Abstract
To assess the clinical utility of targeted Next-Generation Sequencing (NGS) for the diagnosis of Inherited Retinal Dystrophies (IRDs), a total of 109 subjects were enrolled in the study, including 88 IRD affected probands and 21 healthy relatives. Clinical diagnoses included Retinitis Pigmentosa (RP), Leber Congenital Amaurosis (LCA), Stargardt Disease (STGD), Best Macular Dystrophy (BMD), Usher Syndrome (USH), and other IRDs with undefined clinical diagnosis. Participants underwent a complete ophthalmologic examination followed by genetic counseling. A custom AmpliSeq™ panel of 72 IRD-related genes was designed for the analysis and tested using Ion semiconductor Next-Generation Sequencing (NGS). Potential disease-causing mutations were identified in 59.1% of probands, comprising mutations in 16 genes. The highest diagnostic yields were achieved for BMD, LCA, USH, and STGD patients, whereas RP confirmed its high genetic heterogeneity. Causative mutations were identified in 17.6% of probands with undefined diagnosis. Revision of the initial diagnosis was performed for 9.6% of genetically diagnosed patients. This study demonstrates that NGS represents a comprehensive cost-effective approach for IRDs molecular diagnosis. The identification of the genetic alterations underlying the phenotype enabled the clinicians to achieve a more accurate diagnosis. The results emphasize the importance of molecular diagnosis coupled with clinic information to unravel the extensive phenotypic heterogeneity of these diseases.
1. Introduction
Inherited Retinal Dystrophies (IRDs) are a heterogeneous group of eye disorders characterized by rod and/or cone photoreceptor cells degeneration, which include Retinitis Pigmentosa (RP), Leber Congenital Amaurosis (LCA), Stargardt Disease (STGD), Best Macular Dystrophy (BMD), and syndromic forms such as Usher Syndrome (USH). The overall prevalence of these disorders is ~1 in 4,000 individuals for RP, ~1 in 90,000 individuals for LCA and USH, ~1 in 5,000–10,000 individuals for STGD, and 1/5000–1/67000 for BMD (http://www.orpha.net). Classification of IRDs considers the principal site of retinal dysfunction (rod, cone, retinal pigment epithelium, or inner retina), the mode of inheritance, the underlying gene defect, typical age of onset, rate of progression, and association with systemic syndromes. The genetic bases of IRDs are highly heterogeneous, with almost 150 genes currently known [RetNet, https://sph.uth.edu/retnet/] and a wide clinical and genetic overlap among the different disorders, with high phenotypic variability and genes associated with more than one phenotype. The inheritance of these diseases is also complex, with autosomal dominant (AD), autosomal recessive (AR), X-linked (XL), and even digenic patterns [1]. The extensive clinical and genetic heterogeneity in IRD, along with the variable age of onset, the incomplete penetrance, and unclear inheritance, hamper clinical diagnosis.
Recently, Next-Generation Sequencing (NGS) has been used for the genetic diagnosis of retinal diseases [2–6] and has been reported as a cost-effective approach [7, 8] with a wide range of reported mutation detection rates related to differences in number of genes analyzed, NGS platform, and cohort size but above all composition of the study case phenotypes. We therefore present a multidisciplinary approach coupled with a comprehensive NGS amplicon-based strategy to explore IRD genetic complexity and evaluate genotype-phenotype correlations.
2. Patients and Methods
This study was approved by the ethics committee (Comitato Etico di Modena, Modena, Italy). The procedures followed were in accordance with the Helsinki Declaration of 1975, as revised in 2000, and samples were obtained after patients had provided written informed consent.
A total of 109 samples were collected, including 88 IRDs affected probands with unknown molecular diagnosis and 21 healthy family members (Table 1). Subjects were recruited at the Medical Genetics Unit of the University Hospital of Modena (70 samples), at the Medical Genetics Unit of Parma University Hospital (15 samples) and Medical Genetics Unit of Policlinico Sant'Orsola Malpighi, Bologna (24 samples). All subjects underwent a complete ophthalmologic examination (visual acuity, anterior segment and fundus examination, spectral domain-optical coherence tomography, electroretinogram, and/or electrooculogram) followed by genetic counseling. When indicated fundus autofluorescence imaging and visual field were also performed. Clinical information for the patients with identified pathogenic mutations is shown in Supplementary Table 1 (in Supplementary Material available online at http://dx.doi.org/10.1155/2016/6341870). Clinical diagnoses of participating subjects included RP, USH (hearing impairment + RP), LCA, STGD, BMD, and IRDs not otherwise specified or with imprecisely defined clinical diagnosis. Four control patients with known molecular diagnosis were used to validate our method.
Table 1.
Patients cohort.
| Clinical diagnosis | Number of cases | Healthy relatives | Familiar Cases (number of families) | Presumed inheritance in family | Sex | Age at genetic counseling | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sporadic | AD | AR | XL | M | F | Range | Median | ||||
| BMD | 4 | 2 (1) | 4 | 1 | 3 | 12–65 | 58 | ||||
| LCA | 5 | 5 | 1 | 4 | 2 | 3 | 5–85 | 9 | |||
| STGD | 14 | 6 (3) | 14 | 5 | 9 | 8–59 | 28 | ||||
| RP | 45 | 12 | 9 (4) | 14 | 6 | 20 | 5 | 25 | 20 | 2–73 | 47.5 |
| USH | 3 | 3 | 2 | 1 | 33–53 | 51 | |||||
| nd IRD | 17 | 4 | 6 (2) | 6 | 6 | 5 | 13 | 4 | 2–62 | 35 | |
| Total | 88 | 21 | 23 (10) | 21 | 16 | 46 | 5 | 48 | 40 | 2–85 | 37 |
BMD: Best Macular Dystrophy; LCA: Leber Congenital Amaurosis; STGD: Stargardt disease; RP: Retinitis Pigmentosa; USH: Usher syndrome; nd IRD: inherited retinal degeneration not otherwise specified without precisely defined diagnosis; AD: autosomal dominant; AR: autosomal recessive; XL: X-linked; M: male; F: female.
