Abstract
It has been nearly two decades since RNA-interference (RNAi) was first reported. While there are no approved clinical uses, several promising phase II and III clinical trials suggest the great promise of RNAi therapeutics. One challenge for RNAi therapies is the controlled localization and sustained presentation to target tissues, to both overcome systemic toxicity concerns and to enhance in vivo efficacy. One approach that is emerging to address these limitations is the entrapment of RNAi molecules within hydrogels for local and sustained release. In these systems, nucleic acids are either delivered as siRNA conjugates or within nanoparticles. A plethora of hydrogels has been implemented using these approaches, including both traditional hydrogels that have already been developed for other applications and new hydrogels developed specifically for RNAi delivery. These hydrogels have been applied to various applications in vivo, including cancer, bone regeneration, inflammation and cardiac repair. This review will examine the design and implementation of such hydrogel RNAi systems and will cover the most recent applications of these systems.
Keywords: RNAi, hydrogel, gene therapy, nanoparticle, transfection
Graphical abstract
RNA interference therapy delivery from hydrogels has tremendous potential towards translation. This review discusses concepts in designing hydrogels and gives an overview of recent advances in the delivery of siRNA, miRNA and shRNA from hydrogels for various applications both in vitro and in vivo.
1. Introduction
RNA interference (RNAi) was first reported in 1998 when Fire et al. observed that double-stranded RNA molecules (dsRNA) were able to silence the expression of complementary messenger RNA (mRNA) in Caenorhabditis elegans.1 This newfound ability to silence the expression of genes provides the platform for eventual clinical translation, particularly for aberrant protein targets that are difficult to otherwise inhibit therapeutically. Over the past two decades, RNAi has been investigated for a variety of therapeutic purposes, with major advances reported in the fields of infection and cancer.2–6 However, despite the vast therapeutic potential, translation has been limited and there are currently no approved RNAi therapies used clinically.7 One of the major challenges has been the local delivery of RNAi to limit any off-target and undesired outcomes, while enhancing in vivo efficacy. Towards improved clinical translation, biomaterial delivery systems are emerging to facilitate local, sustained, and efficient RNAi-mediated gene silencing. This review will cover recent advances in the use of hydrogels - water-swollen polymer networks - as local RNAi-eluting depots for therapeutic gene silencing.
1.1. RNA-interference Mediated Gene Silencing Mechanisms
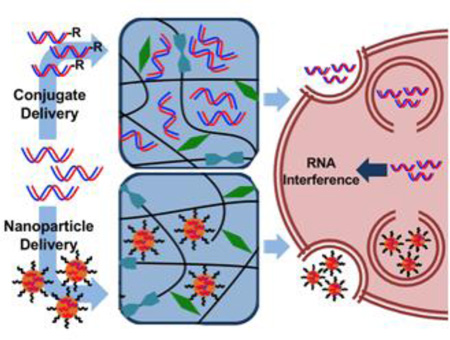
Generally, RNAi occurs through three separate but convergent mechanisms via microRNAs (miRNA), short hairpin RNAs (shRNAs), and small interfering RNAs (siRNAs) (Figure 1). miRNAs are well conserved, endogenously synthesized, non-coding RNA molecules that exert function by silencing expression of one or more complementary messenger RNA molecules.8 Upon transcription, miRNAs are first synthesized as partially complementary hairpins known as primary miRNAs (pri-miRNA). The hairpin structure is recognized by Drosha and cleaved from the rest of the molecule to form precursor miRNA (pre-miRNA). Pre-miRNAs are then exported from the nucleus through a shuttle protein known as Exportin, where they are then cleaved to a dsRNA 21–23 nucleotides in length (miRNA). This same process is coupled to dissociation of dsRNA into single-stranded RNA (ssRNA), in which the single, antisense miRNA molecule is incorporated into a complex of proteins known as the RNA-induced silencing complex (RISC). miRNA/RISC is targeted towards up to hundreds of complementary mRNA sequences, to which it may have incomplete, partial complementarity or complete complementarity through base pairing.9 In the former, binding of miRNA to mRNA localizes complexes to processing bodies, wherein translation is repressed and mRNA is sequestered and eventually destroyed. In the latter, perfect binding of miRNA to mRNA leads to direct target mRNA cleavage. miRNA silencing pathways can be successfully recapitulated exogenously by introducing miRNA “mimics,” which are double-stranded RNA molecules that can associate with RISC in the cytoplasm for gene silencing.10–12
Figure 1.
Roles of miRNA, shRNA and siRNA in RISC binding and RNA interference. Reproduced with permission.186 Copyright 2005, Nature Publishing Group.
Mirroring this endogenous miRNA-mediated RNAi, shRNAs are artificial, exogenously introduced RNAi molecules that are designed as DNA. Typically, shRNAs are introduced as plasmids or through a viral or bacterial vector, which allows for trafficking into the nucleus. shRNA sequences then encode for hairpins known as pre-shRNA that are processed into siRNA. siRNA, in an analogous manner to miRNA, associates with RISC as ssRNA to exert gene silencing. Because shRNAs are usually engineered against a sequence with full complementarity, they exert gene silencing through direct complementary mRNA target cleavage.13 Although shRNAs are a potent form of gene silencing through RNAi, they are limited by the need to introduce DNA into the host genome, which carries the risk of adverse events such as insertional mutagenesis, especially from viral-mediated transfections.14
Exogenously administered siRNA can also lead to gene silencing similar to miRNA mimics. Like miRNA mimics, siRNAs are double-stranded RNA molecules that upon entering the cytoplasm will exert gene silencing against a complementary mRNA target. However, while miRNA may silence a number of genes at once through partial complementarity, siRNAs can be exogenously modified to specifically target a single gene with full complementarity.15 Because of their introduction as RNA, there is no risk of insertional mutagenesis with either siRNA or miRNA mimic delivery. Moreover, siRNA or miRNA only need to get into the cytoplasm, overcoming the need for nuclear trafficking, which is a major barrier associated with shRNA and DNA delivery. However, the major drawbacks to siRNA and miRNA delivery are the relative hydrolytic instability of RNA molecules and poor pharmacokinetics.10,16
1.2. Challenges to Clinical Translation of RNAi
The ability to efficiently silence genes expressed in pathological molecular processes has tremendous potential for therapeutic applications. Currently, clinical trial targets of RNAi-based technology include the eye, liver, gastrointestinal tract, and dermis, which have specifically focused on infectious processes and cancers, with several trials in Phase III development.7,17 However, despite close to two decades of work, RNAi-based therapeutics have yet to be approved by the US Food and Drug Administration.
RNAi molecules in the form of siRNA, miRNA and shRNA are all met with unfavorable physiochemical properties towards delivery. Nucleic acids are negatively charged due to their phosphate backbone and, as such, they are naturally repelled by like-charged cellular membranes.5,18 Moreover, these molecules are all too large and hydrophilic to diffuse passively across cellular membranes that are hydrophobic between their lipid bilayers.19 In the case of siRNA and miRNA, upon internalization, they are also subject to endo-lysosomal fusion and rapid acidification and degradation, which are additional barriers towards efficient delivery. Thus, agents that promote RNAi uptake are needed to both pass through the cellular membrane and allow for endosomal escape for RNAi molecules to complex with RISC machinery. Towards this, there are a plethora of techniques to permit uptake such as through ligand-receptor interactions or endocytosis.20 Traditionally, these approaches include the direct modification of nucleic acids or their complexation with a delivery vehicle.
These intracellular barriers to delivery are further compounded by problems inherent to systemic delivery. As previously discussed, siRNA and miRNA molecules are particularly susceptible to nuclease-mediated hydrolysis through the 2’ hydroxyl group of the pentose ring. When delivered systemically, RNAi molecules are rapidly cleared from circulation (plasma half-life < 10 min)21 and rapidly excreted through the kidneys with accumulation as quickly as 20 minutes after injection.22,23 They can also interact with serum components such as lipoproteins and erythrocytes, causing aggregation and unfavorable accumulation. Moreover, RNA can stimulate the innate immune system through activation of TLR3, TLR7, and retinoic acid-inducible gene I protein. The innate immune response is also characterized by a cytokine response consisting of interleukins, interferons and other pro-inflammatory mediators.20,24 Systemic RNAi delivery also leads to unintended off-target gene silencing, which can be undesirable with accumulation in organs like the liver and lungs.25,26 Although targeting of RNAi can overcome many of these issues through incorporation of antibodies or receptor ligands, there is inherent cytotoxicity to systemic delivery of many non-biodegradable delivery vehicles due to factors like cationic charge and immunogenicity. 27,28
1.3. Hydrogels for RNAi-mediated Gene Silencing
A variety of potential biomaterial drug-eluting depots exist for RNAi therapies, including microparticles,29–32 macroporous scaffolds,33–35 nanofibers,36–40 multilayer films,41,42 and hydrogels. The motivation for the use of biomaterials is their ability to locally deposit and sustain the presentation of RNAi. These drug depot technologies were recently reviewed by Sarett et al. with a focus on controlled, local delivery of siRNA.10 In contrast, this review will focus specifically on the use of hydrogels for RNAi delivery, as hydrogels can be designed with diverse mechanical, chemical and physical properties for this application that can be engineered and tuned specifically for various applications.43
Hydrogels are three-dimensional, water-swollen networks that may be comprised of natural or synthetic polymers. Their various properties, including biodegradability, mechanics, and injectability, may be tailored towards specific applications, which will be reviewed in a later section. For drug delivery, hydrogels overcome many pharmacokinetic and pharmacodynamics concerns. By concentrating and eluting drugs locally, concentrations required for therapeutic efficacy are lowered by orders of magnitude.44,45 When compared to subcutaneous or intramuscular bolus injections, hydrogels further assist in retention and promote sustained drug release. Further, signals can be included into the hydrogel to promote responsiveness to enzymatic activity,46–50 pH,51–53 temperature,54–61 light62–67 or electricity.68,69 Hydrogels also help to overcome problems with undesirable off-target toxicities and accumulation, which are especially relevant to the delivery of RNAi.70 For these reasons, hydrogels have been extensively explored for local, sustained plasmid DNA delivery both in vitro and in vivo,71–79 and RNAi delivery from hydrogels is being increasingly investigated as a means for local and sustained therapies, which is the focus of this review.
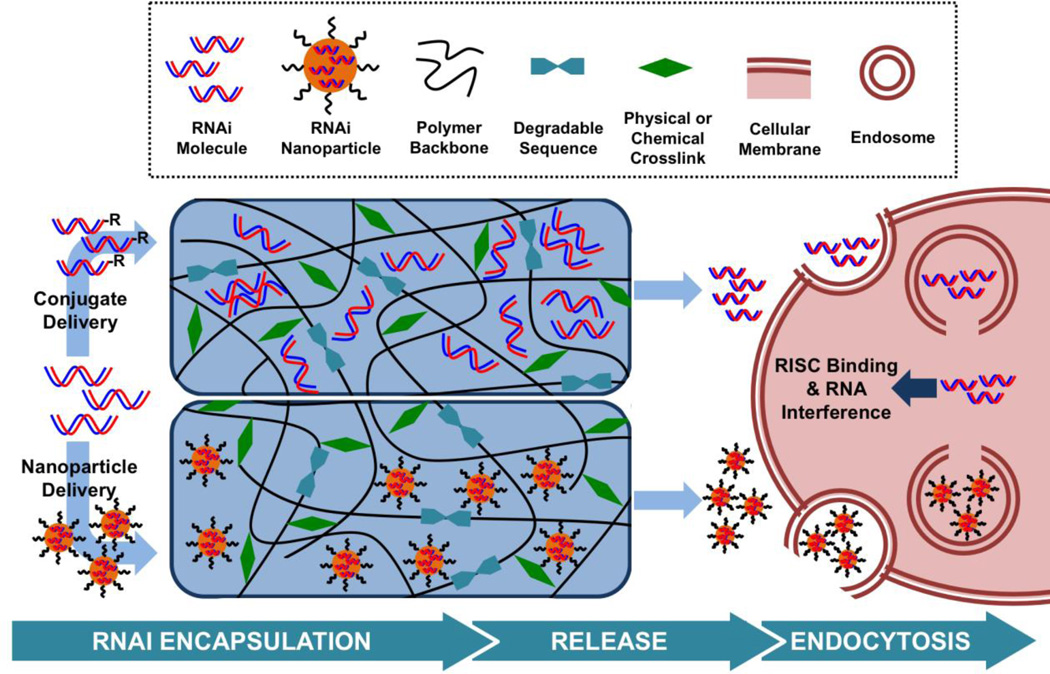
2. Design Considerations in Engineering Hydrogels for RNAi Delivery
Many hydrogel properties (e.g., mechanics, degradation, drug release) are based on the polymers that make up the network and there are numerous design features that should be considered. Towards RNAi delivery, features such as the selection of the polymer that comprises the hydrogel and corresponding properties such as electrostatics are important. Also, the type and composition of hydrogel crosslinks will lead to variations in material properties, such as degradation and whether or not the hydrogel is injectable. Lastly, the incorporation of RNAi nucleic acids may be either direct or with inclusion of nanoparticles (Figure 2). RNAi therapeutics have also been encapsulated along with cells, towards the fabrication of scaffolds to support 3D cellular migration and proliferation towards both regenerative medicine and tissue engineering.80–82 However, this review will focus on the design of hydrogels for local gene silencing without added cells and this section provides an overview of the various design components towards hydrogels for RNAi delivery.
Figure 2.
