Abstract
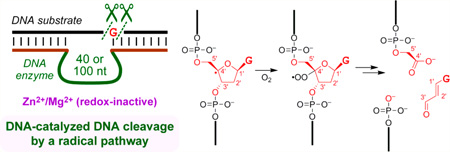
We describe an unprecedented DNA-catalyzed DNA cleavage process in which a radical-based reaction pathway cleanly results in excision of most atoms of a specific guanosine nucleoside. Two new deoxyribozymes (DNA enzymes) were identified by in vitro selection from N40 or N100 random pools initially seeking amide bond hydrolysis, although they both cleave simple single-stranded DNA oligonucleotides. Each deoxyribozyme generates both superoxide (O2-• or HOO•) and hydrogen peroxide (H2O2) and leads to the same set of products (3′-phosphoglycolate, 5′-phosphate, and base propenal) as formed by the natural product bleomycin, with product assignments by mass spectrometry and colorimetric assay. We infer the same mechanistic pathway, involving formation of the C4′ radical of the guanosine nucleoside that is subsequently excised. Consistent with a radical pathway, glutathione fully suppresses catalysis. Conversely, adding either superoxide or H2O2 from the outset strongly enhances catalysis. The mechanism of generation and involvement of superoxide and H2O2 by the deoxyribozymes is not yet defined. The deoxyribozymes do not require redox-active metal ions and function with a combination of Zn2+ and Mg2+, although including Mn2+ increases the activity, and Mn2+ alone also supports catalysis. In contrast to all of these observations, unrelated DNA-catalyzed radical DNA cleavage reactions require redox-active metals and lead to mixtures of products. This study reports an intriguing example of a well-defined, DNA-catalyzed, radical reaction process that cleaves single-stranded DNA and requires only redox-inactive metal ions.
Graphical abstract
INTRODUCTION
Many deoxyribozymes have been identified by in vitro selection1 for a growing reaction scope with biomolecular substrates.2 The first report3 of a deoxyribozyme was for nonhydrolytic RNA cleavage by mediating attack of a 2′-hydroxyl on an adjacent phosphodiester (the ribonuclease mechanism4), and many RNA-cleaving deoxyribozymes were subsequently found.5 While seeking DNA-catalyzed amide bond hydrolysis,6 eventually with success,7 we initially identified deoxyribozymes that hydrolyze specific phosphodiester linkages of single-stranded DNA (ssDNA) substrates8 and RNA substrates.9 We and others also found deoxy-ribozymes that deglycosylate DNA, with subsequent elimination reactions that lead to strand scission.10 Other reported DNA-catalyzed cleavage reactions include oxidative DNA cleavage11,12 and phosphoramidate cleavage.13
Here we report a new, distinct type of DNA-catalyzed ssDNA cleavage. This cleavage process occurs by a super-oxide/H2O2-dependent radical mechanism rather than an ionic mechanism and leads to well-defined reaction products that are identical to those formed by the action of the glycopeptide natural product bleomycin on double-stranded DNA (dsDNA).14 Unlike the reported examples of DNA-catalyzed oxidative DNA cleavage,11,12 the reactions described here do not require redox-active metal ion cofactors such as Cu2+ or Mn2+. Instead, only redox-inactive Zn2+ and Mg2+ are needed. This finding expands the scope of reactions that can be catalyzed by DNA to include a radical pathway that forms well-defined products without obligatory involvement of redox-active metal ions.
RESULTS
Identification of a New DNA-Cleaving Deoxyribozyme by in Vitro Selection
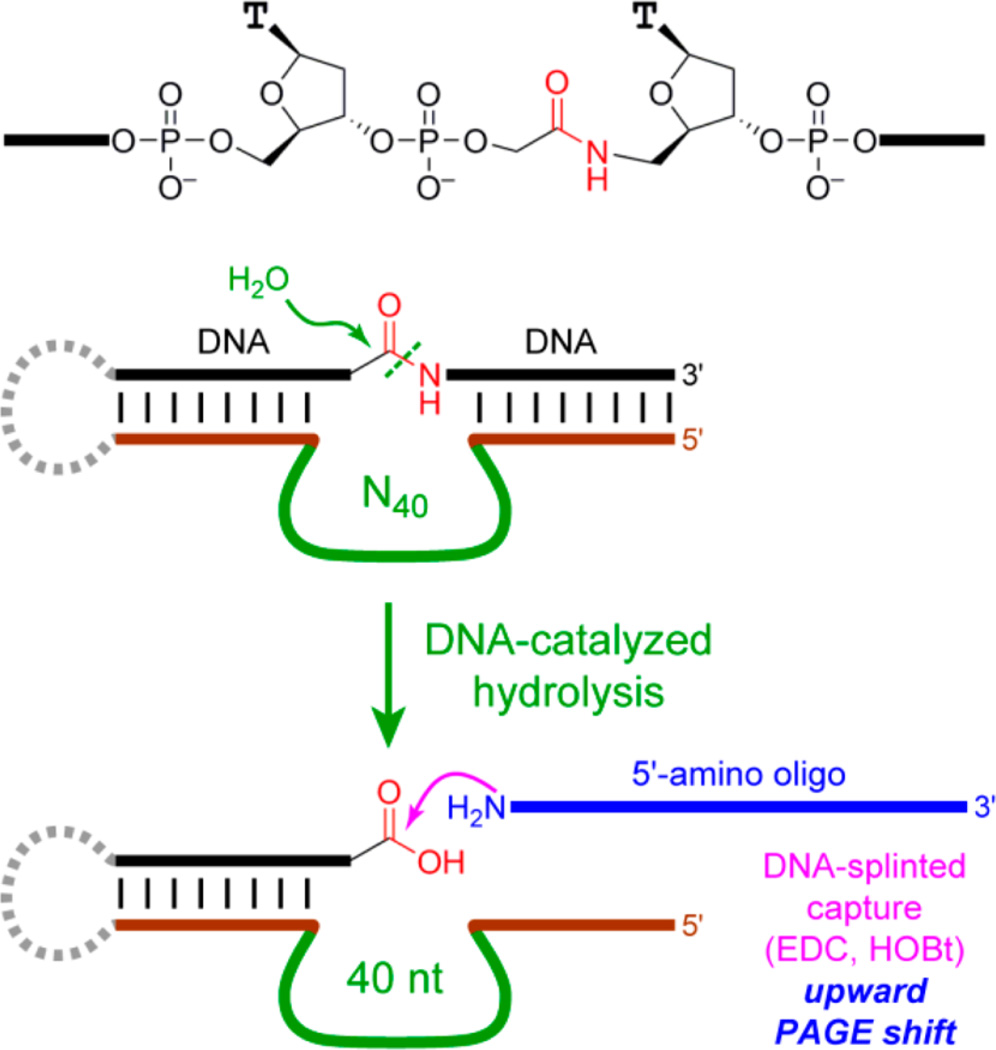
The two new deoxyribozymes in this study were found by in vitro selection, initially seeking DNA-catalyzed amide hydrolysis. A successful in vitro selection experiment often depends upon implementing a suitable “capture reaction”, which is highly selective for a functional group characteristic of the desired reaction product.2g Amide bond hydrolysis leads to a carboxylic acid functional group, which can be captured during each selection round using an amino-modified oligonucleotide and a suitable coupling agent such as the carbodiimide EDC (Figure 1).
