Abstract
PURPOSE
There is a critical clinical need for new predictive and pharmacodynamic biomarkers that evaluate pathway activity in patients treated with targeted therapies. A microscale platform known as VERSA (Versatile Exclusion-based Rare Sample Analysis) was developed to integrate readouts across protein, mRNA and DNA in Circulating Tumor Cells (CTCs) for a comprehensive analysis of the Androgen Receptor (AR) signaling pathway.
EXPERIMENTAL DESIGN
Utilizing exclusion based sample preparation principles, a handheld chip was developed to perform CTC capture, enumeration, quantification and subcellular localization of proteins and extraction of mRNA and DNA. This technology was validated across integrated endpoints in cell lines and a cohort of patients with castrate resistant prostate cancer (CRPC) treated with AR targeted therapies and chemotherapies.
RESULTS
The VERSA was validated in cell lines to analyze AR protein expression, nuclear localization and gene expression targets. When applied to a cohort of patients, radiographic progression was predicted by the presence of multiple AR splice variants and activity in the canonical AR signaling pathway. AR protein expression and nuclear localization identified phenotypic heterogeneity. Next Generation Sequencing with the FoundationOne panel detected copy number changes and point mutations. Longitudinal analysis of CTCs identified acquisition of multiple AR variants during targeted treatments and chemotherapy.
CONCLUSIONS
Complex mechanisms of resistance to AR targeted therapies, across RNA, DNA and protein endpoints, exist in patients with CRPC and can be quantified in CTCs. Interrogation of the AR signaling pathway revealed distinct patterns relevant to tumor progression and can serve as pharmacodynamic biomarkers for targeted therapies.
Keywords: Exclusion-based Sample Preparation, Circulating Tumor Cells, Pharmacodynamic biomarkers, Androgen Receptor, Prostate Cancer
Introduction
The landscape of treatment options for patients with solid tumors has changed dramatically. In the last five years, more than 30 new agents have been approved by the FDA and the need to tailor treatment recommendations to each individual has never been greater (1). However, this personalization of cancer therapies requires biomarkers that predict therapeutic benefit; identify emerging mechanisms of resistance; and tailor subsequent treatment strategies to continually evolving tumors (2). Successful development of predictive and pharmacodynamic biomarkers suitable for these purposes requires frequent sampling cells throughout the course of therapy. This approach is rarely feasible for patients with solid tumors given the invasive nature of tumor biopsies. Circulating tumor cells (CTCs) are shed into peripheral circulation from primary and metastatic tumor sites, and enumeration of CTCs in prostate cancer (PRCA) is prognostic of patient outcomes (1, 3–6). However, enumeration does not identify the underlying mechanisms of resistance that develop during treatment with targeted therapies. Pre-clinical and clinical studies have identified a vast range of resistance mechanisms to therapies targeting the AR signaling pathway that include genomic and functional adaptations. For example, Antonarakis et al (7) identified expression of the splice variant AR-V7 that correlated with primary resistance to both Abiraterone Acetate and Enzalutamide. This group then examined AR-V7 longitudinally in 14 patients and observed conversion of AR-V7 status during the course of treatment, suggesting it may be dynamic biomarker (8). Others have reported heterogeneity in expression and localization of AR protein in CTCs from patients with CRPC (9, 10). However, the molecular analyses from these CTC technologies are limited to a single readout. It is unclear if these biomarkers reflect driver or passenger alterations in resistant PRCA, the extent to which the AR signaling pathway continues to drive disease progression, or if these alterations can be therapeutically targeted. Integrating these distinct endpoints for CTC enumeration, genomic, transcriptomic and protein endpoints may further facilitate discovery endpoints of novel biomarkers or biologic alterations in the development of therapeutic resistance.
Integrating CTC capture with multiplexed molecular analyses such as gene expression or protein analytics presents multiple technological challenges (11). One of the most significant hurdles is analyte loss that occurs during standard cell staining or nucleic acid extraction when isolating rare cell populations (12–14). Most systems are designed for a single endpoint while other multiplexed analyses are add-ons that increase sample loss and impair throughput through transfer of samples or analytes. Over the last decade, sample preparation methods based on traversing an immiscible phase barrier have been developed to minimize analyte minipulation (15–18). We have recently developed Exclusion-based Sample Preparation (ESP) methods that leverage the dominance of surface tension over gravity at the microscale. ESP utilizes microscale constrictions to stabilize the positioning of immiscible fluids side-by-side, creating an immiscible barrier between two aqueous fluids. By pulling paramagnetic particles (PMPs) bound to specific analytes of interest through these immiscible barriers, we isolate the analytes from a general sample without dilution, splitting or perturbation (13, 14, 19–23). The VERSA (Versatile Exclusion-Based Rare Sample Analysis) was designed to use ESP technology for CTC isolation and multi-endpoint analyses (13, 14, 19–22, 24–26) thus achieving modularity without compromising each individual assay.
The VERSA platform integrates CTC capture for any extracellular target of interest, extra- and intracellular staining for enumeration and immunocytochemistry, and mRNA and DNA extraction. We evaluated these integrated endpoints in patients with advanced PRCA for 1. Enumeration(27, 28), 2. Gene expression analysis of multiple AR splice variants; 3) activity in the canonical AR signaling pathway (29, 30); 4. AR protein quantification and subcellular localization(31); and 5. AR genomic alterations(32). We identified expression of multiple AR variants with correspondingly high activity in the canonical AR pathway in patients progressing on targeted therapies. Longitudinal CTC analysis identifies acquisition of multiple AR variants and increased activity in the AR signaling pathway despite AR targeted therapies and chemotherapy. Genomic analysis identifies simultaneous AR amplifications and point mutations in a subset of patients, as predicted in pre-clinical models. These integrated CTC endpoints reveal complex mechanisms of resistance to targeted therapies with corresponding functional alterations in protein and signaling pathways that would direct a subset of patients towards AR independent therapies.
Materials and Methods
VERSA Device Manufacturing
The VERSA device was injection molded in two pieces (Proto Labs, USA). The front and back sides were fabricated separately from polystyrene to a thickness of 2 and 2.5 mm. The fronts side contains the cell capture well (250 µL), the extracellular staining well (30 µL), sieve well (50 µL), oil well (30 µL) and the mRNA extraction well(15 µL). The back side contains the sieve well that mates to the front sieve well (50 µL), oil well (30 µL) and DNA extraction well (15 µL). All wells are connected via trapezoid oil wells that have a 300 mm depth and height that tapers from 2 to 0.8 mm with a channel above for oil loading. The front and back sides were solvent bonded using acetonitrile (Sigma-Aldrich, USA) (33) with an 8 µm microporous membrane (Part PET8025100, Sterlitech, USA) sandwiched in the sieve well. A pressure sensitive adhesive film was applied to each side of the device (MicroAmp, Applied Biosystems, USA).
Cell Culture
The PRCA cell lines 22Rv1 and LNCaPs, were a gift from Dr. Douglas McNeel and were authenticated by short tandem repeat profiling in March 2014 by DDC Medical. Both cell lines were cultured in Corning Cellgro® RPMI 1640 Medium (VWR, USA) containing 10 % fetal bovine serum (FBS), 1 % Pen-Strep, 1 % Sodium Pyruvate and 1 % α-MEM. The PRCA cell line R1-D567 (from the laboratory of Dr. Scott Dehm at the University of Minnesota (34) was cultured in Corning Cellgro®RPMI 1640 Medium and 10 % FBS. The R1-D567 cell line was genetically engineered to remove exon 5, 6 and 7 of the AR LBD. All cells were cultured at 37 °C and maintained under 5 % CO2. Cells were passaged using a 0.05% trypsin/EDTA solution.