2.1. AmpliSeq Panel Design and Ion Torrent™ PGM™ Library Preparation and Sequencing
The Ion AmpliSeq technology (Life Technologies Ltd., Paisley, UK) was used to design a panel of 72 genes (Supplementary Table 2) associated with the following IRD forms: RP, LCA, STGD, BMD, and USH [RetNet, https://sph.uth.edu/retnet/]. The Ion AmpliSeq Designer tool (https://www.ampliseq.com/browse.action) generated an optimized primers design encompassing the coding DNA sequence of the selected genes, for a total of 1.649 amplicons divided into two pools to optimize coverage and multiplex PCR conditions. Libraries were prepared using the Ion AmpliSeq Library Kit 2.0 starting from 15 ng of gDNA/pool according to manufacturer's recommendations. Template preparation was performed using an Ion OneTouch™ 2 System following the latest version of the manufacturer's manuals. The template positive Ion Sphere Particles (ISPs+) were sequenced on an Ion Torrent Personal Genome Machine® (PGM) System (Life Technologies Ltd., Paisley, UK) using the Ion 318™ Chip kit v2 following the Ion PGM Sequencing 200 Kit v2 manual.
2.2. Sanger Sequencing
Sanger sequencing was performed to validate CNGB1 c.875-5_891dup mutation (identified with an anomalous distribution of NGS reads attributable to amplification problems due to the insertion itself located at the end of the target region) and to sequence RPGR ORF15 partially uncovered by the NGS panel. Primers for PCR and sequencing are shown in Supplementary Table 3. The following conditions were used: a 50 μL PCR reaction containing 100 ng of DNA, 100 pmol of forward and reverse primers, 5 μL of buffer, and 0.5 μL of Taq Expand High Fidelity™ DNA Polymerase (Roche). PCR amplification (see Supplementary Table 3) was performed using a Gene Amp PCR System 9700 (Applied Biosystems, California, USA). The resultant amplicons were purified using High Pure PCR Product Purification Kit (Roche). Additional primers for RPGR sequencing were used. The sequencing reactions were performed with BigDye Terminator v1.0 (Life Technologies) and run on ABI PRISM® 3130XL Genetic Analyzer (Life Technologies). Due to sequence composition and technical difficulties, part of RPGR ORF15 (~250 bp, chrX: 38145343–38145593) could not be accurately sequenced with Sanger sequencing.
2.3. Data Analysis
Samples were processed using the Ion Torrent Suite™ (TS) Software for raw data processing and sequence alignment to the human genome reference sequence hg19. The TS Variant Caller was used for the detection of germline variants that were subsequently analyzed using the following optimized filtering and annotation pipeline. Annovar [9] and Variant Effect Predictor (VEP) [10] were used to functionally annotate the detected variants, retrieving RefSeq gene annotation, dbSNP rs identifiers, ClinVar accession, and allele frequency observed in the population (1000-Genome Project, NHLBI GO Exome Sequencing Project ESP6500SI-V2, Exome and Aggregation Consortium ExAC 0.3). Variants with low coverage or low frequency (<30 reads or <30%, resp.) were filtered out. The synonymous variants and variants having an allele frequency greater than 1% reported in the population were discarded as well. In addition, an internal database, built with all variants present in our cohort of processed samples, allowed recognizing and classifying as polymorphisms variants not listed in public databases. Variants were further annotated with conservation scores and functional predictions listed in dbNSFP [11–13], a database which compiles scores from various prediction algorithms, among which are SIFT, Polyphen2, LRT, MutationTaster, MutationAssessor, and FATHMM. Retina International (http://www.retina-international.org/), RPGR database (http://rpgr.hgu.mrc.ac.uk/index.php?select_db=RPGR), CEP290base (http://cep290base.cmgg.be/), and BEST1 LOVD database (http://www-huge.uni-regensburg.de/BEST1_database) were used to explore additional annotations and literature information, if present. Splice-altering predictions were obtained using the online tools Human Splicing Finder (HSF 3.0) [14] and NNSPLICE 0.9 [15] and the databases dbscSNV [16] and SPIDEX [17], which provide predicted effects for all of the potential variants within splicing consensus regions or across the entire genome, respectively. For the prioritization of pathogenetic mutations, the evaluation of inheritance mode was taken into account, along with segregation information coming from the sequencing of healthy family members, if available.
NGS procedure and data analysis were tested on the four control samples with known molecular diagnosis as proof of concept. In all cases the previously identified variants were correctly detected and prioritized as pathogenic variants.
3. Results
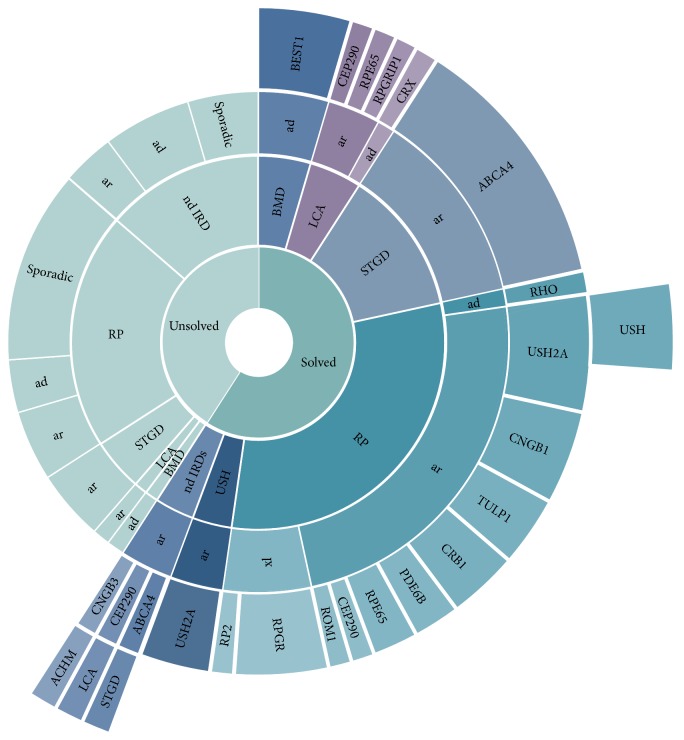
A cohort of 109 samples (Table 1), including 88 IRDs affected probands without molecular diagnosis and 21 unaffected family members, was analyzed by the newly developed system based on NGS and data analysis. A total of 19 sequencing runs were performed (6 samples/Ion Chip 318), obtaining on average a mean coverage of 450 mapped reads, with 92% mean uniformity and 97.6% (SD ± 1.4) of target regions covered at least 30x (96.2% > 50x). For each sample, 242 raw variants were detected on average. Annotation and filtering procedure resulted in the identification of possibly causative mutations in 59.1% of patients (n = 52/88) (Table 2, Figure 1). The majority of the obtained molecular diagnoses were consistent with the subject's clinical presentation and family history.
