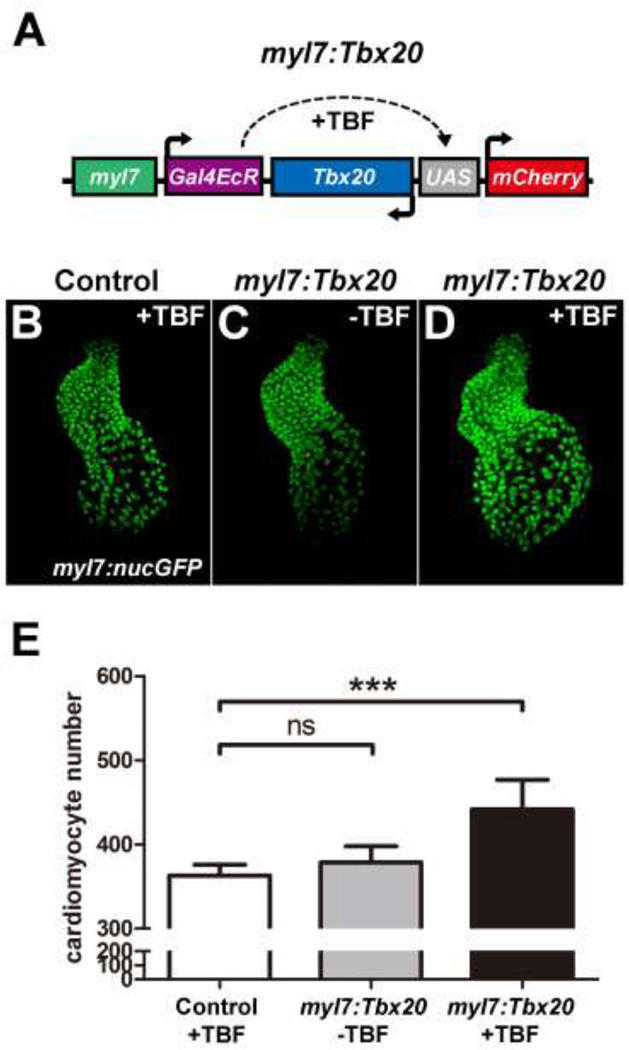
In mammals, overexpression of Tbx20 in differentiated cardiomyocytes is capable of promoting cell proliferation [28]. We thus investigated whether such a mechanism is also present in zebrafish. To do so, we generated the myl7:Gal4EcR-Tbx20-UAS-mCherry construct (hereafter referred to as myl7:Tbx20) in which Tbx20 and mCherry expression is regulated by the Gal4-Ecdysone receptor fusion protein (Gal4-EcR) specifically in myl7-positive cardiomyocytes. In the absence of tebufenozide (TBF), Gal4-EcR is inactive and the expression of Tbx20 and mCherry is not induced. Upon TBF treatment, the activated Gal4-EcR binds to the UAS and drives Tbx20 and mCherry expression [27] (Fig. 7A). We injected myl7:Tbx20 DNA into myl7:nucGFP embryos and treated these embryos with TBF at the 16-somite stage when the myl7 promoter drives strong cardiomyocyte-specific gene expression. There was no difference in cardiomyocyte cell number between control and myl7:Tbx20 embryos in the absence of TBF (362.8±13.1 in control embryos vs 378.5±19.2 in non-treated myl7:Tbx20 embryos, n=6; p>0.05) (Fig. 7B,C and E). Interestingly, TBF-treated myl7:Tbx20 embryos had a significant 22% increase in cardiomyocytes at 2 days post fertilization (dpf) compared to uninjected control embryos (362.8±13.1 in TBF-treated controls vs. 441.5±35.3 in TBF-treated myl7:Tbx20 embryos, n=6, p<0.001) (Fig. 7B,D and E). These findings suggest that Tbx20 is able to cell autonomously promote cardiomyocyte production even after these cells have committed to a cardiac fate.
Fig.7.