Specificity and Sensitivity of Reverse Transcription Polymerase Chain Reaction TaqMan®
Cell dilutions of 100, 10 and 1 for R1-AD1, R1-D567 and VCAP cell lines were created and mRNA extracted as described above. Next, extracted mRNA was reverse transcribed, amplified using TaqMan® PreAmp and used in TaqMan® RT-PCR assays as described above. Threshold cycle (Ct) values were reported.
Clinical Study Design and Statistical Analysis
This was a prospective biomarker study evaluating expression of AR protein and splice variant expression in CTCs from patients with CRPC receiving systemic treatment with chemotherapy, AR targeted therapies or Radium 223. The study was approved by the institutional review board at University of Wisconsin and all patients supplied written informed consent. Patients were required to have histologically confirmed prostate adenocarcinoma, progressive disease despite “castration levels” of serum testosterone (<50 ng per deciliter [1.73 nmol per liter]) with continued androgen-deprivation therapy, and documented metastases, as confirmed on computed tomography (CT) or bone scanning with technetium-99m–labeled methylene diphosphonate. Patients had to have two or more rising serum PSA values obtained 2 or more weeks apart, with the last value being 2.0 ng per milliliter or higher — criteria for PSA progression that are consistent with Prostate Cancer Clinical Trials Working Group 2 (PCWG2) guidelines. Patients with PSA progression underwent restaging radiographic imaging in the form of bone scan and CT scan of the abdomen/pelvis to evaluate for radiographic progression. All the authors vouch for the completeness and integrity of the data and for the fidelity of the study to the clinical protocol. Peripheral blood samples, for analysis of CTCs, were obtained from eligible patients at the time of disease evaluation with serum PSA and radiographic imaging if documented PSA progression. All the clinical investigators were blinded to AR-V7 status of the participants. All the laboratory investigators were blinded to clinical information when determining CTC results. The association between radiographic progression status with gene expression was evaluated using multivariate logistic regression with treatment category as a covariate.
VERSA Operation
Blood specimins were collected in Cellsave (Jansen Diagnostics,fixed) or vacutainer tubes (BD Biosciences, live) with EDTA anticoagulant. Mononuclear cells were isolated with a ficoll gradient. EDTA samples were CD45 depleted to improve purity of live cell capture of CTCs. Cellsave samples were fixed using BD cytofix. CTCs were isolated with VERSA using an antibody to Epcam conjugated to paramagnetic particles. Downstream processes including immunofluorescent staining and extraction of mRNA and DNA are integrated on the VERSA. Details of blood processing, paramagnetic particle preparation, and VERSA operation can be found in the Supplementary Materials and Methods.
AR Nuclear Localization and AR Quantification Analysis
Images were taken with a 10× objective using a Nikon Eclipse Ti-E with a ORCA-Flash 4.0 V2 Digital CMOS camera (Hamamatsu) and NIS-Elements AR Microscope Imaging Software (Nikon, USA). Images were processed using Image J. CTCs were defined as having an intact nuclei, EpCAM or cytokeratin positive and CD45 negative. For AR nuclear localization, a threshold binary image was created for both the nuclear and AR stains to establish regions of interest (ROI). The ROI’s were overlaid on the AR stain and the mean intensity and area measured. Background was subtracted from the mean intensity, which was then multiplied by the area to determine the integrated AR intensity. AR nuclear localization was determined by dividing the corrected AR intensity from the nuclear ROI over the total AR ROI.
Quantatative RT-PCR
The mRNA elution sample containing PMPs was reverse transcribed using a High Capacity cDNA Reverse Transcriptase kit (Life Tech, USA),according to manufacturer’s directions using Bio-Rad C1000 Thermo Cycler (Bio-Rad, USA). The RT reaction (12.5 µL) was then amplified for 10 cycles using TaqMan® PreAmp (Life Tech, USA) according to manufacturer’s directions and diluted 1:5 in 1× TE (10 mM Tris-HCL pH8, 1 mM EDTA). For TaqMan® assays, 5 µL of diluted cDNA template was mixed with 10 µL iTaq® master mix (Bio-Rad, USA), 1 µL TaqMan® Gene Expression Assay (Specified in Table 6, Life Technologies, USA) and 4 µL nuclease free (NF) water. Each reaction was amplified for 45 cycles (denatured at 95 °C for 15 seconds followed by annealing at 60°C for 1 minute) using a CFX Connect® Real-Time PCR System (Biorad, USA). A table of primers used is available in SI (Table S1). Threshold cycle (Ct) values were reported.
Whole Genome Amplification (WGA)
CTC specimens were delivered as extracted DNA. DNA was split and amplified in multiple reactions using Phi29 enzyme at 30°C to allow variant consensus calling across splits. After WGA, the amplified DNA was purified using AMPure XP beads (Agencourt), eluted in EB Buffer (Qiagen), and quantified using Quant-iT PicoGreen dsDNA assay (Invitrogen). Specimens with adequate yield (>200ng) and size profile (200bp-10kb), as determined using the Agilent 2200 TapeStation system, were allowed to proceed for comprehensive genomic profiling.
Comprehensive Genomic Profiling
In samples passing primary quality metrics, amplified DNA (50–200 ng per sample) underwent whole-genome shotgun library construction and solution hybridization using methods previously developed and validated (35), to capture full exons from 315 cancer-related genes, as well as introns from 28 genes frequently rearranged in solid tumors. Hybrid capture libraries meeting yield (>25nM, PicoGreen ds DNA assay) and size specifications (~300bp, Agilent 2200 TapeStation system) were sequenced to a minimum of 300× median coverage, with >88% of exons achieving >100× coverage, using Illumina HiSeq2500 49 × 49 paired-end reads. Sequence data was processed and analyzed using a custom pipeline previously described in detail (35) as well as a secondary pipeline developed specifically for use with the WGA methodologies described above. Base substitutions, short insertions or deletions (indels), and rearrangements were detected by statistical analysis and local re-assembly of mapped reads. Coverage at each target and SNP allele frequencies were used to estimate genome-wide tumor purity and ploidy. Only known somatic oncogenic variants were reported.