Strategies for siRNA delivery from hydrogels include encapsulation within a nanoparticle or as an siRNA conjugate to permit cell transfection. Towards hydrogel design, polymer charges are varied and degradable sequences and crosslinking mechanisms control the rate at which siRNA is released. Upon release, nanoparticles or free siRNA are able to interact with cell membranes and enter the cell, leading to gene silencing.
2.1. Polymer Selection and Electrostatics
Hydrogels are comprised of polymers, which may be of either natural or synthetic origin. Natural polymers include proteins such as collagen and fibrinogen or polysaccharides such as hyaluronic acid (HA), dextran, alginate or chitosan. Traditionally, naturally occurring polymers are biocompatible and possess motifs for cell adhesion and sites for biodegradation, depending on the polymer. However, because they derive from biological sources, these polymers may elicit immune responses, and they may exhibit lower mechanical properties when assembled into hydrogels.83 On the other hand, synthetic polymers are made from monomers such as vinyl acetate, acrylamide, and polyethylene glycol (PEG). Because of their synthetic nature, precise control over properties such as biomechanics and biodegradability is possible. RNAi delivery has been explored from both natural and synthetic polymers.84
With polymer selection comes differences in charge, which may play a major role in the design of hydrogels for RNAi delivery. Because of the anionic charge of nucleic acids and cationic charge of traditional nanoparticle-based nucleic acid delivery approaches, electrostatic interactions will directly influence the release of RNAi from hydrogels. Neutral hydrogels (e.g., comprised of dextran, PEG) are often used because they are electrostatically inert and will not directly interact with charged nucleic acids or nanoparticles.62,85 Anionic hydrogels (e.g., comprised of alginate, HA) have received less attention for RNAi delivery due to rapid release and issues with aggregation. For example, siRNAs were quickly released from alginate hydrogels (e.g., release over 1 week compared to 2 weeks in a collagen hydrogel)86 and it was reported that HA induced aggregation of nucleic acid polyplexes which limited transfection.74,87
Cationic polymers include but are not limited to polyethylenimine (PEI), chitosan, polyamidoamine, poly-β-amino-esters (PBAEs), and poly-L-lysine. Hydrogels from cationic polymers confer several advantages for RNAi delivery. First, cationic polymers alone can condense nucleic acids into polyplexes to promote transfection. As such, the incorporation of cationic polymers as polyplexes into hydrogels promotes the bioactivity of siRNA from hydrogels. Moreover, cationic polymers themselves can be assembled into gels, where the polymer itself can act as the transfection reagent.59,88 Moreover, cationic polymers are able to sequester siRNA for sustained release. Despite the significant advantages of using cationic polymers to formulate hydrogels, the major drawback has been cytotoxicity.89–91 The mechanism for this cytotoxicity has been described as multifaceted and warrants further investigation; however, studies have found that cationic polymers activate cellular necrotic and apoptotic signaling pathways.92 Moreover, polycations are known to induce formation of nanoscale defects in the cellular membrane, including pore formation and membrane thinning.93,94 While these effects may be favorable in the trafficking and permeability of membranes to nucleic acid polyplexes, they ultimately limit cellular viability and potential translation. There have been many advances toward improving the cytotoxicity of cationic polymers, especially PEI, that include modification with amino acids,95,96 sugars,97 lipids,98 and polymers.99
2.2. Crosslinking and Injectability
Various types of crosslinking mechanisms can be employed in hydrogel design, such as Michael addition, radical polymerization, and Schiff-base formation. The mode of hydrogel crosslinking has many important implications for RNAi delivery. For example, the use of free radicals in many chemical crosslinking techniques should be used with care, to avoid any concerns with cell compatibility and the stability of encapsulated cargo.100,101 Crosslinks can also be designed to be biodegradable, which can improve biocompatibility. In addition to hydrolytically degradable hydrogels, polymers can be crosslinked with enzymatically degradable peptide sequences that can accelerate the release of payloads specifically in the presence of proteolytic enzymes,46,48,102 creating responsive hydrogel systems. Other techniques, including photodegradation62,63 or the application of temperature or radiofrequency 103 can be designed into crosslinks for triggered release. Finally, the extent of hydrogel crosslinking can influence hydrogel mechanics and mesh size, which alters the diffusion of any entrapped molecules.104,105
Many hydrogels are also designed to be injectable. Whereas pre-crosslinked hydrogels require surgical implantation, injectable hydrogels can be administered in a minimally invasive fashion.106,107 In addition to alleviating the need for surgical intervention, injectable hydrogels can access otherwise difficult to reach areas.108 These injectable hydrogel systems crosslink through mechanisms such as photoinitiated crosslinking (via application of light and photoinitiator), Michael-type addition or Schiff base reactions (through mixing of two components), or supramolecular interactions (through shear-thinning and self-healing properties). In situ chemical crosslinking may be complicated by poor retention upon injection (e.g., polymer diffusion prior to crosslinking) or premature gelation that can lead to fracture upon injection and delivery failure.109,110 Supramolecularly crosslinked, shear-thinning hydrogels overcome these problems through self-healing,105,111–115 but are softer and have low mechanical properties.116 Thermal transitions can also be used for gelation when injected. For siRNA delivery, some of the most common functionalities introduced here are poly(organophosphazene)s55–57,117 and chitosan/β-glycerophosphates,88,118,119 which undergo sol-gel transitions at 37°C.
2.3. Nanoparticles for RNAi Delivery
Nanoparticle-hydrogel composites are by far the most common approach towards RNAi delivery with hydrogels (Figure 2). Here, RNAi nucleic acids are complexed into nanoparticles (and in the case of polymers, polyplexes) in order to promote cellular uptake and gene silencing. Much of this work builds from previous approaches for DNA transfection.33–35 The traditional approach towards nucleic acid carrier design is to encapsulate the cargo within a vehicle, such as a cationic liposome that mimics the physical characteristics of natural lipids found in the cellular membrane. Neutral lipids also exist as carriers for siRNA. Liposomes are comprised of hydrophobic chains with a positively charged head group that interacts with siRNA to form a lipid bilayer. As siRNA delivery has evolved, new lipid-like materials have been developed, termed “lipidoids”, which have improved transfection120–123 However, lipid preparations and liposomes can be thermodynamically unstable and are prone to aggregation,124 especially in serum or in a charged hydrogel.125 It is likely for these reasons that there have been no studies to date investigating liposomal RNAi transfections from hydrogels.
Cationic polymers have emerged as a more popular approach towards condensing nucleic acids into nanoparticles, which are also commonly referred to as polyplexes. In doing so, they are able to interact favorably with cellular membranes and stimulate endocytosis and endosomal escape. Endosomal escape is thought to occur through the proton-sponge effect, a process in which cationic polymers buffer the endosome pH as it acidifies, leading to accumulation of ions and osmotic pressure that bursts the endosome.126,127 An alternative mechanism for uptake has been proposed that involves fusion between nanoplexes and cell membranes.128 The most commonly used cationic polymer is PEI, which is available in linear and branched forms.129 PEI in its many forms has been used to deliver all types of nucleic acids from DNA to siRNA and has been widely established as a gold-standard in nucleic acid transfections both in vitro and in vivo.130,131 Other cationic polymers that have been used for RNAi delivery from hydrogels include poly-L-lysine,132,133 chitosan,36,119 polyamidoamine134,135 and PBAEs.136–138 As previously discussed, cationic polymers have natural cytotoxicity that should be thoroughly investigated for RNAi therapeutics.
In addition to identifying nanoparticles that promote RNAi gene silencing without affecting cellular viability, there should be strong consideration of the interactions between the hydrogel and the nanoparticle. Naturally, the size and charge of nanoparticles will directly affect these interactions; whereas a cationic nanoparticle will release in a sustained manner from an anionic hydrogel, a negatively charged nanoparticle may be repelled. There have also been previous reports of nucleic acid nanoparticles aggregating and deactivating within hydrogels, emphasizing the importance of nanoparticle/hydrogel interactions in engineering these nucleic acid delivery systems.71,74 The Segura group was able to partially overcome these problems through a method they term “Caged Nanoparticle Encapsulation” in which nanoparticles were coated with agarose to prevent aggregation with charged hydrogels.71,74,87 In some contexts, siRNA nanoparticles have been covalently linked to the hydrogel without loss in activity. For example, the Artzi group covalently tethered arginine-modified PBAE/siRNA nanoparticles to dextran-aldehyde in their hydrogel backbone through Schiff-base formation to sustain nanoparticle release.136
2.4. RNAi Nucleic Acid Conjugates
In addition to or in lieu of using a nanoparticle, siRNA or miRNA can be modified with different functionalities to achieve transfection from hydrogels (Figure 2).139–141 One of the most common modifications is cholesterol, which can be attached to 5’ or 3’ end of the sense, passenger strand, which does not interact with RISC. Many previous reports have shown that cholesterol modifications of siRNA and miRNA facilitate the uptake of RNA into cells with gene silencing, both with and in the absence of nanoparticles.142–147 Commercially, these modified siRNAs are available as Accell siRNAs, which can enter cells without a transfection reagent and have been used with success in vitro and in vivo.85,148–151 Additional modifications such as methylation, especially on the 2’ hydroxyl of the pentose ring where RNA is hydrolytically degraded, can promote RNA stability.5,152 Another common modification is the introduction of phosphothiorate bonds between nucleotides, which can also prevent RNA degradation.153,154 These modifications promote cellular uptake and can silence gene expression. However, the absence of a nanoparticle carrier renders siRNAs and miRNAs especially susceptible to nuclease degradation, and unprotected siRNAs (~7 nm in length and ~2 nm in diameter) may be rapidly released from hydrogels without a nanoparticle sequestrant.86,155,156 Whereas this has not been explored, the use of siRNA conjugates has potential to add additional complexity to siRNA release, where hydrophobic modifications like cholesterol may interact with hydrophobic components of the hydrogel.
3. In Vitro Applications of Hydrogel RNAi Delivery Systems
3.1. Designing Hydrogels for Sustained RNAi Release
In vitro assessment of hydrogel RNAi delivery systems is important towards their development to better understand potential release profiles and the activity of the released RNAi molecules. This area was reviewed in 2010;148 however, there has been significant growth since. The research reviewed in this section details key innovations in hydrogel design towards RNAi delivery.
The first report to investigate and tune the release of siRNA from hydrogels was by the Alsberg group, where they encapsulated Accell siRNA into three different hydrogels: calcium crosslinked alginate, photocrosslinked alginate, and collagen and explored the release of bioactive green fluorescent protein (GFP) siRNA to a GFP-expressing HEK293 line. 86 siRNA was rapidly released over one week from the calcium crosslinked alginate and photocrosslinked alginate hydrogels, and over two weeks for the collagen hydrogel, potentially due to charge interactions with the anionic alginate hydrogels that accelerated release. Incorporation of cationic PEI or chitosan into the alginate systems attenuated release, likely due to differences in cationic charge density. The released siRNA silenced GFP expression by up to 90% by six days. This was a fundamental study pivotal to siRNA delivery from hydrogels for establishing the role of backbone charge in sustaining release.
Recognizing the importance of polymer backbone charge, Nguyen et al. reported a hydrogel in which linear PEI was incorporated into a hydrolyzable and photocrosslinked dextran hydrogel to sustain siRNA release.85 Here, PEI methacrylate was polymerized with hydroxyethyl methacrylate modified dextran, to form hydrogels that were degradable over 17 days. Accell GFP siRNA was encapsulated and release kinetics were tuned with hydrogel weight percent and PEI concentration for up to 17 days, with high knockdown efficiency (~80%) of GFP from GFP-expressing HEK293 cells at 14 days. In a later report, siRNA and miRNA were released from PEG hydrogels (formed through thiol-acrylate reaction) for over 42 days and releasates were able to silence 80% GFP-expression in HEK293 cells.82 Likewise, Noggin siRNA or miR-20a were co-encapsulated with mesenchymal stem cells (MSCs) and promoted osteogenesis through knockdown of Noggin and PPAR-γ with elevation in bone markers including ALP, Runx2 and BSP over a 28 day time period.
Towards spatial control of siRNA delivery within a hydrogel, the Alsberg group engineered siRNA gradients into hydrogels. Here, Hill et al. incorporated a methacrylated dextran into two syringe pumps containing either no siRNA or siRNA with branched PEI polyplexes, respectively.81 With varying flow rates and a mixing unit, they were able to achieve a linear gradient of siRNA/PEI presentation within a dextran hydrogel. Release was sustained for up to 16 days from 4 different sections of the gradient siRNA hydrogel, dependent on the siRNA loading. GFP-knockdown in HEK293 cells was dependent on local presentation of the siRNA/PEI gradient. This study advanced siRNA delivery with complex hydrogel systems.
In another study, Patil et al. synthesized amphiphiles by adding lipopolyamine units to riboflavin.157 Conjugation of lipopolyamines of different lengths gave riboflavin the ability to form hydrogels in water through supramolecular nanofiber formation upon heating and cooling which were stable over four weeks. Moreover, the introduction of cationic polyamines to riboflavin allowed for complexation with anionic siRNA through electrostatic interactions. When hydrogels encapsulated fluorescein-tagged siRNA, there was significant release and uptake of fluorescently labeled VEGF siRNA rivaling that of lipofectamine in HeLa cells. Moreover, the delivery of polyamine-modified riboflavin hydrogels led to 50% knockdown of VEGF expression by ELISA from the hydrogel at 48 hours. The use of protein amphiphiles towards hydrogel formation and siRNA delivery is novel, although this investigation warrants further studies into sustained release and potency of siRNAs.