Cardiomyocyte-specific Tbx20 expression promotes cardiomyocyte production. (A) Schematic representation of the myl7:Tbx20 transgene used for inducible and cardiomyocyte-specific overexpression of Tbx20. The induction of Tbx20 in myl7-expressing cells is controlled by treatment with TBF (tebufenozide). (B–D) Representative confocal projections of myl7:nucGFP hearts from TBF-treated control embryos (B), non-TBF-treated myl7:Tbx20 injected embryos (C) and TBF-treated myl7:Tbx20-injected embryos (D) at 2dpf. (E) Graph of the average number of nucGFP-positive cardiomyocytes in TBF-treated controls, non-TBF-treated myl7:Tbx20-injected embryos and TBF-treated myl7:Tbx20-injected embryos. Cardiomyocyte-specific overexpression of Tbx20 increased cardiomyocyte production. Error bars indicate standard deviations. ns indicates no significant difference compared with control embryos (n=6, p>0.05). Asterisks indicate a significant increase compared with control embryos (n=6, ***p<0.001).
In adult mouse hearts, Tbx20 promotes cardiomyocyte proliferation by modulating cell-cycle gene expression [28]. However, our quantitative RT-PCR analysis detected no significant difference in the expression of cell proliferation activators including ccna2 (cyclin A2), ccnb1 (cyclin B1), ccnd1 (cyclin D1) and ccne1 (cyclin E1) and cell cycle inhibitors including p21, p27 and meis1b between control and Tbx20-overexpressing zebrafish embryos (Fig. S6), indicating that Tbx20 may regulate zebrafish embryonic cardiomyocyte proliferation through a different molecular mechanism than what has been previously observed in mammalian cardiomyocytes.
Tbx20 promotes cardiac expansion primarily through its transcriptional activation activity
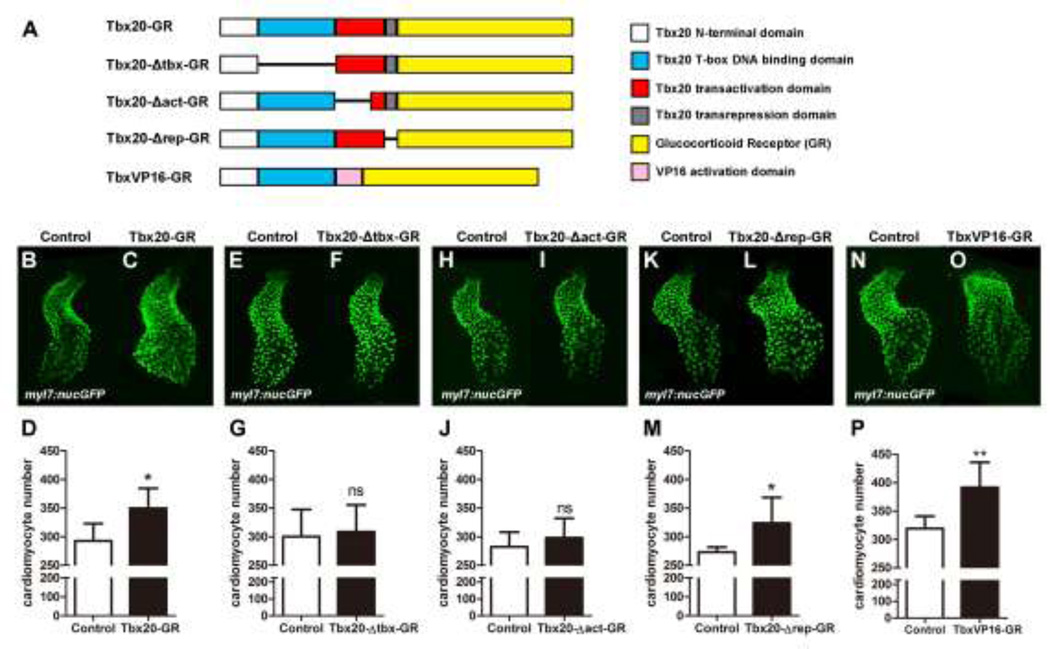
Tbx20 activates and represses the expression of its downstream targets via complex and context-dependent mechanisms [16, 19, 28–31]. We performed a structure-function analysis to identify critical functional domains that execute Tbx20’s cardiac expansion effect. We generated three deletion mutant fusion proteins: Tbx20-Δtbx-GR, Tbx20-Δact-GR, and Tbx20-Δrep-GR which lack the T-box domain, an 86 amino acid portion of the transcription activation domain, and the transcription repression domain of Tbx20, respectively (Fig. 8A). We injected Tbx20-GR, Tbx20-Δtbx-GR, Tbx20-Δact-GR or Tbx20-Δrep-GR mRNA into myl7:nucGFP embryos and treated these embryos with DXS at the 90% epiboly stage to stimulate nuclear translocation of the fusion proteins. Tbx20-GR- and Tbx20-Δrep-GR-injected embryos treated with DXS had significantly more cardiomyocytes than stage-matched uninjected siblings (292.8±30.1 in DXS-treated control embryos vs. 349.8±34.7 in tbx20-GR injected embryos, n=6, p<0.05 and 273.3±8.55 in DXS-treated control embryos vs. 323.8±49.6 in DXS-treated tbx20-Δrep-GR injected embryos, n=6, p<0.05) (Fig. 8B–D and K–M and Fig.S7). Injection of Tbx20-Δtbx-GR or Tbx20-Δact-GR cRNA, on the other hand, did not enhance cardiomyocyte production (296.7.3±20.4 in DXS-treated control embryos vs 279.2±11.3 in DXS-treated Tbx20-Δtbx-GR-injected embryos, n=6, p>0.05 and 282.3±25.8 in DXS-treated control embryos vs. 298.2±33.8 in DXS-treated Tbx20-Δact-GR-injected embryos, n=6, p>0.05) (Fig. 8E–G and H–J and Fig.S7), demonstrating that the T-box domain and the transactivation domain, but not the transrepression domain, are required for Tbx20’s cardiac expansion activity.