Figure 1.
In vitro selection process originally intended to capture the carboxylic acid product of DNA-catalyzed amide hydrolysis. The substrate presents its single amide bond embedded within two DNA oligonucleotide segments, which form Watson-Crick base pairs with the fixed-sequence 5′- and 3′-segments that flank the initially random region, which was either N40 or N100. The N40 selection experiment included modified nucleotides,7 although these modifications are dispensable for activity of the new RadDz3 deoxyribozyme reported here. The N100 selection experiment used unmodified DNA and led to the new RadDz6 deoxyribozyme. Both RadDz3 and RadDz6 lead to DNA cleavage, not amide hydrolysis.
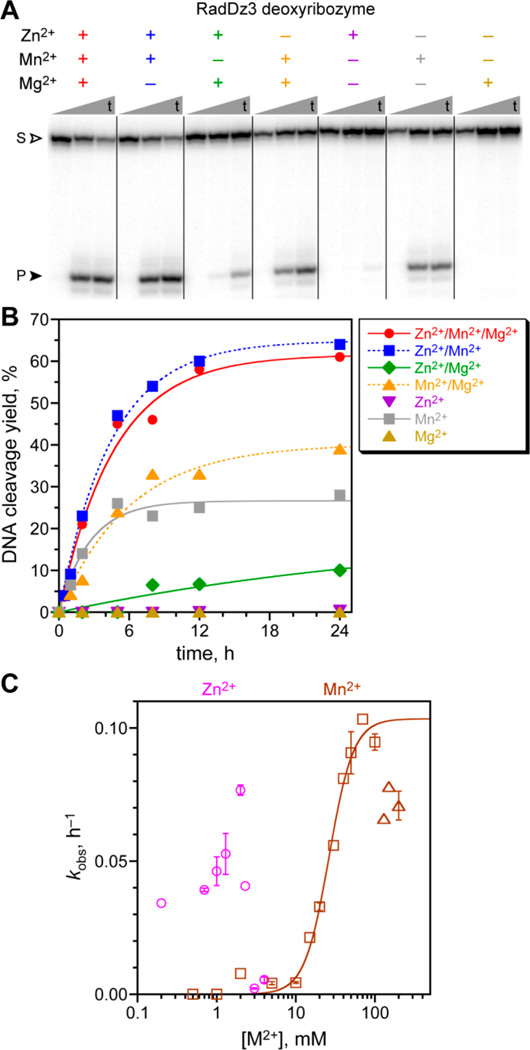
During successful selection experiments for amide hydrolysis in which modified DNA nucleotides were incorporated into the catalytic DNA populations,7 we identified one additional and previously unreported deoxyribozyme, named RadDz3, which cleaves a simple DNA oligonucleotide substrate that lacks any amide bond at all (Figure 2A). From the PAGE analysis, the product is >85% of one species. RadDz3 was identified from an N40 random pool, i.e., DNA sequences with 40 random nucleotides. As originally identified, RadDz3 incorporated several 5-(hydroxymethyl)dU nucleotides, but these modifications were found to be dispensable for catalysis; RadDz3 is catalytically active in its unmodified form as simple DNA, and all experiments in this report used unmodified RadDz3. Under single-turnover conditions, RadDz3 has a kobs value of 0.20 ± 0.02 h−1 (n = 3, mean ± sd) and 50–60% yield in 24 h under the incubation conditions used during its identification, which included each of 1 mM Zn2+, 20 mM Mn2+, and 40 mM Mg2+ (Figure 2B). Zn2+ and Mg2+ together are sufficient for observable RadDz3 activity (although with 50-fold reduced kobs), demonstrating that RadDz3 does not require a redox-active metal ion cofactor. Mn2+ alone also supports substantial catalysis, whereas Mg2+ alone does not, and Zn2+ alone appears to support a trace of activity (~0.5% in 24 h, kobs = 0.0002 h−1). The apparent Kd value was 26 ± 2 mM for Mn2+ (Figure 2C), with some inhibition above 100 mM. The Zn2+ concentration dependence was assayed in the presence of Mn2+, due to the low catalytic activity with Zn2+ alone. Between 0.2 and 2 mM Zn2+, kobs varied by ~2-fold, with inhibition above 2 mM (Figure 2C), as observed with several other deoxyribozymes and perhaps due to precipitation of Zn(OH)2.8a,15 Assays under multiple-turnover conditions did not provide evidence of turnover by RadDz3 (data not shown).
Figure 2.
Assays of the RadDz3 deoxyribozyme that cleaves ssDNA by a radical pathway. Incubation conditions: 70 mM HEPES, pH 7.5, combinations of 1 mM ZnCl2, 20 mM MnCl2, and 40 mM MgCl2 as indicated, and 150 mM NaCl at 37 °C. (A) PAGE analysis of single-turnover DNA cleavage by RadDz3, using 5′-32P-radiolabeled DNA substrate. Representative time points at t = 30 s, 5 h, and 24 h are shown. S = substrate; P = product. (B) Kinetic plots. For this data set, kobs values (h−1): Zn2+/Mn2+/Mg2+ 0.21, Zn2+/Mn2+ 0.22, Zn2+/Mg2+ 0.004, Mn2+/Mg2+ 0.17, Mn2+ 0.38. When Mn2+ was included at 1-300 µM along with 1 mM Zn2+ and 40 mM Mg2+, the DNA cleavage yield was the same as with Zn2+/Mg2+ in the absence of Mn2+ (data not shown), indicating that trace Mn2+ is not responsible for the Zn2+/Mg2+ reactivity. See the Experimental Section for quantitative analysis information on metal ion salts. (C) Determination of metal ion concentration dependence for Zn2+ (in the presence of 20 mM Mn2+) and Mn2+ alone. Data points with error bars were n = 2 (weighted averages; error bars by propagation from curve fit error); data points without error bars were n = 1. For Mn2+ alone, squares were fit, and triangles were not fit. Apparent Kd = 26 ± 2 mM, Hill coefficient n = 2.8 ± 0.4, kmax = 0.10 ± 0.01 h−1.