In an early report, Singh et al. delivered siRNA from hydrogels towards enhancing immune responses via recruitment of antigen presenting cells (APCs).29 Here, they formed poly(lactide-co-glycolide) (PLGA) microparticles with IL-10 siRNA. Branched PEI was conjugated to the surface of these PLGA microparticles to give them a positive surface charge and promote cellular uptake; these microparticles were able to sustain siRNA release for 35 days in a previous report.158 DNA (pgWizLuciferase) was also added to the surface of these microparticles to achieve dual delivery of siRNA with plasmid DNA, upon encapsulation in hydrogels formed via a Michael-addition reaction between a vinylsulfone-dextran and a 4-arm-PEG-thiol. Hydrogels also encapsulated the chemokine MIP3-α to promote APC migration. Primary bone marrow mouse APCs were cultured on top of siRNA-containing hydrogels and IL-10 gene expression was silenced up to 80% in migrated APCs compared to controls at five days. This co-delivery approach has many potential applications, especially in synergistically assisting in RNAi knockdown.
3.2. Stimuli-responsive Hydrogels for siRNA Delivery
As previously discussed, hydrogels can be engineered in ways that permit them to respond to external environmental cues (e.g., light, temperature, enzymes) to promote drug release, including towards RNAi delivery. The research in this section highlights the important role of “smart” hydrogels in tunable RNAi delivery.
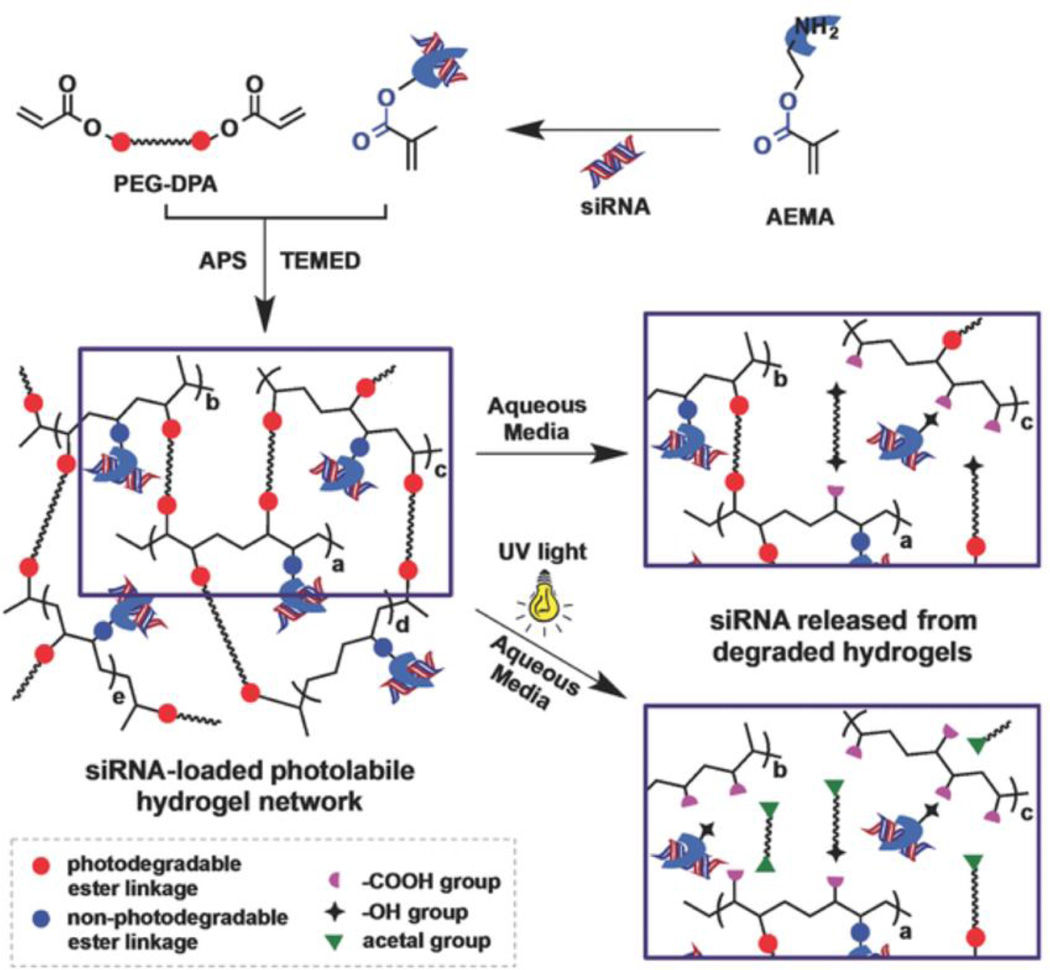
In one report, Hunyh et al. engineered an acrylated PEG hydrogel with photolabile ortho-nitrobenzene groups (PEG-DPA) to permit light-mediated degradation and subsequent siRNA release (Figure 3).62 To incorporate and sustain siRNA release, aminoethyl methacrylate, a cationic molecule, was also crosslinked into the PEG hydrogel. Hydrogels encapsulated Accell GFP siRNA and release was tuned over two weeks by controlling UV intensity and exposure times. Importantly, released GFP siRNA achieved up to 80% knockdown of GFP expression in a GFP-expressing HeLa line at 14 days, which was significantly higher than without UV exposure. This general approach was further applied towards osteogenesis.63 8-arm PEG-thiol was crosslinked to PEG-DPA, and H2O2 was added to form secondary crosslinks between thiols. Noggin siRNAs and miR-20a were co-delivered and were able to transfect hMSCs and induce calcium deposition at early times, with no differences observed with UV-treated and non-UV treated hydrogels.
Figure 3.
Photolabile siRNA delivery through hydrogels formed from PEG-o-nitrobenzene-acrylate permits siRNA release in response to UV light through o-nitrobenzene mediated photo-isomerization. Reproduced with permission.62 Copyright 2015, Wiley.
Radiofrequency has also been used to stimulate siRNA release from a hydrogel.103 N-isopropylacrylamide and acrylamide were crosslinked with bis-acrylamide in the presence of APS/TEMED to form a hydrogel with a critical solution temperature above which it becomes a liquid and releases the cargo. Within these hydrogels, magnetic PEG-diphosphate nanoparticles (80 nm) were co-encapsulated with siRNA, so that radiofrequency heats the particles and increases the hydrogel temperature above the critical solution temperature to stimulate siRNA release. The authors reported siRNA release over seven minutes, wherein radiofrequency induced the release of up to 80% of siRNA, compared to <5% of siRNA without it. They also showed that multiple bursts of radiation could be applied in order to achieve siRNA release. This was the first and only report of a radiofrequency application towards siRNA delivery from a hydrogel, but is limited by extremely short release times and without evidence for bioactivity.
Towards temperature-sensitive siRNA delivery, Liu et al. first formed siRNA polyplexes with poly-L-lysine, 159 which has been previously demonstrated to facilitate siRNA cellular uptake and silencing.132,133 These siRNA polyplexes were encapsulated into a hydrogel consisting of glycol chitosan, a water-soluble chitosan, and a benzaldehyde-capped tri-block copolymer of poly(ethylene oxide)-poly(propylene oxide)-poly(ethylene oxide). siRNA release was measured at two temperatures and a 4-fold increase in siRNA release (122.5 µg siRNA vs. 23.8 µg siRNA) was observed when the hydrogels were incubated at 60°C compared to at 37°C. This demonstrates the role that temperature can play to tune siRNA release; however, this report is limited by the lack of siRNA silencing bioactivity and short release windows over less than a day.
Towards enzymatic control over siRNA release, Saito et al. used cationic gelatin siRNA nanoparticles incorporated into gelatin hydrogels.160 Cationic gelatin was made through the chemical introduction of ethylene diamine to the carboxyl groups of gelatin and was used to form polyplexes with siRNA. The polyplexes were encapsulated with unmodified gelatin and crosslinked through the addition of glutaraldehyde. siRNA release was tuned through the presence of collagenase, which permitted release through hydrogel erosion with 100% release by 48 hours, whereas ~40% release was observed after the first 24 hours without collagenase and negligible release was observed thereafter. Luciferase siRNA was released from hydrogels and added to Colon26-luc cells, a mouse colon carcinoma line, in vitro. In hydrogel/siRNA treated cells, luciferase expression was decreased by up to 40% at nine hours after treatment compared to controls.
In another study with enzymatically-degradable gels, lysozyme was used to release siRNA from chitosan hydrogels.161 Chitosan is a naturally-occurring, cationic polymer that has been used to condense nucleic acids into polyplexes for transfection.36,162 β-glycerophosphate addition allowed chitosan to form a hydrogel at 37°C through hydrophobic interactions for the encapsulation of siRNA, with chitosan improving transfection. siRNA to RANK, a protein that promotes abnormal osteoclast activity in periodontal disease, was released and gene silencing of ~60% observed in RAW264.7 cells by 9 days. Significantly faster release was observed in the presence of lysozyme compared to buffer, wherein lysozyme liberated up to 50% of siRNA by two weeks, compared to less than 20% without.
4. In Vivo Applications of Hydrogel RNAi Delivery Systems
This section covers the progress from in vitro delivery systems that highlight the utility of hydrogel properties for RNAi delivery to applications in vivo. Here, we discuss the most common applications of such systems towards cancer, inflammation, bone regeneration and cardiovascular disease.
4.1. Cancer
The majority of applications of RNAi hydrogel systems have been towards cancer, which is commensurate with a large body of knowledge in siRNAs for cancer therapeutics. The enhanced permeability and retention (EPR) effect and lack of lymphatic drainage have helped to push forward nanoparticle therapeutics in this application, where nanoparticles can accumulate based on increased vasculature.163,164 For these reasons, local targeting to tumor tissues by hydrogels may have increased RNAi specificity towards cancer cells compared to surrounding tissue; as such, several reports of hydrogel/RNAi delivery have shown great promise towards cancer therapies. Many of these systems involve intratumoral injection of hydrogels, which may not be warranted for metastatic, later stage cancers or cancers in sites that are difficult to access.
One of the first applications of hydrogel siRNA delivery to cancer was the treatment of melanoma and breast cancer xenografts in a mouse model.165 Here, chitosan/β-glycerophosphate hydrogels with encapsulated Alexa555-siRNA were injected intratumorally into a melanoma (A375SM)-bearing mouse, displaying a sol-gel transition upon injection. As previously discussed, chitosan is able to effectively condense nucleic acids into nanoparticles to facilitate siRNA uptake and gene silencing.162,166,167 Hydrogel injections helped to localize siRNA fluorescence to tumor cells compared to hydrogel or siRNA alone. siRNA to tissue transglutaminase 2 (TG2) was encapsulated and injected into A375SM mice. Decreased TG2 expression was observed at 10 days and significant decreases in tumor volume, especially when co-administered with docetaxel, a chemotherapy drug, were seen at 30 days. In a similar MDA-MB-231 model of breast cancer, hydrogel/siRNA/docotaxel inhibited tumor growth by up to 92%. Single injections of siRNA alone had no antitumoral effect, suggesting that chitosan was important in facilitating uptake and sustained release.
A report from Guo et al. later demonstrated hydrogel-mediated, local gene silencing through shRNA DNA plasmid transfection for cancer therapy.168 Here, linear PEI was modified with PEG to complex DNA for transfection. PEI-PEG was successful in transfecting luciferase in two breast cancer lines, MDA-MB-231 and MCF-7. shRNA was selected against Akt1, formed into PEI-PEG complexes, and co-encapsulated with paclitaxel, a chemotherapeutic, in an injectable and thermosensitive linoleic acid-modified triblock copolymer of poly(ethylene oxide)-poly(propylene oxide)-poly(ethylene oxide)) hydrogel. In a human tumor xenograft of MDA-MB231, a single hydrogel injection was responsible for a significant reduction in tumor volume of up to 80% with a synergistic effect provided by paclitaxel at 26 days. Akt1 levels were significantly decreased in the tumor and the combination of both drugs increased apoptotic genes, decreased pro-oncogenic genes (Bcl2, Bcl1), and decreased tumor angiogenesis and VEGF levels. This was the first application of a hydrogel/shRNA delivery system towards cancer therapeutics.
In a later report, Ma et al. delivered an shRNA from a hydrogel in osteosarcoma therapy.169 Poly-L-lysine-modified PEI formed nanoparticles with shRNA against polo-like kinase 1 (PLK1), a signaling molecule that is overexpressed in osteosarcoma. Nanoparticles were co-encapsulated with doxorubicin into a PLGA-PEG-PLGA triblock copolymer, a thermosensitive polymer that displays a sol-gel transition at 37°C. Hydrogels were injected in a human osteosarcoma xenograft model of Saos-2 cells. From a single injection, hydrogels with shRNA and doxorubicin significantly reduced tumor size by 16 days, and tissue analysis suggested that PLK1 mRNA levels were significantly lower in hydrogels treated with doxorubicin and shRNA together. A series of markers for cellular proliferation and oncogenesis were also measured, which supported a beneficial role for the PLK1 shRNA co-delivery with doxorubicin in vivo. The two reports by Guo et al.168 and Ma et al.169 paralleled one another in the delivery of a modified PEI from a triblock copolymer forming a thermosensitive hydrogel.