Fig.8.
Tbx20’s cardiac expansion activity requires its DNA binding and transcriptional activation domains. (A) Schematic diagram of the Tbx20-GR, Tbx20-Δtbx-GR (a T-box-defective form of Tbx20), Tbx20-Δact-GR (a transactivation-defective form of Tbx20), Tbx20-Δrep-GR (a transrepression-defective form of Tbx20), and TbxVP16-GR (a transactivation-only form of Tbx20) fusion proteins. (B–D) Tbx20-GR-injected embryos treated with DXS have significantly more cardiomyocytes (n=6). (E–G) Tbx20-Δtbx-GR overexpression did not significantly enhance cardiomyocyte production (n=6). (H–J) Overexpression of Tbx20-Δact-GR did not significantly enhance cardiomyocyte production (n=6). (K–M) Overexpression of Tbx20-Δrep-GR significantly increased cardiomyocyte number (n=6). (N–P) Overexpression of TbxVP16-GR significantly increased cardiomyocyte production (n=6). Error bars indicate standard deviations. ns indicates there is no significant difference compared with control embryos. Asterisks indicate a significant difference (*p<0.05, **p<0.01, ***p<0.001).
We reasoned that if Tbx20 drives cardiogenesis mainly as a transcriptional activator, overexpression of TbxVP16-GR, a chimeric protein consisting of Tbx20’s N-terminal 292 amino acids (including the T-box domain) and the VP16 activation domain (Fig. 8A and Fig S7A), should mimic the effects of Tbx20-GR mRNA. Indeed, direct cell counting indicated a 23% increase in the number of cardiomyocytes in tbxVP16-GR-injected embryos at 40hpf (319.2±21.8 in DXS-treated control embryos vs. 391.5±44.4 in DXS-treated TbxVP16-GR-injected embryos, n=6, p<0.01), an increase comparable to that observed in Tbx20-GR-overexpressing embryos (Fig.8N–P). Together these findings demonstrate that Tbx20’s ability to promote cardiac expansion is mediated by its transcriptional activation activity.
Discussion
Involvement of Tbx20 in cardiac progenitor formation
The differentiation of multi-potent mesodermal cells to a cardiac fate is orchestrated by various signaling events and the cardiac transcriptional program (for review see [32–34]). In this study we showed that Tbx20 is a critical component of the cardiac transcriptional program that drives the formation of cardiac progenitor cells. Loss of Tbx20 activity leads to a reduction in the expression domains of the early cardiac markers nkx2.5, mef2ca, tbx5a and bmp4. This dramatic loss of cardiac gene expression may reflect a failure to activate a subset of the cardiac transcription program, thereby impeding the formation of cardiac progenitor cells. Alternatively, an appropriate number of cardiac progenitors might initially form in the absence of Tbx20 activity but this population may not be able to properly expand or survive in tbx20LA1110 mutant embryos. Given that we did not detect decreased proliferation or increased apoptosis in the heart-forming region of tbx20LA1110 mutants, and because all cardiac progenitor markers we examined were strongly diminished in tbx20LA1110, we favor the interpretation that Tbx20 is directly needed for the initial production of cardiac progenitors from the ALPM. This early requirement for Tbx20 activity in zebrafish cardiogenesis strongly resembles the reported roles of the Tbx20 homologs nmr1 and nmr2 in Drosophila cardiogenesis [14], suggesting that Tbx20 may be a member of a conserved gene program that defines the cardiac population in both invertebrates and vertebrates.
