Abstract
Even in the era of combination antiretroviral therapies used to combat human immunodeficiency virus type 1 (HIV-1) infection, up to 50% of well-suppressed HIV-1-infected patients are still diagnosed with mild neurological deficits referred to as HIV-associated neurocognitive disorders (HAND). The multifactorial nature of HAND likely involves the HIV-1 accessory protein viral protein R (Vpr) as an agent of neuropathogenesis. To investigate the effect of naturally occurring variations in Vpr on HAND in well-suppressed HIV-1-infected patients, bioinformatic analyses were used to correlate peripheral blood-derived Vpr sequences with patient neurocognitive performance, as measured by comprehensive neuropsychological assessment and the resulting Global Deficit Score (GDS). Our studies revealed unique associations between GDS and the presence of specific amino acid changes in peripheral blood-derived Vpr sequences [neuropsychological impairment Vpr (niVpr) variants]. Amino acids N41 and A55 in the Vpr sequence were associated with more pronounced neurocognitive deficits (higher GDS). In contrast, amino acids I37 and S41 were connected to measurably lower GDS. All niVpr variants were also detected in DNA isolated from HIV-1-infected brain tissues. The implication of these results is that niVpr variants alter the genesis and/or progression of HAND through differences in Vpr-mediated effects in the peripheral blood and/or the brain.
Keywords: HIV, Vpr, HAND, neuropathogenesis, GDS, quasispecies, sequence
Introduction
Disease associated with human immunodeficiency virus type 1 (HIV-1) infection includes significant neuropathogenesis subsequent to virus entry into the central nervous system (CNS). Although the availability of effective combination antiretroviral therapy (cART) to infected individuals has both increased life expectancy and reduced the incidence of severe and progressive neuropsychological impairment (NPI) associated with HIV-1 infection (including HIV-1-associated dementia), HIV-1-infected patients continue to develop neuropsychological and neurological deficits (Clifford and Ances, 2013; Heaton et al, 2010; Heaton et al, 2011; Simioni et al, 2010). In the era of cART, up to half of all treated patients are diagnosed with mild to moderate neurological and neuropsychological deficits broadly referred to as HIV-associated neurocognitive disorders (HAND) (Antinori et al, 2007; Clifford and Ances, 2013; Spudich and Gonzalez-Scarano, 2012).
The cellular and molecular basis of HAND has been shown to be complex and multi-factorial. Following HIV-1 entry into the brain, infection of susceptible cells (macrophages, microglia, and, to a lesser extent, astrocytes) causes the aberrant release of numerous cellular neurotoxins, chemokines, and pro-inflammatory cytokines. In addition, productive viral infection results in the release of viral proteins, including gp120, Tat, and Nef, that also function as neurotoxins (Schouten et al, 2011; Spudich and Gonzalez-Scarano, 2012). Collectively, these factors have a detrimental effect on the function and viability of CNS-resident cells, including neurons, astrocytes, and possibly other cell populations (as well as those at the blood-brain barrier). An HIV-1 accessory protein, viral protein R (Vpr), has also been identified as an agent of neuropathogenesis. Vpr has numerous functions and effects (Guenzel et al, 2014), including (i) facilitation of infection in non-dividing cells by participating in pre-integration complex nuclear transport (Hrimech et al, 1999), (ii) trans-activation of the HIV-1 LTR (Hogan et al, 2003; Wang et al, 1995), and (iii) initiation of G2 cell cycle arrest (He et al, 1995). In the context of the brain, Vpr has been shown to cause neuronal apoptosis and cell death (Jones et al, 2007; Patel et al, 2000; Piller et al, 1998; Sabbah and Roques, 2005), inhibition of axonal outgrowth (Kitayama et al, 2008), neuronal dysfunction through microRNA deregulation (Mukerjee et al, 2011), astrocyte necrosis (Huang et al, 2000), induction of astrocyte CCL5 expression (Gangwani et al, 2013), and neuropathic changes that have been associated with memory impairment (Torres and Noel, 2014).
Although many functions of Vpr follow from its intracellular expression, localization with the cell (Ferrucci et al, 2011a), and incorporation into virions in large quantities (Lu et al, 1995), Vpr (like gp120 and Tat) has been shown to function in the extracellular environment (Ferrucci et al, 2011b). Extracellular Vpr has been detected in the sera and cerebrospinal fluids of HIV-1-infected patients (Hoshino et al, 2007; Levy et al, 1994). As an extracellular protein, its effects include astrocyte necrosis (Huang et al, 2000), stunted neuronal axon elongation (Kitayama et al, 2008), cellular transduction resulting in cell cycle arrest and apoptosis (Henklein et al, 2000; Patel et al, 2000; Sabbah and Roques, 2005), activation of signal transduction and transcription in macrophages (Varin et al, 2005), and enhanced HIV-1 replication (Levy et al, 1995; Sherman et al, 2002; Varin et al, 2005). Along these lines, we previously demonstrated that addition of extracellular Vpr to astrocytes caused reductions in adenosine-5’-triphosphate (ATP) and glutathione (GSH), increased caspase activity, increased cytokine and chemokine release, reduced glyceraldehyde-3-phosphate dehydrogenase (GAPDH) activity, and increased reactive oxygen species (Ferrucci et al, 2012; Ferrucci et al, 2013). Furthermore, conditioned media from Vpr-exposed astrocytes caused reductions in neuronal GSH leading to apoptosis (Ferrucci et al, 2013).
The vpr coding sequence, as well as the rest of the HIV-1 proviral genome, is subject to variation during each replication cycle due to the error-prone nature of reverse transcription (Lloyd et al, 2014). Estimates of sequence diversity set the error rate during replication at approximately 1.4 × 10−5 mutations per base pair per replication cycle (Abram et al, 2010). Our recent efforts have turned to investigating potential connections between Vpr sequence variation and HIV-1-associated neuropathogenesis in well-suppressed patients. Given the putative role of Vpr in HIV-1-associated neuropathogenesis, we hypothesized that amino acid changes in Vpr that alter its effects in the brain and/or peripheral blood will result in clinically measurable differences in HAND. Analyses of viral sequences isolated from peripheral blood samples have revealed four amino acids at three positions in Vpr that are associated with significant differences in neuropsychological function as measured by comprehensive neuropsychological assessment. The discovery of these neuropsychological impairment Vpr (niVpr) variants has suggested that Vpr contributes to the onset and/or progression of HAND in HIV-1-infected patients and that those contributions can be altered by specific amino acid changes in Vpr.
Materials and Methods
Collection of PBMCs from HIV-1-infected patients
Patients enrolled in the Drexel Medicine CNS AIDS Research and Eradication Study (CARES) Cohort were recruited from the Partnership Comprehensive Care Practice of the Division of Infectious Disease and HIV Medicine in the Department of Medicine at Drexel University College of Medicine (Philadelphia, Pennsylvania, USA) and the Center for Clinical and Translational Medicine in the Drexel Institute for Molecular Medicine and Infectious Disease. Patients in the Drexel Medicine CARES Cohort were recruited under protocol 1201000748 (Brian Wigdahl, PI), which adheres to the ethical standards of the Helsinki Declaration (1964, amended most recently in 2008), which was developed by the World Medical Association as described (Li et al, 2011). All patients provided written consent upon enrollment.
The CARES Cohort, which was the source of patients involved in this study, is representative of the larger HIV-infected patient population at Drexel, with the majority of patients being black males currently on cART. Key demographics for the patients encompassed by this study, as well as all patients in the Cohort, are provided in Table 1. Forty-five percent of the cohort has never experienced an AIDS-defining illness, while 55% have had at least clinical event. Cohort patients largely use and respond to ART with viral loads and CD4+ T-cell counts consistent with effective viral suppression. Because drug abuse is common in the cohort, patients who showed signs of drug or alcohol intoxication were excluded from the study. A small number of subjects may have been intoxicated at the time of collection with no visible signs of impairment. Patients who later tested positive for drug use but showed no signs of intoxication at the time of collection were not excluded from the study. The presence of blood-borne drug metabolites does not necessarily indicate that a patient was intoxicated during the visit, as the half-lives of these products extend past their neurological effects.