Establishing a Radical Pathway for RadDz3
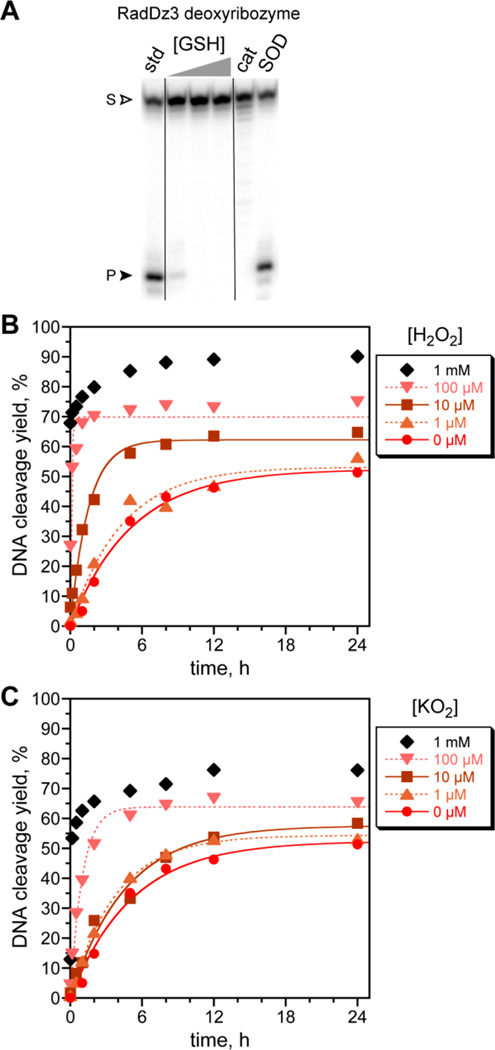
During the in vitro selection process, the survival of RadDz3 cannot be explained by DNA phosphodiester hydrolysis, which would not have led to a capturable product. We therefore considered alternative reaction pathways. RadDz3 was fully inhibited by either glutathione or catalase (Figure 3A), where glutathione quenches radical intermediates and catalase destroys H2O2, suggesting a radical pathway and involvement of H2O2.16 Conversely, adding H2O2 strongly enhanced the catalysis (Figure 3B).18 At 100 µM H2O2, kobs = 10 h−1 (50-fold higher than without H2O2), with even higher kobs (not measurable by manual mixing methods) at 1 mM H2O2. Superoxide dismutase (SOD) did not suppress cleavage product formation, but this outcome is difficult to interpret because superoxide (O2-• or HOO•) could react further immediately after its formation and have no opportunity to be contacted by SOD; also, SOD produces H2O2 from superoxide. Indeed, separately adding potassium superoxide (KO2) also substantially increased the catalysis (Figure 3C), suggesting a pathway in which RadDz3 sequentially forms superoxide and then H2O2.
Figure 3.
Inhibition of RadDz3 catalysis by glutathione or catalase; indifference to superoxide dismutase (SOD); and enhancement of catalysis by H2O2 and potassium superoxide (KO2). All assays used the incubation conditions of Figure 2 with 1 mM Zn2+, 20 mM Mn2+, and 40 mM Mg2+. (A) Assays including glutathione (1, 3, or 10 mM), catalase (1 U/20 µL), or SOD (1 U/20 µL). All time points at 24 h. (B) Assays including H2O2. kobs values (at indicated [H2O2], h−1): 0 µM, 0.19; 1 µM, 0.24; 10 µM, 0.66; 100 µM, 10. The first data point for 1 mM H2O2 is at 30 s. (C) Assays including KO2. kobs values (at indicated [KO2], h−1): 0 µM, 0.19; 1 µM, 0.26; 10 µM, 0.22; 100 µM, 1.0.
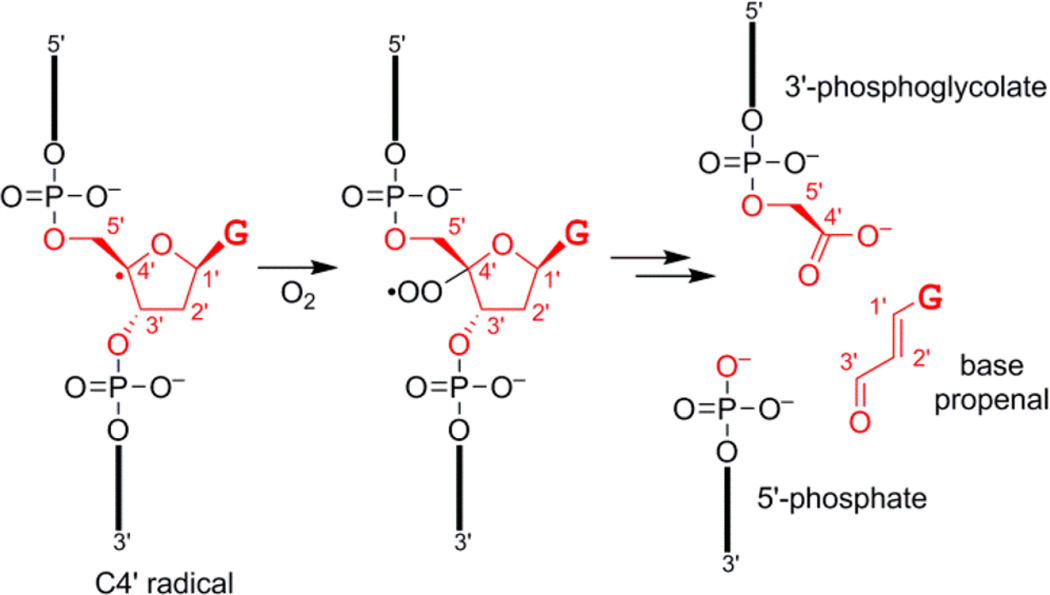
This combination of observations led us to consider the bleomycin radical pathway (Figure 4),14 in which abstraction of a hydrogen atom from the C4′ position of a nucleoside in the presence of O2 leads to a 3′-phosphoglycolate product. Because 3′-phosphoglycolate is a carboxylic acid, this DNA-catalyzed reaction product would be capturable during the in vitro selection process of Figure 1, explaining the survival of RadDz3. Of all 2′-deoxyribose hydrogen atom abstraction intermediates that can be formed from ssDNA, only the C4′ intermediate leads to a carboxylic acid.19 Glutathione is known to quench formation of bleomycin products, presumably by returning a hydrogen atom to the C4′ radical.20 The amide linkage in the originally used substrate would not necessarily be required, and indeed, RadDz3 functions well using an entirely DNA substrate, with 1.7-fold higher kobs than with the amide-containing substrate (data not shown).21
Figure 4.
DNA fragments formed upon reaction with the natural product bleomycin, shown starting with the C4′ radical intermediate and with excision of most atoms of a guanosine (G) nucleoside.14 Carbon atoms C1′-C5′ are marked on each structure. The 3′-phosphoglycolate, 5′-phosphate, and base propenal products are shown. The 3′-phosphoglycolate is a carboxylic acid and therefore capturable by the selection strategy of Figure 1.