The cardiac defects we observed in the zebrafish tbx20 mutant, LA1110, are significantly more severe than the phenotypes reported in previous studies on zebrafish and Xenopus that utilized morpholino knockdown approaches [20, 21]. This discrepancy in cardiac phenotypes may reflect the differences in residual Tbx20 activity between tbx20LA1110 and tbx20 morphants. Transcripts of tbx20LA1110 are unstable and encode a truncated protein lacking all functional domains, leading us to hypothesize that tbx20LA1110 represents a null or a severely hypomorphic allele. If low levels of Tbx20 activity are sufficient for cardiac progenitor specification, then the dramatic loss of Tbx20 activity in tbx20LA1110 could explain our novel findings. The reported mouse tbx20 knockout models represent null alleles but form a relatively normal cardiac crescent [16–18]. It is possible that Tbx20 is not required for the earliest steps of heart formation in the mouse, and has functionally diverged from the ancestral role it plays in Drosophila and zebrafish. Alternatively, compensatory mechanisms or redundancies may exist in mice that enable the formation of cardiac progenitors even in the absence of Tbx20 activity. For example, other transcription factors, including T-box family members, may activate the target genes that are Tbx20-dependent in zebrafish. Further investigation in several model organisms is needed to determine if Tbx20’s regulatory targets differ among vertebrates, and identify any compensatory mechanisms that may have evolved to promote cardiac progenitor formation in Tbx20’s absence.
Tbx20 activity is dispensable for heart tube morphogenesis
Despite the severe defect in cardiomyocyte production present in tbx20-deficient zebrafish embryos, the remaining cardiomyocytes are able to migrate and fuse medially to form the cardiac cone, which later elongates into a linear heart tube within the same timeframe as their wild type siblings. Thus, Tbx20 activity is dispensable for cardiomyocyte migration and primitive heart tube formation in zebrafish. These findings also argue that the threshold of the number of cardiomyocytes needed to drive formation of the cardiac cone and linear heart tube during heart morphogenesis is relatively low. Combining the loss of Tbx20 with that of other factors involved in cardiac progenitor formation may help to clarify how the loss of cardiomyocytes affects the completion or timing of major cardiac developmental events.
Tbx20 regulates the cardiac transcriptional program and cardiomyocyte proliferation
Tbx20 overexpression promotes the proliferation of differentiated embryonic cardiomyocytes in the heart tube between 24 and 40 hpf, resulting in an enlarged heart with significantly more cells. These findings are in line with previous in vitro studies on neonatal rat cardiomyocytes and in vivo studies on mouse fetal hearts showing that Tbx20 overexpression increases cardiomyocyte proliferation and thickens the compact myocardium [15, 35]. While mammalian cardiomyocytes cease to proliferate soon after birth, Tbx20 overexpression is able to induce cardiomyocyte proliferation in adult mouse hearts and leads to the generation of new cardiomyocytes and improved cardiac repair after myocardial infarction [29, 35]. Tbx20’s potent ability to induce cardiomyocyte proliferation in multiple models, along with the finding that Tbx20 expression is reactivated in the regenerating adult zebrafish heart following injury [26, 36], point to the intriguing possibility of utilizing Tbx20 activation as a therapeutic strategy for cardiac repair.
Using a domain dissection approach, we showed that the cardiac expansion effect of Tbx20 in embryonic zebrafish is dependent on its T-box and transcription activation domains. Overexpression of a transactivation-deficient form of Tbx20 (Tbx20-Δact) failed to increase cardiomyocyte production, while overexpression of a TbxVP16 fusion protein composed of the N-terminal portion of Tbx20 and the VP16 activation domain mimicked the effect of wild type Tbx20 and promoted a significant increase in cardiomyocyte number. This suggests that Tbx20 may promote cardiomyocyte production during early zebrafish cardiogenesis by transcriptional activation of its target genes. Previous studies have identified a number of cardiac factors that are activated directly by Tbx20, including nkx2.5, mef2c, and fgf10 [19]. Whether these direct targets, along with the pathways they regulate, are mediators of the cardiac expansion activity of Tbx20 in embryonic zebrafish will require further examination. Interestingly, Tbx20 has been shown to regulate adult mouse cardiomyocyte proliferation by directly repressing cell cycle inhibitors [29], suggesting that the mechanisms by which Tbx20 regulates its direct and downstream targets during development are context and stage-dependent. Future genome-wide analyses will help to reveal the complement of binding partners and gene targets involved in Tbx20-mediated cardiac expansion during zebrafish cardiogenesis.