Table 1.
Demographics of the Drexel Medicine CNS AIDS Research and Eradication Study (CARES) Cohort. Characteristics for the entire patient cohort (as of September 2015) are provided, as are the corresponding demographics for the 112 patients encompassed by this study. Each parameter is accompanied by a 95% confidence interval (in parentheses). Years seropositive is determined by a review of clinical history information.
| Entire Cohort | Patients in this Study | |
|---|---|---|
| Age (years) | 49 (42-45) | 51 (46-56) |
| Current Drug Use | 71.00% | 63.00% |
| Gender | 62% Male | 64% Male |
| Latest CD4 T-Cell Count (cells/mm3) | 521 (328-793) | 641 (428-904) |
| Latest Viral Load (copies/mL) | 48 (22-943) | 20 (13-193) |
| Years on ART | 14 (9-18) | 14 (9-18) |
| Years Seropositive | 16 (11-22) | 19 (14-24) |
Comprehensive Neuropsychological Assessment
A subset of consenting patients recruited into the CARES Cohort undergo yearly Comprehensive Neuropsychological Assessment (CNPA). To date, 191 HIV-1-infected cohort patients have been evaluated using the CNPA. The CNPA includes (i) assessment of global cognition using the Mini-Mental State Examination (MMSE) and premorbid general intellectual functions using the Wide Range Achievement-4 single word reading test and Wechsler Adult Intelligence Scale-IV Information subset; (ii) a self-reported assessment of everyday cognitive and functional status; and (iii) assessment of seven primary neuropsychological domains using tests from the Calibrated Neuropsychological Normative System (CNNS) (Schretlen et al, 2010) and the Spanish and English Neuropsychological Assessment Scales (SENAS) (Mungas et al, 2000) that control for key demographic variables. The seven neuropsychological domains are: (i) working memory/attention, (ii) executive function, (iii) information processing speed, (iv) visuoconstruction, (v) motor function, (vi) verbal/visual learning and memory, and (vii) language.
Approximately 85-90% of our participants are modestly educated African-Americans. The neuropsychological literature clearly underscores the need to use educational/cultural norms in order to obtain an accurate assessment of putative NPI (Antinori et al, 2007). The CNNS and SENAS were both selected because of their strong psychometric properties and because scores may be adjusted for demographic variables, including age, education, race, and gender.
Demographically adjusted scores for each test were converted to deficit scores, which give relatively greater weight to impaired performances than to average or above average performances (Carey et al, 2004). Summary deficit scores for all seven primary neuropsychological domains were averaged to determine a Global Deficit Score (GDS), which indicates an overall level of impairment. The GDS range is 0-5, with 0 indicating no impairment and 5 indicating severe impairment (Blackstone et al, 2012; Carey et al, 2004). The GDS is modeled after the “gold standard” clinical ratings method of diagnosing HAND (Butters et al, 1990; Heaton et al, 1995) and is intended to approximate this approach using a psychometric method that requires less time and expertise. A GDS ≥ 0.5 has demonstrated optimal sensitivity and specificity in detecting HAND as diagnosed by clinical ratings (Carey et al, 2004). Although the present CNPA includes slightly different tests from those included in the initial validation of the GDS, the neuropsychological constructs assessed by both protocols are comparable.
Secondarily, global NPI was assessed during each visit (approximately every six months) using a modified version of the Hopkins Dementia Scale (MHDS) (Li et al, 2011; Power et al, 1995). This exam screens for short-term memory, concentration, and processing speed, and provides a score that ranges from 12 (unaffected) to 0 (severely affected), with scores >10 indicating non-impairment. MHDS scores are used as secondary neurocognitive function indicators between annual CNPA.
Genomic DNA isolation from HIV-1-infected PBMC and brain tissue
A subset of the patients in the Drexel Medicine CARES Cohort who received CNPA (n = 112/191) had whole blood collected and processed for drug screening, plasma analysis, PBMC isolation, and genomic DNA isolation as previously described (Aiamkitsumrit et al, 2014; Li et al, 2011). Brain (head of caudate) tissues and matched spleen samples from three HIV-1-infected patients were acquired from the National NeuroAIDS Tissue Consortium (NNTC) (Morgello et al, 2001). Frozen brain and spleen tissue were punch-biopsied using a 3 mm disposable biopsy punch (Sklar Cat # 96-1105) and placed into a collection tube. DNA was extracted from the sample using the DNeasy® Blood & Tissue procedure (Qiagen Cat # 69504) as described by the manufacturer.
DNA amplification and sequencing – Sanger Sequencing
The vpr coding region was amplified using primers specific for the entire vpr reading frame. HIV-1 vpr was amplified from each patient-derived genomic DNA sample by performing a nested PCR. The first round of this reaction contained Phusion High-Fidelity Polymerase (0.5 U), HF buffer, MgCl2 (1.5 mM), dNTPs (350 μM), primers BA56 (GGAGGAAAAAGAGATATAGCACACAAGTAGACCC) and BA29 (AATAGAGTGGTTGCTTCCTTCC) with 125 ng of genomic DNA in a total volume of 25 μl. The reaction consisted of 98°C for 3 minutes, and then 25 cycles of 98°C for 10 seconds, 58°C for 20 seconds, and 72°C for 30 seconds followed by a 10 minute extension at 72°C. The second round reaction contained Phusion High-Fidelity Polymerase (0.5 U), HF buffer, MgCl2 (1.5 mM), dNTPs (350 μM), primers vprF1 (CAAGTAGACCCTGAACTAGC) and vprR2 (CTTCACTCTCATTGCCACT) with 2.5 μl of round one in a total volume of 50 μl. The reaction included 98°C for 3 minutes, and then 30 cycles of 98°C for 10 seconds, 58°C for 20 seconds, and 72°C for 24 seconds followed by a 10 minute extension at 72°C. PCR products from the second round were then treated with ExoSAP-IT (Affymetrix) at a ratio of approximately 5 μl of PCR reaction to 2 μl of enzyme. Treated product (20 ng) was then sent for Sanger sequencing by Genewiz (South Plainfield, NJ), following their current instructions.
HIV-1 vpr was amplified from each NNTC brain or spleen-derived genomic DNA sample by performing a nested PCR. The first round of this reaction contained Phusion High-Fidelity Polymerase (0.5 U), GC buffer, MgCl2 (1.5 mM), dNTPs (350 μM), primers BA56 and BA29 with 125 ng of genomic DNA in a total volume of 25 μl. Amplification was performed at 98°C for 3 minutes, and then 30 cycles of 98°C for 10 seconds, 58°C for 20 seconds, and 72°C for 35 seconds followed by a 10 minute extension at 72°C. The second round reaction contained Phusion High-Fidelity Polymerase (0.5 U), GC buffer, MgCl2 (1.5 mM), dNTPs (350 μM), primers vprF1 and vprR1 with 2.5 μl of round one in a total volume of 50 μl. The amplification was performed at 98°C for 3 minutes, and then 30 cycles of 98°C for 10 seconds, 51°C for 20 seconds, and 72°C for 30 seconds followed by a 10 minute extension at 72°C. PCR products from the second round were then treated with ExoSAP-IT (Affymetrix) at a ratio of approximately 5 μl of PCR reaction to 2 μl of enzyme. The treated product (20 ng) was then sequenced using Sanger technology ( Genewiz ,South Plainfield, NJ).