Results
VERSA Design and Operation
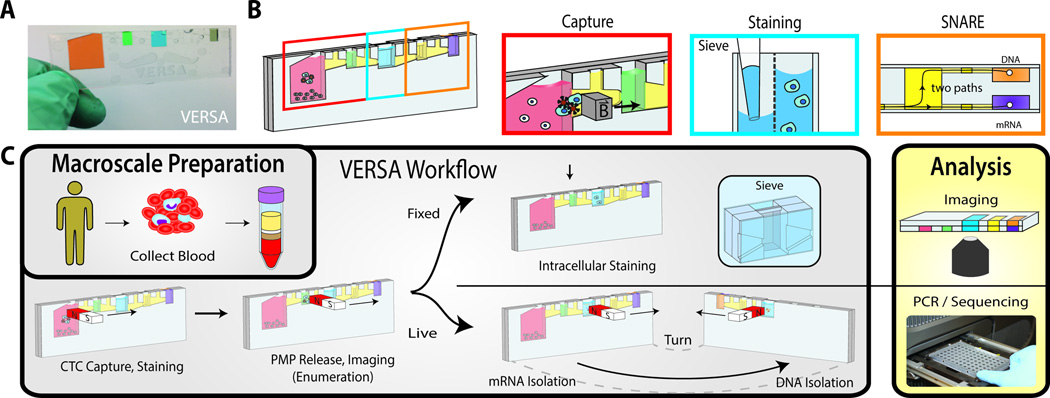
The design of the VERSA and workflow are described in Fig.1. The VERSA device is produced using two pieces of injected molded polystyrene bonded together using solvent bonding (Fig.1A). For CTC capture and isolation from patients with CRPC, functionalized PMPs (3) are added to bind CTCs from the buffy coat fraction and binding occurs on-chip (Fig.1B). CTCs are captured from the residual nucleated cells by moving an external magnet from the input well to the extracellular staining well (Fig.1C). After staining, cells are transferred to the sieve well using the external magnet. The sieve well contains an 8 µm porous membrane, dividing the well into a front and back chamber. The membrane allows low-pressure fluid exchanges to facilitate removal of released and unbound PMPs (2.8 µm) while preventing larger cells (–20 µm) from passing through the porous membrane (21). The sieve well is also used to stain intracellular proteins, enabling cell permeabilization, antibody incubation and fluid exchanges to wash away unbound antibody. The VERSA can be positioned horizontally for on-chip image acquisition. We use the VERSA to identify CTCs as Hoechst+/CD45−/ Cytokeratin+ cells and to measure both intensity and localization of AR. Finally, the VERSA incorporates the SNARE, a highly sensitive ESP nucleic acid extraction method which allows efficient extraction of both mRNA and DNA from a single sample (20) enabling paired genomic/transcriptomic analysis of rare cells such as CTCs (Fig.1B & 1C).
Figure. 1. The VERSA device.
The VERSA integrates efficient cell capture with PMP removal, staining and isolation of mRNA and DNA without dilutive steps. (A) The handheld VERSA is filled with colored dye to differentiate the different chambers. (B) The VERSA is pictured with boxes designating the well used for capture (red), staining (blue) and nucleic acid isolation (orange). (C) A magnet is used to purify PMP-bound CTCs from the input well (pink) through the oil-filled trapezoid into the extracellular staining well (green). After incubation, CTCs are moved into the sieve well (blue), which contains an 8 µm porous membrane, dividing the well into a front and back chamber. The membrane allows low-pressure fluid exchanges to facilitate removal of released and unbound PMPs while preventing cells of interest from passing through. The ability to perform multiple fluid exchanges enables cell permeabilization and incubation with antibodies to intracellular antigens. Cells are imaged in device. mRNA is isolated by lysing cells in device, adding oligo-dT PMPs and moving RNA to the front elution well (orange box, top right). The subsequent addition of silca PMPs with a nuclear lysis buffer enables co-extraction of DNA by magnetic transfer of PMPs to the back well.
Cell Capture and Protein Analysis
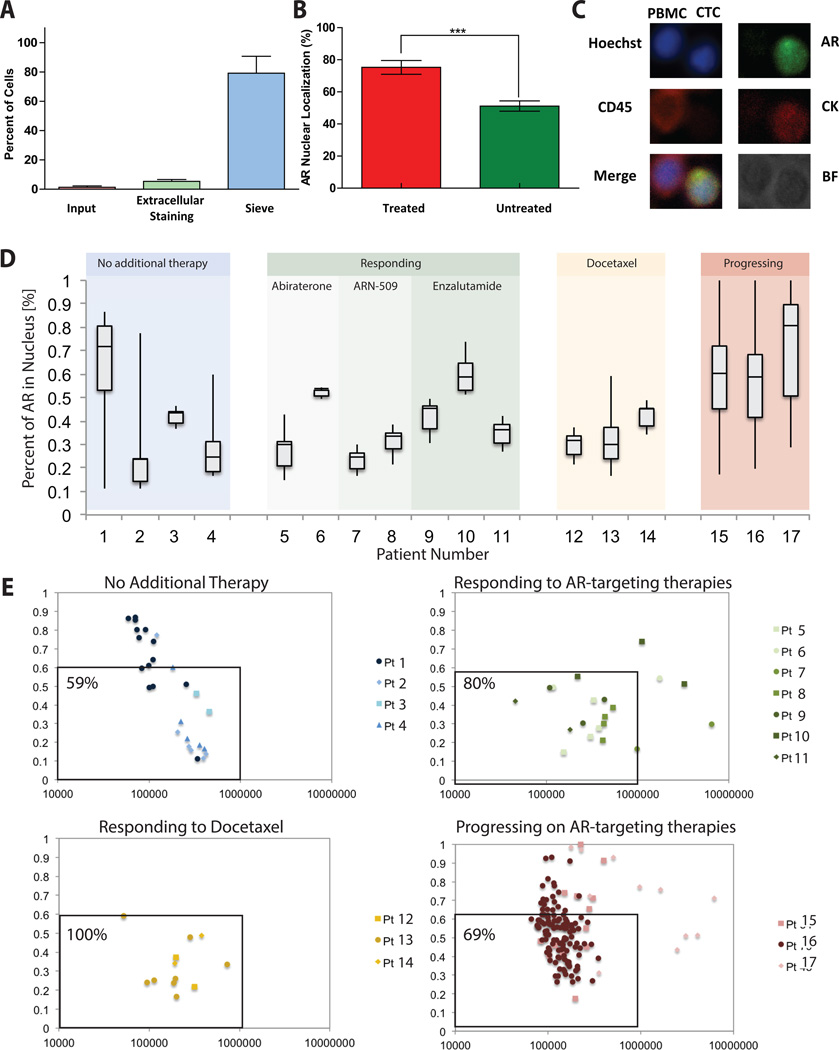
The VERSA captures tumor cells using any capture antibody of interest. Given the clinical relevance of EpCAM in CTCs from patients with PRCA, we validated capture efficiency using an anti-EpCAM antibody. The VERSA demonstrated an average capture efficiency of 79.2±11.6% when isolating ~25 LnCap cells (Fig.2A), similar to prior publications (21). Longitudinal analysis of EpCAM capture demonstrates high reproducibility in capture efficency using VERSA (Fig.S1). Through intracellular staining and imaging of the AR in the VERSA, we can quantify AR nuclear localization and intensity. For initial validation of the image processing to determine AR nuclear localization, AR transfected COS-7 cell lines were treated with an AR agonist or a vehicle control. The treated cells had significantly higher AR nuclear localization as compared to untreated cells (p<0.0004) (Fig.2B).
Figure 2. AR quantification and nuclear localization in CRPC patients.
(A) Approximately 25 calcein-labeled LNCaP cells were spiked into the input well and imaged. Following VERSA procedure, input, extracellular staining and sieve wells were again imaged and LNCaPs counted to determine percent of cells in each well. (B) AR nuclear localization of AR transfected cos-7 cells stimulated with and without mibolerone (AR agonist). Stimulated cells (n=30, mean±SD) showed significantly higher AR nuclear localization as compared to unstimulated cells (p<0.0004). (C) A representative patient PBMC and CTC is shown here stained with CD45 (PBMC) or AR and Cytokeratin (CTC). (D) The percent nuclear localization is shown for 17 CRPC patients grouped by patient treatment and response. Box plots show average and spread (min to max) of the localization percent within CTCs for each patient across different classes of therapy. (E) For each individual CTC within a patient total AR Intensity and AR nuclear localization percentage were plotted for different patient groups.
