Abstract
Introduction. Infections due to extended spectrum β-lactamase producing Enterobacteriaceae are on the rise. They pose serious public health problems due to their resistance to large number of antibiotics. However, little is known about the genotypes of ESBL from Nepal. Therefore, the study presents results of phenotypic and molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. isolated from various clinical specimens in a tertiary care teaching hospital of Nepal. Methods. A total of 172 Enterobacteriaceae clinical isolates recovered from various clinical specimens were analyzed for their antibiotic susceptibility test. Detection of ESBLs was carried out using combination disk test and multiplex PCR for their genotypes (CTX-M, SHV, and TEM). Results. Out of 172 clinical isolates, 70 (40.6%) of them were found ESBL producers. The major source of ESBL producers was urinary tract samples and the highest ESBL production was observed in Escherichia coli (46.5%). Among ESBL genotypes, CTX-M (91.4%) was most predominant, followed by TEM (65.7%) and SHV (11.4%) in both of the isolates. Conclusions. High level of drug resistance and ESBL production was observed among the clinical isolates. There is a need for longitudinal and nationwide surveillance for drug resistance in clinical isolates and antimicrobial stewardship is necessary to guide the appropriate and judicious antibiotic use.
1. General Background
Extended spectrum beta-lactamase producing Enterobacteriaceae (ESBL-E) are among the most important causes of infections in the community and hospital in the recent years [1, 2]. Among them, Klebsiella pneumoniae and Escherichia coli represent the most frequently isolated ESBL producing species worldwide, but ESBLs have also been identified in several other bacterial species [3]. It is global matter of concern since infections associated with ESBL producing clinical isolates are found with higher mortality, length of stay, and health care cost and longer antibiotic therapy in comparison to that with non-ESBL producing pathogens [4]. Furthermore, they pose significant therapeutic challenge in daily management of infectious diseases due to their resistance to additional classes of antibiotics reducing the effectiveness of alternative antimicrobial regimens [5, 6]. Till date, more than 300 types of ESBLs have been described in various members of the Enterobacteriaceae family and other nonenteric organisms; most common of them are TEM, SHV, and CTX-M genotypes [7]. Since their first detection in 1989, CTX-M-type ESBLs are globally disseminating into community and hospital strains in extraordinary extent [1]. In addition, the coexistence of multiple ESBLs in same clinical strain including AmpC beta-lactamases, carbapenemases, and other antibiotic resistance plasmid determinants further creates the therapeutic confrontation [8].
Most studies on prevalence, risk factors, and molecular characterization of ESBL producing organisms have been carried out from developed world but only few data are available concerning the genetic characterization of clinical isolates from Nepal [6, 9, 10]. Rapid and prompt detection of ESBL producing pathogenic Enterobacteriaceae is of utmost importance to promulgate the appropriate antimicrobial therapy and also for preventing cross-transmission to other patients in the vicinity [11]. Several phenotypic methods have been proposed for ESBL detection on isolated Enterobacteriaceae strains such as the double-disk synergy test, the combination disk method, and the ESBL-E tests but their discrepancy in sensitivity has been reported [12]. Moreover, there are specific molecular tests for ESBL detection in bacterial isolates which reduce the time of detection and increase the sensitivity and specificity [13]. In addition, the continuous discovery of new ESBLs and dissemination into various isolates has vitalized the need of molecular approaches [14].
Therefore, in this study, we aimed to determine the burden of multidrug resistant (MDR) enterobacterial strains from various clinical specimens and detection of ESBL production among these isolates by phenotypic and genotypic methods.
2. Materials and Methods
2.1. Study Design and Setup
A hospital based cross-sectional study was conducted at the Department of Microbiology, Manmohan Memorial Medical College and Teaching Hospital (MMTH), a 500-bedded tertiary care hospital in Kathmandu, Nepal. Study hospital is a referral center with medical, surgical, gynecological, pediatric, geriatric, and other specialties. The duration of study was 6 months [from February 2015 to July 2015].
2.2. Bacterial Isolates
A total of 172 consecutive, nonrepetitive Enterobacteriaceae clinical strains were isolated from different clinical specimens such as urine (n = 60), sputum/respiratory secretions (n = 47), pus/wound swab (n = 42), blood (n = 17), body fluids (n = 3), and stool (n = 3) which were received in the clinical bacteriology laboratory, from the patients suspected with infections. The isolation of the significant bacteria and their identification were done according to the standard protocol suggested by American Society for Microbiology [ASM] [15].
