Abstract
Colorectal cancer (CRC), the second leading cause of cancer-related deaths in the US, has been treated with targeted therapies. However, the mechanisms of differential responses and resistance of CRCs to targeted therapies are not well understood. In this study, we found that genetic alterations of FBW7, an E3 ubiquitin ligase and a tumor suppressor frequently mutated in CRCs, contribute to resistance to targeted therapies. CRC cells containing FBW7 inactivating mutations are insensitive to clinically used multi-kinase inhibitors of RAS/RAF/MEK/ERK signaling, including regorafenib and sorafenib. In contrast, sensitivity to these agents is not affected by oncogenic mutations in KRAS, BRAF, PIK3CA, or p53. These cells are defective in apoptosis due to blocked degradation of Mcl-1, a pro-survival Bcl-2 family protein. Deleting FBW7 in FBW7-wild-type CRC cells abolishes Mcl-1 degradation and recapitulates the in vitro and in vivo drug resistance phenotypes of FBW7-mutant cells. CRC cells selected for regorafenib resistance have progressive enrichment of pre-existing FBW7 hotspot mutations, and are cross-resistant to other targeted drugs that induce Mcl-1 degradation. Furthermore, a selective Mcl-1 inhibitor restores regorafenib sensitivity in CRC cells with intrinsic or acquired resistance. Together, our results demonstrate FBW7 mutational status as a key genetic determinant of CRC response to targeted therapies, and Mcl-1 as an attractive therapeutic target.
Keywords: colorectal cancer, targeted therapies, FBW7, Mcl-1, apoptosis
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths in the US 11. CRC progression is driven by a series of well-defined genetic alterations, including mutations in APC, BRAF, KRAS, PIK3CA, p53, and F-box and WD repeat domain-containing 7 (FBW7) 26, 42. FBW7, also known as FBXW7 and CDC4, is a tumor suppressor frequently mutated in human cancers, including ~15-20% of CRCs 12, 35. It functions as an E3 ubiquitin ligase, which binds to phosphorylated substrates to promote their ubiquitination and subsequent proteasomal degradation 12. Heterozygous FBW7 missense mutations are often detected in three arginine residues (R465, R479 and R505) that bind to a conserved CDC4 phosphodegron (CPD) motif of substrates upon phosphorylation 12.
Recent incorporation of targeted therapies has improved efficacy of CRC treatment 10. Regorafenib and sorafenib, multi-kinase inhibitors for targeting RAS/RAF/MEK/ERK signaling, have been shown to increase overall patient survival and approved for treating solid tumors including CRCs 13, 18, 19, 25. Regorafenib and sorafenib inhibit CRAF, BRAF, VEGFRs, PDGFR, c-Kit and other oncogenic kinases 44, 45. Their anticancer effects are associated with induction of apoptosis, inhibition of cell proliferation, and suppression of tumor angiogenesis. The success of targeted therapies is highly dependent on the identification of sensitive tumors 3, exemplified by the use of KRAS mutational status to guide anti-EGFR therapies 1. However, no genetic or epigenetic factors underlying differential responses and resistance to regorafenib or sorafenib have been identified, despite the need for molecular markers to predict responses to these drugs 6.
Killing of tumor cells by apoptosis is a key molecular mechanism of targeted therapies 20. Stress-induced apoptosis in mammalian cells is mediated through mitochondria by the Bcl-2 family proteins, which collectively regulate apoptosis by triggering a cascade of events, including permeabilization of outer mitochondrial membrane, release of the mitochondrial proteins such as cytochrome c, and activation of caspases 5. Myeloid cell leukemia 1 (Mcl-1) is a pro-survival Bcl-2 family member frequently overexpressed in various tumors 39. Distinctive from other Bcl-2 members, it is a very unstable protein 30. Degradation of Mcl-1 can be triggered by various stresses 24, 34, 43, 46, 48, and regulated by kinases such as glycogen synthase kinase 3β (GSK3β) 27. Recent studies identified FBW7 as an E3 ubiquitin ligase that targets phosphorylated Mcl-1 for destruction 22, 43, suggesting that FBW7 mutations may affect responses to targeted therapies through Mcl-1.
To understand the mechanisms of resistance to targeted therapies, we investigated the functional roles of FBW7 mutations and Mcl-1 degradation in determining responses to targeted therapies. Our results suggest that FBW7 mutations mediate intrinsic and acquired resistance of CRCs to targeted agents by blocking Mcl-1 degradation.
Results
CRC cells containing FBW7 mutations are insensitive to targeted drugs
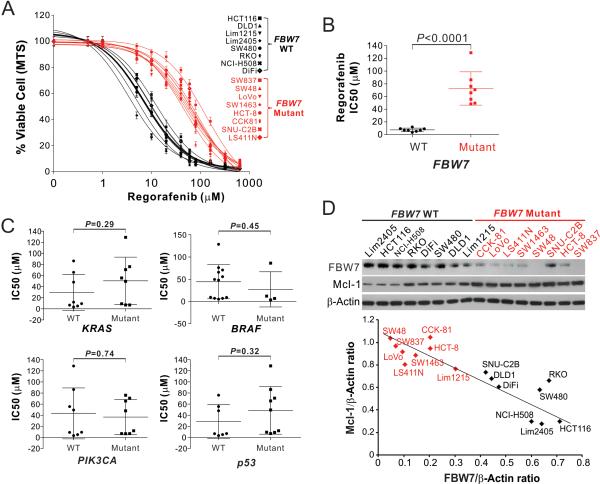
To identify the genetic determinants of CRC response to targeted therapies, we analyzed a panel of 16 CRC cell lines with different mutations in common tumor suppressors and oncogenes, including KRAS, BRAF, PIK3CA, p53 and FBW7 (Table S1). Cells were treated with regorafenib and sorafenib at different concentrations followed by analysis of cell viability using MTS assay. A striking correlation was found between regorafenib sensitivity and FBW7 mutational status (Fig. 1A). All 8 FBW7-mutant CRC cell lines were substantially less sensitive to regorafenib compared to 8 FBW7-wild-type (WT) cell lines, as indicated by higher inhibitory concentration 50 (IC50) (Fig. 1B and Table S1). In contrast, no correlation was found between regorafenib sensitivity and genotypes of KRAS, BRAF, PIK3CA, and p53 (Fig. 1C). The FBW7-mutant cell lines were also significantly less sensitive to sorafenib (Fig. S1, A and B; Table S1). Most (6/8) of the analyzed FBW7-mutant cell lines contain a heterozygous missense mutation inthe substrate-binding arginine or other residues of FBW7, except for SW837 and SW48, which harbor homozygous frame-shift insertion and heterozygous frame-shift deletion, respectively (Table 1 and Fig. S1C). FBW7-mutant cell lines generally express lower levels of FBW7 protein than the WT cell lines (Fig. 1D).