Next, we analyzed CTC number, AR localization and total AR intensity in 17 patients with castrate resistant PRCA. Results are grouped based on PSA response to the patient’s current treatment at the time of the blood draw. Cells were stained using Hoechst, anti-CD45 and anti-Epcam antibodies to distinguish CTCs from hematopoetic cells, as well as an anti-AR antibody to probe expression and nuclear localization (Fig.2C). Patients responding to AR-targeted or chemotherapy treatments showed lower average numbers of CTCs (Table S2) and percentages of AR within the nucleus (37%±12%), compared to patients that have progressed on AR-targeted therapies (60%±8%) (Fig.2D). The low CTC number for patients responding to treatment is consistent with other studies (36), but limits the number of data points available to calculate the average AR localization. Therefore we combined single cell data points from multiple patients in four plots corresponding to different stages in treatment, demonstrating heterogeneity across CTCs with evaluation of AR intensity and AR nuclear localization. (Fig.2E). Patients responding to treatment generally displayed a low total AR intensity and low percent of AR localized to the nucleus. However, patients progressing on AR targeting therapies have more CTCs with higher AR expression/nuclear localization with a unique population of cells expressing low levels of AR, but highly localized to the nucleus (Fig.2E, Pts 15 and Pt 16). Preliminary findings from this initial cohort of patients suggests an emerging mechanism of resistance in AR activity within a subset of CTCs that allows the AR to translocate to the nucleus, despite antagonist therapy. The VERSA thus permits molecular interrogation of phenotypic heterogeneity in CTCs, for prospective evaluation in clinical trials as a pharmacodynamic biomarker.
Gene Expression Analysis of PRCA Cell Lines
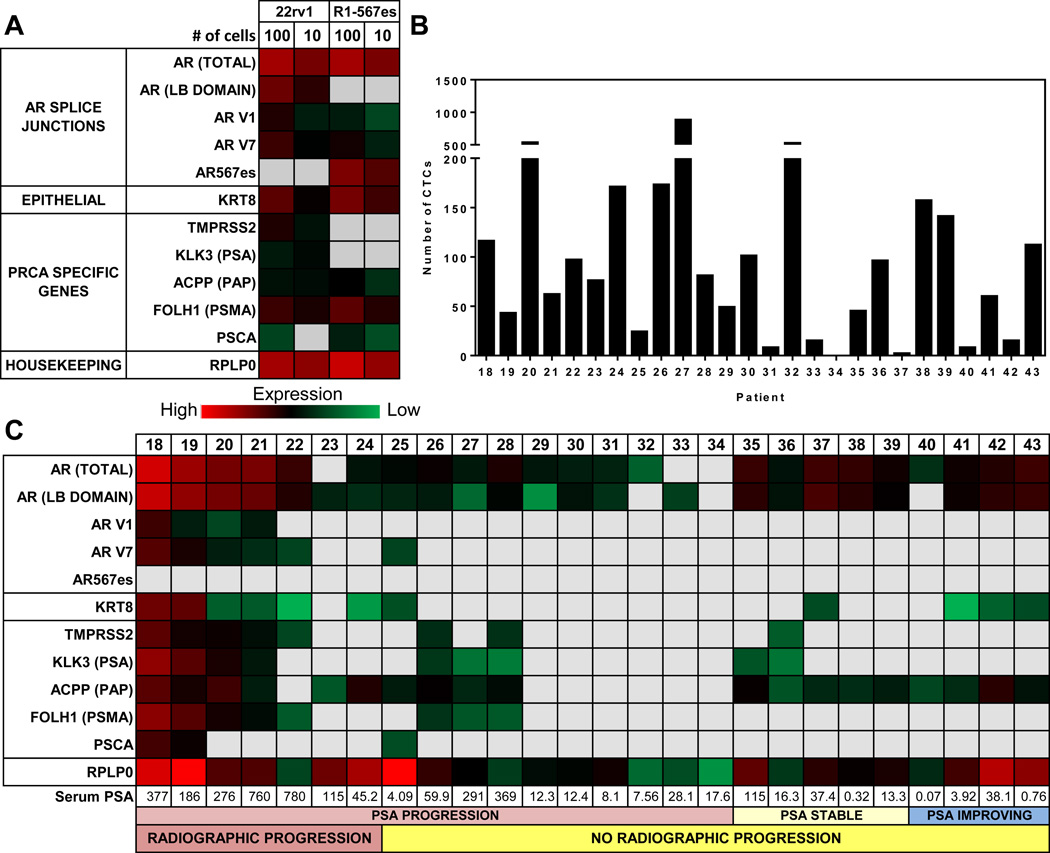
To evaluate the sensitivity and specificity of gene expression using mRNA extracted in the VERSA, we used cell lines which express different AR splice variants (Fig.3A). 22rv1 cells express the variants AR-V1 and AR-V7 while the R1-D567 cell line is engineered to express AR-V567ES (34, 37). mRNA was extracted from 10 or 100 cells reverse transcribed, pre-amplified and probed with the described panel of genes. The AR-V1 and AR-V7 variants are detected in both cell lines at the 10 and 100 cell level. The AR-567es variant is observed only in the R1-D567 cell line. As expected, no detection of the AR LB domain was observed when probing the R1-D567 cells with primers targeting the boundary of exons 4 and 5 of the AR since this cell line has a complete deletion of exons 5, 6, and 7.
Figure 3. Gene expression analysis of the AR signaling pathway.
(A) Results from quantitiative RT-PCR are presented as Ct values represented as a heat map. mRNA was isolated from the indicated number of cells (n=3) from either 22rv1 or R1-D568 cell lines. (B) Enumeration of CTCs (defined as the number of cells with intact nuclei, CK+, CD45−/7.5 mL blood) from a fixed sample run in parallel to gene expression analysis. (C) mRNA was isolated from Epcam positive fraction from15 mL of EDTA anticoagulated blood. mRNA was reversed transcribed, pre –amplified and probed for the AR gene splice junctions including multiple splice variants and PRCA specific genes.
Gene Expression in CTCs
A biomarker clinical trial at the University of Wisconsin Carbone Cancer Center enrolled 26 patients with metastatic PRCA, 19 of whom had received or were currently being treated with AR signaling pathway inhibitors (Table 1, Table S3). CTCs were captured with EpCAM and stained for intact nuclei, cytokeratin, and CD45. CTC enumeration in the VERSA showed detectable CTCs in 25 of 26 patients with a range of 0–1213 CTCs per 7.5 mL of blood (Fig.3B). Patients with both increased serum PSA levels and radiographic evidence of disease progression had detectable expression of full length AR and multiple AR splice variants, with correspondingly high expression of downstream targets in the AR signaling pathway (Fig.3C, Pts 18–22). Detectable expression of the AR-V7 splice variant was significantly different in patients with radiographic progression compared to patients with only PSA progression or PSA response (71% vs 5%, p=0.007). Detectable expression of downstream targets of the AR pathway, FOLH1 (PSMA) (p=0.014) and TMPRSS2 (p=0.030), was also associated with radiographic progression. Importantly, expression of other AR splice variants is found with high coincidence with AR-V7, with detectable expression of the AR-V1 variant in 4 of these 7 patients. Of the two patients without expression of AR variants (Pts 23 and 24), one developed visceral metastases and had no clinical benefit from Abiraterone, suggesting that these integrated biomarkers may identify a subset of patients with progressive disease not dependent on canonical AR signaling. Detectable expression of KLK3 (PSA) and ACPP (PAP) transcripts were not associated with disease response, with patients responding to AR targeted therapies also showing expression of these genes. These results are consistent with recent studies performing single cell transcriptomic analysis which found similar heterogeneity in expression of these targets, with many cells lacking detectable expression of PSMA and PSCA (38, 39). Further prospective clinical trials are needed to determine whether these signatures are solely due to treatment response or a fundamental biologic difference in these tumor cells that have entered circulation.