DNA amplification and sequencing – Next Generation Sequencing
Illumina deep sequencing of PBMC- and brain-derived DNA was performed by using a PCR amplification technique in which the vpr region is flanked by primers that produce an ~1 Kb segment (Henn et al, 2012). Primer sites were selected such that they exactly matched >95% of the subtype B sequences in the Los Alamos National Library (LANL) HIV database. HIV-1 vpr was amplified from patient-derived genomic DNA samples by performing a PCR, which yields a fragment spanning from nucleotide 5516 to nucleotide 6582 with respect to the HXB2 genome. This reaction contained Phusion High-Fidelity Polymerase (0.5 U), HF buffer, MgCl2 (1.5 mM), dNTPs (350 μM), primers Frag-37-L (AAAGCCACCTTTGCCTAGTG) and Frag-37-R (ACACATGGCTTTAGGCTTTG) with 100 ng of genomic DNA in a total volume of 25 μl. The reaction was performed at 95°C for 2 minutes, and then 40 cycles of 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 1 minute followed by a 10 minute extension at 72°C. Samples were then purified using the QIAquick PCR Purification procedure (Qiagen Cat # 28104) as described by the manufacturer and eluted in water. Samples were then prepared using the Nextera XT DNA Library Preparation Guide (Illumina Cat # FC-131-1096) as described by the manufacturer, except where specified below. The amount of input DNA was changed from 0.2 ng/μL to 0.4 ng/μL. The tagmentation reaction was shortened from 5 minutes at 55°C to 4 minutes and 30 seconds to avoid over-fragmenting the shorter PCR fragments. PCR clean-up was completed using 30 μL per sample of the AxyPrep™ Mag PCR Clean-Up procedure (Axygen® Cat # MAG-PCR-CL-5) as described by the manufacturer. Libraries were normalized using the Quant-iT™ dsDNA Assay procedure, High Sensitivity (Invitrogen Cat # Q33120), and validated on an Agilent Technology 2100 Bioanalyzer using the Agilent High Sensitivity DNA procedure as previously described (Agilent Technologies Cat # 5067-4626). Based on the average size of 500 bp, libraries were clustered using the equation 1 ng/μl = 3 nM. Samples were pooled together to reach a final concentration of 1 nM and sequenced using the NextSeq® 500/550 Mid Output Reagent Cartridge v2 300 cycles (Illumina Cat # FC-404-2003) on the Illumina NextSeq 500 Desktop Sequencer.
DNA sequence analyses
Analyses of Vpr sequences obtained using Sanger technology from HIV-1-infected patient blood samples were performed using sequences isolated from 112 CARES Cohort patients with CNPA. Sequences were aligned to consensus B (ConB; 2002) using the MUSCLE alignment tool with default settings (Edgar, 2004). These sequences were translated into the proper reading frame by comparison with HXB2 and all amino acid calls were inferred from the DNA sequence data for statistical analysis.
NGS Vpr sequences were processed by first using Trimmomatic (Bolger et al, 2014) to remove poor quality ends and trim off primer sequences using the default settings. Cleaned paired-end reads were then aligned to the HXB2 genome using the bwa mem tool (Li and Durbin, 2009) and duplicate reads from the same lane were removed using the samblaster tool (Faust and Hall, 2014). Pysam (Li et al, 2009) was used to extract each read with a mapping quality greater than a PHRED score of 60 and Biopython (Cock et al, 2009) was used to translate the read in the proper reading frame and infer the amino acid at each position along the read for downstream statistical analysis.
Combined effect size (CES) analyses
Each of the 112 patient sequences was scored based on the identities of amino acids at positions 37, 41, and 55. A niVpr amino acid at each position was scored using the magnitude and direction of its effect size (Fig. 2), while non-niVpr amino acids were assumed to have no effect on GDS (null) and were assigned a score of 0. GDS scores were grouped and averaged according the total combined effect size (CES), which ranged from −0.250 to +0.516 (12 possible combinations of niVpr and null amino acids). The effects of each site were assumed to be independent.
Statistical analyses
For statistical considerations, we assumed a null model in which neuropsychological function was solely a function of age, gender, ART status, log10 (viral load), and years seropositive. This was implemented as a Least-Squares model using the Python Statsmodels toolbox (Seabold and Perktold, June 28 - July 3, 2010). We then iteratively added each column of the Vpr alignment as a categorical variable to determine whether this sequence variable improved the model. This was determined using a chi-squared distribution to test whether the improved log-likelihood of the model exceeded the expected improvement of adding greater degrees of freedom (Seabold and Perktold, June 28 - July 3, 2010). Neuropsychological function was measured by both the GDS and MHDS at the time of visit. The results of the regression analysis provide the effect of an amino acid change on the neuropsychological function when controlled for age, gender, ART status, log viral load, and years seropositive. Bonferroni multiple-testing correction (p < 0.05/97 = 0.000514) was used to account for potential false discovery.
Results
Cohort patients have a range of HIV-1-associated neurocognitive function
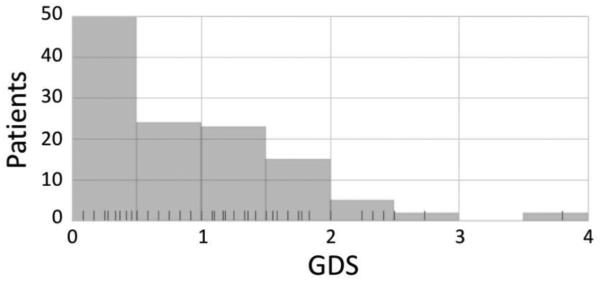
Neuropsychological assessments of HIV-1-infected patients currently enrolled in the CARES Cohort at the Drexel University College of Medicine (Aiamkitsumrit et al, 2014; Dampier et al, 2016; Li et al, 2011; Nonnemacher et al, 2016; Parikh et al, 2014; Shah et al, 2014) revealed a range of neurocognitive function, as indicated by GDS. Most Cohort patients for which Vpr sequences were available had GDS ≤2 (Fig. 1); this is not surprising, since most patients are well-suppressed by effective antiretroviral therapy. Fewer patients had scores in the range of 2-4, and no patients scored >4. To put these results in the context of established measures of HIV-1-associated NPI, ongoing studies with our HIV-1-infected patients using the CNPA (Devlin et al, 2015) have shown 58% of the population meets the Frascati criteria (Antinori et al, 2007) for either Mild Neurocognitive Disorder (MND) or Asymptomatic Neurocognitive Impairment (ANI), a prevalence rate that is comparable to other studies that use different neuropsychological measures (Heaton et al, 2010). Although not yet widely used in the HIV research community, these data suggest that our CNPA protocol can determine the presence of ANI and MND using previously described recommendations (Antinori et al, 2007). In addition, comparative analyses of patient GDS and MHDS results indicated a general correspondence between the two assessment outcomes (data not shown).
Fig. 1.
CARES Cohort patient GDS summary. Rug ticks represent individual GDS scores from patient samples that have Vpr sequences isolated from the same visit as the CNPA exam. Of the 112 patients encompassed by this study, 40.7% were considered unimpaired (GDS < 0.5), 51.7% were mildly impaired (0.5 ≤ GDS < 2), 5.9% were moderately impaired (2 ≤ GDS < 3.5), and 1.7% were severely impaired (GDS ≥ 3.5).