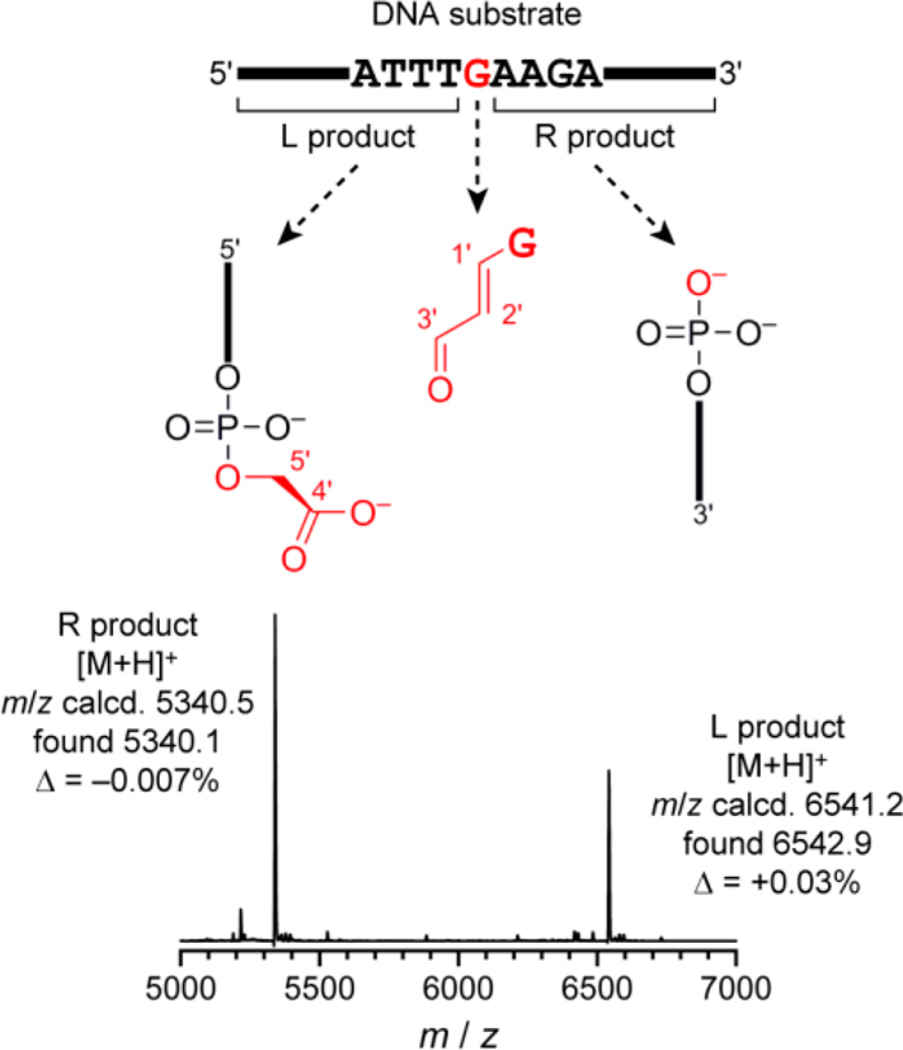
The RadDz3 products, formed either with or without inclusion of H2O2 in the reaction, were analyzed by MALDI mass spectrometry, revealing formation of the 3′-phosphogly-colate and 5′-phosphate products that result from excision of most of a particular guanosine nucleoside of the DNA substrate, located in the middle of the substrate (Figure 5). The small (MW = 205) base propenal fragment formed from the excised mononucleoside was also detected via a colorimetric assay (Figure 6), thereby accounting for all DNA substrate atoms.24 The mfold-predicted secondary structure of RadDz3 is shown in association with its DNA substrate in Figure 7.
Figure 5.
Identification by MALDI mass spectrometry of DNA-catalyzed DNA cleavage products formed by the RadDz3 deoxyribozyme. This spectrum is of the crude RadDz3 products formed in the absence of H2O2 (see the Experimental Section for preparative details). Essentially the same mass spectrum was observed for a sample prepared in the presence of 100 µM H2O2 (data not shown). The illustrated mass spectrum was obtained from cleavage product formed in the presence of all of Zn2+/Mn2+/Mg2+. The same cleavage products were observed upon reaction in the presence of Zn2+/Mg2+ without Mn2+ (data not shown).
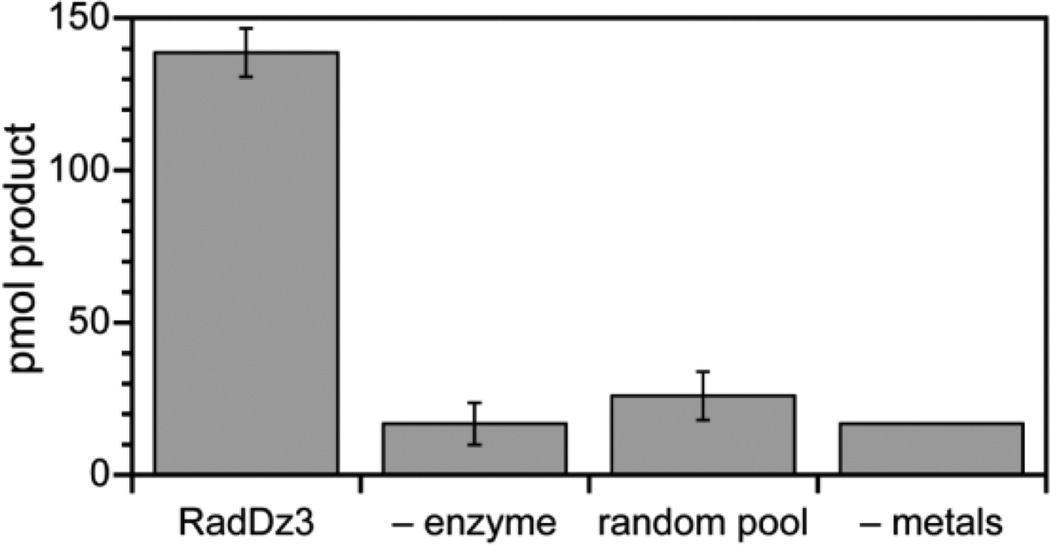
Figure 6.
Detection of the base propenal fragment released upon DNA cleavage by RadDz3 using a colorimetric assay. See the Experimental Section for assay details. The “product” observed for the three negative control experiments corresponds to background signal.