Table 1.
Patient Characteristics stratified by patients Progressing on their current treatment verses patients responding
| Progression (n=7) |
Responding (n=19) |
|
|---|---|---|
| Age, median (range), yrs | 65 (55–78) | 68 (60–83) |
| Time since diagnosis (range), yrs | 7 (1–10) | 9(1–17) |
|
PSA at blood draw, median (range), ng/mL |
276 (45.2–780) | 13.3 (0.07– 369.1) |
| Gleason Score, #, (%) | ||
| ≤7 | 3 (43) | 7 (37) |
| ≥8 | 3 (43) | 10 (53) |
| Poorly differentiated | 1 (14) | 2 (10) |
| Presence of Bone Metastasis,#, (%) | ||
| Yes | 7 (100) | 18 (95) |
| No | 0 (0) | 1 (5) |
| Presence of Visceral Metastasis,#, (%) | ||
| Yes | 2 (29) | 3 (16) |
| No | 5 (71) | 16 (84) |
| Current/Prior use of Enzalutamide / ARN-509,#, (%) | ||
| Yes | 5 (71) | 3 (16) |
| No | 2(29) | 16 (84) |
| Current /Prior use of Abiraterone Acetate / VT-464 / TAK700, #, (%) | ||
| Yes | 5 (71) | 10 (53) |
| No | 2(29) | 9 (47) |
| Current /Prior use of Docetaxel,#, (%) | ||
| Yes | 4 (57) | 9 (47) |
| No | 3 (43) | 10 (53) |
Longitudinal analysis
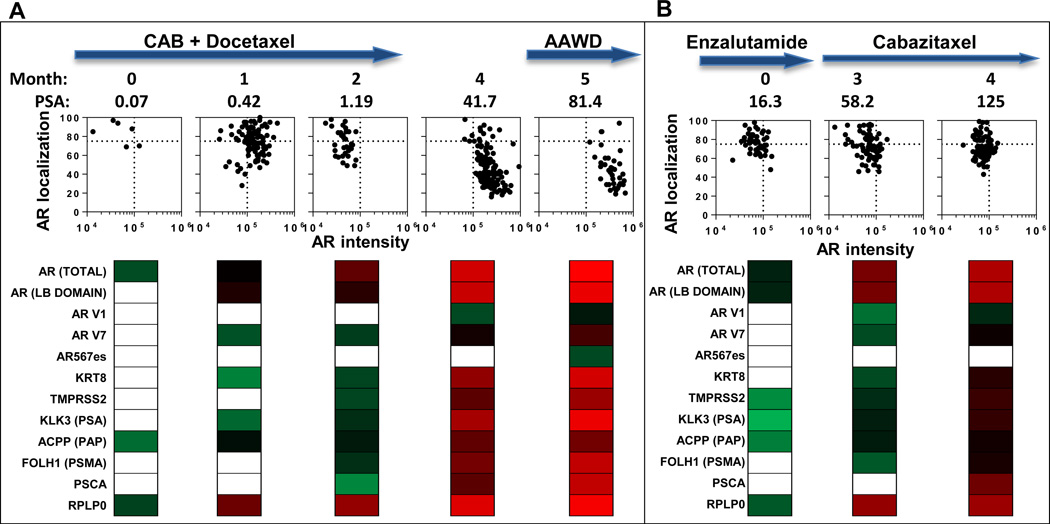
The ability to follow patients longitudinally using CTC analyses is a major potential advantage of the liquid biopsy over traditional biopsies of metastatic sites. Fig.4 shows an example of integrated longitudinal analysis for two patients. Patient 40 (Fig.4A) presented with metastatic PRCA, with lymph node and bone metastases, and a PSA of 35.7. He began chemohormonal therapy as per the CHAARTED protocol (40), with an almost complete resolution in lymphadenopathy and PSA (nadir to 0.07) by cycle 3 (Figure 4A, Month 0). However, his serum PSA began to rise by cycle 4, with a significant increase in CTC number and detectable expression of AR-V7 and PSA genes (Fig.4A, Month 1). By month 2, (cycle 6), detection of PSMA and TMPRSS2 was identified and associated with continued rise in his serum PSA. Restaging radiographic imaging did not show radiographic progression, and the patient elected to take a treatment break given toxicities from docetaxel chemotherapy. However, off therapy, his PSA rapidly rose with corresponding detection of multiple AR splice variants and increasing activity in the canonical AR signaling pathway (Month 4). An anti-androgen withdrawal (AAWD) was performed prior to enrolling on a clinical trial with a PARP inhibitor. AR expression and nuclear localization was highly heterogeneous suggesting a subpopulation of tumor cells retaining sensitivity to targeted therapy. However, there is clearly a resistant population of tumor cells that fluctuated from high nuclear localization to AR overexpression (Fig.4A, graphs), coinciding with AR variant transcript detection. A second patient (Fig.4B, Pt 36) rapidly developed disease progression on Enzalutamide with acquisition of the AR-V7 variant (Month 3). He was subsequently treated with Cabazitaxel chemotherapy with no evidence of a serum PSA response. Corresponding with his lack of response, no phenotypic alterations in AR nuclear localization were identified in this patient, a proposed mechanism by which taxane chemotherapy may exert it’s effect (41). The integrated gene expression data further supports the notion that this patient’s disease is still driven via the AR signaling pathway, with high expression of AR variants, TMPRSS2, and PSMA as the patient developed PSA and radiographic progression (Fig.4B).
Figure 4. Longitudinal analysis of patients with CRPC.