2.2.1. Antimicrobial Susceptibility Testing
The susceptibility of bacterial isolates against different antibiotics was tested by the disk diffusion method [modified Kirby-Bauer method] on Mueller Hinton Agar (Hi-Media Laboratories, India) following standard zone size interpretative criteria recommended by Clinical and Laboratory Standards Institute (CLSI) [16]. Antibiotics that were tested in our study include amoxicillin (AMX 10 μg), amoxicillin clavulanate (AMC 20/10 μg), Gentamycin (GEN 10 μg), Ciprofloxacin (CIP 5 μg), Cotrimoxazole (COT 30 μg), Cefixime (CFM 5 μg), Cefotaxime/Ceftriaxone (CTX/CTR 30 μg), Ceftazidime (CAZ 30 μg), Cefepime (CPM 30 μg), piperacillin tazobactam (PIT 100/10 μg), Imipenem (IMP 10 μg), Meropenem (MEM 10 μg), Polymixin B (PB300 units), and Nitrofurantoin (NI 300 U) (Hi-Media Laboratories, India).
2.3. Screening of Multidrug Resistant (MDR) and Potential ESBL Producers
In this study, if the isolates were resistant to at least one agent of three different classes of commonly used antimicrobial agents, they were regarded as multidrug resistant (MDR) [17]. If the zone of inhibition (ZOI) was ≤25 mm for Ceftriaxone, ≤22 mm for Ceftazidime, and/or ≤27 mm for Cefotaxime, the isolate was considered a potential ESBL producer as recommended by CLSI and further tested by confirmatory methods [16].
2.4. Phenotypic Confirmation of ESBL
Isolates considered potential ESBL producers by initial screening were emulsified with nutrient broth to adjust the inoculum density equal to that of 0.5 McFarland turbidity standards. Combination disk test (CDT), as recommended by the CLSI, was performed in all isolates presumed to be ESBL producers. In this test, Ceftazidime (30 μg) disks alone and in combination with clavulanic acid (Ceftazidime + clavulanic acid, 30/10 μg) disks were applied onto a plate of Mueller Hinton Agar (MHA) which was inoculated with the test strain and then incubated in ambient air for 16–18 hours of incubation at 35 ± 2°C. Isolate that showed increase of ≥5 mm in the zone of inhibition of the combination disks in comparison to that of the Ceftazidime disk alone was considered an ESBL producer [16] (Figure 1). Then molecular analysis was done on all screening positive isolates.
Figure 1.
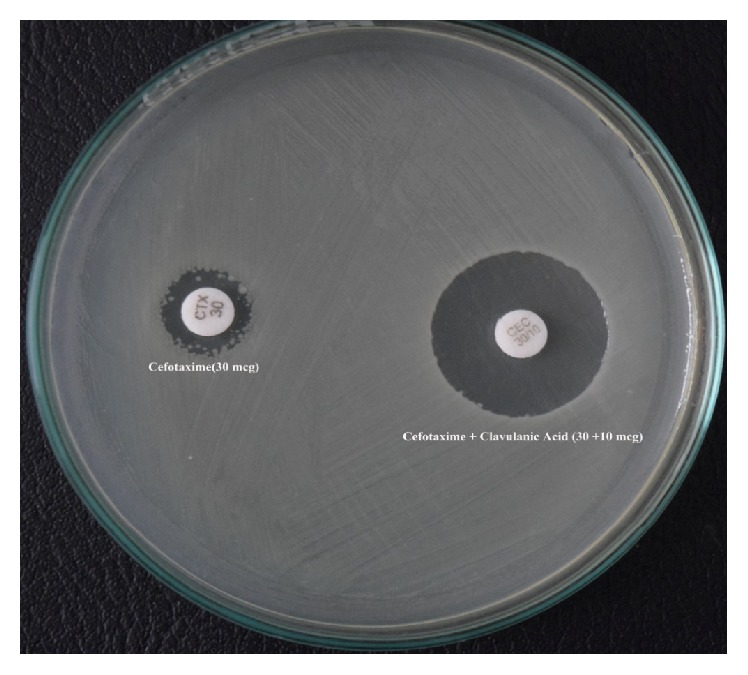
Positive combination disk test (for the phenotypic detection of ESBL).
2.5. Detection of Genes Encoding ESBLs
2.5.1. Plasmid DNA Extraction and Amplification
Crude plasmid DNA was isolated from bacterial cells by using plasmid isolation kit (GeNei™) by using manufacturer instructions. Briefly, the selected bacterial strain (single colony) was grown overnight in Luria-Bertani (LB) broth at 37°C with aeration using an orbital shaker and the plasmid DNA from organisms was extracted by alkaline lysis using sodium dodecyl sulphate (SDS). All phenotypically confirmed potential ESBL positive isolates were analyzed for beta-lactamase genes TEM, SHV, and CTX-M. Primers were obtained from GeNei, India, and they were used for identification of TEM, SHV, and CTX-M. The primer sequence is as shown in Table 1. Multiplex PCR was carried out to detect the plasmid genes for SHV and CTX-M while conventional linear PCR was done for TEM type ESBL genes.