Materials and methods
Zebrafish husbandry and transgenesis
Zebrafish colonies were cared for and bred under standard conditions [37]. Developmental stages of zebrafish embryos were determined using standard morphological features of fish raised at 28.5 °C [37]. The hsp70l:Gal4-Tbx20-UAS-mCherry and myl7:Gal4EcR-Tbx20-UAS-mCherry transgenic constructs were created using the Tol2 kit [38].
Positional Cloning
LA1110 was crossed to the polymorphic WIK strain for mapping. Embryo lysis and PCR-based bulk segregant analysis were performed as previously described [23]. Total RNA was isolated from 1 day post fertilization (dpf) LA1110 homozygous embryos using RNA Wiz (Ambion, TX) and cDNA was synthesized using the Superscript II Kit (Thermo Fisher Scientific, Waltham, MA). Tbx20 cDNA fragments were amplified with Phusion polymerase (New England BioLabs, MA) and cloned into pCR-Blunt II-TOPO (Thermo Fisher Scientific, Waltham, MA) and sequenced.
Whole mount in situ hybridization
Embryos for in situ hybridization were raised in embryo medium supplemented with 0.2 mM 1-phenyl-2-thiourea to maintain optical transparency [37]. Whole-mount in situ hybridization was performed as described previously [39]. The antisense RNA probes used in this study include myl7, nkx2.5, mef2ca, tbx20, scl, tbx5a and bmp4. For fluorescent in situ hybridization, digoxigenin-labeled nkx2.5 probes were then detected by fluorophore deposition using the TSA Plus System (Perkin Elmer, Waltham, MA).
Whole-Mount Immunostaining
Whole mount immunostaining was performed as previously described [22]. Antibodies used include rabbit anti-pHH3 (Santa Cruz Biotechnology, Dallas, TX) and mouse anti-PCNA (Vector Laboratories, Burlingame, CA). For quantification of pHH3-positive cardiac progenitors, immunofluorescence assays were preceded by detection of nkx2.5 by fluorescent in situ hybridization. Proliferating cells were considered cardiac progenitors if they fell within the expression domain of nkx2.5 as determined by examination of confocal image z-stacks in ImageJ.
Whole-Mount Apoptosis Assay
Apoptosis was visualized in situ using the ApopTag Red in situ cell death detection kit (EMD Millipore, Darmstadt, Germany) as previously described [40], preceded by detection of nkx2.5 by fluorescent in situ hybridization. Apoptotic bodies were imaged by confocal microscopy, and were considered to be in the cardiac progenitor field if they fell within the expression domain of nkx2.5 as determined by examination of confocal image z-stacks in ImageJ.
Constructs
Full-length cDNA for tbx20WT and tbx20LA1110 was amplified from 1dpf embryos cDNA using KOD polymerase (EMD Millipore, Darmstadt, Germany) and fused to the glucocorticoid receptor ligand binding domain (GR) (kindly provided by Dr. S. Evans). Tbx20-GR and tbx20LA1110-GR were cloned into pCS2 + 3XFLAG for tagging with the FLAG epitopes. Plasmids were cut with NotI and SP6 RNA polymerase was used to generate mRNA for injection. We generated Tbx20-Δtbx-GR, Tbx20-Δact-GR and Tbx20-Δrep-GR constructs by deleting the entire T-box DNA binding domain (amino acids 97–292) of Tbx20, deleting part of the presumptive transcription activation domain (amino acids 293–378) of Tbx20, and deleting the presumptive transcription repression domain (amino acids 419–446) of Tbx20. The TbxVP16-GR construct was created by replacing the whole C-terminus of Tbx20 (amino acid 293–446) with the VP16 coding region.