(A) Pt 40, and (B) Pt 36 were monitored as they progressed through the indicated treatments. Treatment is indicated at the top. The initial blood draw (Month 0) corresponds to (A) cycle 3 of chemohormonal therapy for Pt 40 and (B) cycle 2 of Enzalutamide treatment for Pt 36. For each patient, and at each time point, we present AR nuclear localization verses intensity (each point representing a single CTC). Below each plot, we show panels with gene expression data (represented as a heat map of Ct values). CAB (Combined Androgen Blockade), AAWD (Anti-Androgen Withdrawl)
Integrated Genomic-Transcriptomic-Phenotypic Analysis of the AR pathway
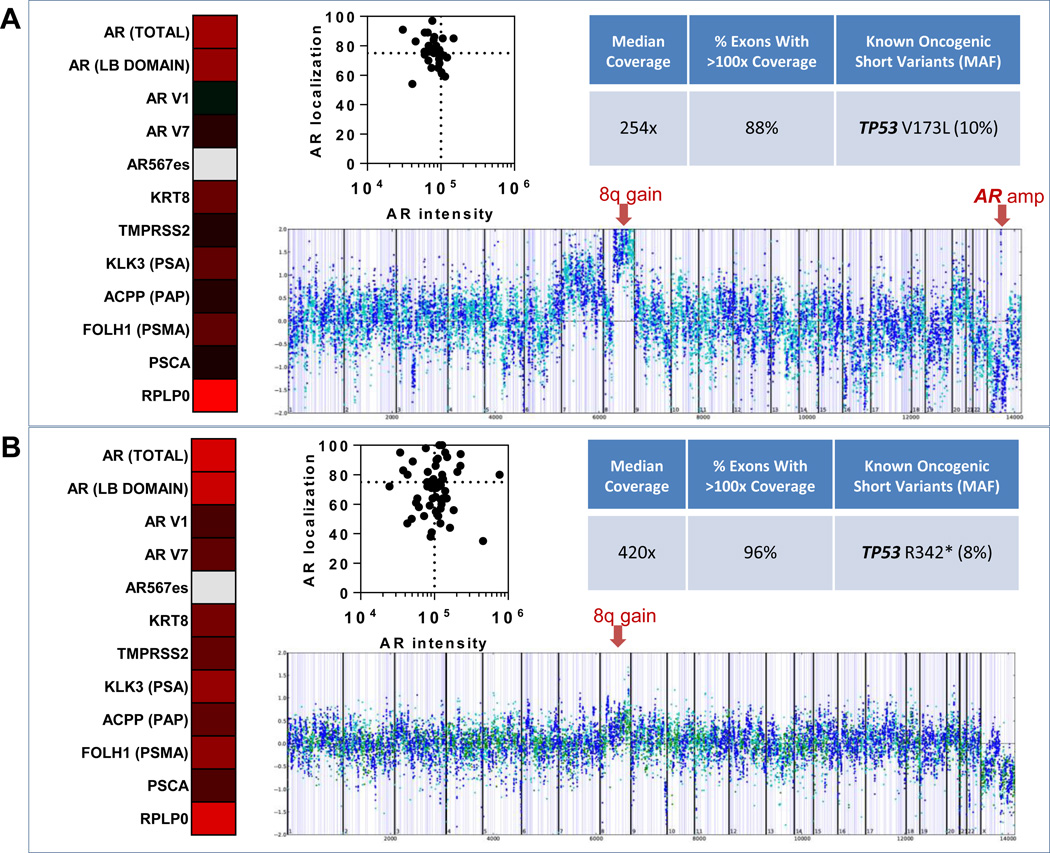
Preclinical and clinical evidence has identified multiple genomic alterations in the AR signaling pathway that contribute to resistance to AR inhibitors, extending beyond AR splice variants (42–45). Using the capability of the VERSA platform to sequentially extract DNA after mRNA isolation, we performed Next Generation Sequencing using a custom genomic assay developed and validated for compatibility with the FoundationOne panel. This analysis was performed in a subset of patients with more than 50 CTCs at greater than 20% purity, and included whole genome amplification (WGA) followed by comprehensive sequencing of full exons from 315 cancer-related genes (including AR), plus introns from 28 additional genes, allowing simultaneous detection of all classes of known oncogenic genomic alterations (base substitutions, short insertions and deletions, copy number changes, and rearrangements). We utilized this assay with the goal of identifying coincident mechanisms of resistance in paired genomic-transcriptomic analyses. CTCs isolated from Pt 19 (Fig.5A), not only expressed multiple AR variants (via mRNA) at the time of progression on Enzalutamide, but also showed genomic evidence for AR amplification (as well as known oncogenic alterations, including a hotspot TP53 mutation (46) and copy number evidence for the 8q gain that is a hallmark of the PRCA genome(47). This contrasts with the CTCs from Pt 28 (Fig.S2), which contained the AR T878A point mutation, a well characterized alteration resulting in a progesterone-activated AR, predicting resistance to AR targeted therapies(44). Pt 18 (Fig.5B) did not have detectable genomic alterations in AR, though again displayed the hallmark 8q gain, as well as a TP53 truncating mutation in the tetramerization domain (48). AR protein analysis revealed broad phenotypic distributions with AR overexpression and AR hyper-nuclear localization, co-occuring in patients with AR splice variant expression and detected genomic AR pathway alterations.
Figure 5. Multi-parametric analysis of gene expression, genomic profiling and AR protein analysis from captured CTCs.
(A) Gene expression and AR protein analysis in a single blood draw from patient 19, and genomic profiling from a subsequent blood draw from this patient. (B) Results shown are from a single blood draw from patient 18. Gene expression data is shown as a heatmap, where red indicates high expression, black median expression, green low expression, and gray no detectable expression. AR protein analysis of CTCs is reported as a graph of AR nuclear localization verses AR intensity. Sequencing coverage and known oncogenic short variants detected using FoundationOne comprehensive genomic profiling are shown along with corresponding copy number plots (each dot represents coverage ratio compared to normal for all exon and intron targets as well as SNPs). Prostate cancer-specific copy number changes are indicated with red arrows (gain of 8q in both patients and focal amplification of AR in patient 19 (A).
Discussion
There is a critical need for predictive and pharmacodynamic biomarkers to guide therapy while simultaneously evaluating emerging mechanisms of resistance. We achieve these multi-endpoint analytics in a microscale platform utilizing ESP concepts that integrate CTC capture with analyte extraction. Termed the VERSA, this handheld device leverages the dominance of surface tension over gravity at the microscale to alternate aqueous and immiscible solutions across distinct operational paths. This enables use of standard reagents and workflows in the VERSA to manipulate and extract captured analytes by hand as all reagents and samples are loaded with standard micropipetters. The flexibility and modularity of the workflow allows collection of live or fixed samples enabling a wide variety of assay endpoints, including visualization of intracellular antigens and extraction of high quality nucleic acids for use in gene expression and comprehensive NGS-based analyses. In this study, magnetic beads conjugated to EpCAM were used for capture, and define CTCs as Hoechst positive, cytokeratin and/or EpCAM positive, and CD45 negative. This platform further permits capture and staining of CTCs for any molecular target of interest, including factors that may contribute to tumor invasion, proliferation and treatment resistance (e.g. epithelial-mesenchymal transitions). The simplicity and cost effectiveness of this platform allows translation to research labs with minimal upfront investment, as VERSA operation is performed in a standard biosafety cabinet and does not require large, expensive machinery.
In this report, we use the VERSA to create a comprehensive CTC assay to evaluate pre-existing and emerging resistance mechanisms to AR targeted therapies including AR amplification, AR point mutations and splice variant expression (49). To better understand the complexity of these resistance mechanisms, we need to evaluate multiple analytes in the same patient. We developed two complementary workflows that leverage the inherent flexibility of the VERSA. First, fixed cell processing allows simultaneous CTC enumeration, and assessment of AR protein expression and localization. Second, the extraction of mRNA and genomic DNA from a matched live sample allows paired analysis of gene expression and NGS-based analysis from the same cells. We have shown that longitudinal analysis across these endpoints can identify emerging biomarkers of resistance, including evidence for persistent activity in the canonical AR signaling pathway as well as expression of multiple AR splice variants. The biomarker evaluations performed in this study are now being evaluated prospectively in clinical trials with Abiraterone Acetate (NCT02025010), Enzalutamide (NCT01942837 and NCT02384382) and VT-464 (NCT02445976). The inherent flexibility of the VERSA creates opportunities to evaluate complex mechanisms of resistance in protein, DNA and mRNA in any solid tumor of interest and is the subject of ongoing studies.