Table 1.
Primers for the bla-CTX-M, bla-TEM, and bla-SHV genes used in this study.
| Gene | Primers (5′-3′) | Amplicon size (bp) |
|---|---|---|
| SHV | F: 5′-GTCAGCGAAAAACACCTTGCC-3′ | 383 bp |
| R: 5′-GTCTTATCGGCGATAAACCAG-3′ | ||
| TEM | F: 5′-GAGACAATAACCCTGGTAAAT-3′ | 459 bp |
| R: 5′-AGAAGTAAGTTGGCAGCAGTG-3′ | ||
| CTX-M | F: 5′-GAAGGTCATCAAGAAGGTGCG-3′ | 560 bp |
| R: 5′-GCATTGCCACGCTTTTCATAG-3′ |
PCR amplification reactions were carried out in a 25 μL volume in which master mix containing 200 μM of dNTPs (dATP, dCTP, dGTP, and dTTP), 120 nM of each primer, 0.5 U/μL of Taq polymerase in 1x PCR buffer, 25 mM MgCl2, and 1 μL of DNA was added. Amplification reactions were performed in a DNA thermal cycler (CG) under following thermal and cycling conditions for the bla-TEM, bla-SHV, and bla-CTX-M genes: initial denaturation at 94°C for 3 minutes, denaturation at 94°C for 45 seconds of 35 cycles, annealing at 60°C for 30 sec of 35 cycles (for bla-SHV and bla-CTX-M) and 55°C for 30 sec of 35 cycles (for bla-TEM), extension at 72°C for 3 minutes of 35 cycles, and final extension at 72°C for 2 minutes.
After PCR amplification, 2.5 μL of each reaction was separated by electrophoresis in 1.5% agarose gel for 30 min at 100 V in 0.5x TBE buffer. DNA was stained with ethidium bromide (1 μg/mL) and the bands were detected using UV-transilluminator [Cleaver Scientific Ltd.] (Figure 2).
Figure 2.
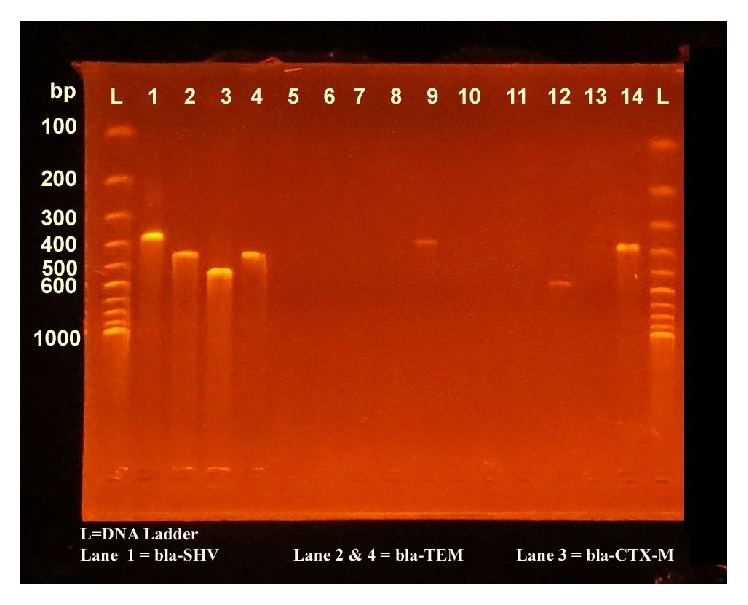
Gel electrophoresis with bands of bla-CTX-M, bla-TEM, and bla-SHV.
2.6. Data Analysis
The information regarding patient's profile, clinical specimens, bacterial isolates, and the results was entered into a computer program. Data analysis was carried out using the Statistical Package for Social Sciences [SPSS™] version 20.0 [IBM, Armonk, NY, USA] and presented in percentage base distribution.
3. Results
3.1. Patient Demographics
During the study period, a total of 172 Enterobacteriaceae clinical strains were recovered from patients suspected with various infections; 75 (43.6%) isolates were from male and 97 (56.4%) from female patients. Majority (69.7%, n = 120) of the isolates were recovered from inpatient department while highest numbers were from age group 31–45 (n = 58). Escherichia coli (n = 138) was the most common organism, next to Klebsiella spp. (n = 20), and the numbers of organisms other than Escherichia coli and Klebsiella were low (n = 14).