Figure 1. FBW7-mutant CRC cells are insensitive to regorafenib.
(A) MTS analysis of cell viability of FBW7-WT (black) and -mutant (red) CRC cell lines treated with regorafenib at different concentrations for 72 hr. Results were expressed as means ± s.d. of three independent experiments. (B) Comparison of regorafenib IC50 of the analyzed FBW7-WT (black) and -mutant (red) CRC cell lines analyzed in (A). (C) Comparison of regorafenib IC50 of the cell lines analyzed in (A) based on the mutational status of KRAS, BRAF, PIK3CA and p53. (D) Upper, western blotting of FBW7 and Mcl-1 in indicated FBW7-WT and -mutant (red) CRC cell lines; lower, plotting of FBW7 and Mcl-1 expression levels quantified by the Image J program and normalized to the loading control β-actin.
Table 1.
FBW7 mutations in CRC cell lines
| Cell lines | Coding sequence mutations | Zygosity | Type of change | Amino acid change |
|---|---|---|---|---|
| SW48 | c.2001delG | Heterozygous | Deletion | p.S668fs*39 |
| SW837 | c.1205_1206insT | Homozygous | Insertion | p.L403fs*34 |
| LoVo | c.1513C>T | Heterozygous | Missense | p.R505C |
| LS411N | c.1514G>A | Heterozygous | Missense | p.R505H |
| CCK81 | c.1393C>T | Heterozygous | Missense | p.R465C |
| SW1463 | c.1436G>A | Heterozygous | Missense | p.R479Q |
| SNU-C2B | c.1735G>T | Heterozygous | Missense | p.G579W |
| HCT-8 | c.1973G>A | Heterozygous | Missense | p.R658Q |
CRC cells containing FBW7 mutations lack Mcl-1 degradation
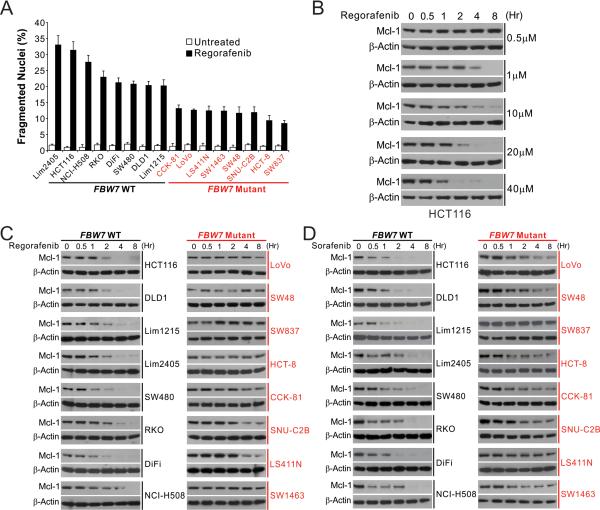
We then investigated the basis of regorafenib and sorafenib sensitivity in CRC cells. Regorafenib suppressed the viability of sensitive cell lines, including HCT116, Lim1215 and RKO cells, at doses that induced caspase activation characteristic of apoptosis induction (Fig. S2A). Regorafenib induced substantially higher levels of apoptosis determined by nuclear fragmentation in FBW7-WT cell lines than in FBW7-mutant cell lines (Fig. 2A), suggesting FBW7 determines regorafenib sensitivity by regulating apoptosis. Analysis of Bcl-2 family members revealed dose- and time-dependent depletion of antiapoptotic Mcl-1 in regorafenib-treated HCT116 cells (Fig. 2B, S2B and S7A), which could be reverted by wash-out of regorafenib (Fig. S2C). Mcl-1 depletion occurred prior to morphological changes of cell death, and required similar doses of regorafenib as those for caspase activation (Fig. 2B and S2A), suggesting a critical role in apoptosis initiation. An inverse correlation between FBW7 and Mcl-1 expression was found in untreated CRC cell lines, with FBW7-mutant cell lines generally expressing higher levels of Mcl-1 than WT cell lines (Fig. 1D and S2D). Upon regorafenib treatment, marked Mcl-1 depletion was detected in all 8 WT cell lines, but not detected or much weaker in 8 FBW7-mutant cell lines (Fig. 2C). Likewise, sorafenib also strongly induced Mcl-1 depletion only in WT cell lines (Fig. 2D). In contrast, the expression of c-Myc and cyclin E, which are also FBW7 substrates 12, were not significantly changed in regorafenib-treated HCT116 cells (Fig. S2E). The striking correlation of regorafenib and sorafenib sensitivity with FBW7 status and Mcl-1 depletion suggests that FBW7 and Mcl-1 are pivotal in determining regorafenib and sorafenib sensitivity of CRC cells.
Figure 2. CRC cells with FBW7 mutations are defective in apoptosis and Mcl-1 degradation.
(A) Indicated FBW7-WT (black) and -mutant (red) CRC cell lines were treated with 40 μM regorafenib for 48 hr. Apoptosis was analyzed by counting condensed and fragmented nuclei after nuclear staining. (B) Western blotting of Mcl-1 in HCT116 cells treated with regorafenib at indicated concentrations and time points. (C) Western blotting of Mcl-1 in FBW7-WT (black) and -mutant CRC (red) cell lines treated with 40 μM regorafenib at indicated time points. (D) Western blotting of Mcl-1 in FBW7-WT (black) and -mutant (red) CRC cell lines treated with 20 μM sorafenib at indicated time points.