The importance of orthogonal endpoints cannot be understated in diseases with complex resistance patterns as occur in PRCA (32). While CTC enumeration or protein signatures identify emerging tumor clones resistant to the current therapy, they do not inform on the underlying mechanisms driving tumor resistance and proliferation. The clear strength of the AR-V7 splice variant analysis pioneered in Antonarakis et al. (7) is the potential link of a biomarker that also acts as the driver of treatment resistance. However, this marker alone does not capture the many complex resistance mechanisms that can lead to progression of CRPC. For example, we have identified patients with radiographic progression that show minimal expression of the AR and other genes in the canonical AR pathway. Genomic evaluation in a subset of patients in this report identified AR amplifications, AR point mutations and other alterations that can contribute to resistance to AR targeted therapies. This integration of orthogonal endpoints, across protein, RNA and DNA readouts, creates opportunities to evaluate the extent that new drugs modulate the AR signaling pathway in the setting of the complex resistance signatures identified in this report.
The last decade has shown a dramatic increase in the number of therapeutic options for patients with advanced cancer. However, the discriminatory power of our available diagnostics does not inform on the relevance or activity of a given pathway to drive cancer progression. One of the most common questions on the relevance of genomic alterations is whether they are driver or passenger events. Integrating transcriptomic and protein analytics in this context will permit pathway specific evaluation that informs on the functional alterations driving disease progression at a given timepoint. It is within the dynamic environment in which resistance develops that longitudinal assessment of pharmacodynamic biomarkers is needed to not only identify tumor clones with emerging resistance, but also evaluate the extent to which these emerging clones impact patient outcomes. For example, in lung carcinoma Sequist et al (50) identified new histological biomarkers on serial biopsies that mandated a change in treatment away from targeted therapies to chemotherapy and radiation. However, disease progression in a subset of these patients was due to the prior lung cancer histology and further benefit was obtained by re-challenging with the prior targeted therapy. It is within this precision therapeutic paradigm that these predictive and pharmacodynamic CTC biomarkers have the greatest potential to improve patient outcomes. In PRCA, the presence of tumor cells with low AR nuclear localization suggests a population of tumor cells that retains sensitivity to AR targeted therapies despite PSA or radiographic progression. Thus a multi-faceted approach for these patients might improve clinical outcomes by combining AR targeted therapies with chemotherapy, immunotherapy, radiation or an investigational agent. These CTC biomarkers may have their greatest clinical benefit as pharmacodynamic biomarkers for targeted therapies, suitable to identify pathway activity in the setting of emerging drug resistance in order to provide greater precision for clinical decision-making.
Development of pharmacodynamic biomarkers may also improve evaluation of pharmacokinetic analyses employed in early phase clinical trials. These trials perform a critical analysis of drug absorption, exposure, and half-life among others. Linking these pharmacokinetic evaluations with pathway-specific pharmacodynamic biomarkers would enable paired analysis on the drug exposure needed to modulate relevant pathways. This integration of pharmacokinetics and pharmacodynamics is a clear route to establishing optimal biologic dosing strategies beyond the current maximally tolerated dosing strategies. For example, evaluation of TMPRSS2 and PSMA expression in CTCs correlates with radiographic progression and reflects increasing activity in the AR signaling pathway despite targeted therapies. It is unknown if increasing drug exposure through dose escalation would be sufficient to regain disease control. However, this therapeutic approach is commonly employed in clinical practice for patients who were previously dose reduced due to excessive toxicity. Clinical trials incorporating pharmacodynamic biomarkers with dose escalation has intriguing potential to drive precision medicine beyond matching a drug to a patient but rather matching drug dose to the individual. It is within this context that novel CTC technologies evaluating orthogonal analytes can drive cancer therapeutics towards precision medical care.
Supplementary Material
Statement of Translational Relevance.
The goal of precision medicine is to tailor treatments to each cancer. Given the complex resistance mechanisms that occur in prostate cancer, it is critical to test protein, DNA and mRNA for resistance signatures that emerge over the course of treatment. Circulating tumor cells may be an ideal source of tumor cells for precision medicine. We report the development of a new handheld chip that leverages the dominance of surface tension over gravity at the microscale to integrate cell capture with protein staining for any target of interest or extraction of both mRNA and DNA. Complex and emerging resistance mechanisms were identified in patients with castrate resistant prostate cancer that can be used to predict benefit and early resistance to targeted therapies. These assays are now being tested in multiple, prospective clinical trials.
Acknowledgments
We would like to thank all patients who participated in this study. We are also grateful for the help of the UWCCC GU clinical research group especially Jamie Wiepz, Jill Kubiak, Mulusew Yayehyirad, Dorothea Horvath, and Mary Jane Staab.
Finanancial Support: This work was supported by grants from the and a Prostate Cancer Foundation Young Investigator award to J.M. Lang and D.J. Beebe, the Bill & Melinda Gates Foundation through the Grand Challenges in Global Health initiative to S.M. Berry and D.J.Beebe, NIH Grant #1R01CA181648 to S.M. Berry and J.M. Lang, DOD PCRP Grant #W81XWH-12-1-0052 to J.M. Lang, NIH Grant #5R33CA137673 to D.J. Beebe, NSF GRFP DGE-0718123 to D.J. Beebe.
Conflict of interest: Lindsay N. Strotman, Benjamin P. Casavant, Scott M. Berry, David J. Guckenberger, David J. Beebe, and Joshua M. Lang hold equity in Salus Discovery LLC, which has licensed some of the technology described in the manuscript. B.P. Casavant holds equity in Tasso, Inc. D. J. Beebe holds equity in Tasso, Inc., Stacks for the Future, LLC, and Bellbrook Labs, LLC.