3.2. Antimicrobial Susceptibility Data
On preliminary antimicrobial susceptibility testing, most of the Escherichia coli and Klebsiella spp. were resistant to third-generation cephalosporins (67% and 71% to Cefixime and more than 60% to Cefotaxime). Similar level of drug resistance was also observed against fluoroquinolones. All isolates of Escherichia coli and 93% of Klebsiella spp. were susceptible to Imipenem and Meropenem. Both isolates were completely susceptible to Polymixin B (Table 2). Isolates other than Escherichia and Klebsiella were not found to be multidrug resistant and were not processed further.
Table 2.
Antimicrobial susceptibility pattern of Escherichia coli and Klebsiella spp. clinical isolates.
| Antibiotics | Escherichia coli (n = 138) | Klebsiella spp. (n = 20) | ||
|---|---|---|---|---|
| R% | S% | R% | S% | |
| Amoxicillin | 92 | 8 | — | — |
| Cephalexin | 92 | 8 | 90 | 10 |
| Cefixime | 67 | 33 | 71 | 29 |
| Cefotaxime | 63 | 37 | 60 | 40 |
| Ciprofloxacin | 55 | 45 | 55 | 45 |
| Piperacillin tazobactam | 28 | 72 | 45 | 55 |
| Gentamycin | 12 | 88 | 25 | 75 |
| Nitrofurantoin | 10 | 90 | 21 | 79 |
| Cotrimoxazole | 49 | 51 | 15 | 85 |
| Amikacin | 1 | 99 | 14 | 86 |
| Imipenem | 0 | 100 | 7 | 93 |
| Meropenem | 0 | 100 | 7 | 93 |
| Polymixin B | 0 | 100 | 0 | 100 |
R = resistant, S = sensitive.
Seventy-one percent of Escherichia coli (n = 98) and 70% of Klebsiella spp. (n = 14) were multidrug resistant. Out of them, 84 (60.8%) Escherichia coli were found to be potential ESBL producer by preliminary screening. However, 7 (63.6%) of the 11 Klebsiella pneumoniae were screen positive ESBL producers and 5 (55.55%) of the 9 Klebsiella oxytoca were found to be potential ESBL producer by preliminary screening test (Table 3). Subsequently, seventy clinical isolates were confirmed as ESBL positive by phenotypic combination disk test (CDT). Escherichia coli were found with the highest prevalence (64, 46.3%) of ESBL production followed by Klebsiella pneumoniae (4, 36.3%) and Klebsiella oxytoca (2, 22.2%).
Table 3.
Preliminary screening and phenotypic confirmation of potential ESBL producers.
| S. N. | Organisms | ESBL screening number (%) | ESBL confirmatory number (%) |
|---|---|---|---|
| 1 | Escherichia coli | 138 | 64 (46.3) |
| Multidrug resistant (MDR) | 98 (71.0) | 64 (65.3) | |
| ESBL positive | 84 (60.8) | 64 (76.1) | |
| ESBL negative | 54 (39.2) | — | |
|
| |||
| 2 | Klebsiella pneumoniae | 11 | 4 (36.3) |
| Multidrug resistant (MDR) | 9 (81.8) | 4 (44.4) | |
| ESBL positive | 7 (63.6) | 4 (57.1) | |
| ESBL negative | 4 (36.4) | — | |
|
| |||
| 3 | Klebsiella oxytoca | 9 | 2 (22.2) |
| Multidrug resistant (MDR) | 5 (55.5) | 2 (40.0) | |
| ESBL positive | 5 (55.5) | 2 (40.0) | |
| ESBL negative | 4 (44.5) | — | |
3.3. β-Lactamase Genes among the Presumptive ESBL Producers
All seventy (70, 44.3%) presumptive ESBL producing isolates were subjected to molecular analysis. Among ESBL producing genes, prevalence of bla-CTX-M (91.4%) was highest followed by bla-TEM (65.7%) and bla-SHV (11.4%). Multiple occurrences of genes were found in some of the isolates. The coexistence of all three genes (bla-SHV, bla-TEM, and bla-CTX-M) was seen in 5 (7.1%) of the isolates, while bla-CTX-M and bla-TEM coexisted in 42 (60%) of the isolates while bla-TEM and bla-SHV coexisted in 7 (10%) of the isolates (Table 4).
Table 4.