FBW7-dependent regorafenib and sorafenib sensitivity is mediated by Mcl-1 depletion
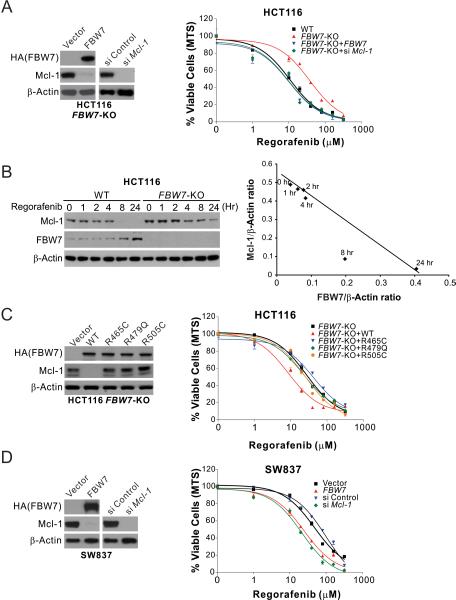
To determine the role of FBW7 in mediating regorafenib and sorafenib sensitivity, we analyzed isogenic FBW7-knockout (KO) HCT116 and DLD1 cells 35. Compared with the WT cells, FBW7-KO cells were substantially more resistant to regorafenib (Fig. 3A and S3A), and also deficient in regorafenib-induced Mcl-1 depletion (Fig. 3B and S3B). Following regorafenib exposure, FBW7 expression was induced when Mcl-1 level started to decline, and showed an inverse correlation with Mcl-1 expression (Fig. 3B and S3B). Transfection of WT FBW7 into FBW7-KO cells depleted Mcl-1 and restored regorafenib sensitivity and apoptosis induction (Fig. 3A; Fig. S3, A, C and D). In contrast to WT FBW7, tumor-derived FBW7 mutants, including R465C, R479Q and R505C, failed to restore regorafenib sensitivity and Mcl-1 depletion in FBW7-KO cells (Fig. 3C and S3E). FBW7 transfection also restored regorafenib sensitivity and apoptosis induction in FBW7-mutant SW837 and SW48 cells (Fig. 3D; Fig. S3, F and G). Treating FBW7-KO and FBW7-mutant cells with sorafenib yielded similar results (Fig. S4, A-D). In contrast to FBW7-KO and FBW7-mutant cells, isogenic CRC cell lines with an engineered change in KRAS, BRAF, PIK3CA, or p53 status did not show any alteration in regorafenib sensitivity compared to the parental cells (Fig. S5, A-D), consistent with lack of correlation with the genotypes of these genes in the cell line panel (Fig. 1C). These results demonstrate that regorafenib and sorafenib sensitivity of CRC cells is dependent on FBW7, whose inactivating mutations cause intrinsic resistance to these drugs, likely through their effect on Mcl-1 depletion.
Figure 3. FBW7 is essential for regorafenib sensitivity and Mcl-1 degradation in CRC cells.
(A) Regorafenib sensitivity of WT and FBW7-KO HCT116 cells with or without HA-tagged FBW7 transfection or Mcl-1 knockdown, which was analyzed by western blotting (left panel). (B) Left, western blotting of Mcl-1 and FBW7 in WT and FBW7-KO HCT116 cells treated with 5 μM regorafenib at indicated time points; right, plotting of FBW7 and Mcl-1 expression quantified by the Image J program and normalized to β-actin. (C) Regorafenib sensitivity of FBW7-KO HCT116 cells transfected with HA-tagged WT FBW7 or indicated mutants (R465C, R479Q or R505C). Transfected FBW7 and endogenous Mcl-1 were analyzed by western blotting (left panel). (D) Regorafenib sensitivity of FBW7-mutant SW837 cells with or without HA-tagged FBW7 transfection or Mcl-1 knockdown, which was analyzed by western blotting (left panel). In (A), (C) and (D), regorafenib sensitivity was analyzed by MTS assay on cells treated with regorafenib at indicated concentrations for 72 hr. Western blotting was performed on untreated cells at 24 hr after transfection. Results were expressed as means ± s.d. of three independent experiments.
CRC cells with acquired regorafenib resistance have FBW7 hotspot mutations
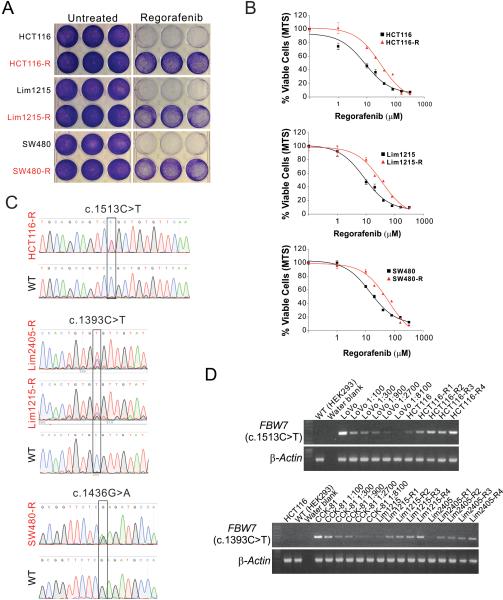
The above observations prompted us to test whether alterations in FBW7 are involved in acquired resistance to regorafenib. We generated regorafenib-resistant cell lines using regorafenib-sensitive and FBW7-WT cell lines, including HCT116, DLD1, SW480, RKO, Lim1215, and Lim2405, by treating cells with 4 consecutive rounds of regorafenib. The survival cells (-R cells) were found to be highly resistant to regorafenib, showing increased cell viability (Fig. 4A, 4B and S6A), and higher IC50 compared to the parental cells (Table 2). The regorafenib-resistant cell lines were found to have reduced apoptosis compared to their parental cell lines (Fig. S6B).
Figure 4. FBW7 mutations are enriched in in CRC cells with acquired regorafenib resistance.
(A) Crystal violet staining of indicated parental and regorafenib-resistant (-R) CRC cells plated at equal density in triplicate after regorafenib (40 μM) treatment for 72 hr. (B) MTS analysis of cell viability of indicated parental (black) and regorafenib-resistant (-R) (red) CRC cells treated with increasing concentrations of regorafenib for 72 hr. Results were expressed as means ± s.d. of three independent experiments. (C) Sequencing of FBW7 genomic region from indicated regorafenib-resistant CRC cell lines highlighting the identified mutations. (D) FBW7 c.1513C>T and c.1393C>T mutations were analyzed by allele-specific PCR in HCT116, Lim1215, and Lim2405 cells after 1-4 rounds (R1-R4) of regorafenib selection. Genomic DNA from LoVo and CCK-81 cells spiked into FBW7-WT HEK293 cells at indicated ratios were used as controls for c.1513C>T and c.1393C>T mutations, respectively.