A PBAE-based nanoparticle hydrogel/system was reported in 2015 by the Artzi group.136 PBAEs condense nucleic acids to form polyplexes.137,138,170 In this report, Segovia et al. formed nanoparticles between siRNA and arginine terminated-PBAEs, which were then encapsulated within a hydrogel from polyamidoamine (PAMAM) dendrimers crosslinked with dextran aldehyde. Arginine modified-PBAEs were able to condense GFP siRNA and silence GFP expression up to 54% relative to controls in GFP-expressing MDA-MB-231 cells. Amines on the nanoparticles reacted with the dextran aldehyde to link to the network. siRNA release was sustained from the nanoparticle hydrogels, with 90% release by 6 days. It was believed that physically entrapped nanoparticles were released first, and then covalently attached nanoparticles later. siRNA to luciferase was encapsulated with the hydrogels and implanted into mice bearing luciferase in MDA-MB-231 breast cancer cells. Injections reduced luciferase expression by as much as 70% at 6 days with hydrogels, whereas a single nanoparticle injection only led to a 20% reduction in luciferase expression.
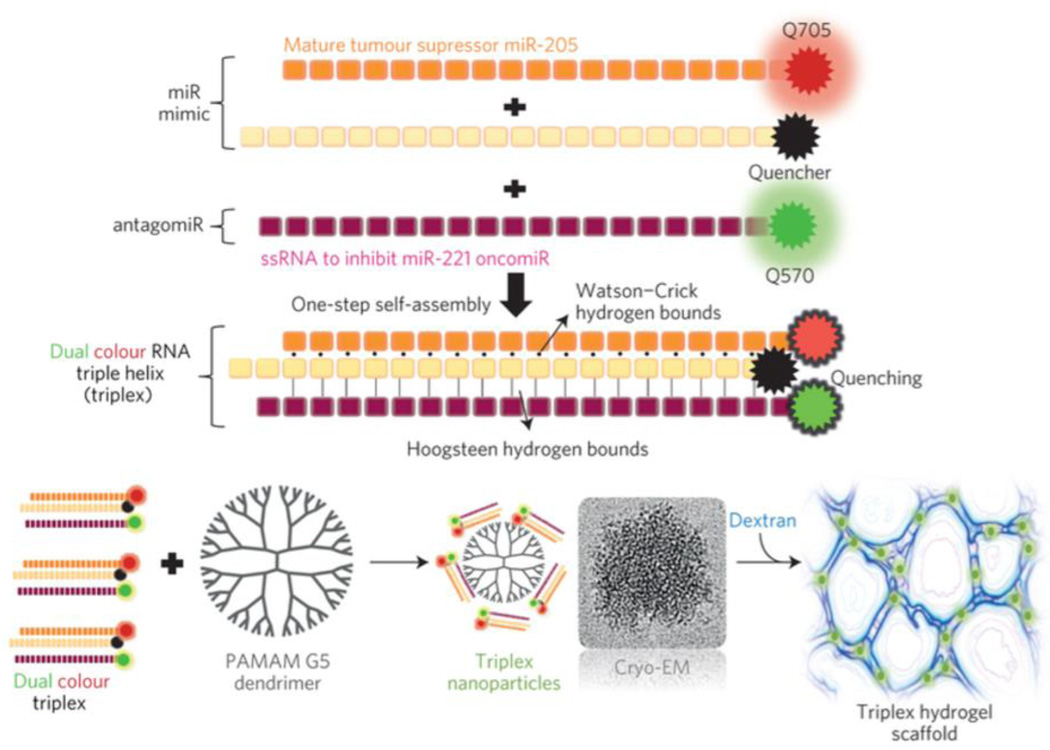
In a subsequent report by the Artzi group, Conde et al. delivered a miRNA triple helix consisting of a miRNA mimic (agomir) and miRNA inhibitor (antagomir) incorporated into the same PAMAM-dextran aldehyde hydrogel they previously used (Figure 4).142 Here, miR-205 (dsRNA) and miR-221 antagomir (ssRNA) were hybridized into a triple helix. Whereas miR-205 has been shown to have anticancer properties, miR-221 has been shown to have tumorigenic effects necessitating antagonism. The miR-205 sense strand was also modified with a 3’ cholesterol to facilitate cellular uptake and protection from endonuclease degradation. RNA triple helices consisting of fluorescently-labeled miR-205 mimics and a ssRNA inhibitor of miR-221 formed triple helix nanoparticles 50 nm in size and were complexed with PAMAM through electrostatic interactions to form aggregates approximately three microns in size. Triple helices were uptaken into cells significantly more (99.8%) than double helices (38%), likely due to micropinocytosis. Hydrogels were implanted adjacent to tumors in mammary fat pads of SCID hairless congenic mice. Thirteen days following hydrogel implantation, the mice tumor xenografts were reduced by 90% in size compared to just 50% from miR administration separately and only 25% and 35% in doxorubicin or paclitaxel-loaded hydrogels, respectively. This was the first report of the co-delivery of a miR agomir and antagomir within the same system, demonstrating synergism in their capacity to form a triple helix. Robust transfection and uptake both in vitro and in vivo from their PAMAM/dextran hydrogel suggests strong translational potential.
Figure 4.
miR-205 mimic can interact with miR-221 antagomiR via Hoogsteen interactions to form RNAi triple helices. These interact with polyamidoamine (PAMAM) dendrimers to form nanoparticles that can be further crosslinked into a hydrogel by addition of dextran aldehyde. Reproduced with permission.142 Copyright 2015, Nature Publishing Group.
Towards the treatment of gastric cancer, Peng et al. developed a type 1 collagen hydrogel encapsulating a commercial linear PEI/siRNA complex.171 Id1 siRNA, which is thought to inhibit proliferation and migration of cancer cells, was encapsulated with PEI and silenced up to 60% of gene expression in SGC-7901 gastric cancer cells. Downstream targets, including Cyclin D1 and p-AKt, were also downregulated, consistent with siRNA activity. Cellular proliferation, the direct target of Id1 siRNA, was significantly inhibited at 14 days as well when compared to controls. In a gastric cancer xenograft, hydrogels were injected hypodermally, which decreased tumor size at 4 weeks. Moreover, immunostaining of cellular targets of Id1 siRNA were consistent with gene silencing activity, with less cyclin D1 and more P21, indicating suppression of proliferation. Proliferating cell nuclear antigen also decreased in siRNA/PEI/collagen treated samples, suggesting inhibition of proliferation.
As an alternate approach to encapsulation in hydrogels, the Song group directly assembled polyplexes into hydrogels for siRNA release.55,57,59,117 In their first report, Kim et al. engineered an injectable PEI-based thermosensitive hydrogel with this approach (Figure 5).59 PEI was modified with poly(organophosphazene) and a biodegradable ester linkage to enable thermogelation at 37°C into a hydrogel. At room temperature, modified PEI-poly(organophosphazene)s were able to form polyplexes and then assemble into hydrogels for sustained release of a Cy5.5-labeled siRNA for up 28 days. Towards anticancer therapy, siRNA was selected against Cyclin B1, a regulator of the cell cycle and encapsulated into hydrogels. Cy5.5-siRNA was sustained for 21 days intratumorally and Cyclin B1 siRNA reduced tumor growth at 30 days. This report was incredibly novel not only for its anticancer response and sustained release but also for its engineering design principle. The idea of using PEI as a member of the backbone and as a transfection reagent for condensing siRNA into polyplexes was previously undescribed.
Figure 5.
Modification of PEI with poly(organophosphazene) to form polyplexes with siRNAs. Upon gelation at 37°C, hydrogels form that dissociate into siRNA polyplexes over time. Reproduced with permission.59 Copyright 2012, American Chemical Society.
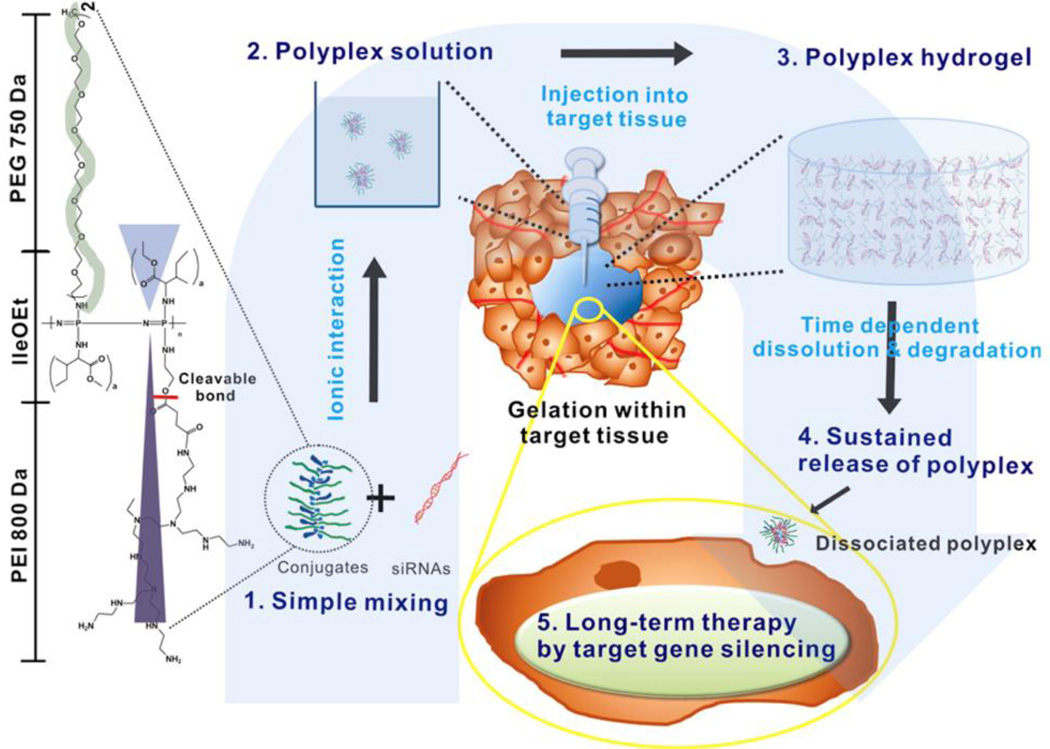
Following this initial report, a series of studies used this poly(organophosphazene) system towards other cancer applications. Hydrophobic isoleucine ethylester and hydrophilic PEG were introduced to PEI-poly(organophosphazene) to form an amphiphile to co-deliver an siRNA to BCL-2 with docetaxel, a hydrophobic drug that assisted in polyplex condensation.55 siRNA release was sustained over one month in vitro, and Cy3-siRNA signal was present up to 41 days in vivo. Hydrogel injections of BCL-2 siRNA with doxotaxel were able to sustain MDA-MB-231 breast cancer tumor inhibition in vivo at 30 days, ~80% compared to a PBS control. In another study, protamine, a potent transfection reagent,172,173 was conjugated to poly(organophosphazene) to increase electrostatic interactions.117 Protamine/siRNA polyplexes were assembled into hydrogels. Gels were observed up to 24 days intratumorally upon injection and significantly decreased tumor growth with siRNA to vascular endothelial growth factor (VEGF).
The PEI-poly(organophosphazene) was later modified with folate to form polyplexes that specifically targeted the folate receptor (FR).57 Modification by folate allowed for condensation of siRNA into polyplexes. When encapsulated with VEGF siRNA, VEGF was silenced to 50% from siRNA micelleplexes. When injected in a mouse tumor xenograft model, hydrogels with Cy3-siRNAs persisted for up to 10 days with the hydrogel. With siRNA to VEGF, the hydrogel significantly reduced tumor volume over 21 days up to 50% and reduced VEGF expression in the tumor by 50%. Moreover, by doubling the amount of siRNA from 100 µg to 200 µg, they were able to achieve a more robust gene silencing effect demonstrated by relative tumor volume growth inhibition (>80%) at 30 days. The idea of a transfection reagent being used as part of a hydrogel backbone has interesting implications towards siRNA delivery, especially in being able to deliver and transfect cells with unmodified, naked siRNA. It is likely that previous applications of chitosan hydrogels in which siRNAs were also encapsulated naked119,161,165 were behaving through a similar idea, and further investigation should look into the release of biologically active polyplexes as the mechanism through which siRNA activity was potentiated with these systems.
4.2. Inflammation
RNAi delivery from hydrogels has been used to mediate inflammation, including towards the treatment of hypertrophic scarring, a process characterized by excessive collagen deposition. Zhao et al. delivered a TGFβ-337 siRNA complexed with Entranster-R, a commercially available transfection reagent. TGFβ-337 is a protein that when silenced can inhibit cellular proliferation, arrest cells in G1, and induce apoptosis of fibroblasts in hypertrophic scars. Polyvinylpyrrolidone, polyvinyl alcohol, glycerol and water were mixed under vacuum for hydrogel formation and polyplex encapsulation, coated onto a polyethylene glycol terephthalate film for adhesion, and then applied transdermally to a rat. Compared to controls, hydrogel/Entranster penetrated significantly more into the skin at 24 hours, with increased siRNA fluorescence in the dermis. In mice with hypertrophic scars treated with hydrogel/siRNA combinations, there was a significant decrease in the volume change of hypertrophic scars, and the orientation of collagen fibers was more similar to that of normal dermis, with more type 1 collagen. 174 When applied topically, this system promoted both sustained and local delivery.