Distribution of ESBL genotypes among bacterial strains.
| Isolates | ESBL genotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TEM | SHV | CTX-M | TEM + SHV | TEM + CTX-M | TEM + SHV + CTX-M | |||||||
| N | % | N | % | N | % | N | % | N | % | N | % | |
| E. coli (n = 64) | 40 | 62.5 | 5 | 7.8 | 58 | 90.6 | 4 | 6.2 | 36 | 56.2 | 3 | 4.6 |
| Klebsiella spp. (n = 6) | 6 | 100 | 3 | 50 | 6 | 100 | 3 | 50 | 6 | 100 | 2 | 33.3 |
|
| ||||||||||||
| Total | 46 | 65.7 | 8 | 11.4 | 64 | 91.4 | 7 | 10 | 42 | 60.0 | 5 | 7.1 |
Among 64 ESBL positive strains of E. coli, all the three genes, that is, bla-TEM, bla-SHV, and bla-CTX-M, were present in 3 strains of E. coli while only two genes, that is, bla-TEM and bla-SHV, were present in 4 strains, and bla-TEM and bla-CTX-M were present in 36 strains of E. coli. The prevalence of bla-CTX-M, bla-TEM, and bla-SHV for Escherichia coli was 90.6%, 62.5%, and 7.8%, respectively. The distribution of the genes based on the organisms isolated is as shown in Table 4.
Among 6 presumptive ESBL positive strains of Klebsiella spp., all three genes, that is, bla-TEM, bla-SHV, and bla-CTX-M, were present only in two strains of Klebsiella spp. while two genes, that is, bla-TEM and bla-SHV, were present only in three strains, and bla-TEM and bla-CTX-M were present in all 6 ESBL positive strains. The prevalence of bla-CTX-M, bla-TEM, and bla-SHV for Klebsiella spp. was 100%, 100%, and 50%, respectively. Concurrently, urinary isolates were the most common ESBL producers (31, 51.6%) followed by respiratory isolates and others (Table 5).
Table 5.
Distribution of ESBL among various clinical specimens and strains.
| S. N. | Specimens (n = 172) | Organisms | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli (n = 138) | Klebsiella spp. (n = 20) | ESBL Escherichia coli | ESBL Klebsiella spp. | Total ESBL (n = 70) | |||||
| Number | % | Number | % | Number | % | ||||
| 1 | Urine (60) | 46 | 8 | 27 | 58.6 | 4 | 50.0 | 31 | 51.6 |
| 2 | Sputum (47) | 38 | 4 | 14 | 36.8 | 2 | 50.0 | 16 | 34.0 |
| 3 | Pus (42) | 36 | 3 | 15 | 41.6 | 0 | 0 | 15 | 35.7 |
| 4 | Blood (17) | 12 | 5 | 6 | 50.0 | 0 | 0 | 6 | 35.2 |
| 5 | Body fluid (3) | 3 | 0 | 1 | 33.3 | 0 | 0 | 1 | 33.3 |
| 6 | Stool (3) | 3 | 0 | 1 | 33.3 | 0 | 0 | 1 | 33.3 |
|
| |||||||||
| Total | 172 | 138 | 20 | 64 | 46.3 | 6 | 30.0 | 70 | 40.6 |
4. Discussions
Increased global prevalence and dissemination of ESBL genes among pathogenic microorganisms are a serious peril for medical fraternity. This brings new demands on routine clinical microbiology laboratories to investigate the potential of MDR strains and perform ESBL typing on every suspected isolate. This study has examined the common enterobacterial strains from clinical specimens, their antibiogram, and production of ESBL among MDR strains with phenotypic and genotypic approaches. To the best of our knowledge, this is probably the first effort from Nepal to characterize the ESBL strains genotypically from clinical specimens of routine care.
In the present study, ESBL production among Enterobacteriaceae clinical isolates was found to be 40.6%. This rate of ESBL prevalence is highest from Nepal among the published literature till date. Previous studies from our country have reported the prevalence of the ESBL producing bacteria ranging from 13.5% to 33.2%. Chander and Shrestha from nearby hospital reported the ESBL prevalence rate to be 13.5% [18] whereas, recently, Neupane et al. have reported 33.2% of ESBL producers in their study [19]. Similarly, Ansari et al. [20], Khanal et al. [21], Pokhrel et al. [10], Yadav et al. [22], and Pant et al. [23] have reported 24%, 25%, 25.8%, 26.8%, and 31.3% ESBL producers, respectively. These findings correlated well with those of our study. The prevalence of ESBL production is high in the referral hospitals and the intensive care units where the patients are referred from the peripheral centers reference laboratories and hospitals with huge arsenal of antibiotic use. The relatively high prevalence of ESBLs recorded in this study might be due to the extreme empirical use of third-generation cephalosporins in primary care. Moreover, higher rates (52.49%) of ESBL production among Enterobacteriaceae clinical isolates have been reported from an Indian study [13]. However, extremely lower rates of ESBL production have been documented from Japan, Korea, and United States [7, 24]. The differences in the ESBL rates may be attributable to the geographic difference, antimicrobial stewardship programme, and infection control practices.