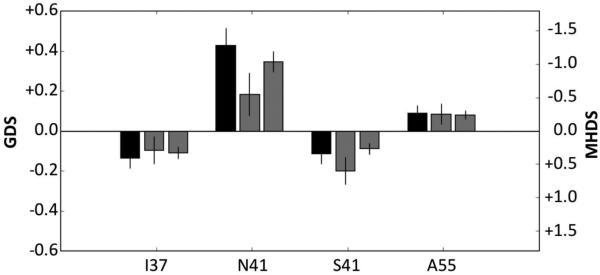
Specific amino acids in Vpr are significantly associated with differences in neurocognitive function in HIV-1-infected Cohort patients
Correlations were examined between Vpr sequences from 112 Cohort patients (one Sanger-derived sequence per patient) and corresponding CNPA results. Results of the analyses, which passed Bonferroni multiple-testing correction (p < 0.05/97 = 0.000514), had an R2 > 0.5, and an effect size greater than 0.15 (Fig. 2). Effect size (left y axis) is the difference in GDS associated with each amino acid position when compared to the average GDS. Positive and negative effect sizes indicate higher or lower GDS, respectively, relative to the patient average. These analyses indicated significant associations between the presence of four amino acids at three specific sites (niVpr variants) within the primary Vpr sequence (positions 37, 41, and 55) and altered neurocognitive function in HIV-1-infected patients. Specifically, amino acids N41 and A55 were associated with higher GDS (“detrimental”), while amino acids I37 and S41 were associated with reduced GDS (“beneficial”). Strikingly, MHDS testing results were in general agreement with the magnitudes and directions of differences in NPI indicated by the GDS effect sizes. While significant GDS effect sizes were indicated at other positions (positive effect sizes for V37, Y84, S89, and I93; negative effect size for T55), those changes were not completely correlated with changes in MHDS (data not shown), possibly as a consequence of the greater sensitivity of the CNPA relative to the MHDS.
Fig. 2.
Vpr amino acid variants are associated with differences in GDS. The change in GDS (left y-axis and black bars) in 112 CNPA-evaluated patients is plotted with respect to the position and identity of the amino acid. The middle gray bars show the effect sizes on MHDS (right y-axis) for the same 112 patients. The right gray bars show the effect sizes on MHDS in all patients (543 patients who were evaluated with both CNPA and MHDS or MHDS only). Error bars indicate 95% confidence intervals on estimated effect sizes.
Further analyses of amino acids found at each niVpr position revealed a discrete number of variants at each position (Table 2). At position 37, niVpr variant I37 was one of three prevalent amino acids at that position, and one of 10 residues found at that site. At position 41, the N41 niVpr variant was found at a relatively low frequency out of four amino acids; this position was dominated by glycine, which was not associated with a change in GDS. The A55 variant, however, was the dominant residue out of four amino acids coded at this position.
Table 2.
Amino acid presence and prevalence at each niVpr position. Amino acids found at each niVpr position are shown in conjunction with their frequency of appearance and relative effect on GDS. A zero effect indicates that the amino acid was not significantly associated with a change in GDS.
| Position 37 | Position 41 | Position 55 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid | P | I | V | M | T | A | L | E | H | S | G | S | N | A | A | T | E | V |
| frequency | 33 | 32 | 28 | 7 | 4 | 3 | 2 | 2 | 1 | 1 | 68 | 34 | 13 | 1 | 59 | 46 | 9 | 2 |
| GDS effect | 0 | (−) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | (−) | (+) | 0 | (+) | 0 | 0 | 0 |
niVpr variant combinations result in a continuum of effect sizes
As each niVpr variant was associated with an effect size of a specific magnitude and direction, we hypothesized that the combined effects of the niVpr variants on GDS would be additive. In analyses of the combined effect sizes (CES), a positive correlation between CES (the cumulative effect size of amino acids at the three sites within a patient Vpr sequence) and average GDS was evident (Fig. 3). In well-suppressed patients with GDS ≤ 2, combined effects of niVpr variants could contribute significantly to NPI.
Fig. 3.
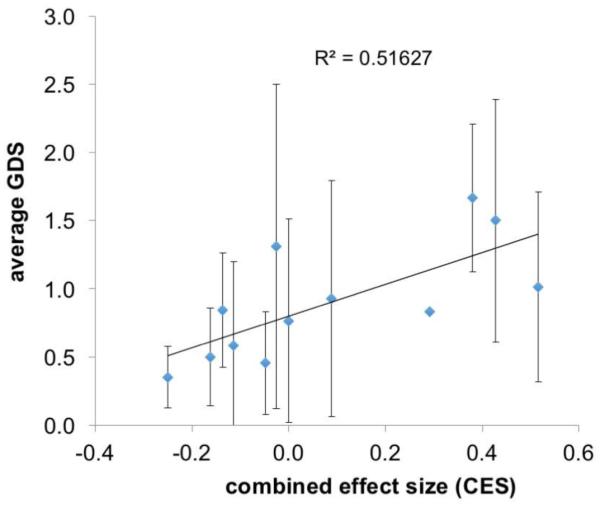
Average patient GDS is positively correlated with the combined effect size (CES). The summed effect sizes for each patient sequence are plotted against average GDS for each of the 12 possible combined effect size groups.
Deep sequencing reveals that niVpr variant amino acids appear with varied frequencies in cohort patients
Since Sanger sequencing only yields what is usually the predominant or highly prevalent sequence within the population of sequences, sequencing was also performed using Illumina deep sequencing. Through next generation sequencing (NGS), variant sequences at all frequencies can be documented. Applying NGS to a select number of patient DNA samples, a depth of coverage was obtained that ranged from 9231 to 48,028 reads, with a mean of 13,931 across all samples and was not correlated with clinical parameters or biological compartment. This approach demonstrated that niVpr variants and non-niVpr amino acids appeared at varied frequencies in individual patients (Table 3). In seven of the nine patients included in these analyses, at least one niVpr amino acid was present in 100% of the sequence reads, suggesting that the effects of these variants in HIV-1-infected patients could be considerable. These results also confirm in individual patients that different combinations of niVpr amino acids and “null” residues can be found at these three positions (Table 3). These results, in conjunction with the CES analyses (Fig. 3), support the hypothesis that Vpr variants can have a range of effects on GDS dependent on the identities of the amino acids at positions 37, 41, and 55.
Table 3.
Prevalence of amino acids at positions 37, 41, and 55 in peripheral blood Vpr sequences collected using deep sequencing. Samples from seven HIV-1-infected patients were sequenced using NGS focused on the vpr open reading frame in genomic DNA. The prevalence of each amino acid at a site is shown as a fraction of the total. Bold text highlights the niVpr variants identified in Figure 2.
| Patient | E37 | G37 | I37 | M37 | P37 | V37 | G41 | N41 | S41 | A55 | E55 | T55 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A0103 | 0.25 | 0.75 | 1.00 | 1.00 | ||||||||
| A0432 | 1.00 | 1.00 | 1.00 | |||||||||
| A0438 | 0.07 | 0.51 | 0.41 | 0.88 | 0.12 | 0.71 | 0.29 | |||||
| A0451 | 1.00 | 1.00 | 1.00 | |||||||||
| A0457 | 1.00 | 1.00 | 1.00 | |||||||||
| A0460 | 1.00 | 1.00 | 1.00 | |||||||||
| A0461 | 1.00 | 1.00 | 1.00 | |||||||||
| A0467 | 1.00 | 1.00 | 1.00 | |||||||||
| A0469 | 1.00 | 1.00 | 1.00 |
niVpr variants are found in brain samples from HIV-1-infected patients
Any model of niVpr-associated neuropathogenesis must address mechanistic connections between niVpr variants found in the peripheral blood and the onset of NPI in the brain. The simplest explanation is that niVpr variants arise in the blood and are seeded into the brain, where they act to increase or decrease clinically apparent NPI. To begin to address this point, vpr sequences were collected using NGS from matched HIV-1-infected brain and spleen tissues banked through the NNTC (Morgello et al, 2001). While niVpr amino acids were clearly detected in both brain and spleen tissue, they were not universally present nor were they always the most predominant residue (Table 4). In addition, these results suggest niVpr variant compartmentalization between brain and spleen, since some residues were present in one compartment but absent in the other (e.g., patient T56766, N41), while some residues appeared in both compartments but differed considerably in their prevalence (e.g., patient T56766, I37). Although the detection of an niVpr variant in the brain but not the spleen (patient T56766, N41) appears counter to the identification of niVpr variants in peripheral blood cells, this particular finding may be the result of other factors, including the timing of tissue collection (post-mortem sampling from patients potentially in end-stage disease versus samples from well-suppressed living patients). Relevant to tissue origin, niVpr variants were also detected using Sanger sequencing in other areas of the brain, including cerebellum, deep white matter, parietal lobe, and thalamus (data not shown).