Table 2.
FBW7 mutations in regorafenib-resistant CRC cell lines
| Cell lines | IC50 (μM) Resistant vs. Parental | *50% Mcl-1 reduction (hr) Resistant vs. Parental | Coding sequence mutations | Zygosity | Amino acid change |
|---|---|---|---|---|---|
| HCT116-R | 30.4 vs.13.7 | >8 vs. 1.8 | c.1513C>T | Heterozygous | p.R505C |
| Lim1215-R | 35.2 vs. 5.1 | >8 vs. 2.2 | c.1393C>T | Heterozygous | p.R465C |
| Lim2405-R | 39.1 vs. 3.5 | >8 vs. 1.2 | c.1393C>T | Heterozygous | p.R465C |
| SW480-R | 61.1 vs. 9.1 | >8 vs. 1.0 | c.1436G>A | Heterozygous | p.R479Q |
| DLD1-R | 32.8 vs. 6.9 | >8 vs. 1.7 | Not detected | ||
| RKO-R | 39.0 vs. 6.8 | >8 vs. 2.8 | Not detected |
Time for 50% Mcl-1 reduction analyzed in cells treated with 40 μM regorafenib
Remarkably, FBW7 mutations were identified in 4 out of 6 resistant cell lines, including R505C (c.1513C>T) in HCT116-R cells, R465C (c.1393C>T) in Lim1215-R and Lim2405-R cells, and R479Q (c.1436G>A) in SW480-R cells (Fig. 4C and Table 2). These mutations account for 25%-43% of FBW7 alleles in the resistant cells, determined by sequencing of individual clones of PCR products from genomic DNA (Fig. S6C). They are the same hotspot mutations found in FBW7-mutant CRC cell lines and tumors (Table 1) 35. Using allele-specific PCR assays that can detect rare FBW7 mutations in FBW7-WT cells, we found that the R505C mutation pre-existed in ~0.03-0.1% of HCT116 cells, but became enriched upon each successive round of regorafenib treatment (Fig. 4D). Similarly, the R465C mutation also pre-existed in Lim1215 and Lim2405 cells and was selected by regorafenib treatment (Fig. 4D). Consistent with the mutations, FBW7 mRNA expression, which was elevated in the parental cells upon regorafenib treatment, could not be induced by regorafenib in the resistant cell lines (Fig. S6D).
Mcl-1 knockdown or inhibition restores regorafenib sensitivity in CRC cells
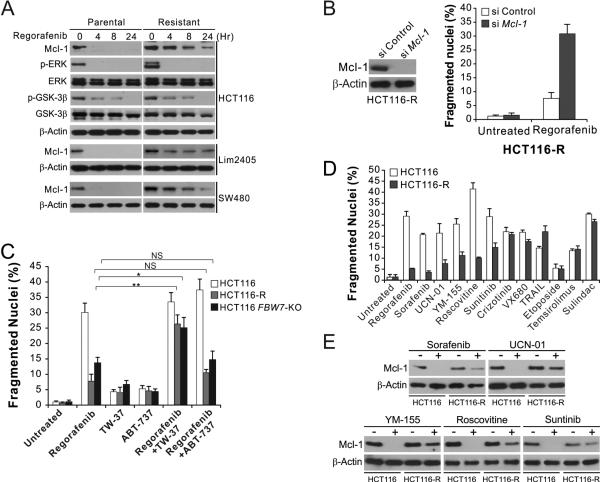
Upon regorafenib treatment, all of the resistant cell lines showed delayed and/or attenuated Mcl-1 degradation than the parental cell lines (Fig. 5A and Table 2), whereas other regorafenib-induced changes, including inhibition of ERK phosphorylation, de-phosphorylation of GSK3β, and induction of PUMA and other Bcl-2 family proteins 8, were intact in the resistant cell lines (Fig. 5A and S7A), suggesting acquired regorafenib resistance is mediated by blocked Mcl-1 degradation. Indeed, knockdown of Mcl-1 restored regorafenib sensitivity in each of the 6 resistant lines (Fig. 5B and S7B).
Figure 5. Regorafenib-resistant cells are re-sensitized by Mcl-1 inhibition, and cross-resistant to other anticancer agents that induce Mcl-1 degradation.
(A) Western blotting of indicated proteins in parental and regorafenib-resistant HCT116, Lim2405 and SW480 cells treated with 40 μM regorafenib at indicated time points. p-ERK: Thr202/Tyr204; p-GSK3β: Ser9. (B) HCT116-R cells transfected with control or Mcl-1 siRNA were treated with 40 μM regorafenib for 48 hr. Left, western blot analysis of Mcl-1 knockdown; right, analysis of apoptosis by counting condensed and fragmented nuclei after nuclear staining. (C) HCT116-R cells were treated for 48 hr with 40 μM regorafenib alone or in combination with 1 μM of the Mcl-1 inhibitor TW-37 or the Bcl-2/Bcl-XL inhibitor ABT-737. Apoptosis was analyzed as in (B). (D) Parental and regorafenib-resistant HCT116 cells were treated with 40 μM regorafenib, 20 μM sorafenib, 1 μM UCN-01, 1 μM YM-155, 10 μM roscovitine, 15 μM sunitinib, 10 μM crizotinib, 10 nM TRAIL, 10 μM VX680, 20 μM etoposide, 20 μM temsirolimus, or 120 μM sulindac sulfide for 48 hr. Apoptosis was analyzed as in (B). (E) Western blotting of Mcl-1 in parental and regorafenib-resistant HCT116 cells treated with indicated agents as in (D) for 24 hr. Results in (B)-(D) represent the means ± s.d. of three independent experiments. NS, P>0.05; *, P<0.05; **, P<0.01.