In the present study, urinary isolates were the most common ESBL producing strains (51.6%) followed by respiratory tract samples, pus/swabs, blood, body fluid, and stool. Similar findings have also been documented in other studies [10, 20, 25].
In the current study, we observed that 46.3% Escherichia coli and 30% Klebsiella isolates were ESBL producers. Although Klebsiella spp. were more often reported as ESBL producers in other studies [18, 26], we observed that the ESBL production was more common in the Escherichia coli isolates. Similar predominance of ESBL producing Escherichia coli has been reported by Pant et al. [23] from Nepal and Sharma et al. [13] and Shanthi and Sekar [25] from India.
Regarding the high diversity ESBL types obtained in our study, we find it interesting to compare with neighbor regions. Majority of phenotypic ESBL positive strains carried multiple bla genes, where CTX-M-type was the most predominant (91.4%) in this study. Majority of Escherichia coli (90.6%) and Klebsiella spp. (100%) harbored CTX-M gene while sixty-two percent of Escherichia coli and all Klebsiella spp. were found with bla-TEM. Our finding corroborates with the reports of Pokhrel et al. [10] where bla-CTX-M-type ESBL was most common (23, 95.8%) followed by bla-TEM (7, 29.2%) and bla-SHV (3, 12.5%) among Escherichia coli clinical isolates. Moreover, the higher prevalence of CTX-M gene among ESBL producing isolates in our study is also in concordance with study of Moses et al. [27] and Sharma et al. [13]. A study from Thailand by Kiratisin et al. reported 99.6% of ESBL producing Escherichia coli and 99.2% of ESBL producing Klebsiella spp. isolates carried bla-CTX-M genotype. The bla-TEM and bla-SHV groups were detected in 77.0% and 3.8% of ESBL producing Escherichia coli and 71.7% and 87.4% of ESBL producing Klebsiella spp., respectively [28]. However, in another study by Bali et al., TEM type ESBLs were found in 72.72% of Escherichia coli and 75% of Klebsiella spp., while they reported 9.09% of SHV gene and 22.72% of CTX-M gene in their study which is quite low as compared to our study [29]. Bonnet and Livermore also reviewed the dominance of CTX-M in European and Asian countries [30, 31].
In the present study, it was observed that there were multiple occurrences of genes in some of the isolates. It has been observed that 54 out of 70 (77.1%) isolates carried more than one type of β-lactamase genes. This is exactly corroborating the previous findings of Sharma et al. from India [13]. However, lower coproduction of various genotypes has been documented by Goyal et al. (57.3%) [32] and Bali et al. (19.2%) [29]. The significance of the finding may correlate with the ability of organisms to hydrolyse broader substrates and more likely to have the propensity for widespread nosocomial transmission.
Multidrug resistance mediated by ESBL producing bacteria is a global problem. The genetic mobile elements carrying the resistant determinants are easily transferable to other clinical strains. Prompt and precise detection of these strains in microbiology laboratories is very important. The lack of understanding or limited resources are responsible for rapid worldwide dissemination of the pathogens which possess these β-lactamases. Therefore, the regular detection of ESBLs by conventional methods and further confirmation with molecular methods should be included in routine care laboratories [7, 30].
5. Conclusions
This study concluded that there is high prevalence of multidrug resistant pathogenic enterobacterial strains in our hospital. Higher rates of ESBL genotypes among clinical strains of Escherichia coli and Klebsiella spp. have vitalized the importance of appropriate antimicrobial regimen selection for empirical therapy. CTX-M genotypes are disseminating at the alarming rate among pathogenic strains; therefore molecular detection and identification of beta-lactamases would be essential for a reliable epidemiological investigation of antimicrobial resistance. Regular national-wide surveillance of multidrug resistance seems necessary step to combat the severity caused by ESBL producing bugs.
Acknowledgments
The authors are deeply thankful to all the patients participating in this study. Their special thanks go to all the laboratory staffs, management, and officials of Manmohan Memorial Teaching Hospital, Kathmandu, for providing the opportunity to carry out this research work.
Abbreviations
- CLSI:
Clinical and Laboratory Standard Institute
- ESBLs:
Extended spectrum beta-lactamases
- AmpC:
AmpC beta-lactamase
- ASM:
American Society for Microbiology
- CAZ:
Ceftazidime
- CTR:
Ceftriaxone
- CTX:
Cefotaxime
- CTX-M:
Cefotaximase-Munich
- SHV:
Sulfhydryl variant
- TEM:
Temoniera
- bla:
β-Lactamases
- PCR:
Polymerase chain reaction
- CDT:
Combined disk test.