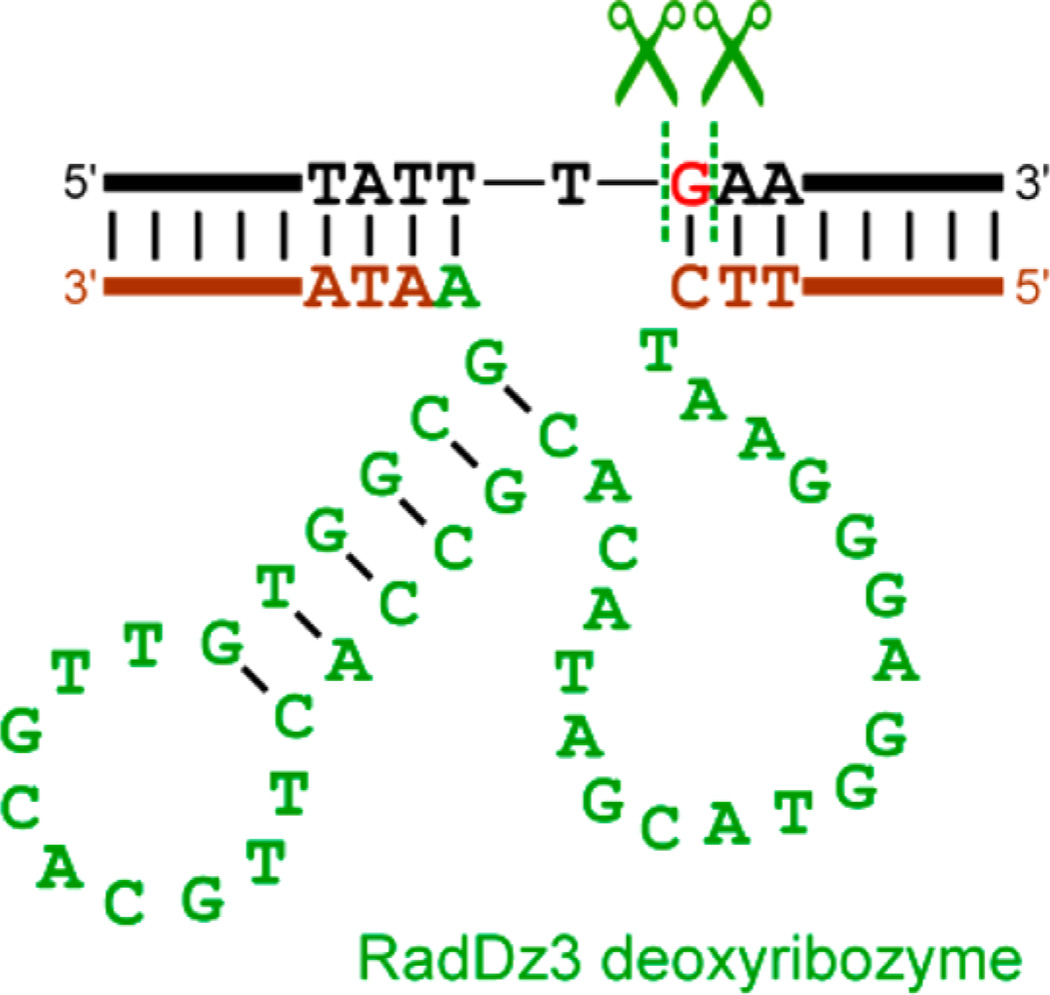
Figure 7.
Mfold-predicted25 secondary structure of the RadDz3 deoxyribozyme (green and brown), shown in association with the DNA substrate that RadDz3 cleaves (black; the excised G nucleoside is red). The RadDz6 deoxyribozyme has numerous alternative predicted secondary structures within 2 kcal/mol, none of which are shown here. See the Experimental Section for full sequences of the fixed segments (brown) that flank the initially random N40 region of RadDz3 (green) as well as the full RadDz6 sequence. The predicted 9 nt loop (TTGCACGTT) of RadDz3 was changed to 4 nt TTTT or GCGA. In each case, the shortened deoxyribozyme retained full activity (data not shown), consistent with but not proving the stem-loop structure. Further experiments to evaluate the RadDz3 secondary structure have not yet been performed.
A Second, N100 Deoxyribozyme That Cleaves DNA by the Same Radical Pathway
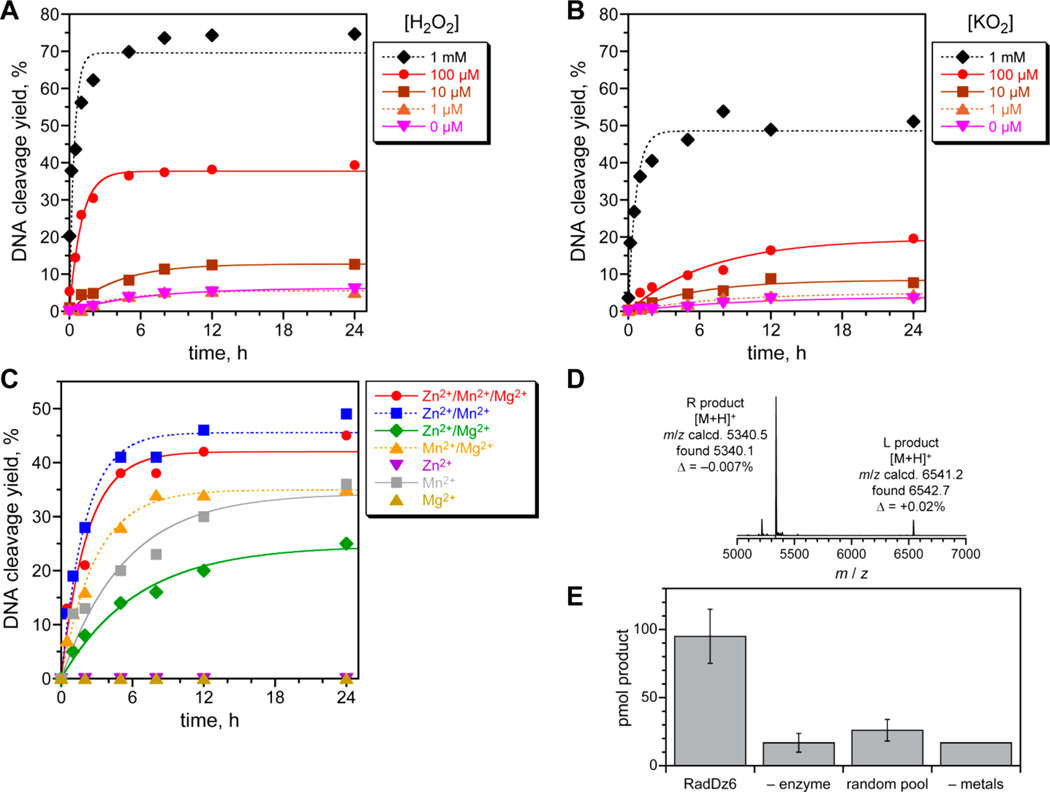
In parallel with the above experiments, we performed a separate, new in vitro selection experiment using an N100 random sequence population, considering that, in general, the length of the initially random region can be an important experimental variable.26 This selection experiment used the amide substrate of Figure 1 along with solely unmodified DNA and resulted in one new deoxyribozyme, named RadDz6, that cleaves a simple DNA oligonucleotide substrate. Under single-turnover conditions and in the absence of H2O2, RadDz6 has only 6% yield in 24 h, whereas, in the presence of 100 µM H2O2, kobs is 1.0 h−1 with 39% yield in 24 h; both kobs and yield are even higher at 1 mM H2O2 (Figure 8A). As was observed for RadDz3, inclusion of KO2 also increased the catalytic ability of RadDz6 (Figure 8B). Mn2+ supported the greatest activity by RadDz6, either alone or in combination with Zn2+ or Mg2+ (Figure 8C). Significantly, the combination of 1 mM Zn2+ and 40 mM Mg2+ in the absence of Mn2+ also supported substantial catalysis, demonstrating that RadDz6, like RadDz3, does not require a redox-active metal ion cofactor for its catalytic activity. As for RadDz3, glutathione quenches product formation (data not shown). Analysis of the RadDz6 DNA cleavage products by MALDI mass spectrometry revealed the same 3′-phosphoglycolate and 5′-hydroxyl products as formed by RadDz3 (Figure 8D), and the base propenal product was detected colorimetrically (Figure 8E). The combined data strongly suggest that RadDz3 and RadDz6 form their products through the common Figure 4 mechanistic pathway.