Table 2.
Diagnostic yields for the clinical subtypes of this study.
| Clinical diagnosis |
Cases (n) |
Genetic diagnosis (n) |
Unsolved cases (n) |
Clinical reassessment (final diagnosis) |
Diagnostic yield (%) |
|---|---|---|---|---|---|
| BMD | 4 | 4 | — | 100 | |
| LCA | 5 | 4 | 1 | 80 | |
| STGD | 14 | 11 | 3 | 78.5 | |
| RP | 45 | 27 | 18 | 2 (USH) | 60.0 |
| USH | 3 | 3 | — | 100 | |
| nd IRD | 17 | 3 | 14 | 3 (ACHM, LCA, STGD) | 17.6 |
| Total | 88 | 52 | 36 | 5 | 59.1 |
BMD: Best Macular Dystrophy; LCA: Leber Congenital Amaurosis; STGD: Stargardt Disease; RP: Retinitis Pigmentosa; USH: Usher Syndrome; nd IRD: inherited retinal degeneration not otherwise specified without precisely defined diagnosis; ACHM: Achromatopsia.
Figure 1.
The chart summarizes the diagnostic yields obtained for the clinical subtypes of this study. The different levels of circles (from inner to outside) specify clinical diagnoses, inheritance mode, mutated genes, and clinical reassessment.
We found pathogenic mutations in 16 genes, with the most recurrent being ABCA4 for STGD and USH2A for RP/USH patients. The majority of the mutated genes were inherited with an AR pattern (78.9%), followed in order by AD (11.5%) and XL (9.6%) inheritance. The majority of cases displaying recessive inheritance were compound heterozygous of two different pathogenic variants, in line with the low frequency of consanguineous marriages in Italy
Identified candidate pathogenic mutations are shown in Table 3. Overall, 63 different mutations were identified: 62.5% of variants were already reported in previous studies, while 37.5% were novel. Among the list of novel variants, 56% were missense predicted to have deleterious protein functional effect by the prediction algorithms described in the Patients and Methods (predicted to be damaging by at least three of the applied algorithms), and 44% were frameshift, nonsense, or splice-site mutations that might severely affect protein function. Notably, 12% of identified variants were located within splicing consensus regions, and additional 12% were exonic variants predicted to alter splicing through enhancer/silencer motif modification or the creation of new potential donor/acceptor sites.
Table 3.
| Patient ID | Family | Clinical diagnosis | Clinical reassessment | Genotype | Inheritance | Gene | Mutation type | Region | cds change |
|
| |||||||||
| IRD027 | STGD | Comp Het | ar | ABCA4 | Splice_region | INTRON_40 | c.5714+5G>A | ||
| ABCA4 | Frameshift | EXON_11 | c.1375delA | ||||||
| IRD036 | Familiar case | STGD | Comp Het | ar | ABCA4 | Stop_gained | EXON_14 | c.2099G>A | |
| ABCA4 | Splice_region syn | EXON_6 | c.768G>T | ||||||
| IRD037 | STGD | Comp Het | ar | ABCA4 | Stop_gained | EXON_14 | c.2099G>A | ||
| ABCA4 | Splice_region syn | EXON_6 | c.768G>T | ||||||
| IRD042 | Familiar case | STGD | Comp Het | ar | ABCA4 | Missense | EXON_42 | c.5882G>A | |
| ABCA4 | Missense | EXON_6 | c.634C>T | ||||||
| IRD043 | STGD | Comp Het | ar | ABCA4 | Missense | EXON_42 | c.5882G>A | ||
| ABCA4 | Missense | EXON_12 | c.1622T>C | ||||||
| IRD050 | STGD | Comp Het | ar | ABCA4 | Missense | EXON_16 | c.2461T>A | ||
| ABCA4 | Missense | EXON_15 | c.2300T>A | ||||||
| IRD054 | STGD | Comp Het | ar | ABCA4 | Stop_gained | EXON_47 | c.6445C>T | ||
| ABCA4 | Missense | EXON_42 | c.5882G>A | ||||||
| IRD055 | STGD | Comp Het | ar | ABCA4 | Missense | EXON_19 | c.2842C>T | ||
| ABCA4 | Missense | EXON_15 | c.2300T>A | ||||||
| IRD061 | STGD | Comp Het | ar | ABCA4 | Missense | EXON_42 | c.5882G>A | ||
| ABCA4 | Missense | EXON_28 | c.4139C>T | ||||||
| IRD062 | STGD | Comp Het | ar | ABCA4 | Missense | EXON_42 | c.5882G>A | ||
| ABCA4 | Missense | EXON_16 | c.2549A>G | ||||||
| IRD073 | nd IRD | STGD | Hom | ar | ABCA4 | Missense | EXON_19 | c.2894A>G | |
| IRD077 | STGD | Comp Het | ar | ABCA4 | Missense | EXON_37 | c.5285C>A | ||
| ABCA4 | Missense | EXON_15 | c.2300T>A | ||||||
| IRD047 | BMD | Het | ad | BEST1 | Missense | EXON_2 | c.73C>T | ||
| IRD057 | Familiar case | BMD | Het | ad | BEST1 | Missense | EXON_2 | c.80G>C | |
| IRD058 | BMD | Het | ad | BEST1 | Missense | EXON_2 | c.80G>C | ||