Consent
This research was approved by the Institutional Review Committee of Manmohan Memorial Institute of Health Sciences, Kathmandu, Nepal. Letter of approval was obtained after submitting and presenting the proposal to the committee. All the patients were asked verbally to include them as a sample source in this study.
Competing Interests
There are no competing interests to disclose.
Authors' Contributions
Govardhan Joshi and Puspa Raj Khanal are equal contributors.
References
- 1.Pitout J. D., Laupland K. B. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. The Lancet Infectious Diseases. 2008;8(3):159–166. doi: 10.1016/s1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 2.Doi Y., Park Y. S., Rivera J. I., et al. Community-associated extended-spectrum beta-lactamase-producing Escherichia coli infection in the United States. Clinical Infectious Diseases. 2013;56(5):641–648. doi: 10.1093/cid/cis942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brolund A. Overview of ESBL-producing Enterobacteriaceae from a Nordic perspective. Infection Ecology & Epidemiology. 2014;4:1–9. doi: 10.3402/iee.v4.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwaber M. J., Carmeli Y. Mortality and delay in effective therapy associated with extended-spectrum β-lactamase production in Enterobacteriaceae bacteraemia: a systematic review and meta-analysis. Journal of Antimicrobial Chemotherapy. 2007;60(5):913–920. doi: 10.1093/jac/dkm318. [DOI] [PubMed] [Google Scholar]
- 5.Karisik E., Ellington M. J., Pike R., Warren R. E., Livermore D. M., Woodford N. Molecular characterization of plasmids encoding CTX-M-15 β-lactamases from Escherichia coli strains in the United Kingdom. Journal of Antimicrobial Chemotherapy. 2006;58(3):665–668. doi: 10.1093/jac/dkl309. [DOI] [PubMed] [Google Scholar]
- 6.Livermore D. M. Current epidemiology and growing resistance of Gram-negative pathogens. Korean Journal of Internal Medicine. 2012;27(2):128–142. doi: 10.3904/kjim.2012.27.2.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paterson D. L., Bonomo R. A. Extended-spectrum β-lactamases: a clinical update. Clinical Microbiology Reviews. 2005;18(4):657–686. doi: 10.1128/cmr.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruppé É., Woerther P.-L., Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Annals of Intensive Care. 2015;5, article 21 doi: 10.1186/s13613-015-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherchan J. B., Hayakawa K., Miyoshi-Akiyama T., et al. Clinical epidemiology and molecular analysis of extended-spectrum-β-lactamase-producing Escherichia coli in Nepal: characteristics of sequence types 131 and 648. Antimicrobial Agents and Chemotherapy. 2015;59(6):3424–3432. doi: 10.1128/aac.00270-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pokhrel R. H., Thapa B., Kafle R., Shah P. K., Tribuddharat C. Co-existence of beta-lactamases in clinical isolates of Escherichia coli from Kathmandu, Nepal. BMC Research Notes. 2014;7, article 694 doi: 10.1186/1756-0500-7-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livermore D. M., Andrews J. M., Hawkey P. M., et al. Are susceptibility tests enough, or should laboratories still seek ESBLs and carbapenemases directly? Journal of Antimicrobial Chemotherapy. 2012;67(7):1569–1577. doi: 10.1093/jac/dks088. [DOI] [PubMed] [Google Scholar]
- 12.Drieux L., Brossier F., Sougakoff W., Jarlier V. Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clinical Microbiology and Infection. 2008;14(1):90–103. doi: 10.1111/j.1469-0691.2007.01846.x. [DOI] [PubMed] [Google Scholar]
- 13.Sharma M., Pathak S., Srivastava P. Prevalence and antibiogram of Extended Spectrum β-lactamase (ESBL) producing Gram negative bacilli and further molecular characterization of ESBL producing Escherichia coli and Klebsiella spp. Journal of Clinical and Diagnostic Research. 2013;7(10):2173–2177. doi: 10.7860/jcdr/2013/6460.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaur M., Aggarwal A. Occurrence of the CTX-M, SHV and the TEM genes among the extended spectrum β-lactamase producing isolates of enterobacteriaceae in a tertiary care hospital of north India. Journal of Clinical and Diagnostic Research. 2013;7(4):642–645. doi: 10.7860/jcdr/2013/5081.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isenberg H. D. Clinical Microbiology Procedure Handbook. 2nd. Washington, DC, USA: ASM Press; 2004. [Google Scholar]
- 16.Performance Standards for Antimicrobial Disk Susceptibility Tests 2012. Wayne, Pa, USA: Clinical and Laboratory Standards Institute; 2012. (M02-A11(Approved Standard—Eleventh Edition)). [Google Scholar]