Figure 8.
Assays of the RadDz6 deoxyribozyme that cleaves ssDNA. (A) Kinetic results from PAGE analysis of single-turnover DNA cleavage. Incubation conditions: 70 mM HEPES, pH 7.5, 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2, 150 mM NaCl, and [H2O2] as indicated at 37 °C. (B) Kinetic results in the presence of KO2. Incubation conditions as in panel A, with [KO2] as indicated. (C) Kinetic results in the presence of various divalent metal ions. Incubation conditions as in panel A, with combinations of 1 mM ZnCl2, 20 mM MnCl2, and 40 mM MgCl2 as indicated, each at 100 µM H2O2. For this data set, kobs values (h−1): Zn2+/Mn2+/Mg2+ 0.46, Zn2+/Mn2+ 0.50, Zn2+/Mg2+ 0.16, Mn2+/Mg2+ 0.35, Mn2+ 0.19. (D) MALDI mass spectrometry of DNA-catalyzed DNA cleavage products, formed with Zn2+/Mn2+/Mg2+ and 100 µM H2O2. The same DNA cleavage site as for RadDz3 is indicated by the data (cf. Figure 5). (E) Detection of the base propenal fragment released upon DNA cleavage using a colorimetric assay (cf. Figure 6). See the Experimental Section for assay details. The “product” observed for the three negative control experiments corresponds to background signal.
DISCUSSION
In this study, we describe a new DNA-catalyzed reaction in which a deoxyribozyme, accompanied by redox-inactive metal ion cofactors, cleaves a single-stranded DNA substrate with excision of most atoms of a specific guanosine nucleoside. The biochemical and mass spectrometry data fully support a radical reaction pathway directly analogous to the O2-dependent bleomycin pathway, in which creation of a C4′ radical at the guanosine leads to formation of 3′-phosphoglycolate and 5′-phosphate DNA termini, along with loss of the remainder of the guanosine nucleoside as the base propenal fragment. The present findings constitute the first report of DNA-catalyzed oxidative DNA cleavage that does not require redox-active metals such as Cu2+ or Mn2+,11,12 as both the RadDz3 and RadDz6 deoxyribozymes can form their DNA cleavage products using only redox-inactive Zn2+ and Mg2+ as the cofactors. In contrast, the Cu2+/Mn2+-dependent oxidatively cleaving deoxyribozymes do not lead to well-defined products, instead forming a variety of fragments via “oxidative destruction” of one or more nucleosides.11,12
The new DNA-catalyzed cleavage reaction appears to represent a new mechanistic class for nucleic acid enzymes, considering the combination of radical-based chemistry and selectivity in product formation, in contrast to the myriad of oxidative DNA cleavage products formed by other deoxy-ribozymes.11,12 The current mechanistic model for the RadDz3/RadDz6 reaction pathway has unresolved aspects. The homogeneity of the reaction products indicates that the C4′ radical is formed at a specific guanosine nucleoside of the substrate, presumably as directed by the deoxyribozyme, but the details of C4′ radical generation are undefined by the current data. Metal ion redox activity is not required, because each deoxyribozyme is active in the presence of Zn2+/Mg2+. Additional work is required to understand formation of the C4′ radical and the detailed roles of superoxide and H2O2 in the reaction pathway. No damage of the deoxyribozymes themselves was detected by postreaction isolation and piperidine treatment (data not shown), indicating no obligatory redox participation of any of the deoxyribozyme nucleotides.
EXPERIMENTAL SECTION
Oligonucleotides
DNA oligonucleotides (including deoxyribozymes) were obtained from Integrated DNA Technologies (Coralville, IA) or prepared by solid-phase synthesis on an ABI 394 instrument using reagents from Glen Research. All oligonucleotides were purified by 7 M urea denaturing PAGE with running buffer 1× TBE (89 mM each Tris and boric acid and 2 mM EDTA, pH 8.3), extracted from the polyacrylamide with TEN buffer (10 mM Tris, pH 8.0, 1 mM EDTA, 300 mM NaCl), and precipitated with ethanol as described previously.27 The DNA substrate sequence was 5′-GGATAATACGACTCACTATTTGAAGAGATGGCGACTTCG-3′, where most of the underlined G is excised by RadDz3 and RadDz6.
In Vitro Selection and Identification of RadDz3 and RadDz6 Deoxyribozymes
RadDz3 was identified in our recent study that sought DNA-catalyzed amide hydrolysis, specifically from the in vitro selection experiment in which the HOdU modification was included.7 The incubation conditions used during identification of RadDz3 were 70 mM HEPES, pH 7.5, 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2, and 150 mM NaCl at 37 °C. The RadDz3 sequence is 5-CGAAGTCGCCATCTCTTCTAAGGGAGGTAC-GATACACGCCACTTGCACGTTGTGGCGAATAGTGAGTCGT-ATTA-3’ (74 nt), where the initially random N40 region is underlined.
RadDz6 was identified in a new in vitro selection experiment in which only unmodified DNA was used, along with an initially random N100 region. This experiment used the amide-containing substrate from Figure 1 (same substrate as was used in ref 7) and our earlier selection procedure, which uses the same capture strategy as depicted in Figure 1. The same incubation conditions as in identification of RadDz3 were used. After eight selection rounds, the capture yield was 8%, compared to 60–70% capture yield for the capture standard reaction, and individual deoxyribozymes were cloned from round 8. The RadDz6 sequence is 5-CGAAGTCG-CCATCTCTTCTAAGATAAACACACCGGCCGTCACGCTG-CCTCAGACGGTGAGTGACCCTAAAATAAGGGGATAACCCA-AGGCCGATATCCCAATTTGAGATGGTCGGGGAATAGTGAG-TCGTATTA-3’ (134 nt), where the initially random N100 region is underlined. No homology is evident between the RadDz3 and RadDz6 sequences.