| IRD064 | BMD | Het | ad | BEST1 | Missense | EXON_2 | c.80G>C | ||
| IRD010 | LCA | Comp Het | ar | CEP290 | Missense | EXON_33 | c.4237G>C | ||
| CEP290 | Frameshift | EXON_23 | c.2390delA | ||||||
| IRD066 | RP | Comp Het | ar | CEP290 | Stop_gained | EXON_48 | c.6640A>T | ||
| CEP290 | Frameshift | EXON_14 | c.1219_1220delAT | ||||||
| IRD072 | nd IRD | LCA | Comp Het | ar | CEP290 | Missense | EXON_14 | c.1298A>G | |
| CEP290 | Frameshift | EXON_3 | c.164_167delCTCA | ||||||
| IRD039 | RP | Hom | ar | CNGB1 | Frameshift | EXON_13 | c.875-5_891dup | ||
| IRD052 | RP | Comp Het | ar | CNGB1 | Missense | EXON_29 | c.2957A>T | ||
| CNGB1 | Frameshift | EXON_13 | c.875-5_891dup | ||||||
| IRD068 | RP | Comp Het | ar | CNGB1 | Splicing, syn | EXON_26 | c.2526C>T | ||
| CNGB1 | Missense | EXON_21 | c.2153G>C | ||||||
| IRD085 | RP | Hom | ar | CNGB1 | Missense | EXON_23 | c.2284C>T | ||
| IRD032 | nd IRD | ACHM | Comp Het | ar | CNGB3 | Splice_donor | INTRON_13 | c.1578+1G>A | |
| CNGB3 | Frameshift | EXON_10 | c.1148delC | ||||||
| IRD029 | Familiar case | RP | Hom | ar | CRB1 | Missense | EXON_5 | c.2200G>A | |
| IRD030 | RP | Hom | ar | CRB1 | Missense | EXON_5 | c.2200G>A | ||
| IRD031 | RP | Hom | ar | CRB1 | Missense | EXON_5 | c.2200G>A | ||
| IRD035 | LCA | Het | ad | CRX | Frameshift | EXON_4 | c.514delC | ||
| IRD008 | RP | Hom | ar | PDE6B | Splice_region | EXON_18 | c.2193+1delG | ||
| IRD013 | RP | Comp Het | ar | PDE6B | Missense | EXON_4 | c.794G>A | ||
| ar | PDE6B | Intron | INTRON_8 | c.1108-10G>A | |||||
| IRD026 | RP | Het | ad | RHO | Missense | EXON_3 | c.568G>T | ||
| IRD016 | RP | Comp Het | ar | ROM1 | Missense | EXON_1 | c.178C>A | ||
| ROM1 | Missense | EXON_1 | c.323C>T | ||||||
| IRD033 | RP | Hem | xl | RP2 | Frameshift | EXON_2 | c.382_383delTT | ||
| IRD076 | RP | Hom | ar | RPE65 | Missense | EXON_2 | c.65T>C | ||
| IRD001 | RP | Comp Het | ar | RPE65 | Missense | EXON_2 | c.65T>C | ||
| RPE65 | Frameshift | EXON_9 | c.893delA | ||||||
| IRD074 | LCA | Hom | ar | RPE65 | Missense | EXON_5 | c.430T>G | ||
| IRD002 | LCA | Comp Het | ar | RPGRIP1 | Frameshift | EXON_15 | c.2225_2226delGA | ||
| RPGRIP1 | Frameshift | EXON_17 | c.2795_2796insT | ||||||
| IRD012 | RP | Hem | xl | RPGR | Missense | EXON_8 | c.785C>G | ||
| IRD067 | RP | Hem | xl | RPGR | Missense | EXON_8 | c.814G>T | ||
| IRD075 | RP | Hem | xl | RPGR | Missense, Splice_region | EXON_2 | c.154G>A | ||
| IRD017 | RP | Hem | De novo | RPGR | Frameshift | EXON_2 | c.89delT | ||
| IRD059 | Familiar case | RP | Comp Het | ar | TULP1 | Missense | EXON_15 | c.1590C>G | |
| TULP1 | Missense | EXON_13 | c.1255C>T | ||||||
| IRD060 | RP | Comp Het | ar | TULP1 | Missense | EXON_15 | c.1590C>G | ||
| TULP1 | Missense | EXON_13 | c.1255C>T | ||||||
| IRD041 | RP | Comp Het | ar | TULP1 | Splice_region | INTRON_14 | c.1496-6C>A | ||
| TULP1 | Missense | EXON_14 | c.1445G>A | ||||||
| IRD007 | USH | Comp Het | ar | USH2A | Missense | EXON_63 | c.12420T>G | ||
| USH2A | splice_region, syn | EXON_28 | c.5775A>T | ||||||
| IRD009 | USH | Comp Het | ar | USH2A | Missense | EXON_63 | c.13546G>T | ||
| USH2A | splice_region, Missense | EXON_10 | c.1645T>C | ||||||
| IRD021 | RP | Comp Het | ar | USH2A | Missense | EXON_69 | c.14995A>G | ||
| USH2A | Missense | EXON_8 | c.1481A>G | ||||||
| IRD023 | Familiar case | RP | USH | Comp Het | ar | USH2A | Missense | EXON_13 | c.2296T>C |
| USH2A | Frameshift | EXON_3 | c.545_548delAAGA | ||||||
| IRD024 | RP | USH | Comp Het | ar | USH2A | Missense | EXON_13 | c.2296T>C | |
| USH2A | Frameshift | EXON_3 | c.545_548delAAGA | ||||||
| IRD038 | RP | Comp Het | ar | USH2A | Missense | EXON_13 | c.2296T>C | ||
| USH2A | Missense | EXON_13 | c.2276G>T | ||||||
| IRD084 | USH | Hom | ar | USH2A | Frameshift | EXON_69 | c.14977_14978delTT | ||
| IRD034 | RP | Hom | ar | USH2A | Missense | EXON_63 | c.12574C>T | ||
|
| |||||||||
| Patient ID | Protein change | Frequency (%) | Coverage (# reads) |
Segregation and unaffected siblings | Functional predictions (dbNSFP) | Splicing predictions | Reference | ||
| Human Splicing Finder | dbscSNV | SPIDEX | |||||||