For the rescue experiments, 100pg of tbx20-GR or tbx20LA1110-GR mRNA was injected into one-cell stage zebrafish embryos collected from tbx20 heterozygous crosses, and 100μM dexamethasone was added to activate the GR protein at the 90% epiboly stage. The embryos were fixed at the 16S stage for in situ hybridization followed by genotyping. Genotyping of tbx20LA1110 embryos was performed using two PCR primers that amplify a fragment of tbx20 that spans the nonsense mutation. The PCR products were then purified and sequenced. Primers used were 5’- GTGGGGTTGTCCTTAACTTAGG -3’ and 5’- AGGTAAAGGTGGATCTGCCTTTC-3’.
For the overexpression experiments, 100pg of each mRNA (tbx20-GR, Tbx20-Δtbx-GR, Tbx20-Δact-GR, Tbx20-Δrep-GR or TbxVP16-GR) was injected into one-cell stage myl7:nucGFP embryos. 100µM dexamethasone was added to activate the GR protein at the 90% epiboly stage, while control embryos (uninjected) were treated in parallel with an equivalent dosage of dexamethasone.
Western Blotting
Embryos for Western analysis were lysed at day 1 of development in Rubinfeld’s Lysis Buffer (20 mM Tris pH 8.0, 140 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EGTA, 1.5 mM MgCl2, 1 mM Na3VO4, 50 mM NaF, 1 mM DTT) with protease inhibitors (Sigma-Aldrich, St. Louis, MO). Western Blotting analysis was performed following standard procedures. Antibodies used include mouse anti-Flag (Sigma-Aldrich, St. Louis, MO) and mouse anti-β-actin (Sigma-Aldrich, St. Louis, MO).
Tbx20 induction
The hsp70l:tbx20; tbx20LA1110 and hsp70l:tbx20; myl7:nucGFP embryos were exposed to 38.5°C for one hour at the dome stage. Embryos were fixed at 10S and 16S for gene expression analysis or at 40hpf for cell counting.
The myl7:Gal4EcR-Tbx20-UAS-mCherry transgene was injected into myl7:nucGFP embryos at the one-cell stage. 1µM TBF was added to the embryo media at the 16-somite stage to induce the production of Tbx20. The number of cardiomyocytes was measured at 2dpf.
Imaging and cardiomyocyte cell counting
Tg(myl7:nucGFP) embryos were used to facilitate cardiomyocyte cell counting [41]. The Tg(myl7:nucGFP) fish line was kindly provided by Dr. John Mably. Embryos were embedded in 1% low-melt agarose and imaged on a Zeiss LSM510 confocal microscope equipped with a 20X water objective. The number of green fluorescent nuclei was measured using ImageJ.
BrdU incorporation and immunostaining
Embryos were exposed to BrdU from 24–40 hpf (5 mg/mL) and fixed at 40hpf with 4% paraformaldehyde/PBS solution overnight at 4°C. Fixed embryos were rinsed with PBTX and acid treated (2N HCl for 30min at room temperature) before primary antibody incubation (rat anti-BrdU, Abcam). Anti-rat IgG-Alexa Fluor 647-conjugated antibody was used as the secondary antibody (Invitrogen). BrdU staining was imaged on a Zeiss LSM510 confocal microscope and BrdU and myl7-positive nuclei were counted using ImageJ.
Quantitative RT-PCR
RNA samples of non-transgenic and Tg (hsp70l:Tbx20) embryos were extracted using Trizol RNA isolation reagents (Thermo Fisher Scientific, Waltham, MA). cDNA was synthesized using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). Real-time quantitative PCR was carried out using the Roche LightCycler 480 Real-Time PCR System. Primers used are:
Tbx20-F: 5’- ACTCTGGGAGAAGAAGGACATTCAGC -3’
Tbx20-R: 5’- GTGCTGAAAGCCAGAGAAACCAGATG -3’
Ccna2-F: 5’- GCTTTTGGCTTCGAAGTTTGA -3’
Ccna2-R: 5’-GTGGGTGGTCTTCAGGTTTG-3’
Ccnb1-F: 5’- GCTTATGCCCTGACCCTGAA -3’
Ccnb1-R: 5’- GCATCACAGGAACCAGCTCAT -3’
Ccnd1-F: 5’- TTCCTTGCCAAACTGCCTAT -3’
Ccnd1-R: 5’- GGTGAGGTTCTGGGATGAGA -3’
Ccne1-F: 5’- CGCAGTATGCATCAGAAAGCA -3’
Ccne1-R: 5’- GAGCAGGTTGTTCCAAACTTCAT -3’
P21-F: 5’- CCGCATGAAGTGGAGAAAAC -3’
P21-R: 5’- ACGCTTCTTGGCTTGGTAGA -3’
P27-F: 5’- GTCCGACACCCACATAAACACA -3’
P27-R: 5’- CATCGAAGCGACGACAATGA -3’
Meis1b–F: 5’- CCCGCACACAGCCCACAC -3’
Meis1b–R: 5’- CCGGGTTCTCTAGGTGTGCAAG -3’
eef1a1l1-F: 5’- CCTCTTTCTGTTACCTGGCAA -3’
eef1a1l1-R: 5’- CTTTTCCTTTCCCATGATTGA -3’
Statistics
All values are expressed as mean ± SD. Significance values were calculated using an unpaired Student's t-test.