Towards the treatment of colonic inflammation in inflammatory bowel disease, Laroui et al. targeted CD98, a protein highly expressed on epithelial and immune cells that promotes crucial roles in homeostasis and the innate immune response in the gut.175 CD98 siRNA was complexed with PEI and polyvinyl alcohol and encapsulated into an alginate/chitosan hydrogel crosslinked with calcium and sulfate. Release was pH dependent, such as in the colon. When colitis mice were gavaged with hydrogel-encapsulated siRNA nanoparticles daily for 8 days, the nanoparticles trafficked across the mucosal layer and invaded submucosal epithelium. Weight loss, myeloperoxidase, a measure of inflammation, and pro-inflammatory cytokines and markers all decreased with treatment. In a later study, TNF-α siRNA/PEI complexes with PLA-PEG nanoparticles were targeted to macrophages by adding the Fab portion of an antibody targeting F4/80, a glycoprotein expressed by murine macrophages.176 These nanoparticles decreased TNF-α expression from RAW macrophages and then decreased markers of inflammation in vivo when delivered with a hydrogel. As an siRNA delivery system, this hydrogel is a “smart” material in the ability to release payload only in the pH in the colon. Moreover, the ability to deliver a hydrogel through an oral gavage has not been used previously for siRNA delivery.
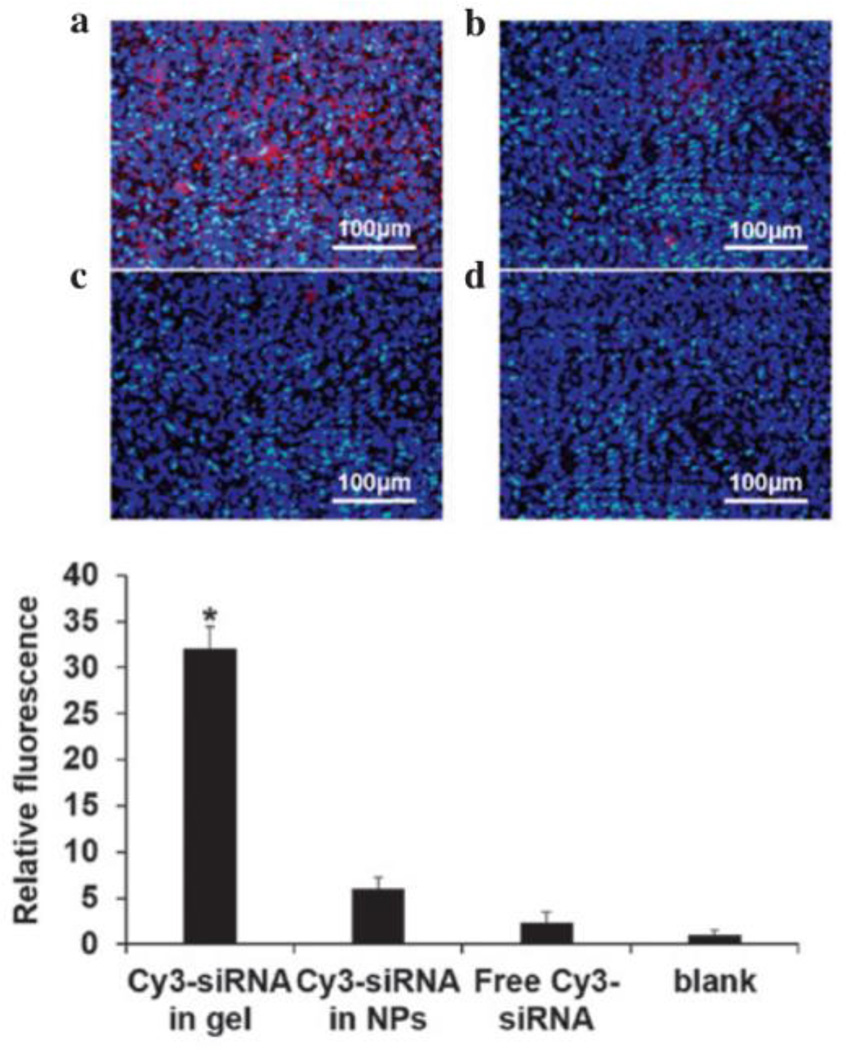
Another application of hydrogel RNAi delivery was toward mucosal inflammation in sinusitis (Figure 6).119 Here, Cao et al. engineered an injectable hydrogel from chitosan/β-glycerophosphate. siRNA release was pH dependent - acidified PBS led to accelerated release (70% by two weeks) compared to moderate release in neutral pH (40%). VEGF siRNA released to seeded bronchial epithelial cells silenced up to 40% of VEGF over one week. siRNA was delivered to mucosal cells in a chronic rhinutis rat model, and hydrogels/siRNA were injected bilaterally into the maxillary sinus. After 2 weeks, there was a significant decrease in mucosal thickness in the sinus mucosa, suggesting inhibited inflammation by silencing VEGF.
Figure 6.
Hydrogel/Cy3-siRNA (a) significantly enhanced retention upon injection compared to nanoparticles (b) or free siRNA (c) alone compared to untreated (d), as quantified via fluorescence intensity. Reproduced with permission.119 Copyright 2015, Cao et al.
Kanazawa et al. reported on a hydrogel for RNAi delivery towards inflammation, where they engineered a hydrogel that encapsulated RelA siRNA, a gene thought to improve the pathogenesis of atopic dermatitis through silencing of NFkB.177 siRNA was complexed with TAT peptide and AT1002 into nanoparticles and further encapsulated into hydrogels made from sericin, a protein created in the production of silk. Sericin contains many polar side chains, exhibiting hydrophobic characteristics that drive β-sheet structures in the hydrated, hydrogel state. TAT peptide functions as a cell penetrating peptide and transfection vehicle and AT1002 is a peptide that functions in opening tight junctions to permit transdermal siRNA delivery. In a model for dermatitis, hydrogels were applied topically over the ears of mice. When hydrogel/RelA siRNA was applied three times a week for two weeks after sensitization for atopic dermatitis in mice, improvements in ear thickness and clinical skin severity were seen. Immunohistochemistry also demonstrated the absence of parakeratosis, eosinophils, and mononuclear cells in the hydrogel/siRNA treated groups.
Towards a different inflammatory outcome, Browne et al. engineered a system to reduce the foreign body response in organs and transplants.178 Here, they co-delivered IL-6 siRNA with a plasmid encoding for endothelial nitric oxide synthase (eNOS). IL-6 can be a pro-inflammatory cytokine, especially in recruiting monocytes and neutrophils. eNOS has been shown to inhibit IL-6 expression and is pro-angiogenic. Here, they encapsulated siRNA and DNA with commercially available polyamidoamine dendrimers (Superfect) to form polyplexes and then further into collagen microspheres and then into collagen hydrogels. Hydrogels were implanted subcutaneously in rats and decreased the volume fraction of inflammatory cells compared to controls. By combining with eNOS DNA, they were able to decrease the volume fraction of inflammatory cells, found at 7 days, and significantly increased blood vessel density of host tissue at 14 days. Protein expression revealed an overall reduction in inflammatory cytokines with increased expression of angiogenic factors in treatment groups. Together, this system suggests that hydrogels could be used to enhance the foreign body response and promote the vascularization of transplanted organs while decreasing inflammation.
4.3. Bone Regeneration
RNAi delivery for bone regeneration is still relatively new and there are only a few reports in this area. Manaka et al. encapsulated Noggin siRNA into a biodegradable hydrogel consisting of a poly-d,L-lactic acid-p-dioxanone-polyethylene glycol block copolymer (PLA-DX-PEG).179 Hydrogels were implanted with rhBMP-2 and either with or without noggin siRNA into the dorsal muscle pouch of mice to control ectopic bone formation. At two weeks, hydrogels in which Noggin siRNA was delivered with BMP-2 led to formation of new bone with increased mineral content. An interesting aspect of this study was the absence of any vehicle for siRNA internalization. For these reasons, the authors argue for the internalization by endocytosis of PLA-PEG polymers in complex with siRNA that are formed from the hydrogel. This mechanism has not been previously described and should be further characterized. As a delivery system, this hydrogel is novel in delivering a growth factor (BMP2) with an siRNA to an antagonist of that growth factor (Noggin). The ability to achieve functional outcomes in vivo warrants further experiments to improve therapeutic efficacy and a better understanding of the role that the hydrogel plays towards sustained delivery.
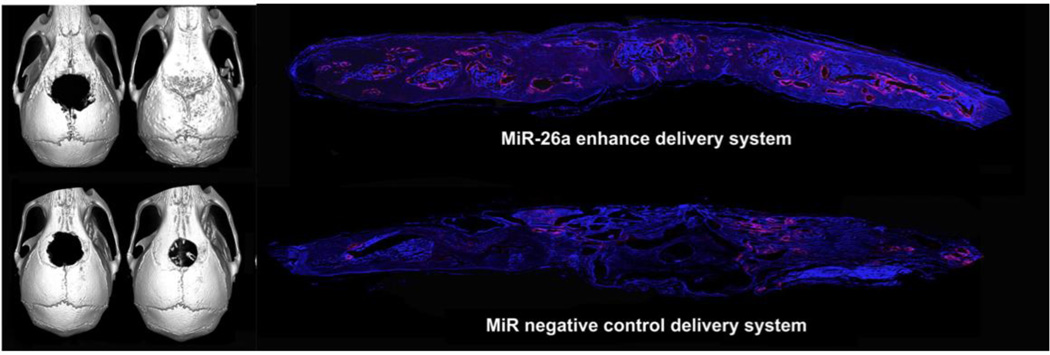
A report by Li et al. delivered miR-26a through a hydrogel in vivo to promote bone healing through angiogenesis and is one of few reports to deliver microRNAs by hydrogel.180 In this report, they identified miR-26a as promoting the coordination of angiogenesis with osteogenesis during bone regeneration. Using a miR-26a mimic or inhibitor on MSCs, they showed that microRNA transfection by siPORT NeoFX (Ambion) promoted the expression of VEGF and Ang1 concurrently with BMP2, Runx2, osteocalcin and collagen over two weeks. hMSCs transfected with miR-26a were found to promote vascular and bone tissue formation. Subsequently, they incorporated Cy3-labeled, 3’ cholesterol-modified, 2’ O-methyl modified, single-stranded miR-26a mimic into a biodegradable hydrogel consisting of thiolated heparin, thiolated HA, and PEG crosslinked through Michael addition chemistry. The cholesterol-modified microRNAs led to therapeutic, endogenous gene silencing in vivo without the need for a transfection reagent. To test the hydrogel-miR system in vivo, a 5-mm sized calvarial bone defect was created in nude mice. Hydrogel-miR or controls were implanted into the defect, and bone formation and vascularization was assessed after 12 weeks. From histological examination, newly formed bone coordinated with vessel size and area density, which was significantly higher in hydrogel-miR groups compared to controls. By micro-CT, hydrogel-miR groups led to complete repair by 12 weeks, wherein controls, including a hydrogel-MSC group only led to moderate regeneration (Figure 7). The ability to completely repair bone in the calvarial defect model suggests that this is a powerful platform.
Figure 7.
Complete regeneration of calvarial bone defects was observed with hydrogel/miR-26a treatment compared to hydrogel/miR controls, including increased vascularization by immunohistochemistry at 12 weeks. Reproduced with permission. 180 Copyright 2013, Elsevier.
4.4. Cardiovascular Disease
One of the first examples of an siRNA being delivered through a hydrogel was towards MMP2 inhibition in hydrogel-coated carotid stents to prevent atherosclerosis by Lie et al.181 Here, pullulan, a naturally occurring polysaccharide, was functionalized with diaminoethylamine so that it would complex siRNA through electrostatic interactions into nanoparticles. These nanoparticles were mixed with unmodified pullulans and dextrans, which were physically crosslinked with sodium-trimetaphosphate. Cationic pullulan was able to sustain siRNA release from hydrogels of ~20% over two hours compared to neutral hydrogels, where 100% was released within 10 minutes. siRNA was then swollen into the stent-covered hydrogels. A balloon catheter was used to induce right carotid abrasion in a rabbit model, and stents were implanted after 15 days. After 24 hours, Tamra-tagged siRNA showed clear uptake of siRNA upon implantation into rabbit carotids. siMMP2 was able to silence MMP2 activity of about 30% in cationized pullulan groups.