Several small-molecule inhibitors of the antiapoptotic Bcl-2 family members have been identified, among which TW-37 was shown to be the most effective Mcl-1 inhibitor 41. Treating cells with TW-37, but not ABT-737, a Bcl-2/Bcl-XL inhibitor that does not inhibit Mcl-1 38, restored regorafenib-induced apoptosis in regorafenib-resistant and FBW7-KO HCT116 cells (Fig. 5C and S7C), suggesting that inhibiting Mcl-1 can overcome regorafenib resistance in CRC cells.
Regorafenib-resistant CRC cells are cross-resistant to other agents that induce Mcl-1 degradation
To determine whether the FBW7/Mcl-1 axis has a broad functional role in drug resistance, we analyzed the response of regorafenib-resistant cells to other anticancer agents including targeted agents (Fig. 5D). Analysis of over 30 anticancer agents identified several additional targeted agents that can induce Mcl-1 degradation, including the multi-kinase inhibitors UCN-01 and Sunitinib, the CDK inhibitor Roscovitine, and the survivin inhibitor YM-155 (Fig. 5E). In response to each of these agents, apoptosis and Mcl-1 degradation were found to be suppressed in regorafenib-resistant HCT116, SW480 and Lim 2405 cells compared to the parental cells (Fig. 5D, 5E, S8B, and S8C). In contrast, no change was detected in the sensitivity of regorafenib-resistant cells to the agents that did not induce Mcl-1 degradation, including the Met/Alk inhibitor crizotinib, the death receptor ligand TRAIL, the aurora kinase inhibitor VX680, the DNA damaging agent etoposide, the mTOR inhibitor temsirolimous, and the non-steroidal anti-inflammatory drug sulindac sulfide (Fig. 5D, S8B and S8C). These results suggest that deficiency in Mcl-1 degradation, frequently caused by FBW7 inactivating mutations, is widely involved in intrinsic and acquired resistance to different classes of anticancer agents in CRC cells.
FBW7 and Mcl-1 depletion mediate the in vivo antitumor effects of regorafenib
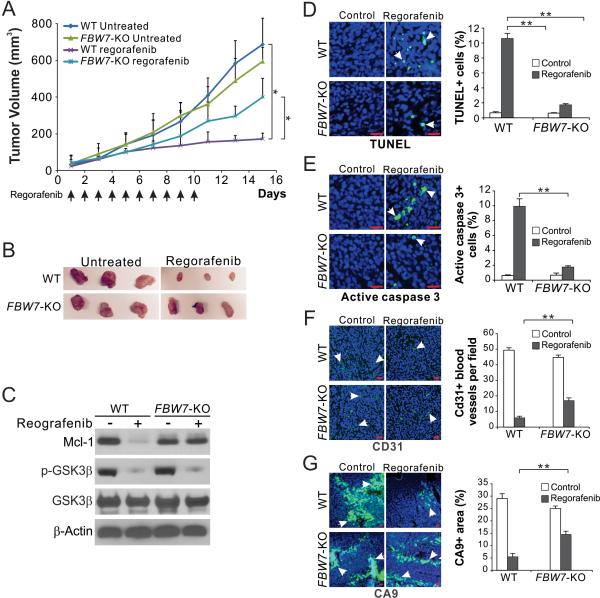
To determine the role of FBW7 in mediating the in vivo effects of regorafenib, WT and FBW7-KO HCT116 cells were injected subcutaneously into nude mice to establish xenograft tumors. Mice were then treated with 30 mg/kg regorafenib or the control vehicle by oral gavage for 10 consecutive days. In contrast to WT tumors, FBW7-KO HCT116 tumors were substantially less sensitive to regorafenib treatment (Fig. 6, A and B). Blocked Mcl-1 depletion but intact GSK3β de-phosphorylation was found in FBW7-KO tumors (Fig. 6C). Apoptosis analyzed by TUNEL and active caspase 3 staining was significantly reduced in the FBW7-KO tumors compared to WT tumors (Fig. 6, D and E). Analysis of tumor vasculature by CD31 staining showed that the antiangiogenic effect of regorafenib was reduced in the FBW7-KO tumors (Fig. 6F). Suppression of tumor hypoxia, analyzed by Carbonic Anhydrase 9 (CA9) staining, by regorafenib was also decreased in the FBW7-KO tumors compared to WT tumors (Fig. 6G). These findings demonstrate a pivotal role of FBW7 and Mcl-1 degradation in mediating the antitumor effects of regorafenib.
Figure 6. FBW7 and Mcl-1 degradation contribute to the in vivo antitumor effects of regorafenib.
(A) Nude mice were injected s.c. with 4 × 106 WT or FBW-KO HCT116 cells. After 1 week, mice were treated with 30 mg/kg regorafenib daily by oral gavage (indicated by arrows), or the vehicle control cremephor EL/ethanol for 10 consecutive days. Tumor volume at indicated time points after treatment was calculated and plotted with statistical significance for indicated comparisons (n=7 in each group). (B) Representative tumors at the end of the experiment. (C) Nude mice with WT or FBW-KO HCT116 tumors were treated with regorafenib as in (A) for 4 consecutive days. Indicated proteins in randomly selected tumors were analyzed by western blotting. (D)-(G) Tissue sections from WT or FBW-KO HCT116 tumors treated as in (C) were analyzed for apoptosis by TUNEL (D) and active caspase 3 (E) staining, tumor angiogenesis by CD31 staining (F), and tumor hypoxia by CA9 staining (G). Left, representative staining pictures with arrows indicating example cells with positive staining;right, quantification of cells with positive staining. Results were expressed as means ± s.d. of three independent experiments. Scale bars, 25 μm. *, P <0.05, **, P <0.01.
Discussion
Metastatic CRC is one of the most deadly cancers, characterized by poor prognosis and low survival rate. Incorporation of targeted agents, including the anti-VEGF antibody bevacizumab and the anti-EGFR antibodies cetuximab and panitumumab, has improved efficacy of CRC treatment 10. However, anticancer therapies, especially targeted therapies, often generate highly heterogeneous patient responses. A major goal of precision medicine is to prevent unnecessary treatments and therapy-associated adverse effects through patient stratification. For example, KRAS status has been used to guide anti-EGFR therapy on a CRC patient 1. Our findings elucidate a critical functional role of FBW7 and Mcl-1 in differential sensitivity and resistance of CRC cells to targeted therapies.