Supplementary Material
Highlights.
Tbx20 is required for cardiac progenitor cell formation.
Tbx20 deficiency results in diminished cardiac progenitor population.
Tbx20 promotes cardiomyocyte proliferation.
Tbx20 overexpression causes cardiomegaly.
The cardiac expansion activity of Tbx20 requires its T-box and transactivation domains.
Acknowledgments
We thank the members of the Chen lab for helpful discussions. Fei Lu is supported by Graduate Student Fellowships from the China Scholarship Council and the Philip Whitcome Training Program at UCLA. This work is funded by grants from the American Heart Association (15GRNT25850032) and the National Institute of Health (HL096980 and HL129052) to JNC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- 1.Qian L, et al. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc Natl Acad Sci U S A. 2008;105(50):19833–19838. doi: 10.1073/pnas.0808705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen T, et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121(12):4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirk EP, et al. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet. 2007;81(2):280–291. doi: 10.1086/519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C, et al. T-box transcription factor TBX20 mutations in Chinese patients with congenital heart disease. Eur J Med Genet. 2008;51(6):580–587. doi: 10.1016/j.ejmg.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Hammer S, et al. Characterization of TBX20 in human hearts and its regulation by TFAP2. J Cell Biochem. 2008;104(3):1022–1033. doi: 10.1002/jcb.21686. [DOI] [PubMed] [Google Scholar]
- 6.Posch MG, et al. A gain-of-function TBX20 mutation causes congenital atrial septal defects, patent foramen ovale and cardiac valve defects. J Med Genet. 2010;47(4):230–235. doi: 10.1136/jmg.2009.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Y, et al. TBX20 loss-of-function mutation contributes to double outlet right ventricle. Int J Mol Med. 2015;35(4):1058–1066. doi: 10.3892/ijmm.2015.2077. [DOI] [PubMed] [Google Scholar]
- 8.Zhao C-M, et al. TBX20 loss-of-function mutation associated with familial dilated cardiomyopathy. Clinical Chemistry and Laboratory Medicine (CCLM) 2016:325. doi: 10.1515/cclm-2015-0328. [DOI] [PubMed] [Google Scholar]
- 9.Yu LW, et al. Mild decrease in TBX20 promoter activity is a potentially protective factor against congenital heart defects in the Han Chinese population. Sci Rep. 2016;6:23662. doi: 10.1038/srep23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahn DG, et al. tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech Dev. 2000;95(1–2):253–258. doi: 10.1016/s0925-4773(00)00346-4. [DOI] [PubMed] [Google Scholar]
- 11.Griffin KJ, et al. A conserved role for H15-related T-box transcription factors in zebrafish and Drosophila heart formation. Dev Biol. 2000;218(2):235–247. doi: 10.1006/dbio.1999.9571. [DOI] [PubMed] [Google Scholar]
- 12.Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse Tbox gene tbx20. Mech Dev. 2001;100(1):87–91. doi: 10.1016/s0925-4773(00)00499-8. [DOI] [PubMed] [Google Scholar]
- 13.Brown DD, et al. Developmental expression of the Xenopus laevis Tbx20 orthologue. Dev Genes Evol. 2003;212(12):604–607. doi: 10.1007/s00427-002-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian L, Liu J, Bodmer R. Neuromancer Tbx20-related genes (H15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Dev Biol. 2005;279(2):509–524. doi: 10.1016/j.ydbio.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Chakraborty S, Yutzey KE. Tbx20 regulation of cardiac cell proliferation and lineage specialization during embryonic and fetal development in vivo. Dev Biol. 2012;363(1):234–246. doi: 10.1016/j.ydbio.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai CL, et al. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132(10):2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh MK, et al. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132(12):2697–2707. doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- 18.Stennard FA, et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132(10):2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- 19.Takeuchi JK, et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132(10):2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 20.Szeto DP, Griffin KJ, Kimelman D. HrT is required for cardiovascular development in zebrafish. Development. 2002;129(21):5093–5101. doi: 10.1242/dev.129.21.5093. [DOI] [PubMed] [Google Scholar]