- 17.Magiorakos A.-P., Srinivasan A., Carey R. B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 18.Chander A., Shrestha C. D. Prevalence of extended spectrum beta lactamase producing Escherichia coli and Klebsiella pneumoniae urinary isolates in a tertiary care hospital in Kathmandu, Nepal. BMC Research Notes. 2013;6(1, article no. 487) doi: 10.1186/1756-0500-6-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neupane S., Pant N. D., Khatiwada S., Chaudhary R., Banjara M. R. Correlation between biofilm formation and resistance toward different commonly used antibiotics along with extended spectrum beta lactamase production in uropathogenic Escherichia coli isolated from the patients suspected of urinary tract infections visiting Shree Birendra Hospital, Chhauni, Kathmandu, Nepal. Antimicrobial Resistance and Infection Control. 2016;5, article 5 doi: 10.1186/s13756-016-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ansari S., Nepal H. P., Gautam R., et al. Community acquired multi-drug resistant clinical isolates of Escherichia coli in a tertiary care center of Nepal. Antimicrobial Resistance and Infection Control. 2015;4(1, article no. 15) doi: 10.1186/s13756-015-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanal S., Joshi D. R., Bhatta D. R., Devkota U., Pokhrel B. M. β-lactamase-producing multidrug-resistant bacterial pathogens from tracheal aspirates of intensive care unit patients at national institute of neurological and allied sciences, Nepal. ISRN Microbiology. 2013;2013:5. doi: 10.1155/2013/847569.847569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yadav K. K., Adhikari N., Khadka R., Pant A. D., Shah B. Multidrug resistant Enterobacteriaceae and extended spectrum β-lactamase producing Escherichia coli: a cross-sectional study in National Kidney Center, Nepal. Antimicrobial Resistance and Infection Control. 2015;4, article 42 doi: 10.1186/s13756-015-0085-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pant N. D., Bhandari R., Poudel A., Sharma M. Assessment of the effectiveness of three different cephalosporins/clavulanate combinations for the phenotypic confirmation of extended spectrum beta lactamases producer bacterial isolates from urine samples at National Public Health Laboratory, Kathmandu, Nepal. BMC Research Notes. 2016;9, article 390 doi: 10.1186/s13104-016-2192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Y.-Z., Sun K.-D., Pan L.-H., et al. A screening strategy for phenotypic detection of carbapenemase in the clinical laboratory. Canadian Journal of Microbiology. 2014;60(4):211–215. doi: 10.1139/cjm-2013-0692. [DOI] [PubMed] [Google Scholar]
- 25.Shanthi M., Sekar U. Extended spectrum beta lactamase producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Journal of Association of Physicians of India. 2010;58(supplement):41–44. [PubMed] [Google Scholar]
- 26.Babypadmini S., Appalaraju B. Extended spectrum β-lactamases in urinary isolates of Escherichia coli and Klebsiella pneumoniae—prevalence and susceptibility pattern in a tertiary care hospital. Indian Journal of Medical Microbiology. 2004;22(3):172–174. [PubMed] [Google Scholar]
- 27.Moses A., Bwanga F., Boum Y., Bazira J. Prevalence and genotypic characterization of extended- spectrum beta-lactamases produced by gram negative bacilli at a tertiary care hospital in Rural South Western Uganda. British Microbiology Research Journal. 2014;4(12):1541–1550. doi: 10.9734/bmrj/2014/9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiratisin P., Apisarnthanarak A., Laesripa C., Saifon P. Molecular characterization and epidemiology of extended-spectrum-β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrobial Agents and Chemotherapy. 2008;52(8):2818–2824. doi: 10.1128/aac.00171-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bali E. B., Açik L., Sultan N. Phenotypic and molecular characterization of SHV, TEM, CTX-M and extended-spectrum β-lactamase produced by Escherichia coli, Acinobacter baumannii and Klebsiella isolates in a Turkish hospital. African Journal of Microbiology Research. 2010;4(8):650–654. [Google Scholar]
- 30.Livermore D. M., Canton R., Gniadkowski M., et al. CTX-M: changing the face of ESBLs in Europe. Journal of Antimicrobial Chemotherapy. 2007;59(2):165–174. doi: 10.1093/jac/dkl483. [DOI] [PubMed] [Google Scholar]
- 31.Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrobial Agents and Chemotherapy. 2004;48(1):1–14. doi: 10.1128/aac.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goyal A., Prasad K. N., Prasad A., Gupta S., Ghoshal U., Ayyagari A. Extended spectrum β-lactamases in Escherichia coli & Klebsiella pneumoniae & associated risk factors. Indian Journal of Medical Research. 2009;129(6):695–700. [PubMed] [Google Scholar]