Most of the FBW7 mutations in CRCs are heterozygous point mutations. It has been controversial as to how heterozygous FBW7 mutations abolish the protein function in cancer cells. Mutant FBW7 may have altered protein stability 28, or act as dominant negative proteins upon hetero-dimerization with WT FBW7 12. The lower levels of FBW7 in FBW7-mutant CRC cells (Fig. 1D) suggest a gene dosage effect and altered protein stability. FBW7 mutations are found in various human cancers and likely play a broad functional role in determining therapeutic responses of cancer cells 12. They have been shown to affect responses to γ secretase inhibitors in leukemia cells 31, 40, to HDAC inhibitors in squamous tumor cells 21, and to antimitotic drugs in CRC cells 43. FBW7 is an F-box protein that functions as a substrate receptor for SKP1-CUL1-F-box (SCF)-type of ubiquitin ligase complexes to promote polyubiquitination 14. In addition to Mcl-1, dozens of other FBW7 substrates have been identified, including Jun, Myc, cyclin E, and Notch, which contain CPD motifs of various binding affinities for FBW7 12. These substrates can be differentially affected by FBW7 mutations, underlying the agent- and cell-type-dependent effects on therapeutic response.
FBW7 determines regorafenib and sorafenib sensitivity through proteasomal degradation of Mcl-1. It has been shown that upon GSK3β-dependent phosphorylation, phosphorylated Mcl-1 binds to FBW7 and recruited to the SCF ubiquitin ligase complex formed 43, which can covalently link ubiquitin chains to Mcl-1, leading to its degradation in the 26S proteasome 4. Several other kinases have also been implicated in regulating Mcl-1 turnover, including p38, JNK, CDK1, and casein kinase II 22, 27, 43. FBW7 induction, which correlates with Mcl-1 degradation (Fig. 3B) and is likely responsible for FBW7 activation, may be related to transcriptional regulators of FBW7, such as microRNA 27a and C/EBPδ 2, 23. Alternatively, the stability of FBW7 itself can be regulated by the deubiquitinase Usp28 36. In addition to FBW7, Mcl-1 stability can be regulated by other E3 ubiquitin ligases such as Mule and β-TrCP 16, 48, and by the deubiquitinase USP9X 37. These proteins may also be involved in drug resistance mediated by Mcl-1 stabilization. Furthermore, a recent study showed that Mcl-1 stability is regulated by cyclin E/cdk2-dependent phosphorylation at specific sites, suggesting indirect regulation of Mcl-1 stability by FBW7 through cyclin E 9.
A major challenge in anticancer therapies is emergence of drug resistance. However, little is known about why initial therapeutic responses are short lived, with re-growing tumors insensitive to further treatment 3. Acquired resistance to the anti-EGFR therapy has been associated with enrichment of KRAS-mutant CRC cells 15, 29, suggesting a critical role of preexisting or de novo oncogenic mutations in acquired resistance. Our data show that acquired regorafenib resistance can be due to enrichment of a small fraction of FBW7-mutant cells (Fig. 4). The nature of genomic instability that is responsible for generating these mutant cells remain to be identified. FBW7 genotype does not seem to be the sole determinant of regorafenib sensitivity, suggested by residual Mcl-1 degradation in some FBW7-mutant CRC cells (Fig. 2C) and lack of FBW7 mutations in 2 out of 6 regorafenib-resistant cell lines (Table 2). Other tumor suppressors or oncogenes that regulate the expression and protein stability of FBW7 and Mcl-1 may be involved in regorafenib and sorafenib resistance. The role of Mcl-1 in the intrinsic and acquired regorafenib resistance suggests that Mcl-1 inhibition is an effective approach for overcoming such resistance. The cell line and resistance models generated in this study can be useful for analysis of specific Mcl-1 targeting, which has been difficult to assess 41.
In parallel with ongoing clinical studies, our studies may establish FBW7 mutational status and Mcl-1 stability as key determinants of regorafenib and sorafenib sensitivity. They may provide a rationale for effective combinations of regorafenib and sorafenib with other drugs, for establishing the broad functional roles of FBW7 and Mcl-1 in targeted anticancer therapies, and for overcoming therapeutic resistance caused by genomic instability in cancer cells.
Materials and Methods
Cell culture
The human CRC cell lines (Table S1), including HCT116, RKO, DLD1, LoVo, Lim1215, Lim2405, SW480, SNU-C2B, LS411N, SW48, SW1463, SW837 and HCT-8 were obtained from the American Type Culture Collection (Manassas, VA). CCK-81, DiFi and NCI-H508 cells were obtained from Dr. Alberto Bardelli at University of Torino in Italy. Isogenic p53-KO, FBW7-KO, KRAS-KO (WT or G13D mutant allele), PIK3CA-KO (WT or H1047R or E545K mutant allele) HCT116 or DLD1 cell lines, as well as BRAF-KO (WT or V600E mutant allele) RKO and VACO432 cells, were obtained either from Dr. Bert Vogelstein at Johns Hopkins, or from Horizon Discovery (Cambridge, UK). The cell lines were last tested and authenticated for genotypes, drug response, morphology, and absence of mycoplasma in Feb, 2016. Loss of expression of targeted proteins was confirmed by western blotting and Mycoplasma testing was performed routinely by PCR. Regorafenib-resistant cell lines were generated by exposing regorafenib-sensitive HCT116, DLD1, RKO, SW480, Lim1215 and Lim2405 cells to 40 µM regorafenib for 3 days, followed by recovery for 5 days, and then repeated treatment/recovery for a total of 4 cycles.
All cell lines were maintained at 37°C in 5% CO2 and cultured in McCoy's 5A modified media (Invitrogen) supplemented with 10% defined FBS (HyClone), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). For drug treatment, cells were plated in 12-well plates at 20% to 30% density 24 hr before treatment. The DMSO (Sigma) stocks of agents used, including regorafenib, sorafenib, TW-37, ABT-737, UCN-01, YM-155, roscovitine, sunitinib, crizotinib, VX680, etoposide, temsirolimus, and sulindac (Selleck Chemicals), were diluted to appropriate concentrations with the cell culture medium. TRAIL (XcessBio, San Diego, CA) was diluted with distilled water.