- 21.Brown DD, et al. Tbx5 and Tbx20 act synergistically to control vertebrate heart morphogenesis. Development. 2005;132(3):553–563. doi: 10.1242/dev.01596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langenbacher AD, et al. The PAF1 complex differentially regulates cardiomyocyte specification. Dev Biol. 2011;353(1):19–28. doi: 10.1016/j.ydbio.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen CT, et al. The PAF1 complex component Leo1 is essential for cardiac and neural crest development in zebrafish. Dev Biol. 2010;341(1):167–175. doi: 10.1016/j.ydbio.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolm PJ, Sive HL. Efficient hormone-inducible protein function in Xenopus laevis. Dev Biol. 1995;171(1):267–272. doi: 10.1006/dbio.1995.1279. [DOI] [PubMed] [Google Scholar]
- 25.Tada M, Reilly MA, Smith JC. Analysis of competence and of Brachyury autoinduction by use of hormone-inducible Xbra. Development. 1997;124(11):2225–2234. doi: 10.1242/dev.124.11.2225. [DOI] [PubMed] [Google Scholar]
- 26.Schindler YL, et al. Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development. 2014;141(16):3112–3122. doi: 10.1242/dev.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esengil H, et al. Small-molecule regulation of zebrafish gene expression. Nat Chem Biol. 2007;3(3):154–155. doi: 10.1038/nchembio858. [DOI] [PubMed] [Google Scholar]
- 28.Singh R, et al. Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ Res. 2009;105(5):442–452. doi: 10.1161/CIRCRESAHA.109.196063. [DOI] [PubMed] [Google Scholar]
- 29.Xiang F-l, Guo M, Yutzey KE. Overexpression of Tbx20 in Adult Cardiomyocytes Promotes Proliferation and Improves Cardiac Function After Myocardial Infarction. Circulation. 2016 doi: 10.1161/CIRCULATIONAHA.115.019357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stennard FA, et al. Cardiac T-box factor Tbx20 directly interacts with Nkx2–5, GATA4, and GATA5 in regulation of gene expression in the developing heart. Dev Biol. 2003;262(2):206–224. doi: 10.1016/s0012-1606(03)00385-3. [DOI] [PubMed] [Google Scholar]
- 31.Plageman TF, Jr, Yutzey KE. Differential expression and function of Tbx5 and Tbx20 in cardiac development. J Biol Chem. 2004;279(18):19026–19034. doi: 10.1074/jbc.M314041200. [DOI] [PubMed] [Google Scholar]
- 32.Später D, et al. How to make a cardiomyocyte. Development. 2014;141(23):4418–4431. doi: 10.1242/dev.091538. [DOI] [PubMed] [Google Scholar]
- 33.Paige SL, et al. Molecular Regulation of Cardiomyocyte Differentiation. Circulation Research. 2015;116(2):341–353. doi: 10.1161/CIRCRESAHA.116.302752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu F, Langenbacher AD, Chen JN. Transcriptional Regulation of Heart Development in Zebrafish. J Cardiovasc Dev Dis. 2016;3(2) doi: 10.3390/jcdd3020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chakraborty S, Sengupta A, Yutzey KE. Tbx20 promotes cardiomyocyte proliferation and persistence of fetal characteristics in adult mouse hearts. J Mol Cell Cardiol. 2013;62:203–213. doi: 10.1016/j.yjmcc.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 36.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127(3):607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 37.Westerfield M. The zebrafish book. The University of Oregon Press; 2000. [Google Scholar]
- 38.Kwan KM, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236(11):3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 39.Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122(12):3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- 40.Mouillesseaux K, Chen J-N. Mutation in utp15 Disrupts Vascular Patterning in a p53-Dependent Manner in Zebrafish Embryos. PLoS ONE. 2011;6(9):e25013. doi: 10.1371/journal.pone.0025013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavanaugh AM, Huang J, Chen JN. Two developmentally distinct populations of neural crest cells contribute to the zebrafish heart. Dev Biol. 2015;404(2):103–112. doi: 10.1016/j.ydbio.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.