Table 4.
Prevalence of amino acids at positions 37, 41, and 55 in matched brain and spleen Vpr sequences collected using deep sequencing. Matched samples from three HIV-1-infected patients were sequenced using NGS focused on vpr in genomic DNA. The prevalence of each amino acid at a site is shown as a fraction of the total. Bold text highlights the niVpr variants (Fig. 2). Abbreviations: HOC, head of caudate; SPL, spleen.
Discussion
Through patient-based bioinformatic studies, we have identified three positions in the primary sequence of HIV-1 Vpr where select amino acids are specifically and significantly linked to differences in patient neurocognitive status. From these results, several important conclusions can be formulated. First, Vpr appears to have a causative role in HIV-1-associated neuropathogenesis. While this conclusion has been inferred by previous in vitro and animal model studies (Ferrucci et al, 2012; Ferrucci et al, 2013; Gangwani et al, 2013; Huang et al, 2000; Jones et al, 2007; Kitayama et al, 2008; Mukerjee et al, 2011; Patel et al, 2000; Piller et al, 1998; Sabbah and Roques, 2005; Torres and Noel, 2014), we believe that this is the first study that shows an association in humans between different Vpr variants and the extent of clinically apparent neurocognitive function. Second, the contribution of Vpr to viral neuropathogenesis can be altered by changes in the amino acid sequence of Vpr that occur during the course of infection. This conclusion is consistent with investigations that explored the functional consequences of Vpr variation and mutation (Pandey et al, 2009), as well as studies that suggest clinical outcomes associated with distinct changes amino acids within Vpr (Hadi et al, 2014; Jacquot et al, 2009; Mologni et al, 2006).
Third, the appearance of niVpr variants in well-suppressed patients suggests a level of replication sufficient to support viral evolution and, specifically, Vpr sequence diversity. This conclusion is consistent with our more global analyses of longitudinal HIV-1 genomic diversity, which revealed a low but measureable level of quasispecies development in patients with infections well-suppressed by effective antiretroviral combination therapies (Dampier et al, 2016). Viral replication in the face of highly effective antiretroviral therapy is also consistent with recent studies that suggest ongoing viral replication in the lymph nodes despite chemotherapeutic reductions in viral loads in the peripheral circulation (Cohen, 2016; Lorenzo-Redondo et al, 2016). The inability to fully suppress replication and therefore negate the generation of diversity in Vpr and other parts of the viral genome may be a consequence of insufficient levels of antiretroviral drugs penetrating into the lymphatic tissues (Cohen, 2011; Fletcher et al, 2014) and other tissue compartments, such as the brain and/or gastrointestinal tract.
Fourth and finally, the identification of niVpr variants in the blood (Fig. 2) and brain (Table 4) suggests one of two scenarios: trafficking of cells harboring niVpr variants into the brain from the peripheral circulation or virus reservoirs (including the lymph nodes or gastrointestinal tract); or the genesis of niVpr variants in infected cells within the CNS (e.g., perivascular macrophages or microglial cells) and subsequent trafficking of those cells from the brain into the peripheral circulation. The generally accepted role of infected macrophages as cells that seed the CNS, as well as demonstrated similarities between proviral sequences isolated from bone marrow-derived monocytes and infected cells in the brain (Liu et al, 2000), suggest the possibility that niVpr variants are harbored in monocytic cells that pass between the peripheral circulation and brain. Conversely, it is more probable that the blood-derived niVpr sequences were derived from infected T lymphocytes, since this cell type is in much greater abundance relative to cells of monocytic lineage, and since Sanger sequencing yields sequences that are predominant in proviral genomes isolated from infected cells. Upcoming studies will focus on localizing niVpr variants to specific cell populations in the peripheral blood and brain.
The identification of four discrete amino acid changes in Vpr associated with measurable differences in neurocognition provides a starting point for structure-function studies focused on determining the molecular and cellular mechanisms that underlie the clinically apparent differences in neurological function. Along those lines, previously published studies suggest that amino acid changes at niVpr positions 37 and 41 may affect Vpr oligomerization, which has been implicated in Vpr incorporation into nascent virions (Fritz et al, 2010; Venkatachari et al, 2010). Amino acids in this region of the protein (residues 36-46) have been shown to be critical for Vpr oligomerization (Zhao et al, 1994) and Vpr incorporation during virus assembly (Singh et al, 2000). Specifically, an R36W polymorphism was associated with rapid progression of HIV-1-associated disease, higher levels of oligomerization, and increased levels of virus replication (Hadi et al, 2014). The effects of niVpr variants on the functions of Vpr, including oligomerization and virus assembly, facilitation of pre-integration complex nuclear transport (Hrimech et al, 1999), trans-activation of the HIV-1 LTR (Wang et al, 1995), and initiation of G2 cell cycle arrest (He et al, 1995), will need to be explored in future studies. Since the pathogenic effects of niVpr variants may be manifested in the peripheral blood and brain, the impact of Vpr variants will need to be assessed in cell populations from both compartments.
As an extension of our previous investigations (Ferrucci et al, 2012; Ferrucci et al, 2013), preliminary in vitro experiments of this nature suggest that substitution of an asparagine for a serine at position 41 results in greater Vpr-associated reductions in cellular glutathione in astrocytes (data not shown). Because Vpr has been shown to cause adverse changes in astrocyte and neuron metabolism and viability (Ferrucci et al, 2012; Ferrucci et al, 2013), increased oxidative stress associated with expression of the 41N variant suggests one possible mechanism through which niVpr variants can affect the degree of HIV-1-associated neuropathogenesis.
Structure-function experiments will also yield insights into the selective pressures that influence the appearance and prevalence of niVpr variants in HIV-1-infected patients. Viruses that harbor specific amino acid changes at niVpr positions may have advantages over other viruses due to alterations in replicative capacity, persistence in viral reservoirs, or the release of extracellular Vpr. Advantages conferred by amino acid changes at niVpr positions may also be linked to the identity of the host cell, since we have shown that Vpr sequence diversity and divergence at positions 37, 41, and others are linked to co-receptor phenotype (Antell et al., submitted for publication, 2016). Advantages associated with specific changes at niVpr positions may then indirectly or directly affect HIV-1-associated neuropathogenesis.
Future studies will also examine associations between localized expression of niVpr variants in the brain and specific neurocognitive deficits (as indicated by specific CNPA domain scores). The CNPA results preliminarily suggest that N41 is associated with increased deficits in processing speed, executive functions, and verbal/visual memory (data not shown). In this regard, niVpr variants were detected in areas of the brain that are well-known to be associated with previously described patterns of NPI involving motor, executive, and memory deficits (Devlin et al, 2015). Deficits in specific neuropsychological domains could indicate that a specific, underlying neuropsychological network within the brain is affected by contributions of niVpr variants to neuropathogenesis. This information may provide new information about the mechanisms through which regionally expressed niVpr variants contribute to specific neuropsychological syndromes.
The demonstration of associations between specific amino acids in Vpr and significant, measurable differences in the neurocognitive abilities of HIV-1-infected patients provides new insights into the roles of Vpr during HIV-1 infection and the contributions of Vpr to HIV-1-associated neuropathogenesis and HAND. Because of the links between vpr sequence variation and neuropathogenesis, these studies also suggest the possible use of niVpr variants as diagnostic or predictive biomarkers for HAND. Since many studies currently show no relation between neurocognitive function and classic measures of HIV-1 infection (e.g., viral load and CD4 T-cell counts), Vpr may be a meaningful biomarker for HIV-1-associated NPI. Finally, a more comprehensive understanding of pathogenic mechanisms arising from Vpr expression in the periphery and brain may point toward new therapeutic strategies effective against HIV-1-associated neuropathogenesis.