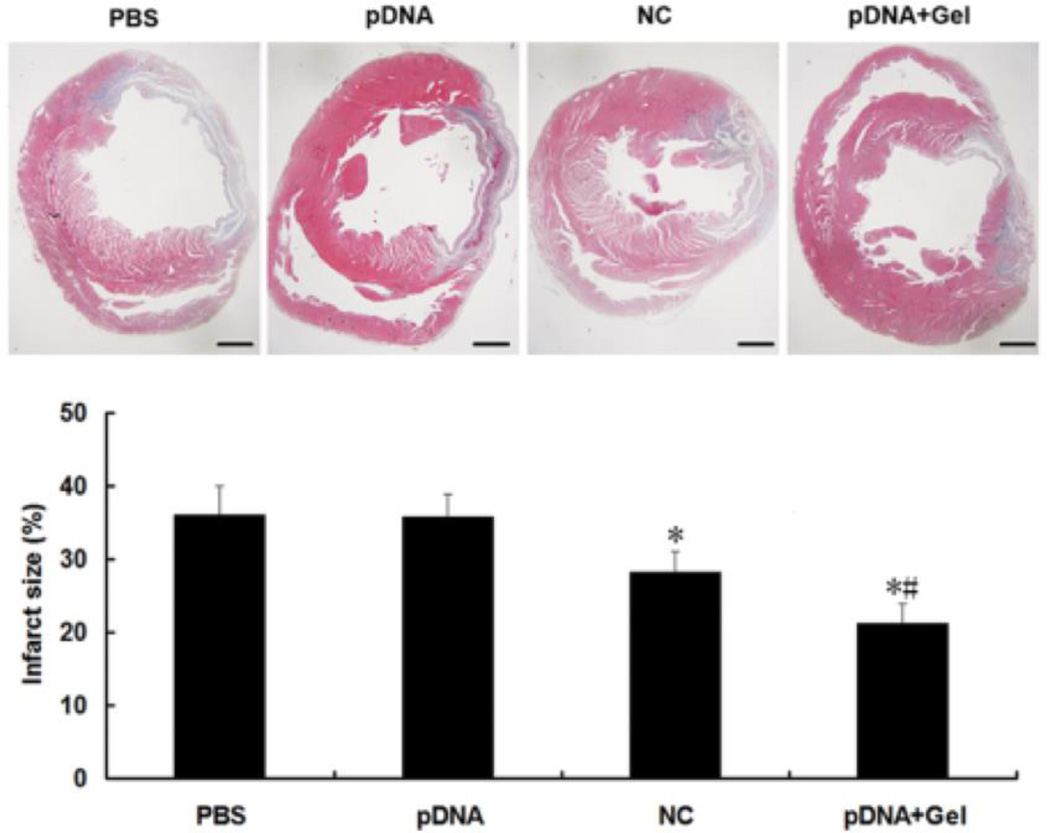
Wan et al. reported a hydrogel/RNAi combination towards a cardiovascular application, where they delivered an shRNA DNA plasmid against angiotensin converting enzyme (ACE).182 ACE has been shown to enhance apoptosis, increase infarct size, and lead to heart failure after myocardial infarction.183,184 A biodegradable, thermoresponsive dextran hydrogel modified with a hydrophobic poly(e-caprolactone)-2-hydroxyethylmethacrylate and a poly(N-isopropylacrylamide) thermoresponsive group was used. Within this hydrogel, ACE shRNA plasmids complexed to Lipofectamine 2000 were encapsulated. In a rat model of infarction, the left coronary artery was ligated and rats were injected with 4 × 25 uL of gel/shRNA into the infarcted region. From their plasmid, they co-expressed GFP, which was present 30 days after injection. Moreover, hydrogel/shRNA reduced infarct size 21%, apoptotic index 11.4%, and ACE mRNA by 50% at 30 days, suggesting a cardioprotective effect in rats (Figure 8). This is the only example of RNAi delivery from a hydrogel leading to improved cardiac function after myocardial infarction. In this regard, the ability to achieve such outcomes suggests that additional targets should be explored and a better understanding of hydrogel behavior in a contractile environment is warranted.
Figure 8.
Reduction in infarct size was observed with hydrogel+shRNA delivery when compared to controls after MI. Reproduced with permission.182 Copyright 2013, Wiley.
5. Summary
As outlined here, there is tremendous potential in the ability to achieve local and sustained gene silencing through the use of hydrogels. The evidence of this technology has been promising, showing antitumorigenic effects, improved bone healing and cardiac function, and modulation of immune responses over long periods in vivo. The variety of systems mentioned demonstrate how hydrogels can be tailored with various properties to address specific, unique needs for each application. Great promise was especially evident from hydrogel/RNAi systems in cancer and bone regeneration.
While hydrogels demonstrate potential for pushing RNAi therapies forward, there are limitations that should be acknowledged. First, hydrogels can sustain the delivery of a single amount of RNAi over time periods of weeks. This can be a disadvantage, as it decreases the concentration that cells are exposed to at any given time window and may not achieve the physiological response that serial, systemic injections (or serial local injections) may achieve. Thus, with hydrogel delivery there is a trade-off between sustained delivery and bioactivity. Second, hydrogels themselves carry a host of their own potential issues, including degradability, clearance and a foreign body response. These should be further investigated in in vivo settings. Finally, hydrogels are unable to target multiple organs at once and thus may be inferior to systemic delivery systems for processes like metastatic cancer.
Compared to other bulk delivery systems, hydrogels may have unique advantages in tunability of properties, ease of cell adhesion and migration, and injectability. Their high water content also improves the tissue response. Despite these advantages, other bulk systems should also be considered as RNAi delivery vehicles, which may have improved properties such as towards mechanical stability and representing the 3D structure of tissues.
Towards the development of therapies, an improved fundamental understanding of release and uptake from hydrogels is warranted. Specifically, in vitro characterization of how bulk material properties affect drug release will permit improved tuning of release properties in vivo. The structure and size of nanoparticles should also be better explored in the context of these hydrogel delivery approaches. Micropatterning approaches can potentially be used to as another mechanism of controlling and understanding cellular responses to RNAi.185 Lastly, an improved understanding of different methods of cellular trafficking can help in the design and characterization of novel polymeric methods towards in vivo application. These should be explored by cell type and organ system to promote the application of hydrogel RNAi therapeutics in a range of tissues that are previously unexplored.
Towards sustained delivery and improved patient adherence, hydrogels should be explored to various endpoints in vivo and RNAi molecule release should be characterized spatially and temporally. Further experiments should assess the manner by which hydrogels either become incorporated or are degraded by the body. For improved translation, injectable hydrogels should be considered over implantable ones that require invasive surgical interventions, especially in organs such as the heart, which can be accessed percutaneously by catheter. Future technologies will also strongly benefit from the ability to externally control RNAi release to achieve on-demand and temporal control over gene silencing.
Acknowledgments
The authors would like to acknowledge funding to LLW via a predoctoral fellowship from the American Heart Association and the National Institutes of Health (F30 HL134255).
Biographies

Jason A. Burdick is currently a Professor in the Department of Bioengineering at the University of Pennsylvania. His current research interests are in the design of biomaterials, particularly hydrogels, for applications in drug delivery and regenerative medicine. These hydrogels are typically designed from natural polysaccharides and are processed for injection and with techniques such as 3D printing and electrospinning. Applications include controlling stem cell fate, musculoskeletal tissue regeneration, and cardiac repair.

Leo L. Wang graduated from the University of Pennsylvania in 2012 with his B.A. in Biochemistry and M.S. in Chemistry, where he studied platelet biology with a focus on gene editing of megakaryocytes. He stayed at the University of Pennsylvania to pursue his M.D./Ph.D. in a dual-degree program through the School of Medicine and School of Engineering. In 2014, he joined the Burdick Polymeric Biomaterials Laboratory as a Ph.D. student to study injectable hydrogels for cardiac RNAi therapeustic delivery.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Devi GR. Cancer Gene Ther. 2006;13(9):819–829. doi: 10.1038/sj.cgt.7700931. [DOI] [PubMed] [Google Scholar]
- 3.Mollaie HR, Monavari SHR, Arabzadeh SAM, Shamsi-Shahrabadi M, Fazlalipour M, Afshar RM. Asian Pac. J. Cancer Prev. 2013;14(12):7045–7056. doi: 10.7314/apjcp.2013.14.12.7045. [DOI] [PubMed] [Google Scholar]
- 4.Mansoori B, Sandoghchian Shotorbani S, Baradaran B. Adv. Pharm. Bull. 2014;4(4):313–321. doi: 10.5681/apb.2014.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitehead KA, Langer R, Anderson DG. Nat. Rev. Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okumura A, Pitha PM, Harty RN. Proc. Natl. Acad. Sci. U. S. A. 2008;105(10):3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobbin ML, Rossi JJ. Annu. Rev. Pharmacol. Toxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]
- 8.Hannon GJ. Nature. 2002;418(6894):244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 9.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 10.Sarett SM, Nelson CE, Duvall CL. J. Control. Release. 2015;218:94–113. doi: 10.1016/j.jconrel.2015.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rooij E, Olson EN. Nat. Rev. Drug Discov. 2012;11(11):860–872. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Rooij E, Kauppinen S. EMBO Mol. Med. 2014;6(7):851–864. doi: 10.15252/emmm.201100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore CB, Guthrie EH, Huang MT-H, Taxman DJ. Methods Mol. Biol. 2010;629:141–158. doi: 10.1007/978-1-60761-657-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanya S, Kim S-S, Manjunath N, Shankar P. Expert Opin. Biol. Ther. 2010;10(2):201–213. doi: 10.1517/14712590903448158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lam JKW, Chow MYT, Zhang Y, Leung SWS. Mol. Ther. Acids. 2015;4(9):e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu Y-L, Rana TM. RNA. 2003;9(9):1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozcan G, Ozpolat B, Coleman RL, Sood AK, Lopez-Berestein G. Adv. Drug Deliv. Rev. 2015;87:108–119. doi: 10.1016/j.addr.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokatlian T, Segura T. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010;2(3):305–315. doi: 10.1002/wnan.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu C, Wang J. Asian J. Pharm. Sci. 2015;10(1):1–12. [Google Scholar]
- 20.Kanasty R, Dorkin JR, Vegas A, Anderson D. Nat. Mater. 2013;12(11):967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 21.van de Water FM, Boerman OC, Wouterse AC, Peters JGP, Russel FGM, Masereeuw R. Drug Metab. Dispos. 2006;34(8):1393–1397. doi: 10.1124/dmd.106.009555. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Lu Z, Wientjes MG, Au JL-S. AAPS J. 2010;12(4):492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santel A, Aleku M, Keil O, Endruschat J, Esche V, Fisch G, Dames S, Löffler K, Fechtner M, Arnold W, Giese K, Klippel A, Kaufmann J. Gene Ther. 2006;13(16):1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- 24.Kanasty RL, Whitehead KA, Vegas AJ, Anderson DG. Mol. Ther. 2012;20(3):513–524. doi: 10.1038/mt.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenzer C, Dirin M, Winkler A-M, Baumann V, Winkler J. J. Control. Release. 2015;203:1–15. doi: 10.1016/j.jconrel.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Tian Y, Liu Y, Wang T, Zhou N, Kong J, Chen L, Snitow M, Morley M, Li D, Petrenko N, Zhou S, Lu M, Gao E, Koch WJ, Stewart KM, Morrisey EE. Sci. Transl. Med. 2015;7(279) doi: 10.1126/scitranslmed.3010841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 28.Parhamifar L, Larsen AK, Hunter AC, Andresen TL, Moghimi SM. Soft Matter. 2010;6(17):4001. [Google Scholar]
- 29.Singh A, Suri S, Roy K. Biomaterials. 2009;30(28):5187–5200. doi: 10.1016/j.biomaterials.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pradhan P, Qin H, Leleux JA, Gwak D, Sakamaki I, Kwak LW, Roy K. Biomaterials. 2014;35(21):5491–5504. doi: 10.1016/j.biomaterials.2014.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mountziaris PM, Sing DC, Chew SA, Tzouanas SN, Lehman ED, Kasper FK, Mikos AG. Pharm. Res. 2011;28(6):1370–1384. doi: 10.1007/s11095-010-0354-9. [DOI] [PubMed] [Google Scholar]
- 32.Mountziaris PM, Tzouanas SN, Sing DC, Kramer PR, Kasper FK, Mikos AG. Acta Biomater. 2012;8(10):3552–3560. doi: 10.1016/j.actbio.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson CE, Gupta MK, Adolph EJ, Guelcher SA, Duvall CL. Adv. wound care. 2013;2(3):93–99. doi: 10.1089/wound.2011.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson CE, Gupta MK, Adolph EJ, Shannon JM, Guelcher SA, Duvall CL. Biomaterials. 2012;33(4):1154–1161. doi: 10.1016/j.biomaterials.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson CE, Kim AJ, Adolph EJ, Gupta MK, Yu F, Hocking KM, Davidson JM, Guelcher SA, Duvall CL. Adv. Mater. 2014;24(4):607–614. doi: 10.1002/adma.201303520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen M, Gao S, Dong M, Song J, Yang C, Howard KA, Kjems J, Besenbacher F. ACS Nano. 2012;6(6):4835–4844. doi: 10.1021/nn300106t. [DOI] [PubMed] [Google Scholar]