MTS assay
CRC cells were seeded in 96-well plates at a density of 1×104 cells/well. After overnight incubation, cells were treated with regorafenib for 72 hr. 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay was performed using the MTS assay kit (Promega) according to the manufacturer's instructions. Luminescence was measured with a Wallac Victor 1420 Multilabel Counter (Perkin Elmer). Each assay was conducted in triplicate and repeated three times.
Western blotting
Western blotting was performed as previously described 32, with antibodies for PUMA 47, Akt, phospho-Akt (S473), ERK, phospho-ERK (T202/Y204), GSK3β, phospho-GSK3β (S9) (Cell Signaling), Bak (Millipore), Bax, cyclin E, c-Myc, Mcl-1 (Santa Cruz), Bcl-2 (Dako), Bim, Bid, Noxa, and β-actin (EMD Biosciences), HA and Bcl-XL (BD Biosciences).
Transfection and siRNA knockdown
FBW7 expression construct was a gift from Dr. Wenyi Wei at Harvard Medical School. Mutations were introduced into FBW7 using QuickChange XL site-directed mutagenesis kit (Agilent Technologies). Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Knockdown experiments were performed 24 hr before regorafenib or sorafenib treatment using 200 pmol of siRNA. The control scrambled siRNA and siRNA for human Mcl-1 (CGCCGAATTCATTAATTTATT-dTdT) was from GE Dharmacon.
Genomic and reverse transcriptase (RT) PCR
To detect FBW7 hotspot mutations in parental and regorafenib-resistant cells, genomic DNA was isolated from 5-10×104 cells by using ZR-96 Quick-gDNA Kit (ZYMO Research) according to the manufacturer's instructions. One μl out of 50 μl genomic DNA preparation was used for PCR using previously described conditions 17 and primers listed in Table S2. Cycle conditions are available upon request. For analysis of FBW-7 mRNA expression, total RNA was isolated from cells using the Mini RNA Isolation II kit (ZYMO Research, Irvine, CA) according to the manufacturer's protocol. One μg of total RNA was used to generate cDNA using SuperScript II reverse transcriptase (Invitrogen). Real-time PCR was carried out using the primer pairs listed in Table S2 as described 17.
Analysis of apoptosis
Adherent and floating cells were harvested, stained with Hoechst 33258 (Invitrogen), and analyzed for apoptosis by nuclear staining assay. A minimum of 300 cells were analyzed for each treatment. Caspase activity was measured using the SensoLyte Homogeneous AMC Caspase-3/7 Assay Kit (Anaspec) as previously described 7.
Xenograft tumor experiments
All animal experiments were approved by the University of Pittsburgh Institutional Animal Care and Use Committee. Female 5- to 6-week-old Nu/Nu mice (Charles River) were housed in a sterile environment with micro isolator cages and allowed access to water and chow ad libitum. Mice were injected subcutaneously with 4×106 WT or FBW7-KO HCT116 cells. After tumor growth for 7 days, mice were treated daily with regorafenib at 30 mg/kg by oral gavage for 10 consecutive days. Regorafenib was dissolved in Cremephor EL/95% ethanol (50:50) as a 4× stock solution. Tumor growth was monitored by calipers, and tumor volumes were calculated according to the formula 0.5 × length × width2. Mice were euthanized when tumors reached 1.0 cm3 in size. Tumors were dissected and fixed in 10% formalin and embedded in paraffin. Terminal deoxynucleotidyl transferase mediated dUTP Nick End Labeling (TUNEL; Millipore), active caspase 3 (Cell Signaling), CD31 (Spring Bioscience, Pleasanton, CA), and Carbonic Anhydrase 9 (CA9; Santa Cruz) immunostaining was performed on 5 μM paraffin-embedded tumor sections as previously described 33, by using an AlexaFluor 488-conjugated secondary antibody (Invitrogen) for detection.
Statistical Analysis
Statistical analyses were carried out using GraphPad Prism IV software. P values were calculated by the student's t-test and were considered significant if p < 0.05. The means ± one standard deviation (s.d.) were displayed in the figures.
Supplementary Material
Acknowledgements
The authors would like to thank our lab members for critical reading. This work is supported by U.S. National Institute of Health grants (CA106348, CA172136, and CA203028 to L.Z.; U01DK085570 and AI068021to J.Y.) and American Cancer Society grant (RGS-10-124-01-CCE to J.Y.). This project used the UPCI shared facilities that were supported in part by award P30CA047904.
Abbreviations
- CA9
Carbonic Anhydrase 9
- CPD
CDC4 phosphodegron
- CRC
colorectal cancer
- DAPI
4’ 6-Diamidino-2-phenylindole
- ERK
extracellular signal-regulated kinase
- FBW7
F-box and WD repeat domain-containing 7
- 5-FU
5-fluorouracil
- GSK3β
glycogen synthase kinase 3β
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IC50
inhibitory concentration 50
- KO
knockout
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- Mcl-1
myeloid cell leukemia 1
- RT-PCR
reverse transcriptase polymerase chain reaction
- siRNA
small interfering RNA
- TUNEL
terminal deoxynucleotidyl transferase mediated dUTP nick end labeling
- WT
wild-type
Footnotes
Conflicts of interest statement
None declared.