Acknowledgments
We would like to thank all patients who are part of the Drexel Medicine CNS AIDS Research and Eradication Study (CARES) Cohort (studies of which were approved under Institutional Review Board protocol # 1201000748, Dr. Brian Wigdahl, Principal Investigator). We would also like to thank the clinical staff within the Division of Infectious Diseases and HIV Medicine at the Drexel University College of Medicine who were involved in the recruitment, enrollment, obtaining consent, obtaining clinical history, venipuncture, and delivery of peripheral blood to the research laboratories in the Center for Molecular Virology and Translational Neuroscience in the Institute for Molecular Medicine and Infectious Disease. In addition, we want to recognize Carrie Lamberson, Elizabeth Schell, and Jessica Kurczewski (Temple University) for their participation in and contributions to patient neuropsychological assessment. These studies were funded in part by the Public Health Service National Institutes of Health through grants from the National Institute of Neurological Disorders and Stroke (NS32092 and NS46263, Dr. Brian Wigdahl, PI; NS089435, Dr. Michael Nonnemacher, PI), the National Institute of Drug Abuse (DA19807, Dr. Brian Wigdahl, PI), the National Institute of Mental Health Comprehensive NeuroAIDS Center (CNAC) (P30 MH092177, Dr. Kamel Khalili, PI, and Dr. Brian Wigdahl, PI of the CNAC Clinical and Translational Research Support Core), a CNAC developmental grant (Dr. Fred Krebs, PI), and the Ruth L. Kirschstein National Research Service Award (T32 MH079785, Dr. Jay Rappaport, PI, and Dr. Brian Wigdahl, PI of the Drexel component). This publication was also made possible by NIH/NIMH funding of the Texas NeuroAIDS Research Center grant (U24 MH100930, Dr. Benjamin B. Gelman, PI). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NNTC or NIH. Drs. Nonnemacher, Dampier, and Krebs were also supported by faculty development funds provided by the Department of Microbiology and Immunology and the Institute for Molecular Medicine and Infectious Disease.
Footnotes
Author Contributions
WD, GA, MRN, JJ, VP, KND, TG, DJL, BW, and FCK conceived and designed the studies. BA, WZ, KK, SP, JWW, and TJ performed sample preparation. WD and GA performed the bioinfomatic analyses. KND, TG, and DJL performed the neuropsychological assessment. WD, GA, MRN, VP, JJ, TG, DJL, BW, and FCK contributed to analyses and discussion of the results. WD, MRN, DJL, and FCK wrote the paper. All authors proofed and edited the paper.
Conflicts of Interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
DNA Sequence Sharing
Sequences used for these analyses have been submitted to GenBank and attached to NCBI BioProject PRJNA325815.
References
- Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J Virol. 2010;84:9864–78. doi: 10.1128/JVI.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiamkitsumrit B, Dampier W, Antell G, Rivera N, Martin-Garcia J, Pirrone V, Nonnemacher MR, Wigdahl B. Bioinformatic analysis of HIV-1 entry and pathogenesis. Curr HIV Res. 2014;12:132–61. doi: 10.2174/1570162x12666140526121746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, Gisslen M, Grant I, Heaton RK, Joseph J, Marder K, Marra CM, McArthur JC, Nunn M, Price RW, Pulliam L, Robertson KR, Sacktor N, Valcour V, Wojna VE. Updated research nosology for HIV-associated neurocognitive disorders. Neurology. 2007;69:1789–99. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackstone K, Moore DJ, Franklin DR, Clifford DB, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, Ellis RJ, Atkinson JH, Grant I, Heaton RK. Defining neurocognitive impairment in HIV: deficit scores versus clinical ratings. Clin Neuropsychol. 2012;26:894–908. doi: 10.1080/13854046.2012.694479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butters N, Grant I, Haxby J, Judd LL, Martin A, McClelland J, Pequegnat W, Schacter D, Stover E. Assessment of AIDS-related cognitive changes: recommendations of the NIMH Workshop on Neuropsychological Assessment Approaches. J Clin Exp Neuropsychol. 1990;12:963–78. doi: 10.1080/01688639008401035. [DOI] [PubMed] [Google Scholar]
- Carey CL, Woods SP, Gonzalez R, Conover E, Marcotte TD, Grant I, Heaton RK, Group H. Predictive validity of global deficit scores in detecting neuropsychological impairment in HIV infection. J Clin Exp Neuropsychol. 2004;26:307–19. doi: 10.1080/13803390490510031. [DOI] [PubMed] [Google Scholar]
- Clifford DB, Ances BM. HIV-associated neurocognitive disorder. Lancet Infect Dis. 2013;13:976–86. doi: 10.1016/S1473-3099(13)70269-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJ, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJ. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–3. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. HIV/AIDS research. Tissue says blood is misleading, confusing HIV cure efforts. Science. 2011;334:1614. doi: 10.1126/science.334.6063.1614. [DOI] [PubMed] [Google Scholar]
- Cohen J. INFECTIOUS DISEASE. Researchers claim to find HIV sanctuaries. Science. 2016;351:434. doi: 10.1126/science.351.6272.434. [DOI] [PubMed] [Google Scholar]
- Dampier W, Nonnemacher MR, Mell J, Earl J, Ehrlich G, Pirrone V, Aiamkitsumrit B, Zhong W, Kercher K, Passic S, Williams J, Jacobson JM, Wigdahl B. HIV-1 genetic variation resulting in the development of new quasispecies continues to be encountered in the peripheral blood of well-suppressed patients. PLoS One. 2016 doi: 10.1371/journal.pone.0155382. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin KN, Giovannetti T, Dampier W, Pirrone V, Nonnemacher MR, Schell E, Lamberson C, Kurczewski JR, Jacobson JM, Libon DJ, Wigdahl B. Mild cognitive impairment in a clinically latent HIV-1 patient population; 7th International Workshop on HIV Persistence during Therapy; Miami, FL. 2015. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust GG, Hall IM. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics. 2014;30:2503–5. doi: 10.1093/bioinformatics/btu314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci A, Nonnemacher MR, Cohen EA, Wigdahl B. Extracellular human immunodeficiency virus type 1 viral protein R causes reductions in astrocytic ATP and glutathione levels compromising the antioxidant reservoir. Virus Res. 2012;167:358–69. doi: 10.1016/j.virusres.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci A, Nonnemacher MR, Wigdahl B. Cellular phenotype impacts human immunodeficiency virus type 1 viral protein R subcellular localization. Virol J. 2011a;8:397. doi: 10.1186/1743-422X-8-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci A, Nonnemacher MR, Wigdahl B. Human immunodeficiency virus viral protein R as an extracellular protein in neuropathogenesis. Adv Virus Res. 2011b;81:165–99. doi: 10.1016/B978-0-12-385885-6.00010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrucci A, Nonnemacher MR, Wigdahl B. Extracellular HIV-1 viral protein R affects astrocytic glyceraldehyde 3-phosphate dehydrogenase activity and neuronal survival. J Neurovirol. 