- 37.Cao H, Jiang X, Chai C, Chew SY. J. Control. Release. 2010;144(2):203–212. doi: 10.1016/j.jconrel.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 38.Diao HJ, Low WC, Milbreta U, Lu QR, Chew SY. J. Control. Release. 2015;208:85–92. doi: 10.1016/j.jconrel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rujitanaroj P, Jao B, Yang J, Wang F, Anderson JM, Wang J, Chew SY. Acta Biomater. 2013;9(1):4513–4524. doi: 10.1016/j.actbio.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rujitanaroj P, Wang Y-C, Wang J, Chew SY. Biomaterials. 2011;32(25):5915–5923. doi: 10.1016/j.biomaterials.2011.04.065. [DOI] [PubMed] [Google Scholar]
- 41.Castleberry S, Wang M, Hammond PT. ACS Nano. 2013;7(6):5251–5261. doi: 10.1021/nn401011n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartmann H, Hossfeld S, Schlosshauer B, Mittnacht U, Pêgo AP, Dauner M, Doser M, Stoll D, Krastev R. J. Control. Release. 2013;168(3):289–297. doi: 10.1016/j.jconrel.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, Marchant RE. Expert Rev. Med. Devices. 2011;8(5):607–626. doi: 10.1586/erd.11.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Laporte L, Shea LD. Adv. Drug Deliv. Rev. 2007;59(4–5):292–307. doi: 10.1016/j.addr.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shea LD, Smiley E, Bonadio J, Mooney DJ. Nat. Biotechnol. 1999;17(6):551–554. doi: 10.1038/9853. [DOI] [PubMed] [Google Scholar]
- 46.Purcell BP, Lobb D, Charati MB, Dorsey SM, Wade RJ, Zellars KN, Doviak H, Pettaway S, Logdon CB, Shuman JA, Freels PD, Gorman JH, Gorman RC, Spinale FG, Burdick JA. Nat. Mater. 2014;13(6):653–661. doi: 10.1038/nmat3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wade RJ, Bassin EJ, Rodell CB, Burdick JA. Nat. Commun. 2015;6:6639. doi: 10.1038/ncomms7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodell CB, Wade RJ, Purcell BP, Dusaj NN, Burdick JA. ACS Biomater. Sci. Eng. 2015;1(4):277–286. doi: 10.1021/ab5001673. [DOI] [PubMed] [Google Scholar]
- 49.Van Hove AH, Beltejar M-JG, Benoit DSW. Biomaterials. 2014;35(36):9719–9730. doi: 10.1016/j.biomaterials.2014.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Hove AH, Antonienko E, Burke K, Brown E, Benoit DSW. Adv. Healthc. Mater. 2015;4(13):2002–2011. doi: 10.1002/adhm.201500304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Tang Y, Narain R, Lewis AL, Armes SP. Langmuir. 2005;21(22):9946–9954. doi: 10.1021/la050356u. [DOI] [PubMed] [Google Scholar]
- 52.Wang K, Fu Q, Chen X, Gao Y, Dong K. RSC Adv. 2012;2(20):7772. [Google Scholar]
- 53.Mahkam M. Drug Deliv. 2010;17(3):158–163. doi: 10.3109/10717541003604908. [DOI] [PubMed] [Google Scholar]
- 54.Qiu Y, Park K. Adv. Drug Deliv. Rev. 2001;53(3):321–339. doi: 10.1016/s0169-409x(01)00203-4. [DOI] [PubMed] [Google Scholar]
- 55.Kim Y-M, Kim C-H, Song S-C. ACS Macro Lett. 2016;5(3):297–300. doi: 10.1021/acsmacrolett.6b00008. [DOI] [PubMed] [Google Scholar]
- 56.Kim Y-M, Park M-R, Song S-C. Biomaterials. 2013;34(18):4493–4500. doi: 10.1016/j.biomaterials.2013.02.050. [DOI] [PubMed] [Google Scholar]
- 57.Kim Y-M, Song S-C. Biomaterials. 2014;35(27):7970–7977. doi: 10.1016/j.biomaterials.2014.05.070. [DOI] [PubMed] [Google Scholar]
- 58.Kim Y-M, Song S-C. Biomaterials. 2014;35(27):7970–7977. doi: 10.1016/j.biomaterials.2014.05.070. [DOI] [PubMed] [Google Scholar]
- 59.Kim Y-M, Park M-R, Song S-C. ACS Nano. 2012;6(7):5757–5766. doi: 10.1021/nn300842a. [DOI] [PubMed] [Google Scholar]
- 60.Fang J-Y, Chen J-P, Leu Y-L, Hu J-W. Eur. J. Pharm. Biopharm. Off. J. Arbeitsgemeinschaft für Pharm. Verfahrenstechnik e.V. 2008;68(3):626–636. [Google Scholar]
- 61.Ruel-Gariépy E, Leroux J-C. Eur. J. Pharm. Biopharm. 2004;58(2):409–426. doi: 10.1016/j.ejpb.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 62.Huynh CT, Nguyen MK, Tonga GY, Longé L, Rotello VM, Alsberg E. Adv. Healthc. Mater. 2016;5(3):305–310. doi: 10.1002/adhm.201500778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huynh CT, Nguyen MK, Naris M, Tonga GY, Rotello VM, Alsberg E. Nanomedicine (Lond) 2016 doi: 10.2217/nnm-2016-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Griffin DR, Kasko AM. J. Am. Chem. Soc. 2012;134(31):13103–13107. doi: 10.1021/ja305280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Science. 2009;324(5923):59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong DY, Griffin DR, Reed J, Kasko AM. Macromolecules. 2010;43(6):2824–2831. [Google Scholar]
- 67.Kloxin AM, Tibbitt MW, Anseth KS. Nat. Protoc. 2010;5(12):1867–1887. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murdan S. J. Control. Release. 2003;92(1–2):1–17. doi: 10.1016/s0168-3659(03)00303-1. [DOI] [PubMed] [Google Scholar]
- 69.Kulkarni RV, Sa B. J. Bioact. Compat. Polym. 2009;24(4):368–384. [Google Scholar]
- 70.Jackson AL, Linsley PS. Nat. Rev. Drug Discov. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 71.Lei Y, Huang S, Sharif-Kashani P, Chen Y, Kavehpour P, Segura T. Biomaterials. 2010;31(34):9106–9116. doi: 10.1016/j.biomaterials.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Keeney M, Onyiah S, Zhang Z, Tong X, Han L-H, Yang F. Biomaterials. 2013;34(37):9657–9665. doi: 10.1016/j.biomaterials.2013.08.050. [DOI] [PubMed] [Google Scholar]
- 73.Li Y, Yang C, Khan M, Liu S, Hedrick JL, Yang Y-Y, Ee PLR. Biomaterials. 2012;33(27):6533–6541. doi: 10.1016/j.biomaterials.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 74.Lei Y, Rahim M, Ng Q, Segura T. J. Control. Release. 2011;153(3):255–261. doi: 10.1016/j.jconrel.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lei P, Padmashali RM, Andreadis ST. Biomaterials. 2009;30(22):3790–3799. doi: 10.1016/j.biomaterials.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lei Y, Segura T. Biomaterials. 2009;30(2):254–265. doi: 10.1016/j.biomaterials.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meilander NJ, Pasumarthy MK, Kowalczyk TH, Cooper MJ, Bellamkonda RV. J. Control. Release. 2003;88(2):321–331. doi: 10.1016/s0168-3659(03)00007-5. [DOI] [PubMed] [Google Scholar]
- 78.Segura T, Chung PH, Shea LD. Biomaterials. 2005;26(13):1575–1584. doi: 10.1016/j.biomaterials.2004.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ta HT, Dass CR, Larson I, Choong PFM, Dunstan DE. Biomaterials. 2009;30(27):4815–4823. doi: 10.1016/j.biomaterials.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 80.Yang H, van Ee RJ, Timmer K, Craenmehr EGM, Huang JH, Öner FC, Dhert WJA, Kragten AHM, Willems N, Grinwis GCM, Tryfonidou MA, Papen-Botterhuis NE, Creemers LB. Acta Biomater. 2015;23:214–228. doi: 10.1016/j.actbio.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 81.Hill MC, Nguyen MK, Jeon O, Alsberg E. Adv. Healthc. Mater. 2014 doi: 10.1002/adhm.201400458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nguyen MK, Jeon O, Krebs MD, Schapira D, Alsberg E. Biomaterials. 2014;35(24):6278–6286. doi: 10.1016/j.biomaterials.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Asti A, Gioglio L. Int. J. Artif. Organs. 2014;37(3):187–205. doi: 10.530/ijao.5000307. [DOI] [PubMed] [Google Scholar]
- 84.Vorhies JS, Nemunaitis JJ. Methods Mol. Biol. 2009;480:11–29. doi: 10.1007/978-1-59745-429-2_2. [DOI] [PubMed] [Google Scholar]
- 85.Nguyen K, Dang PN, Alsberg E. Acta Biomater. 2013;9(1):4487–4495. doi: 10.1016/j.actbio.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krebs MD, Jeon O, Alsberg E. J. Am. Chem. Soc. 2009;131(26):9204–9206. doi: 10.1021/ja9037615. [DOI] [PubMed] [Google Scholar]
- 87.Tokatlian T, Cam C, Siegman SN, Lei Y, Segura T. Acta Biomater. 2012;8(11):3921–3931. doi: 10.1016/j.actbio.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma Z, Yang C, Song W, Wang Q, Kjems J, Gao S. J. Nanobiotechnology. 2014;12:23. doi: 10.1186/1477-3155-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rezvani Amin Z, Rahimizadeh M, Eshghi H, Dehshahri A, Ramezani M. Iran. J. Basic Med. Sci. 2013;16(2):150–156. [PMC free article] [PubMed] [Google Scholar]
- 90.Fröhlich E. Int. J. Nanomedicine. 2012;7:5577–5591. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lv H, Zhang S, Wang B, Cui S, Yan J. J. Control. Release. 2006;114(1):100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 92.Wei X, Shao B, He Z, Ye T, Luo M, Sang Y, Liang X, Wang W, Luo S, Yang S, Zhang S, Gong C, Gou M, Deng H, Zhao Y, Yang H, Deng S, Zhao C, Yang L, Qian Z, Li J, Sun X, Han J, Jiang C, Wu M, Zhang Z. Cell Res. 2015;25(2):237–253. doi: 10.1038/cr.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Verma A, Stellacci F. Small. 2010;6(1):12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- 94.Cai J, Yue Y, Rui D, Zhang Y, Liu S, Wu C. Macromolecules. 2011;44(7):2050–2057. [Google Scholar]
- 95.Zhang Q-F, Luan C-R, Yin D-X, Zhang J, Liu Y-H, Peng Q, Xu Y, Yu X-Q. Polymers (Basel) 2015;7(12):2316–2331. [Google Scholar]
- 96.Hashemi M, Parhiz BH, Hatefi A, Ramezani M. Cancer Gene Ther. 2011;18(1):12–19. doi: 10.1038/cgt.2010.57. [DOI] [PubMed] [Google Scholar]
- 97.Pun SH, Bellocq NC, Liu A, Jensen G, Machemer T, Quijano E, Schluep T, Wen S, Engler H, Heidel J, Davis ME. Bioconjug. Chem. 2004;15(4):831–840. doi: 10.1021/bc049891g. [DOI] [PubMed] [Google Scholar]
- 98.Song H, Wang G, He B, Li L, Li C, Lai Y, Xu X, Gu Z. Int. J. Nanomedicine. 2012;7:4637–4648. doi: 10.2147/IJN.S33923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu L, Zheng M, Librizzi D, Renette T, Merkel OM, Kissel T. Mol. Pharm. 2016;13(1):134–143. doi: 10.1021/acs.molpharmaceut.5b00575. [DOI] [PubMed] [Google Scholar]
- 100.Mironi-Harpaz I, Wang DY, Venkatraman S, Seliktar D. Acta Biomater. 2012;8(5):1838–1848. doi: 10.1016/j.actbio.2011.12.034. [DOI] [PubMed] [Google Scholar]
- 101.El-Sherbiny IM, Yacoub MH. Glob. Cardiol. Sci. Pract. 2013;2013(3):316–342. doi: 10.5339/gcsp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wade RJ, Bassin EJ, Rodell CB, Burdick JA. Nat. Commun. 2015;6:6639. doi: 10.1038/ncomms7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Babincova M, Novotny J, Rosenecker J, Babinec P. Optoelectron. Adv. Mater. 2007;1(11):644–647. [Google Scholar]
- 104.Hoare TR, Kohane DS. Polymer (Guildf) 2008;49(8):1993–2007. [Google Scholar]
- 105.Rodell CB, Kaminski AL, Burdick JA. Biomacromolecules. 2013;14(11):4125–4134. doi: 10.1021/bm401280z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Burdick JA, Prestwich GD. Adv. Mater. 2011;23(12):H41–H56. doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nguyen QV, Huynh DP, Park JH, Lee DS. Eur. Polym. J. 2015;72:602–619. [Google Scholar]
- 108.Nguyen MK, Lee DS. Macromol. Biosci. 2010;10(6):563–579. doi: 10.1002/mabi.200900402. [DOI] [PubMed] [Google Scholar]
- 109.Martens TP, Godier AFG, Parks JJ, Wan LQ, Koeckert MS, Eng GM, Hudson BI, Sherman W, Vunjak-Novakovic G. Cell Transplant. 2009;18(3):297–304. doi: 10.3727/096368909788534915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hillel AT, Unterman S, Nahas Z, Reid B, Coburn JM, Axelman J, Chae JJ, Guo Q, Trow R, Thomas A, Hou Z, Lichtsteiner S, Sutton D, Matheson C, Walker P, David N, Mori S, Taube JM, Elisseeff JH. Sci. Transl. Med. 2011;3(93) doi: 10.1126/scitranslmed.3002331. 93ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang Z, He Z, Liang R, Ma Y, Huang W, Jiang R, Shi S, Chen H, Li X. Biomacromolecules. 2016;17(3):798–807. doi: 10.1021/acs.biomac.5b01526. [DOI] [PubMed] [Google Scholar]
- 112.Rodell CB, Mealy JE, Burdick JA. Bioconjug. Chem. 2015;26(12):2279–2289. doi: 10.1021/acs.bioconjchem.5b00483. [DOI] [PubMed] [Google Scholar]
- 113.Voorhaar L, Hoogenboom R. Chem. Soc. Rev. 2016 doi: 10.1039/c6cs00130k. [DOI] [PubMed] [Google Scholar]
- 114.Highley CB, Kim M, Lee D, Burdick JA. Nanomedicine. 2016 doi: 10.2217/nnm-2016-0070. nnm-2016-0070. [DOI] [PubMed] [Google Scholar]
- 115.Mealy JE, Rodell CB, Burdick JA. J. Mater. Chem. B. 2015;3(40):8010–8019. doi: 10.1039/C5TB00981B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rodell CB, MacArthur JW, Dorsey SM, Wade RJ, Wang LL, Woo YJ, Burdick JA. Adv. Funct. Mater. 2014;25(4):636–644. doi: 10.1002/adfm.201403550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim Y-M, Park M-R, Song S-C. Biomaterials. 2013;34(18):4493–4500. doi: 10.1016/j.biomaterials.2013.02.050. [DOI] [PubMed] [Google Scholar]