|
| |||||||||
| IRD027 | 44.9 | 514 | Broken WT Donor Site | 0.999|0.988 | −3.21 | PMID: 15494742 | |||
| p.Thr459GlnfsX2 | 47.7 | 1179 | PMID: 21911583 | ||||||
| IRD036 | p.Trp700X | 48.2 | 303 | .|..|N|A|.|.|.|.|.|D | PMID: 11702214 | ||||
| p.Val256Val | 47.2 | 53 | Broken WT Donor Site | 1.000|0.952 | −2.43 | PMID: 12037008 | |||
| IRD037 | p.Trp700X | 44.5 | 110 | .|..|N|A|.|.|.|.|.|D | New Acceptor Site | −5.41 | PMID: 11702214 | ||
| p.Val256Val | 48.3 | 29 | Broken WT Donor Site | 1.000|0.952 | −2.43 | PMID: 12037008 | |||
| IRD042 | p.Gly1961Glu | 47.1 | 1325 | D|DD|D|D|N|D|D|D|D|D | PMID: 9295268 | ||||
| p.Arg212Cys | 49.1 | 432 | D|DD|D|A|M|D|D|D|D|D | PMID: 11726554 | |||||
| IRD043 | p.Gly1961Glu | 46.9 | 796 | D|DD|D|D|N|D|D|D|D|D | PMID: 9295268 | ||||
| p.Leu541Pro | 51.9 | 727 | D|DD|D|A|M|D|D|D|D|D | PMID: 11527935 | |||||
| IRD050 | p.Trp821Arg | 43.8 | 309 | D|DD|D|D|H|T|D|D|D|D | PMID: 11527935 | ||||
| p.Val767Asp | 46.3 | 452 | D|BB|D|D|M|D|T|D|D|D | PMID: 15494742 | |||||
| IRD054 | p.Arg2149X | 49.1 | 422 | .|..|D|A|.|.|.|.|.|D | New ESS site | −58.3 | PMID: 12202497 | ||
| p.Gly1961Glu | 49.4 | 1448 | D|DD|D|D|N|D|D|D|D|D | PMID: 9295268 | |||||
| IRD055 | p.Arg948Cys | 52.0 | 175 | T|BB|N|D|L|D|T|T|N|N | This study | ||||
| p.Val767Asp | 51.5 | 437 | D|BB|D|D|M|D|T|D|D|D | PMID: 15494742 | |||||
| IRD061 | p.Gly1961Glu | 50.0 | 729 | D|DD|D|D|N|D|D|D|D|D | PMID: 9295268 | ||||
| p.Pro1380Leu | 55.8 | 437 | D|DP|N|A|M|D|D|D|D|D | New ESS site | −5.44 | PMID: 11726554 | |||
| IRD062 | p.Gly1961Glu | 100 | 787 | D|DD|D|D|N|D|D|D|D|D | PMID: 9295268 | ||||
| p.Tyr850Cys | 49.4 | 176 | D|DD|D|D|M|T|D|D|D|D | PMID: 23096905 | |||||
| IRD073 | p.Asn965Ser | 100 | 225 | D|DD|D|D|L|D|D|D|D|D | PMID: 9054934 | ||||
| IRD077 | p.Ala1762Asp | 50.8 | 259 | D|DD|D|A|M|D|D|D|D|D | PMID: 15192030 | ||||
| p.Val767Asp | 51.4 | 752 | D|BB|D|D|M|D|T|D|D|D | PMID: 15494742 | |||||
| IRD047 | p.Arg25Trp | 56.0 | 348 | D|DD|U|D|M|D|D|D|D|D | New Donor Site, New ESS site | PMID: 10798642 | |||
| IRD057 | p.Ser27Thr | 46.8 | 344 | D|DD|U|D|H|D|D|D|D|D | This study | ||||
| IRD058 | p.Ser27Thr | 45.5 | 317 | D|DD|U|D|H|D|D|D|D|D | This study | ||||
| IRD064 | p.Ser27Thr | 47.1 | 453 | D|DD|U|D|H|D|D|D|D|D | This study | ||||
| IRD010 | p.Asp1413His | 49.2 | 413 | D|BB|D|D|N|T|T|T|N|D | ClinVar: RCV000082249.5 | ||||
| p.Lys797SerfsX2 | 30.1 | 163 | This study | ||||||
| IRD066 | p.Lys2214X | 47.5 | 705 | .|..|D|A|.|.|.|.|.|D | ESE Site Broken | −86.6 | This study | ||
| p.Met407GlufsX14 | 51.1 | 225 | PMID: 17724218 | ||||||
| IRD072 | p.Asp433Gly | 53.4 | 116 | T|DP|D|D|L|T|T|T|D|D | New ESS site, New donor site | This study | |||
| p.Thr55SerfsX3 | 43.2 | 243 | PMID: 20690115 | ||||||
| IRD039 | p.Gly298CysfsX13 | 100 ∗ | This study | ||||||
| IRD052 | p.Asn986Ile | 51.7 | 471 | D|DD|D|D|M|D|D|D|D|D | PMID: 21147909 | ||||
| p.Gly298CysfsX13 | 26,7∗ | 258 | This study | ||||||
| IRD068 | Thr842Thr | 52.1 | 431 | ESE Site Broken | This study | ||||
| p.Gly718Ala | 47.1 | 153 | D|PP|D|D|M|T|T|T|D|D | This study | |||||
| IRD085 | p.Arg762Cys | 100 | 57 | D|DD|D|D|H|D|D|D|D|D | This study | ||||
| IRD032 | 47.8 | 907 | Broken WT Donor Site | −8.56 | PMID: 15657609 | ||||
| p.Thr383IlefsX13 | 46.5 | 588 | PMID: 15657609 | ||||||
| IRD029 | p.Gly734Arg | 100 | 397 | D|DD|.|D|M|T|D|D|D|D | This study | ||||
| IRD030 | p.Gly734Arg | 100 | 397 | D|DD|.|D|M|T|D|D|D|D | This study | ||||
| IRD031 | p.Gly734Arg | 100 | 397 | D|DD|.|D|M|T|D|D|D|D | This study | ||||
| IRD035 | p.Pro172LeufsX15 | 50.5 | 521 | This study | |||||
| IRD008 | 100 | 395 | Brother: Het | Broken WT Donor Site | This study | ||||
| IRD013 | p.Arg265Gln | 51.7 | 319 | n.a. | T|DD|D|D|L|T|T|T|N|D | ClinVar: RCV000178068.1 | |||
| 54.7 | 75 | Mother: Het | 0.001|0.096 | PMID: 8698075 | |||||
| IRD026 | p.Asp190Tyr | 44.6 | 168 | D|DD|D|D|M|T|T|T|D|D | PMID: 8401533 | ||||
| IRD016 | p.Pro60Thr | 56.1 | 278 | T|BB|N|N|L|T|T|T|N|N | PMID: 8595413 | ||||
| p.Thr108Met | 52.8 | 108 | T|PB|N|D|L|T|T|T|N|D | PMID: 8595413 | |||||
| IRD033 | p.Leu129ValfsX9 | 100 | 392 | This study | |||||