2013;19:239–53. doi: 10.1007/s13365-013-0170-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CV, Staskus K, Wietgrefe SW, Rothenberger M, Reilly C, Chipman JG, Beilman GJ, Khoruts A, Thorkelson A, Schmidt TE, Anderson J, Perkey K, Stevenson M, Perelson AS, Douek DC, Haase AT, Schacker TW. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A. 2014;111:2307–12. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JV, Dujardin D, Godet J, Didier P, De Mey J, Darlix JL, Mely Y, de Rocquigny H. HIV-1 Vpr oligomerization but not that of Gag directs the interaction between Vpr and Gag. J Virol. 2010;84:1585–96. doi: 10.1128/JVI.01691-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwani MR, Noel RJ, Jr., Shah A, Rivera-Amill V, Kumar A. Human immunodeficiency virus type 1 viral protein R (Vpr) induces CCL5 expression in astrocytes via PI3K and MAPK signaling pathways. J Neuroinflammation. 2013;10:136. doi: 10.1186/1742-2094-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenzel CA, Herate C, Benichou S. HIV-1 Vpr-a still "enigmatic multitasker". Front Microbiol. 2014;5:127. doi: 10.3389/fmicb.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadi K, Walker LA, Guha D, Murali R, Watkins SC, Tarwater P, Srinivasan A, Ayyavoo V. Human immunodeficiency virus type 1 Vpr polymorphisms associated with progressor and nonprogressor individuals alter Vpr-associated functions. J Gen Virol. 2014;95:700–11. doi: 10.1099/vir.0.059576-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–11. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Clifford DB, Franklin DR, Jr., Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier AC, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan JA, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I, Group C. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75:2087–96. doi: 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Franklin DR, Ellis RJ, McCutchan JA, Letendre SL, Leblanc S, Corkran SH, Duarte NA, Clifford DB, Woods SP, Collier AC, Marra CM, Morgello S, Mindt MR, Taylor MJ, Marcotte TD, Atkinson JH, Wolfson T, Gelman BB, McArthur JC, Simpson DM, Abramson I, Gamst A, Fennema-Notestine C, Jernigan TL, Wong J, Grant I, Group C, Group H. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. J Neurovirol. 2011;17:3–16. doi: 10.1007/s13365-010-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Grant I, Butters N, White DA, Kirson D, Atkinson JH, McCutchan JA, Taylor MJ, Kelly MD, Ellis RJ, et al. The HNRC 500--neuropsychology of HIV infection at different disease stages. HIV Neurobehavioral Research Center. J Int Neuropsychol Soc. 1995;1:231–51. doi: 10.1017/s1355617700000230. [DOI] [PubMed] [Google Scholar]
- Henklein P, Bruns K, Sherman MP, Tessmer U, Licha K, Kopp J, de Noronha CM, Greene WC, Wray V, Schubert U. Functional and structural characterization of synthetic HIV-1 Vpr that transduces cells, localizes to the nucleus, and induces G2 cell cycle arrest. J Biol Chem. 2000;275:32016–26. doi: 10.1074/jbc.M004044200. [DOI] [PubMed] [Google Scholar]
- Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S, Zody MC, Erlich RL, Green LM, Berical A, Wang Y, Casali M, Streeck H, Bloom AK, Dudek T, Tully D, Newman R, Axten KL, Gladden AD, Battis L, Kemper M, Zeng Q, Shea TP, Gujja S, Zedlack C, Gasser O, Brander C, Hess C, Gunthard HF, Brumme ZL, Brumme CJ, Bazner S, Rychert J, Tinsley JP, Mayer KH, Rosenberg E, Pereyra F, Levin JZ, Young SK, Jessen H, Altfeld M, Birren BW, Walker BD, Allen TM. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan TH, Nonnemacher MR, Krebs FC, Henderson A, Wigdahl B. HIV-1 Vpr binding to HIV-1 LTR C/EBP cis-acting elements and adjacent regions is sequence-specific. Biomed Pharmacother. 2003;57:41–8. doi: 10.1016/s0753-3322(02)00333-5. [DOI] [PubMed] [Google Scholar]
- Hoshino S, Sun B, Konishi M, Shimura M, Segawa T, Hagiwara Y, Koyanagi Y, Iwamoto A, Mimaya J, Terunuma H, Kano S, Ishizaka Y. Vpr in plasma of HIV type 1-positive patients is correlated with the HIV type 1 RNA titers. AIDS Res Hum Retroviruses. 2007;23:391–7. doi: 10.1089/aid.2006.0124. [DOI] [PubMed] [Google Scholar]
- Hrimech M, Yao XJ, Bachand F, Rougeau N, Cohen EA. Human immunodeficiency virus type 1 (HIV-1) Vpr functions as an immediate-early protein during HIV-1 infection. J Virol. 1999;73:4101–9. doi: 10.1128/jvi.73.5.4101-4109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MB, Weeks O, Zhao LJ, Saltarelli M, Bond VC. Effects of extracellular human immunodeficiency virus type 1 vpr protein in primary rat cortical cell cultures. J Neurovirol. 2000;6:202–20. doi: 10.3109/13550280009015823. [DOI] [PubMed] [Google Scholar]
- Jacquot G, Le Rouzic E, Maidou-Peindara P, Maizy M, Lefrere JJ, Daneluzzi V, Monteiro-Filho CM, Hong D, Planelles V, Morand-Joubert L, Benichou S. Characterization of the molecular determinants of primary HIV-1 Vpr proteins: impact of the Q65R and R77Q substitutions on Vpr functions. PLoS One. 2009;4:e7514. doi: 10.1371/journal.pone.0007514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GJ, Barsby NL, Cohen EA, Holden J, Harris K, Dickie P, Jhamandas J, Power C. HIV-1 Vpr causes neuronal apoptosis and in vivo neurodegeneration. J Neurosci. 2007;27:3703–11. doi: 10.1523/JNEUROSCI.5522-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama H, Miura Y, Ando Y, Hoshino S, Ishizaka Y, Koyanagi Y. Human immunodeficiency virus type 1 Vpr inhibits axonal outgrowth through induction of mitochondrial dysfunction. J Virol. 2008;82:2528–42. doi: 10.1128/JVI.02094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DN, Refaeli Y, MacGregor RR, Weiner DB. Serum Vpr regulates productive infection and latency of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1994;91:10873–7. doi: 10.1073/pnas.91.23.10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DN, Refaeli Y, Weiner DB. Extracellular Vpr protein increases cellular permissiveness to human immunodeficiency virus replication and reactivates virus from latency. J Virol. 1995;69:1243–52. doi: 10.1128/jvi.69.2.1243-1252.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Aiamkitsumrit B, Pirrone V, Nonnemacher MR, Wojno A, Passic S, Flaig K, Kilareski E, Blakey B, Ku J, Parikh N, Shah R, Martin-Garcia J, Moldover B, Servance L, Downie D, Lewis S, Jacobson JM, Kolson D, Wigdahl B. Development of co-selected single nucleotide polymorphisms in the viral promoter precedes the onset of human immunodeficiency virus type 1-associated neurocognitive impairment. J Neurovirol. 2011;17:92–109. doi: 10.1007/s13365-010-0014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tang XP, McArthur JC, Scott J, Gartner S. Analysis of human immunodeficiency virus type 1 gp160 sequences from a patient with HIV dementia: evidence for monocyte trafficking into brain. J Neurovirol. 2000;6(Suppl 1):S70–81. [PubMed] [Google Scholar]