- 118.Han HD, Mora EM, Roh JW, Nishimura M, Lee SJ, Stone RL, Bar-Eli M, Lopez-Berestein G, Sood AK. Cancer Biol. Ther. 2011;11(9):839–845. doi: 10.4161/cbt.11.9.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cao C, Yan C, Hu Z, Zhou S. Mol. Med. Rep. 2015;12(5):6688–6694. doi: 10.3892/mmr.2015.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Whitehead Ka, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, Fenton OS, Zhang Y, Olejnik KT, Yesilyurt V, Chen D, Barros S, Klebanov B, Novobrantseva T, Langer R, Anderson DG. Nat. Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Akinc A, Goldberg M, Qin J, Dorkin JR, Gamba-Vitalo C, Maier M, Jayaprakash KN, Jayaraman M, Rajeev KG, Manoharan M, Koteliansky V, Röhl I, Leshchiner ES, Langer R, Anderson DG. Mol. Ther. 2009;17(5):872–879. doi: 10.1038/mt.2009.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sun S, Wang M, Knupp SA, Soto-Feliciano Y, Hu X, Kaplan DL, Langer R, Anderson DG, Xu Q. Bioconjug. Chem. 2012;23(1):135–140. doi: 10.1021/bc200572w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cho S-W, Goldberg M, Son SM, Xu Q, Yang F, Mei Y, Bogatyrev S, Langer R, Anderson DG. Adv. Funct. Mater. 2009;19(19):3112–3118. doi: 10.1002/adfm.200900519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Toh M-R, Chiu GNC. Asian J. Pharm. Sci. 2013;8(2):88–95. [Google Scholar]
- 125.Thoniyot P, Tan MJ, Karim AA, Young DJ, Loh XJ. Adv. Sci. 2015;2(1–2) doi: 10.1002/advs.201400010. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dominska M, Dykxhoorn DM. J. Cell Sci. 2010;123(Pt 8):1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 127.Akinc A, Thomas M, Klibanov AM, Langer R. J. Gene Med. 2005;7(5):657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 128.Hsu CYM, Uludağ H. Biomaterials. 2012;33(31):7834–7848. doi: 10.1016/j.biomaterials.2012.06.093. [DOI] [PubMed] [Google Scholar]
- 129.Xiong MP, Forrest ML, Ton G, Zhao A, Davies NM, Kwon GS. Biomaterials. 2007;28(32):4889–4900. doi: 10.1016/j.biomaterials.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 130.Höbel S, Aigner A. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 5(5):484–501. doi: 10.1002/wnan.1228. [DOI] [PubMed] [Google Scholar]
- 131.Neuberg P, Kichler A. Advances in Genetics. 2014;88:263–288. doi: 10.1016/B978-0-12-800148-6.00009-2. [DOI] [PubMed] [Google Scholar]
- 132.Inoue Y, Kurihara R, Tsuchida A, Hasegawa M, Nagashima T, Mori T, Niidome T, Katayama Y, Okitsu O. J. Control. Release. 2008;126(1):59–66. doi: 10.1016/j.jconrel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 133.Watanabe K, Harada-Shiba M, Suzuki A, Gokuden R, Kurihara R, Sugao Y, Mori T, Katayama Y, Niidome T. Mol. Biosyst. 2009;5(11):1306–1310. doi: 10.1039/b900880b. [DOI] [PubMed] [Google Scholar]
- 134.Zhang Y, Zhou C, Kwak KJ, Wang X, Yung B, Lee LJ, Wang Y, Wang PG, Lee RJ. Pharm. Res. 2012;29(6):1627–1636. doi: 10.1007/s11095-012-0676-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arima H, Tsutsumi T, Yoshimatsu A, Ikeda H, Motoyama K, Higashi T, Hirayama F, Uekama K. Eur. J. Pharm. Sci. 2011;44(3):375–384. doi: 10.1016/j.ejps.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 136.Segovia N, Pont M, Oliva N, Ramos V, Borrós S, Artzi N. Adv. Healthc. Mater. 2015;4(2):271–280. doi: 10.1002/adhm.201400235. [DOI] [PubMed] [Google Scholar]
- 137.Tzeng SY, Hung BP, Grayson WL, Green JJ. Biomaterials. 2012;33(32):8142–8151. doi: 10.1016/j.biomaterials.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee J-S, Green JJ, Love KT, Sunshine J, Langer R, Anderson DG. Nano Lett. 2009;9(6):2402–2406. doi: 10.1021/nl9009793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sarett SM, Werfel TA, Chandra I, Jackson MA, Kavanaugh TE, Hattaway ME, Giorgio TD, Duvall CL. Biomaterials. 2016;97:122–132. doi: 10.1016/j.biomaterials.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sarett SM, Kilchrist KV, Miteva M, Duvall CL. J. Biomed. Mater. Res. A. 2015;103(9):3107–3116. doi: 10.1002/jbm.a.35413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Muratovska A, Eccles MR. FEBS Lett. 2004;558(1–3):63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 142.Conde J, Oliva N, Atilano M, Song HS, Artzi N. Nat. Mater. 2015;15(3):353–363. doi: 10.1038/nmat4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ambardekar VV, Han HY, Varney ML, Vinogradov SV, Singh RK, Vetro JA. Biomaterials. 2011;32(5):1404–1411. doi: 10.1016/j.biomaterials.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Röhl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher H-P. Nature. 2004;432(7014):173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 145.De Paula D, Bentley MVLB, Mahato RI. RNA. 2007;13(4):431–456. doi: 10.1261/rna.459807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.He X-X, Chang Y, Meng F-Y, Wang M-Y, Xie Q-H, Tang F, Li P-Y, Song Y-H, Lin J-S. Oncogene. 2012;31(28):3357–3369. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 147.Zhou Q, Anderson C, Zhang H, Li X, Inglis F, Jayagopal A, Wang S. Mol. Ther. 2014;22(2):378–389. doi: 10.1038/mt.2013.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Krebs MD, Alsberg E. Chemistry. 2011;17(11):3054–3062. doi: 10.1002/chem.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nakajima H, Kubo T, Semi Y, Itakura M, Kuwamura M, Izawa T, Azuma Y-T, Takeuchi T. J. Biotechnol. 2012;157(2):326–333. doi: 10.1016/j.jbiotec.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 150.Larsen HØ, Roug AS, Nielsen K, Søndergaard CS, Hokland P. Exp. Hematol. 2011;39(11):1081–1089. doi: 10.1016/j.exphem.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 151.Gupta AK, Eshraghi Y, Gliniak C, Gosain AK. Plast. Reconstr. Surg. 2010;125(2):494–501. doi: 10.1097/PRS.0b013e3181c82df1. [DOI] [PubMed] [Google Scholar]
- 152.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, Marshall W, Khvorova A, Linsley PS. RNA. 2006;12(7):1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Behlke MA. Oligonucleotides. 2008;18(4):305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 154.Winkler J. Ther. Deliv. 2013;4(7):791–809. doi: 10.4155/tde.13.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Segovia N, Pont M, Oliva N, Ramos V, Borrós S, Artzi N. Adv. Healthc. Mater. 2015;4(2):271–280. doi: 10.1002/adhm.201400235. [DOI] [PubMed] [Google Scholar]
- 156.Wang J, Lu Z, Wientjes MG, Au JL-S. AAPS J. 2010;12(4):492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Patil SP, Jeong HS, Kim BH. Chem. Commun. 2012;48(71):8901. doi: 10.1039/c2cc34466a. [DOI] [PubMed] [Google Scholar]
- 158.Singh A, Nie H, Ghosn B, Qin H, Kwak LW, Roy K. Mol. Ther. 2008;16(12):2011–2021. doi: 10.1038/mt.2008.206. [DOI] [PubMed] [Google Scholar]
- 159.Liu J, Xue W-R, Zhao L-L, Qu X-Z, Yang Z-Z, Li R, Deng S-Z, Xiao F, Hu X-P, Bai W-J. Natl. J. Androl. 2013;19(9):771–775. [PubMed] [Google Scholar]
- 160.Saito T, Tabata Y. J. Drug Target. 2012;20(10):864–872. doi: 10.3109/1061186X.2012.725170. [DOI] [PubMed] [Google Scholar]
- 161.Ma Z, Yang C, Song W, Wang Q, Kjems J, Gao S. J. Nanobiotechnology. 2014;12(1):23. doi: 10.1186/1477-3155-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Louw AM, Kolar MK, Novikova LN, Kingham PJ, Wiberg M, Kjems J, Novikov LN. Nanomedicine. 2016;12(3):643–653. doi: 10.1016/j.nano.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 163.Kobayashi H, Watanabe R, Choyke PL. Theranostics. 2013;4(1):81–89. doi: 10.7150/thno.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jain RK, Stylianopoulos T. Nat. Rev. Clin. Oncol. 2010;7(11):653–664. doi: 10.1038/nrclinonc.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Han HD, Mora EM, Roh JW, Nishimura M, Lee SJ, Stone RL, Bar-Eli M, Lopez-Berestein G, Sood AK. Cancer Biol. Ther. 2011;11(9):839–845. doi: 10.4161/cbt.11.9.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yu K, Wu S, Li H. J. Exp. Nanosci. 2015:1–16. [Google Scholar]
- 167.Zhao Q-Q, Chen J-L, Lv T-F, He C-X, Tang G-P, Liang W-Q, Tabata Y, Gao J-Q. Biol. Pharm. Bull. 2009;32(4):706–710. doi: 10.1248/bpb.32.706. [DOI] [PubMed] [Google Scholar]
- 168.Guo D-D, Hong S-H, Jiang H-L, Kim J-H, Minai-Tehrani A, Kim J-E, Shin J-Y, Jiang T, Kim Y-K, Choi Y-J, Cho C-S, Cho M-H. Biomaterials. 2012;33(7):2272–2281. doi: 10.1016/j.biomaterials.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 169.Ma H, He C, Cheng Y, Li D, Gong Y, Liu J, Tian H, Chen X. Biomaterials. 2014;35(30):8723–8734. doi: 10.1016/j.biomaterials.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 170.Kozielski KL, Green JJ. Methods Mol. Biol. 2016;1364:79–87. doi: 10.1007/978-1-4939-3112-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Peng H, Yang H, Song L, Zhou Z, Sun J, Du Y, Lu K, Li T, Yin A, Xu J, Wei S. J. Exp. Clin. Cancer Res. 2016;35(1):57. doi: 10.1186/s13046-016-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hauser PV, Pippin JW, Kaiser C, Krofft RD, Brinkkoetter PT, Hudkins KL, Kerjaschki D, Reiser J, Alpers CE, Shankland SJ. PLoS One. 2010;5(3):e9463. doi: 10.1371/journal.pone.0009463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Shukla RS, Tai W, Mahato R, Jin W, Cheng K. Mol. Pharm. 2013;10(12):4534–4545. doi: 10.1021/mp400355q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhao R, Yan Q, Huang H, Lv J, Ma W. Matrix Biol. 2013;32(5):265–276. doi: 10.1016/j.matbio.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 175.Laroui H, Geem D, Xiao B, Viennois E, Rakhya P, Denning T, Merlin D. Mol. Ther. 2014;22(1):69–80. doi: 10.1038/mt.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Laroui H, Viennois E, Xiao B, Canup BSB, Geem D, Denning TL, Merlin D. J. Control. Release. 2014;186:41–53. doi: 10.1016/j.jconrel.2014.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kanazawa T, Shizawa Y, Takeuchi M, Tamano K, Ibaraki H, Seta Y, Takashima Y, Okada H. Pharmaceutics. 2015;7(3):294–304. doi: 10.3390/pharmaceutics7030294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Browne S, Monaghan MG, Brauchle E, Berrio DC, Chantepie S, Papy-Garcia D, Schenke-Layland K, Pandit A. Biomaterials. 2015;69:133–147. doi: 10.1016/j.biomaterials.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 179.Manaka T, Suzuki A, Takayama K, Imai Y, Nakamura H, Takaoka K. Biomaterials. 2011;32(36):9642–9648. doi: 10.1016/j.biomaterials.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 180.Li Y, Fan L, Liu S, Liu W, Zhang H, Zhou T, Wu D, Yang P, Shen L, Chen J, Jin Y. Biomaterials. 2013;34(21):5048–5058. doi: 10.1016/j.biomaterials.2013.03.052. [DOI] [PubMed] [Google Scholar]
- 181.Lie A, Juan S, Bala M, Hlawaty H, Portes P, Vranckx R, Feldman LJ, Letourneur D. doi: 10.1021/bm900740g. [DOI] [PubMed] [Google Scholar]
- 182.Wan W-G, Jiang X-J, Li X-Y, Zhang C, Yi X, Ren S, Zhang X-Z, W-g W, X-j J, X-y L, X-z Z. J Biomed Mater Res Part A. 2014;102:3452–3458. doi: 10.1002/jbm.a.35014. [DOI] [PubMed] [Google Scholar]
- 183.Pfeffer MA. Circulation. 1998;97(22):2192–2194. doi: 10.1161/01.cir.97.22.2192. [DOI] [PubMed] [Google Scholar]
- 184.Kobara M, Tatsumi T, Kambayashi D, Mano A, Yamanaka S, Shiraishi J, Keira N, Matoba S, Asayama J, Fushiki S, Nakagawa M. J. Cardiovasc. Pharmacol. 2003;41(6):880–889. doi: 10.1097/00005344-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 185.LeDuc P, Ostuni E, Whitesides G, Ingber D. Methods Cell Biol. 2002;69:385–401. doi: 10.1016/s0091-679x(02)69024-7. [DOI] [PubMed] [Google Scholar]
- 186.Cullen BR. Nat. Genet. 2005;37(11):1163–1165. doi: 10.1038/ng1105-1163. [DOI] [PubMed] [Google Scholar]