References
- 1.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti- epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 2.Balamurugan K, Wang JM, Tsai HH, Sharan S, Anver M, Leighty R, et al. The tumour suppressor C/EBPdelta inhibits FBXW7 expression and promotes mammary tumour metastasis. EMBO J. 2010;29:4106–4117. doi: 10.1038/emboj.2010.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254–1261. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15:122–133. doi: 10.1038/nrm3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhola PD, Letai A. Mitochondria-Judges and Executioners of Cell Death Sentences. Mol Cell. 2016;61:695–704. doi: 10.1016/j.molcel.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan SL, Ma BB. An update on the safety and efficacy of regorafenib in the treatment of solid cancers. Expert opinion on drug metabolism & toxicology. 2014;10:1607–1614. doi: 10.1517/17425255.2014.970169. [DOI] [PubMed] [Google Scholar]
- 7.Chen D, Ming L, Zou F, Peng Y, Van Houten B, Yu J, et al. TAp73 promotes cell survival upon genotoxic stress by inhibiting p53 activity. Oncotarget. 2014;5:8107–8122. doi: 10.18632/oncotarget.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, Wei L, Yu J, Zhang L. Regorafenib inhibits colorectal tumor growth through PUMA-mediated apoptosis. Clin Cancer Res. 2014;20:3472–3484. doi: 10.1158/1078-0432.CCR-13-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhary GS, Tat TT, Misra S, Hill BT, Smith MR, Almasan A, et al. Cyclin E/Cdk2-dependent phosphorylation of Mcl-1 determines its stability and cellular sensitivity to BH3 mimetics. Oncotarget. 2015;6:16912–16925. doi: 10.18632/oncotarget.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu E. An update on the current and emerging targeted agents in metastatic colorectal cancer. Clinical colorectal cancer. 2012;11:1–13. doi: 10.1016/j.clcc.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010;375:1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 12.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demetri GD, Reichardt P, Kang YK, Blay JY, Rutkowski P, Gelderblom H, et al. Efficacy and safety of regorafenib for advanced gastrointestinal stromal tumours after failure of imatinib and sunitinib (GRID): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:295–302. doi: 10.1016/S0140-6736(12)61857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual review of biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 15.Diaz LA, Jr., Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudgeon C, Peng R, Wang P, Sebastiani A, Yu J, Zhang L. Inhibiting oncogenic signaling by sorafenib activates PUMA via GSK3beta and NF-kappaB to suppress tumor cell growth. Oncogene. 2012;31:4848–4858. doi: 10.1038/onc.2011.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 19.Grothey A, Van Cutsem E, Sobrero A, Siena S, Falcone A, Ychou M, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013;381:303–312. doi: 10.1016/S0140-6736(12)61900-X. [DOI] [PubMed] [Google Scholar]
- 20.Hata AN, Engelman JA, Faber AC. The BCL2 Family: Key Mediators of the Apoptotic Response to Targeted Anticancer Therapeutics. Cancer discovery. 2015;5:475–487. doi: 10.1158/2159-8290.CD-15-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He L, Torres-Lockhart K, Forster N, Ramakrishnan S, Greninger P, Garnett MJ, et al. Mcl-1 and FBW7 control a dominant survival pathway underlying HDAC and Bcl-2 inhibitor synergy in squamous cell carcinoma. Cancer discovery. 2013;3:324–337. doi: 10.1158/2159-8290.CD-12-0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lerner M, Lundgren J, Akhoondi S, Jahn A, Ng HF, Akbari Moqadam F, et al. MiRNA-27a controls FBW7/hCDC4-dependent cyclin E degradation and cell cycle progression. Cell Cycle. 2011;10:2172–2183. doi: 10.4161/cc.10.13.16248. [DOI] [PubMed] [Google Scholar]
- 24.Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res. 2006;66:11851–11858. doi: 10.1158/0008-5472.CAN-06-1377. [DOI] [PubMed] [Google Scholar]
- 25.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 26.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Min SH, Lau AW, Lee TH, Inuzuka H, Wei S, Huang P, et al. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol Cell. 2012;46:771–783. doi: 10.1016/j.molcel.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mojsa B, Lassot I, Desagher S. Mcl-1 ubiquitination: unique regulation of an essential survival protein. Cells. 2014;3:418–437. doi: 10.3390/cells3020418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng R, Tong JS, Li H, Yue B, Zou F, Yu J, et al. Targeting Bax interaction sites reveals that only homo-oligomerization sites are essential for its activation. Cell Death Differ. 2013;20:744–754. doi: 10.1038/cdd.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu W, Wang X, Leibowitz B, Liu H, Barker N, Okada H, et al. Chemoprevention by nonsteroidal anti-inflammatory drugs eliminates oncogenic intestinal stem cells via SMAC-dependent apoptosis. Proc Natl Acad Sci U S A. 2010;107:20027–20032. doi: 10.1073/pnas.1010430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahmani M, Davis EM, Bauer C, Dent P, Grant S. Apoptosis induced by the kinase inhibitor BAY 43-9006 in human leukemia cells involves down-regulation of Mcl-1 through inhibition of translation. J Biol Chem. 2005;280:35217–35227. doi: 10.1074/jbc.M506551200. [DOI] [PubMed] [Google Scholar]
- 35.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 36.Schulein-Volk C, Wolf E, Zhu J, Xu W, Taranets L, Hellmann A, et al. Dual regulation of Fbw7 function and oncogenic transformation by Usp28. Cell Rep. 2014;9:1099–1109. doi: 10.1016/j.celrep.2014.09.057. [DOI] [PubMed] [Google Scholar]
- 37.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 38.Tahir SK, Yang X, Anderson MG, Morgan-Lappe SE, Sarthy AV, Chen J, et al. Influence of Bcl-2 family members on the cellular response of small-cell lung cancer cell lines to ABT-737. Cancer Res. 2007;67:1176–1183. doi: 10.1158/0008-5472.CAN-06-2203. [DOI] [PubMed] [Google Scholar]
- 39.Thomas LW, Lam C, Edwards SW. Mcl-1; the molecular regulation of protein function. FEBS Lett. 2010;584:2981–2989. doi: 10.1016/j.febslet.2010.05.061. [DOI] [PubMed] [Google Scholar]
- 40.Thompson BJ, Buonamici S, Sulis ML, Palomero T, Vilimas T, Basso G, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varadarajan S, Vogler M, Butterworth M, Dinsdale D, Walensky LD, Cohen GM. Evaluation and critical assessment of putative MCL-1 inhibitors. Cell Death Differ. 2013;20:1475–1484. doi: 10.1038/cdd.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 43.Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 44.Wilhelm S, Carter C, Lynch M, Lowinger T, Dumas J, Smith RA, et al. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5:835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 45.Wilhelm SM, Dumas J, Adnane L, Lynch M, Carter CA, Schutz G, et al. Regorafenib (BAY 73-4506): a new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int J Cancer. 2011;129:245–255. doi: 10.1002/ijc.25864. [DOI] [PubMed] [Google Scholar]
- 46.Yu C, Bruzek LM, Meng XW, Gores GJ, Carter CA, Kaufmann SH, et al. The role of Mcl-1 downregulation in the proapoptotic activity of the multikinase inhibitor BAY 43-9006. Oncogene. 2005;24:6861–6869. doi: 10.1038/sj.onc.1208841. [DOI] [PubMed] [Google Scholar]
- 47.Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci U S A. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.