| IRD076 | p.Leu22Pro | 100 | 495 | T|BB|D|D|M|D|D|D|N|D | PMID: 9801879 | ||||
| IRD001 | p.Leu22Pro | 46.3 | 257 | Brother: wt | T|BB|D|D|M|D|D|D|N|D | PMID: 9801879 | |||
| p.Lys298SerfsX27 | 98 | 150 | Brother: wt | PMID: 11462243 | |||||
| IRD074 | p.Tyr144Asp | 100 | 430 | Father: Het | D|DD|D|D|M|D|D|D|D|D | PMID: 11462243 | |||
| IRD002 | p.Glu743ArgfsX24 | 48.8 | 570 | Father: Het | This study | ||||
| p.Glu933X | 48.8 | 400 | Mother: Het | This study | |||||
| IRD012 | p.Ala262Gly | 100 | 280 | T|BB|N|N|L|D|T|T|N|N | This study | ||||
| IRD067 | p.Gly272Cys | 100 | 155 | D|DD|D|D|H|D|D|D|D|D | This study | ||||
| IRD075 | p.Gly52Arg | 100 | 348 | D|DP|U|D|M|T|T|T|D|D | Broken WT Donor Site | PMID: 15364249 | |||
| IRD017 | p.Phe30SerfsX38 | 100 | 113 | Brother: wt Female twin: wt |
This study | ||||
| IRD059 | p.Ile530Met | 50.6 | 682 | D|DD|D|D|H|D|D|D|D|N | This study | ||||
| p.Arg419Trp | 49.5 | 645 | D|DD|D|D|H|D|D|D|D|D | PMID: 25342620 | |||||
| IRD060 | p.Ile530Met | 51.0 | 655 | D|DD|D|D|H|D|D|D|D|N | This study | ||||
| p.Arg419Trp | 45.3 | 575 | D|DD|D|D|H|D|D|D|D|D | PMID: 25342620 | |||||
| IRD041 | 54.1 | 727 | Father: Het | 0.005|0.419 | PMID: 9660588 | ||||
| p.Arg482Gln | 48.5 | 485 | Mother: Het | D|DD|D|D|H|D|D|D|D|D | New Acceptor Site | −1.28 | PMID: 22665969 | ||
| IRD007 | p.Cys4140Trp | 50.5 | 214 | D|DD|D|D|M|T|T|T|D|D | This study | ||||
| 49.5 | 398 | Broken WT Donor Site | 0.998|0.986 | −4.24 | This study | ||||
| IRD009 | p.Gly4516Trp | 53.8 | 239 | D|DD|U|D|H|T|D|D|D|D | This study | ||||
| p.Cys549Arg | 49.2 | 566 | D|DD|U|D|H|D|D|D|D|D | 0.417|0.520 | This study | ||||
| IRD021 | p.Thr4999Ala | 51.0 | 400 | D|DD|U|D|M|T|T|T|D|D | This study | ||||
| p.Tyr494Cys | 49.0 | 400 | D|DD|N|D|L|T|T|T|D|D | This study | |||||
| IRD023 | p.Cys766Arg | 39.0 | 82 | D|DD|D|D|H|D|D|D|D|D | PMID: 23591405 | ||||
| p.Lys182ArgfsX9 | 61.4 | 202 | This study | ||||||
| IRD024 | p.Cys766Arg | 43.5 | 124 | D|DD|D|D|H|D|D|D|D|D | PMID: 23591405 | ||||
| p.Lys182ArgfsX9 | 48.0 | 225 | This study | ||||||
| IRD038 | p.Cys766Arg | 47.2 | 89 | D|DD|D|D|H|D|D|D|D|D | PMID: 23591405 | ||||
| p.Cys759Phe | 51.1 | 90 | D|DD|D|A|H|D|D|D|D|D | PMID: 10775529 | |||||
| IRD084 | p.Phe4993ProfsX7 | 100 | 483 | PMID: 24944099 | |||||
| IRD034 | p.Arg4192Cys | 100 | 515 | D|DP|N|D|M|D|D|D|D|D | PMID: 24498627 | ||||
ACHM: Achromatopsia; ad: autosomal dominant; ar: autosomal recessive; BMD: best macular disease; Comp Het: compound heterozygous; ESE: exonic splicing enhancer; ESS: exonic splicing silencer; Hem: Hemizygous; Het: heterozygous; Hom: homozygous; LCA: Leber Congenital Amaurosis; nd IRD: inherited retinal degeneration not otherwise specified without precisely defined diagnosis; RP: Retinitis Pigmentosa; STGD: Stargardt Disease; USH: Usher Syndrome; wt: wild-type; xl: X-linked. For nonsynonymous variants, predictions from dbNSFP are reported, comprising scores from the following alghoritms: SIFT | Polyphen2HDIV Polypehn2HVAR | LRT | MutationTaster | MutationAssessor | FATHMM | MetaSVM | MetaLR | PROVEAN | fathmm-MKL. For splicing variants, predictions from Human Splicing Finder, dbscSNV (ada_score|rf_score) and SPIDEX are reported. For SPIDEX, max dPSI is shown if lower than −1 (maximum mutation − induced change in the percentage of transcripts with the exon spliced in). Familiar case: the patients were from the same family. *Sanger sequencing was performed to confirm mutation frequency.
Table 2 summarizes the mutation detection rates obtained for the different clinical subtypes of our study cohort. The highest diagnostic yields were achieved for BMD, LCA, USH, and STGD patients with well-defined clinical diagnosis, where the number of known genes associated with each disease is relatively limited.
For BMD cases, all diagnosed patients were heterozygous for mutations on BEST1. Three patients (mother and son) were found to harbour a novel BEST1 missense mutation c.80G>C (p.Ser27Thr) located in the immediate N-terminus, in one of the four mutational hotspots regions in the highly conserved N-terminal half of the protein [18] and predicted to be deleterious by all interrogated algorithms.