References
- 1.Lang JM, Casavant BP, Beebe DJ. Circulating tumor cells: getting more from less. Science translational medicine. 2012;4:141ps13. doi: 10.1126/scitranslmed.3004261. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong AJ, Tannock IF, de Wit R, George DJ, Eisenberger M, Halabi S. The development of risk groups in men with metastatic castration-resistant prostate cancer based on risk factors for PSA decline and survival. Eur J Cancer. 2010;46:517–525. doi: 10.1016/j.ejca.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 3.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:6302–6309. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 4.Danila DC, Anand A, Sung CC, Heller G, Leversha MA, Cao L, et al. TMPRSS2-ERG status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with abiraterone acetate. European urology. 2011;60:897–904. doi: 10.1016/j.eururo.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scher HI, Morris MJ, Larson S, Heller G. Validation and clinical utility of prostate cancer biomarkers. Nature reviews Clinical oncology. 2013;10:225–234. doi: 10.1038/nrclinonc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Molecular cancer research : MCR. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. The New England journal of medicine. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakazawa M, Lu C, Chen Y, Paller CJ, Carducci MA, Eisenberger MA, et al. Serial blood-based analysis of AR-V7 in men with advanced prostate cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2015;26:1859–1865. doi: 10.1093/annonc/mdv282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes EE, VanderWeele DJ, Isikbay M, Duggan R, Campanile A, Stadler WM, et al. Quantitative characterization of androgen receptor protein expression and cellular localization in circulating tumor cells from patients with metastatic castration-resistant prostate cancer. Journal of translational medicine. 2014;12:313. doi: 10.1186/s12967-014-0313-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crespo M, van Dalum G, Ferraldeschi R, Zafeiriou Z, Sideris S, Lorente D, et al. Androgen receptor expression in circulating tumour cells from castration-resistant prostate cancer patients treated with novel endocrine agents. British journal of cancer. 2015;112:1166–1174. doi: 10.1038/bjc.2015.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plaks V, Koopman CD, Werb Z. Cancer. Circulating tumor cells. Science. 2013;341:1186–1188. doi: 10.1126/science.1235226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. Journal of oncology. 2010;2010:617421. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry SM, Alarid ET, Beebe DJ. One-step purification of nucleic acid for gene expression analysis via Immiscible Filtration Assisted by Surface Tension (IFAST) Lab on a chip. 2011;11:1747–1753. doi: 10.1039/c1lc00004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berry SM, Strotman LN, Kueck JD, Alarid ET, Beebe DJ. Purification of cell subpopulations via immiscible filtration assisted by surface tension (IFAST) Biomedical microdevices. 2011;13:1033–1042. doi: 10.1007/s10544-011-9573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shikida MTK, Inouchi K, Honda H, Sato K. Using wettability and interfacial tension to handle droplets of magnetic beads in a micro-chemical-analysis system. Sensors and Actuators B: Chemical. 2006;113:563–569. [Google Scholar]
- 16.Sur K, McFall SM, Yeh ET, Jangam SR, Hayden MA, Stroupe SD, et al. Immiscible phase nucleic acid purification eliminates PCR inhibitors with a single pass of paramagnetic particles through a hydrophobic liquid. The Journal of molecular diagnostics : JMD. 2010;12:620–628. doi: 10.2353/jmoldx.2010.090190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Stanley E, Jin S, Zirkin BR. Stem Leydig cells: from fetal to aged animals. Birth defects research Part C, Embryo today : reviews. 2010;90:272–283. doi: 10.1002/bdrc.20192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bordelon H, Adams NM, Klemm AS, Russ PK, Williams JV, Talbot HK, et al. Development of a low-resource RNA extraction cassette based on surface tension valves. ACS applied materials & interfaces. 2011;3:2161–2168. doi: 10.1021/am2004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strotman LN, Lin G, Berry SM, Johnson EA, Beebe DJ. Facile and rapid DNA extraction and purification from food matrices using IFAST (immiscible filtration assisted by surface tension) The Analyst. 2012;137:4023–4028. doi: 10.1039/c2an35506j. [DOI] [PubMed] [Google Scholar]
- 20.Strotman L, O'Connell R, Casavant BP, Berry SM, Sperger JM, Lang JM, et al. Selective nucleic acid removal via exclusion (SNARE): capturing mRNA and DNA from a single sample. Analytical chemistry. 2013;85:9764–9770. doi: 10.1021/ac402162r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casavant BP, Guckenberger DJ, Berry SM, Tokar JT, Lang JM, Beebe DJ. The VerIFAST: an integrated method for cell isolation and extracellular/intracellular staining. Lab on a chip. 2013;13:391–396. doi: 10.1039/c2lc41136a. [DOI] [PubMed] [Google Scholar]
- 22.Casavant BP, Guckenberger DJ, Beebe DJ, Berry SM. Efficient sample preparation from complex biological samples using a sliding lid for immobilized droplet extractions. Analytical chemistry. 2014;86:6355–6362. doi: 10.1021/ac500574t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beebe DJ, Mensing GA, Walker GM. Physics and applications of microfluidics in biology. Annual review of biomedical engineering. 2002;4:261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- 24.Casavant BP, Strotman LN, Tokar JJ, Thiede SM, Traynor AM, Ferguson JS, et al. Paired diagnostic and pharmacodynamic analysis of rare non-small cell lung cancer cells enabled by the VerIFAST platform. Lab on a chip. 2013;14:99–105. doi: 10.1039/c3lc50912e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berry SM, Regehr KJ, Casavant BP, Beebe DJ. Automated operation of immiscible filtration assisted by surface tension (IFAST) arrays for streamlined analyte isolation. Journal of laboratory automation. 2013;18:206–211. doi: 10.1177/2211068212462023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas PC, Strotman LN, Theberge AB, Berthier E, O'Connell R, Loeb JM, et al. Nucleic acid sample preparation using spontaneous biphasic plug flow. Analytical chemistry. 2013;85:8641–8646. doi: 10.1021/ac4012914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armstrong AJ, Carducci MA. Novel therapeutic approaches to advanced prostate cancer. Clin Adv Hematol Oncol. 2005;3:271–282. [PubMed] [Google Scholar]
- 28.Ramiah V, George DJ, Armstrong AJ. Clinical endpoints for drug development in prostate cancer. Curr Opin Urol. 2008;18:303–308. doi: 10.1097/MOU.0b013e3282fb7807. [DOI] [PubMed] [Google Scholar]
- 29.Page ST, Lin DW, Mostaghel EA, Marck BT, Wright JL, Wu J, et al. Dihydrotestosterone administration does not increase intraprostatic androgen concentrations or alter prostate androgen action in healthy men: a randomized-controlled trial. The Journal of clinical endocrinology and metabolism. 2011;96:430–437. doi: 10.1210/jc.2010-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine reviews. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 32.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou P, Young L, Chen Z. Weak solvent based chip lamination and characterization of on-chip valve and pump. Biomedical microdevices. 2010;12:821–832. doi: 10.1007/s10544-010-9436-z. [DOI] [PubMed] [Google Scholar]
- 34.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA, et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17492–17497. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nature biotechnology. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scher HI, Heller G, Molina A, Attard G, Danila DC, Jia X, et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:1348–1355. doi: 10.1200/JCO.2014.55.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer research. 2013;73:483–489. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Science translational medicine. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sweeney CJ, Chen YH, Carducci M, Liu G, Jarrard DF, Eisenberger M, et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer. The New England journal of medicine. 2015;373:737–746. doi: 10.1056/NEJMoa1503747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darshan MS, Loftus MS, Thadani-Mulero M, Levy BP, Escuin D, Zhou XK, et al. Taxane-induced blockade to nuclear accumulation of the androgen receptor predicts clinical responses in metastatic prostate cancer. Cancer research. 2011;71:6019–6029. doi: 10.1158/0008-5472.CAN-11-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korpal M, Korn JM, Gao X, Rakiec DP, Ruddy DA, Doshi S, et al. An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide) Cancer discovery. 2013;3:1030–1043. doi: 10.1158/2159-8290.CD-13-0142. [DOI] [PubMed] [Google Scholar]
- 43.Joseph JD, Lu N, Qian J, Sensintaffar J, Shao G, Brigham D, et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer discovery. 2013;3:1020–1029. doi: 10.1158/2159-8290.CD-13-0226. [DOI] [PubMed] [Google Scholar]
- 44.Chen EJ, Sowalsky AG, Gao S, Cai C, Voznesensky O, Schaefer R, et al. Abiraterone treatment in castration-resistant prostate cancer selects for progesterone responsive mutant androgen receptors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:1273–1280. doi: 10.1158/1078-0432.CCR-14-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romanel A, Tandefelt DG, Conteduca V, Jayaram A, Casiraghi N, Wetterskog D, et al. Plasma AR and abiraterone-resistant prostate cancer. Science translational medicine. 2015;7:312re10. doi: 10.1126/scitranslmed.aac9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker DR, Bond JP, Tarone RE, Harris CC, Makalowski W, Boguski MS, et al. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene. 1999;18:211–218. doi: 10.1038/sj.onc.1202298. [DOI] [PubMed] [Google Scholar]
- 47.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chene P. The role of tetramerization in p53 function. Oncogene. 2001;20:2611–2617. doi: 10.1038/sj.onc.1204373. [DOI] [PubMed] [Google Scholar]
- 49.Karantanos T, Evans CP, Tombal B, Thompson TC, Montironi R, Isaacs WB. Understanding the mechanisms of androgen deprivation resistance in prostate cancer at the molecular level. European urology. 2015;67:470–479. doi: 10.1016/j.eururo.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Science translational medicine. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.