- Lloyd SB, Kent SJ, Winnall WR. The high cost of fidelity. AIDS Res Hum Retroviruses. 2014;30:8–16. doi: 10.1089/aid.2013.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Redondo R, Fryer HR, Bedford T, Kim EY, Archer J, Kosakovsky Pond SL, Chung YS, Penugonda S, Chipman JG, Fletcher CV, Schacker TW, Malim MH, Rambaut A, Haase AT, McLean AR, Wolinsky SM. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016;530:51–6. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YL, Bennett RP, Wills JW, Gorelick R, Ratner L. A leucine triplet repeat sequence (LXX)4 in p6gag is important for Vpr incorporation into human immunodeficiency virus type 1 particles. J Virol. 1995;69:6873–9. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mologni D, Citterio P, Menzaghi B, Zanone Poma B, Riva C, Broggini V, Sinicco A, Milazzo L, Adorni F, Rusconi S, Galli M, Riva A, rHoPe SG. Vpr and HIV-1 disease progression: R77Q mutation is associated with long-term control of HIV-1 infection in different groups of patients. AIDS. 2006;20:567–74. doi: 10.1097/01.aids.0000210611.60459.0e. [DOI] [PubMed] [Google Scholar]
- Morgello S, Gelman BB, Kozlowski PB, Vinters HV, Masliah E, Cornford M, Cavert W, Marra C, Grant I, Singer EJ. The National NeuroAIDS Tissue Consortium: a new paradigm in brain banking with an emphasis on infectious disease. Neuropathol Appl Neurobiol. 2001;27:326–35. doi: 10.1046/j.0305-1846.2001.00334.x. [DOI] [PubMed] [Google Scholar]
- Mukerjee R, Chang JR, Del Valle L, Bagashev A, Gayed MM, Lyde RB, Hawkins BJ, Brailoiu E, Cohen E, Power C, Azizi SA, Gelman BB, Sawaya BE. Deregulation of microRNAs by HIV-1 Vpr protein leads to the development of neurocognitive disorders. J Biol Chem. 2011;286:34976–85. doi: 10.1074/jbc.M111.241547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungas D, Reed BR, Marshall SC, Gonzalez HM. Development of psychometrically matched English and Spanish language neuropsychological tests for older persons. Neuropsychology. 2000;14:209–23. doi: 10.1037//0894-4105.14.2.209. [DOI] [PubMed] [Google Scholar]
- Nonnemacher MR, Pirrone V, Feng R, Moldover B, Passic S, Aiamkitsumrit B, Dampier W, Wojno A, Kilareski E, Blakey B, Ku TS, Shah S, Sullivan NT, Jacobson JM, Wigdahl B. HIV-1 Promoter Single Nucleotide Polymorphisms Are Associated with Clinical Disease Severity. PLoS One. 2016;11:e0150835. doi: 10.1371/journal.pone.0150835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RC, Datta D, Mukerjee R, Srinivasan A, Mahalingam S, Sawaya BE. HIV-1 Vpr: a closer look at the multifunctional protein from the structural perspective. Curr HIV Res. 2009;7:114–28. doi: 10.2174/157016209787581508. [DOI] [PubMed] [Google Scholar]
- Parikh N, Dampier W, Feng R, Passic SR, Zhong W, Frantz B, Blakey B, Aiamkitsumrit B, Pirrone V, Nonnemacher MR, Jacobson JM, Wigdahl B. Cocaine alters cytokine profiles in HIV-1-infected African American individuals in the DrexelMed HIV/AIDS genetic analysis cohort. J Acquir Immune Defic Syndr. 2014;66:256–64. doi: 10.1097/QAI.0000000000000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CA, Mukhtar M, Pomerantz RJ. Human immunodeficiency virus type 1 Vpr induces apoptosis in human neuronal cells. J Virol. 2000;74:9717–26. doi: 10.1128/jvi.74.20.9717-9726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piller SC, Jans P, Gage PW, Jans DA. Extracellular HIV-1 virus protein R causes a large inward current and cell death in cultured hippocampal neurons: implications for AIDS pathology. Proc Natl Acad Sci U S A. 1998;95:4595–600. doi: 10.1073/pnas.95.8.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C, Selnes OA, Grim JA, McArthur JC. HIV Dementia Scale: a rapid screening test. J Acquir Immune Defic Syndr Hum Retrovirol. 1995;8:273–8. doi: 10.1097/00042560-199503010-00008. [DOI] [PubMed] [Google Scholar]
- Sabbah EN, Roques BP. Critical implication of the (70-96) domain of human immunodeficiency virus type 1 Vpr protein in apoptosis of primary rat cortical and striatal neurons. J Neurovirol. 2005;11:489–502. doi: 10.1080/13550280500384941. [DOI] [PubMed] [Google Scholar]
- Schouten J, Cinque P, Gisslen M, Reiss P, Portegies P. HIV-1 infection and cognitive impairment in the cART era: a review. AIDS. 2011;25:561–75. doi: 10.1097/QAD.0b013e3283437f9a. [DOI] [PubMed] [Google Scholar]
- Schretlen DJ, Testa SM, Pearlson GD. Calibrated Neuropsychological Normative System (CNNS) Psychological Assessment Resources; Lutz, F.L.: 2010. [Google Scholar]
- Seabold JS, Perktold J. In: van der Walt S, Millman J, editors. Statsmodels: Econometric and Statistical Modeling with Python; Proceedings of the 9th Python in Science Conference; 2010. June 28 - July 3. [Google Scholar]
- Shah S, Alexaki A, Pirrone V, Dahiya S, Nonnemacher MR, Wigdahl B. Functional properties of the HIV-1 long terminal repeat containing single-nucleotide polymorphisms in Sp site III and CCAAT/enhancer binding protein site I. Virol J. 2014;11:92. doi: 10.1186/1743-422X-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MP, Schubert U, Williams SA, de Noronha CM, Kreisberg JF, Henklein P, Greene WC. HIV-1 Vpr displays natural protein-transducing properties: implications for viral pathogenesis. Virology. 2002;302:95–105. doi: 10.1006/viro.2002.1576. [DOI] [PubMed] [Google Scholar]
- Simioni S, Cavassini M, Annoni JM, Rimbault Abraham A, Bourquin I, Schiffer V, Calmy A, Chave JP, Giacobini E, Hirschel B, Du Pasquier RA. Cognitive dysfunction in HIV patients despite long-standing suppression of viremia. AIDS. 2010;24:1243–50. doi: 10.1097/QAD.0b013e3283354a7b. [DOI] [PubMed] [Google Scholar]
- Singh SP, Tomkowicz B, Lai D, Cartas M, Mahalingam S, Kalyanaraman VS, Murali R, Srinivasan A. Functional role of residues corresponding to helical domain II (amino acids 35 to 46) of human immunodeficiency virus type 1 Vpr. J Virol. 2000;74:10650–7. doi: 10.1128/jvi.74.22.10650-10657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich S, Gonzalez-Scarano F. HIV-1-related central nervous system disease: current issues in pathogenesis, diagnosis, and treatment. Cold Spring Harb Perspect Med. 2012;2:a007120. doi: 10.1101/cshperspect.a007120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres L, Noel RJ., Jr. Astrocytic expression of HIV-1 viral protein R in the hippocampus causes chromatolysis, synaptic loss and memory impairment. J Neuroinflammation. 2014;11:53. doi: 10.1186/1742-2094-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varin A, Decrion AZ, Sabbah E, Quivy V, Sire J, Van Lint C, Roques BP, Aggarwal BB, Herbein G. Synthetic Vpr protein activates activator protein-1, c-Jun N-terminal kinase, and NF-kappaB and stimulates HIV-1 transcription in promonocytic cells and primary macrophages. J Biol Chem. 2005;280:42557–67. doi: 10.1074/jbc.M502211200. [DOI] [PubMed] [Google Scholar]
- Venkatachari NJ, Walker LA, Tastan O, Le T, Dempsey TM, Li Y, Yanamala N, Srinivasan A, Klein-Seetharaman J, Montelaro RC, Ayyavoo V. Human immunodeficiency virus type 1 Vpr: oligomerization is an essential feature for its incorporation into virus particles. Virol J. 2010;7:119. doi: 10.1186/1743-422X-7-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Mukherjee S, Jia F, Narayan O, Zhao LJ. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J Biol Chem. 1995;270:25564–9. doi: 10.1074/jbc.270.43.25564. [DOI] [PubMed] [Google Scholar]
- Zhao LJ, Wang L, Mukherjee S, Narayan O. Biochemical mechanism of HIV-1 Vpr function. Oligomerization mediated by the N-terminal domain. J Biol Chem. 1994;269:32131–7. [PubMed] [Google Scholar]