For STGD patients, genetic diagnosis was achieved in 11 out of 14 (78.5% of the cases). All diagnosed patients in our cohort carried mutations on ABCA4. In 75% of the unsolved cases at least one ABCA4 pathogenic allele was identified, suggesting the presence of disease-causing mutations lying outside the coding sequence covered by our panel, as reported in a previous study [19].
In LCA patients, causative mutations were identified in CEP290, RPE65, RPGRIP1, and CRX genes, and only one case remained unsolved (20% of the total LCA cases), whereas all Usher 2 syndrome cases were found to carry mutations in USH2A gene.
For RP patients, genetic diagnosis was achieved in 27 out of 45 (60% of the cases), involving mutations in 11 different genes: confirming that these phenotypes are genetically heterogeneous (Figure 1). Dominant mutations were identified in RHO gene, whereas USH2A, CNGB1, and TULP1 were the most recurrently mutated genes in ARRP. X-linked inheritance was established for 5 RP male patients (4 probands had mutations in RPGR, whereas one had a mutation in RP2). The identification of USH2A as the defective gene in patients with initial clinical diagnosis of RP was followed by audiometric testing to establish if there were any hearing deficiencies. A hearing impairment was found in 2 cases out of 5 leading to clinical reassessment and final diagnosis of USH (Table 2).
For patients with IRD without a defined clinical diagnosis or with unclear disease manifestations, we identified causative mutations in 7 out of 17 probands (23.5% of the total IRD cases). In two cases the molecular results allowed a refined clinical diagnosis: a compound heterozygosity of two mutations in CEP290 led to a genetic diagnosis of LCA in a patient with initial diagnosis of North Carolina or Stargardt macular dystrophy, whereas a homozygous pathogenic variant in ABCA4 was found in a patient with tapetoretinal degeneration.
In 36 patients (12 familiar and 24 sporadic) the molecular analysis did not achieve any definitive result, even after the analysis of the healthy family members, which was performed in 8 cases. Half of the cases with a negative test result (18 out of 36) were affected by RP. The additional analysis of the RPGR ORF15 (a mutational hotspot which was nonsufficiently covered in our panel) for the male patients with a sporadic or suspected X-linked pattern of inheritance (10 patients) by Sanger sequencing yielded no additional mutations.
4. Discussion
The results of the present study confirm that high-throughput Next-Generation Sequencing represents a comprehensive cost-effective approach for the molecular diagnosis of Inherited Retinal Dystrophies (IRDs), achieving a molecular diagnosis for 59.1% of the studied cases. More specifically, among the different clinical phenotypes, the highest detection rates were achieved for BMD, LCA, USH, and STGD patients, in whom the genetic test clearly confirmed the clinical diagnoses (Table 2). The results of the RP and of the not defined IRD cohorts, instead, demonstrated the high genetic heterogeneity of this diseases and the essential contribution of our NGS analysis to achieving an accurate diagnosis, with the involvement of 12 different genes in 28 sporadic cases. Revision of the initial diagnosis, performed for 9.6% of the genetically diagnosed patients, further emphasizes the importance of a comprehensive genotype/phenotype analysis to unravel the extensive heterogeneity of these diseases. Notably, a remarkable fraction of identified variants are splice-altering mutations (25% of the total mutation burden, 16 out of 64), located within splicing consensus regions, or exonic variants predicted to cause enhancer/silencer motif modification or the creation of new potential donor/acceptor, which are amenable to the antisense-mediated splicing-correction approaches, as recently reported for several genetic diseases, including CEP290-caused LCA [20, 21].
The prevalence of IRD and most importantly the frequency of gene mutations causing those diseases are not well characterized in Italy and only few data have been reported [22–24]. RPE65, CRB1, and GUCY2D were identified as the most prevalent mutated genes in Italian LCA patients [22] and RHO was reported to be the gene most commonly responsible for ADRP [23] and EYS the most recurrent for nonsyndromic ARRP and sporadic cases [24]. Our study contributes only partially to the knowledge of the gene mutation frequencies, since each IRD type is represented by small cohorts of cases (i.e., the LCA and dominant RP phenotypes were accounted for by 5 and 6 cases, resp.), and some probands of other ethnicities have been included too. Indeed, regarding LCA, we identified mutations in CEP290, RPE65, CRX, and RPGRIP1 genes.
For ADRP, RHO was identified to be responsible for the phenotype in one case, whereas, in ARRP and sporadic RP, USH2A, CNGB1, and TULP1 were the most recurrently mutated genes. RPE65 mutations were found in two ARRP cases: in one more case, still unsolved, a single RPE65 heterozygous pathogenic variant was found. ROM1 compound heterozygosity was established in one RP proband, suggesting a mechanism of recessive inheritance for this gene associated with dominant and digenic forms. X-linked inheritance was established for 5 RP affected probands, with RPGR and RP2 identified as the disease-causing gene in 4 cases and 1 case, respectively. All BMD diagnosed patients were heterozygous for mutations on BEST1 gene, the major gene responsible for Best's juvenile form [25], whereas the 78.5% of patients with clinically diagnosed STGD carried pathogenic variants on ABCA4 [26].
Similarly to a recent study [6], the clinical sensitivity of our NGS analysis was not uniform, with the highest diagnostic yields obtained in conditions where the disease-causing genes have been nearly all identified.
Direct comparison of our findings with other recently published NGS studies [2–6, 27] is not straightforward, due to differences in the number of genes analyzed but especially due to composition and relative representation of the different phenotypes in the patients cohorts. However, the finding of USH2A and ABCA4 as the most mutated genes for RP/USH and STGD patients is consistent with previous reports [27–29]. In our RP cohort, USH2A is followed by CNGB1 and RPGR. These two genes, already reported among the most frequently mutated genes in IRD patients [29], were not highly frequently altered in the Saudi population [6] or in a large cohort of Western European and South Asian individuals [27]. Also, we did not find any alteration in EYS, one of the top three genes contributing to IRD in other populations [28, 29].
The different gene alterations identified in our LCA cohort (CEP290, RPE65, RPGRIP1, and CRX genes) were consistent with the different disease manifestations of the analyzed patients, in accordance with the specific clinical features described for each of the LCA-associated genes [30, 31]. Less direct is the correlation between the genes involved and the phenotypic features in RP, due to the known contribution of environmental factors to late-childhood- and adult-onset-diseases.