Single-Turnover Deoxyribozyme Assay Procedure
The DNA substrate was 5’-32P-radiolabeled using γ-32P-ATP and polynucleotide kinase. A 10 µL sample containing 5 pmol of DNA substrate (of which 0.2 pmol was 5’-32P-radiolabeled) and 15 pmol of deoxyribozyme was annealed in 5 mM HEPES, pH 7.5, 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. The DNA-catalyzed cleavage reaction was initiated by bringing the sample to 20 µL total volume containing 70 mM HEPES, pH 7.5, 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2, and 150 mM NaCl. ZnCl2 was 99.999% (Alfa), where 0.001% of 1 mM is 10 nM. MnCl2 was 99.999% (Alfa). MgCl2 was >99.0% with ≤5 ppm Mn (EMD), where 5 ppm of 40 mM is 200 nM. H2O2 was added from a 100× stock solution, which was prepared from 30% H2O2 (8.82 M). KO2 was added from a 100× stock solution in water. Glutathione was added from a 10× stock solution in water (Sigma, cat. no. G4251). Catalase was added from a 200× stock solution in water (1.6 mg in 54.4 µL = 100 U/µL; catalase from bovine liver from Sigma, cat. no. C9322, 3390 U/mg). Superoxide dismutase (SOD) was added from a 200× stock solution in water (0.8 mg in 30.2 µL = 100 U/µL; Cu/Zn SOD from bovine erythrocytes from Sigma, cat. no. S7571, 3780 U/mg).
The sample was incubated at 37 °C. At each time point, a 2 µL aliquot was quenched with 5 µL stop solution (80% formamide, 1× TBE [89 mM each Tris and boric acid and 2 mM EDTA, pH 8.3], 50 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol).
Before PAGE for RadDz3 assays, to each quenched sample was added 100 pmol of a “decoy oligonucleotide”, which was a 60-mer complementary to the deoxyribozyme’s initially random region (40 nt) along with 10 nt of binding arm on either side. This decoy oligonucleotide was added to displace the deoxyribozyme from the substrate and cleavage product. When the decoy was omitted, gel bands were noticeably smeared, which inhibited proper quantification.
Samples were separated by 20% PAGE and quantified with a PhosphorImager. Values of kobs were obtained by fitting the yield versus time data directly to first-order kinetics; i.e., yield = Y(1 − e−kt), where k = kobs and Y is the final yield. When kobs was sufficiently low such that an exponential fit was not meaningful, the initial points were fit to a straight line, and kobs was taken as the slope of the line.
Mass Spectrometry Procedure
To prepare the cleavage products using a deoxyribozyme, a 50 µL sample containing 1.0 nmol of DNA substrate and 1.1 nmol of deoxyribozyme was annealed in 5 mM HEPES, pH 7.5, 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. The DNA-catalyzed cleavage reaction was initiated by bringing the sample to 100 µL total volume containing 70 mM HEPES, pH 7.5, 1.0 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2, 150 mM NaCl, and (when included) 100 µM H2O2. The sample was incubated at 37 °C for 24 h, precipitated with ethanol, desalted by Millipore C18 ZipTip, and analyzed by MALDI mass spectrometry. Data were acquired on a Bruker UltrafleXtreme MALDI-TOF mass spectrometer with matrix 3-hydroxypicolinic acid in positive ion mode at the UIUC School of Chemical Sciences Mass Spectrometry Laboratory.
Base Propenal Colorimetric Assay
Upon heating in aqueous solution, base propenal hydrolyzes to malondialdehyde, which may be detected by colorimetric assay upon condensation with 2 equiv of thiobarbituric acid.28 A 120 µL sample containing 1.0 nmol of DNA substrate and 1.1 nmol of deoxyribozyme was annealed in 5 mM HEPES, pH 7.5, 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. The DNA-catalyzed DNA cleavage reaction was initiated by bringing the sample to 300 µL total volume containing 70 mM HEPES, pH 7.5, 1 mM ZnCl2, 20 mM MnCl2, 40 mM MgCl2, 150 mM NaCl, and 100 µM H2O2. The sample was incubated at 37 °C for 2 h; longer incubation times could not be used due to malondialdehyde degradation. After addition of 500 µL of 50 mM thiobarbituric acid in water, the sample was incubated at 95 °C for 20 min and diluted by addition of 200 µL of water to a total of 1 mL volume. The absorbance at 532 nm was measured in a quartz cuvette with 1 cm path length (Thermo Scientific NanoDrop; 600 nm baseline subtraction). A calibration curve was created using malondialdehyde, which was prepared as follows. A sample containing 100 µL of 50 mM aqueous malondialdehyde bis(dimethyl acetal) (1,1,3,3-tetramethoxypropane; Aldrich), 100 µL of 1 M HCl, and 800 µL of water was heated at 55 °C for 1 h, cooled to room temperature, and diluted by addition of 4 mL of water, providing a 10 mM solution of malondialdehyde. A 0.1 mM stock solution of malondialdehyde was prepared by diluting 100 µL of the 10 mM solution to 10 mL. The calibration curve was created using 50–2000 pmol of malondialdehyde (0.5–20 µL of the 0.1 mM stock solution) in the above DNA cleavage procedure. The slope of the calibration plot was 0.145 nmol−1 .
Acknowledgments
This work was supported by a grant to S.K.S. from the National Institutes of Health (R01GM065966). Y.L. was partially supported by a Grant-In-Aid of Research from Sigma Xi. B.M.B. was partially supported by NIH T32GM070421.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.(a) Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]; (b) Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; (c) Robertson DL, Joyce GF. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]; (d) Joyce GF. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]; (e) Joyce GF. Angew. Chem., Int. Ed. 2007;46:6420–6436. doi: 10.1002/anie.200701369. [DOI] [PubMed] [Google Scholar]
- 2.(a) Silverman SK. Chem. Commun. 2008:3467–3485. doi: 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]; (b) Schlosser K, Li Y. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]; (c) Silverman SK. Acc. Chem. Res. 2009;42:1521–1531. doi: 10.1021/ar900052y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Zhang XB, Kong RM, Lu Y. Annu. Rev. Anal. Chem. 2011;4:105–128. doi: 10.1146/annurev.anchem.111808.073617. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Gong L, Zhao Z, Lv YF, Huan SY, Fu T, Zhang XB, Shen GL, Yu RQ. Chem. Commun. 2015;51:979–995. doi: 10.1039/c4cc06855f. [DOI] [PubMed] [Google Scholar]; (g) Silverman SK. Acc. Chem. Res. 2015;48:1369–1379. doi: 10.1021/acs.accounts.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Hollenstein M. Molecules. 2015;20:20777–20804. doi: 10.3390/molecules201119730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breaker RR, Joyce GF. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 4.Cuchillo CM, Nogues MV, Raines RT. Biochemistry. 2011;50:7835–7841. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Faulhammer D, Famulok M. Angew. Chem., Int. Ed. Engl. 1996;35:2837–2841. [Google Scholar]; (b) Santoro SW, Joyce GF. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Li J, Zheng W, Kwon AH, Lu Y. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Schlosser K, Li Y. ChemBioChem. 2010;11:866–879. doi: 10.1002/cbic.200900786. [DOI] [PubMed] [Google Scholar]; (e) Silverman SK. Nucleic Acids Res. 2005;33:6151–6163. doi: 10.1093/nar/gki930. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Torabi S-F, Wu P, McGhee CE, Chen L, Hwang K, Zheng N, Cheng J, Lu Y. Proc. Natl. Acad. Sci. U. S. A. 2015;112:5903–5908. doi: 10.1073/pnas.1420361112. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Saran R, Liu J. Anal. Chem. 2016;88:4014–4020. doi: 10.1021/acs.analchem.6b00327. [DOI] [PubMed] [Google Scholar]
- 6.Brandsen BM, Hesser AR, Castner MA, Chandra M, Silverman SK. J.. Am. Chem. Soc. 2013;135:16014–16017. doi: 10.1021/ja4077233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou C, Avins JL, Klauser PC, Brandsen BM, Lee Y, Silverman SK. J. Am. Chem. Soc. 2016;138:2106–2109. doi: 10.1021/jacs.5b12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Chandra M, Sachdeva A, Silverman SK. Nat. Chem. Biol. 2009;5:718–720. doi: 10.1038/nchembio.201. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xiao Y, Wehrmann RJ, Ibrahim NA, Silverman SK. Nucleic Acids Res. 2012;40:1778–1786. doi: 10.1093/nar/gkr860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker DJ, Xiao Y, Aguilar JM, Silverman SK. J. Am. Chem. Soc. 2013;135:8472–8475. doi: 10.1021/ja4032488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Sheppard TL, Ordoukhanian P, Joyce GF. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7802–7807. doi: 10.1073/pnas.97.14.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hobartner C, Pradeepkumar PI, Silverman SK. Chem. Commun. 2007:2255–2257. doi: 10.1039/b704507g. [DOI] [PubMed] [Google Scholar]; (c) Dokukin V, Silverman SK. Chem. Sci. 2012;3:1707–1714. doi: 10.1039/C2SC01067D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Carmi N, Shultz LA, Breaker RR. Chem. Biol. 1996;3:1039–1046. doi: 10.1016/s1074-5521(96)90170-2. [DOI] [PubMed] [Google Scholar]; (b) Carmi N, Balkhi SR, Breaker RR. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2233–2237. doi: 10.1073/pnas.95.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Carmi N, Breaker RR. Bioorg. Med. Chem. 2001;9:2589–2600. doi: 10.1016/s0968-0896(01)00035-9. [DOI] [PubMed] [Google Scholar]
- 12.(a) Wang M, Zhang H, Zhang W, Zhao Y, Yasmeen A, Zhou L, Yu X, Tang Z. Nucleic Acids Res. 2014;42:9262–9269. doi: 10.1093/nar/gku592. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang M-Q, Dong J, Zhang H, Tang Z. Org. Biomol. Chem. 2016;14:2347–2351. doi: 10.1039/c6ob00148c. [DOI] [PubMed] [Google Scholar]
- 13.Burmeister J, von Kiedrowski G, Ellington AD. Angew. Chem., Int. Ed. Engl. 1997;36:1321–1324. [Google Scholar]
- 14.(a) Sugiyama H, Xu C, Murugesan N, Hecht SM, van der Marel GA, van Boom JH. Biochemistry. 1988;27:58–67. doi: 10.1021/bi00401a011. [DOI] [PubMed] [Google Scholar]; (b) Burger RM. Chem. Rev. 1998;98:1153–1170. doi: 10.1021/cr960438a. [DOI] [PubMed] [Google Scholar]; (c) Claussen CA, Long EC. Chem. Rev. 1999;99:2797–2816. doi: 10.1021/cr980449z. [DOI] [PubMed] [Google Scholar]; (d) Hecht SMJ. Nat. Prod. 2000;63:158–168. doi: 10.1021/np990549f. [DOI] [PubMed] [Google Scholar]
- 15.(a) Cuenoud B, Szostak JW. Nature. 1995;375:611–614. doi: 10.1038/375611a0. [DOI] [PubMed] [Google Scholar]; (b) Dokukin V, Silverman SK. Chem. Commun. 2014;50:9317–9320. doi: 10.1039/c4cc04253k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ma L, Liu B, Huang PJ, Zhang X, Liu J. Langmuir. 2016;32:5672–5680. doi: 10.1021/acs.langmuir.6b00906. [DOI] [PubMed] [Google Scholar]
- 16.Chelation of metal ions by glutathione cannot explain the observed inhibition. Even if 1 mM glutathione chelates all 1 mM Zn2+, 17 we would still observe considerable activity with the remaining Mn2+/Mg2+, as shown in Figure 2.
- 17.Kręzel A, Maret W. Arch. Biochem. Biophys. 2016;611:3–19. doi: 10.1016/j.abb.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adding H2O2 increases the activity of unrelated Cu2+/Mn2+-dependent deoxyribozymes that oxidatively cleave DNA.12
- 19.Dedon PC. Chem. Res. Toxicol. 2008;21:206–219. doi: 10.1021/tx700283c. [DOI] [PubMed] [Google Scholar]
- 20.Giese B, Dussy A, Elie C, Erdmann P, Schwitter U. Angew. Chem., Int. Ed. Engl. 1994;33:1861–1863. [Google Scholar]
- 21.We made several unsuccessful attempts to exclude O2. Use of the glucose oxidase oxygen scavenging system22 was precluded by its direct production of H2O2, and catalase could not be added as usual for this system because the deoxyribozyme itself generates and requires H2O2. Use of the protocatechuate dioxygenase oxygen scavenging system23 was thwarted because the PCD enzyme itself, in the absence of its protocatechuic acid substrate, was found to inhibit the deoxyribozyme. Finally, physical purging of O2 using argon had no measurable effect.
- 22.Benesch RE, Benesch R. Science. 1953;118:447–448. doi: 10.1126/science.118.3068.447. [DOI] [PubMed] [Google Scholar]
- 23.Aitken CE, Marshall RA, Puglisi JD. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A distinct deoxyribozyme identified alongside RadDz3 formed analogous products but with excision of most of the T nucleoside immediately to the 5′-side of the G that is excised by RadDz3 (data not shown).
- 25.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velez TE, Singh J, Xiao Y, Allen EC, Wong O, Chandra M, Kwon SC, Silverman SK. ACS Comb. Sci. 2012;14:680–687. doi: 10.1021/co300111f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(a) Flynn-Charlebois A, Wang Y, Prior TK, Rashid I, Hoadley KA, Coppins RL, Wolf AC, Silverman SK. J. Am. Chem. Soc. 2003;125:2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]; (b) Wang Y, Silverman SK. Biochemistry. 2003;42:15252–15263. doi: 10.1021/bi0355847. [DOI] [PubMed] [Google Scholar]
- 28.(a) Waravdekar VS, Saslaw LD. J. Biol. Chem. 1959;234:1945–1950. [PubMed] [Google Scholar]; (b) Burger RM, Berkowitz AR, Peisach J, Horwitz SB. J. Biol. Chem. 1980;255:11832–11838. [PubMed] [Google Scholar]; (c) Zhou X, Taghizadeh K, Dedon PC. J. Biol. Chem. 2005;280:25377–25382. doi: 10.1074/jbc.M503079200. [DOI] [PubMed] [Google Scholar]