Allelic heterogeneity, with different mutations in the same gene causing different phenotypes, is evident also in USH2A-related retinal disease. Genotype-phenotype correlations observed in our cohort were in accordance with the allelic hierarchy proposed in a recent study [32], supporting the model that USH represents the null phenotype consequent upon severe USH2A defects, whereas milder mutations in at least one allele result in a pure retinal phenotype associated with normal auditory function.
IRD genetic heterogeneity, reflected in the identification of mutations in many genes with a considerable number of previously undescribed alterations, supported the conclusion that molecular diagnosis of these disorders should rely on massive parallel multigene sequencing. Nevertheless, for 36 probands, including 12 familiar cases and 24 unrelated probands, our NGS procedure did not result in the identification of a clear genetic cause of the disease. Some subjects may have mutations that cannot be detected by our amplicon-based approach, such as deep intronic mutations, copy-number variations, or large deletions. In the perspective of the design of a more complete new version of the panel, additional deep intronic regions reported in the literature as carrying disease-causing mutations [19, 33, 34] or a higher exon padding (5 bp in our design, up to 100 bp available in the current pipeline version of the Ion AmpliSeq Designer tool) could be implemented. Moreover, technical limitations, including the difficult amplification of RPGR ORF15, a mutational hotspot for X-linked RP, may have accounted for some of the missed diagnosis (our panel is presently covering only 30% of this critical exon), but the addition of the specific analysis by Sanger sequencing of the ORF15 of the RPGR gene in 10 males patients, with sporadic/X-linked RP and previously testing negative for pathogenic mutations using our NGS panel, did not reveal any mutation in the analyzed region. Finally, as an improvement to further support the pathogenicity of novel mutations identified in probands, the analysis of both affected and unaffected family member should be performed, when possible.
In some of the patients who tested negative we however identified single potentially pathogenic heterozygous mutations in recessive genes or novel heterozygous missense variants in dominant genes with unknown significance, lacking the appropriate level of evidence to classify them as disease-causing or not in concordance with patients' clinical presentations or family data. The contribution of these variants in combination with deep intronic mutations or large deletions is suspected but could not be demonstrated with the present technique.
Database incompleteness further complicates variant interpretation. Two probands with BMD phenotype and BEST1 mutation were found to harbour also heterozygous mutation in RHO (c.578C>T, p.Thr193Met), which was predicted to be damaging and listed as associated with ADRP in a public database [http://www.retina-international.org/sci-news/databases/mutation-database] but in our cohort was carried also by healthy subject, reinforcing the need of a critical interpretation of the molecular findings in view of the phenotypic features of the patients with IRD until a more thorough knowledge of the frequency of the variants and a critical amount of data present in the public disease databases are reached.
In conclusion, by presenting profoundly different mutation rates varying according to the clinical diagnosis and by reporting 9.61% of cases of reassessment of the initial diagnosis on the basis of the results of the test, our study reinforces the need of a multidisciplinary work-up before and after the genetic testing, due to the implications of the results in terms of risk assessment for family members and inclusion in gene-based clinical trials.
Supplementary Material
Clinical information for the patients with identified pathogenic mutations is reported in Supplementary Table1. Supplementary Table2 lists the genes included in the designed NGS panel and their associated retinal disease. Primers used for PCR and Sanger sequencing are reported in Supplementary Table 3.
Acknowledgments
This study was supported by Regione Emilia Romagna “RARER,” Area1 (E35E09000880002). The authors thank Professor Sandro Banfi for kindly providing the control samples used to validate their procedure. Programma di Ricerca Regione-Università 2010–2012 “Next-Generation Sequencing and Gene Therapy to Diagnose and Cure Rare Diseases in Regione Emilia Romagna (RARER),” Area 1, Strategic Programmes (E35E09000880002), is acknowledged. The funding organization participated in the design of the study.
Abbreviations/Acronyms
- AD:
Autosomal dominant
- AR:
Autosomal recessive
- BMD:
Best Macular Dystrophy
- IRDs:
Inherited Retinal Dystrophies
- LCA:
Leber Congenital Amaurosis
- NGS:
Next-Generation Sequencing
- RP:
Retinitis Pigmentosa
- STGD:
Stargardt Disease
- USH:
Usher Syndrome
- XL:
X-linked.
Competing Interests
No conflicting relationship exists for any author.
Authors' Contributions
Isabella Bernardis and Laura Chiesi contributed equally.
References
- 1.Dryja T. P., Hahn L. B., Kajiwara K., Berson E. L. Dominant and digenic mutations in the peripherin/RDS and ROM1 genes in retinitis pigmentosa. Investigative Ophthalmology and Visual Science. 1997;38(10):1972–1982. [PubMed] [Google Scholar]
- 2.Neveling K., Collin R. W. J., Gilissen C., et al. Next-generation genetic testing for retinitis pigmentosa. Human Mutation. 2012;33(6):963–972. doi: 10.1002/humu.22045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eisenberger T., Neuhaus C., Khan A. O., et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: the example of retinal dystrophies. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0078496.e78496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J., Zhang V. W., Feng Y., et al. Dependable and efficient clinical utility of target capture-based deep sequencing in molecular diagnosis of retinitis pigmentosa. Investigative Ophthalmology & Visual Science. 2014;55(10):6213–6223. doi: 10.1167/iovs.14-14936. [DOI] [PubMed] [Google Scholar]
- 5.Consugar M. B., Navarro-Gomez D., Place E. M., et al. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genetics in Medicine. 2015;17(4):253–261. doi: 10.1038/gim.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel N., Aldahmesh M. A., Alkuraya H., et al. Expanding the clinical, allelic, and locus heterogeneity of retinal dystrophies. Genetics in Medicine. 2015;18(6):554–556. doi: 10.1038/gim.2015.127. [DOI] [PubMed] [Google Scholar]
- 7.Chiang J. P.-W., Lamey T., McLaren T., Thompson J. A., Montgomery H., De Roach J. Progress and prospects of next-generation sequencing testing for inherited retinal dystrophy. Expert Review of Molecular Diagnostics. 2015;15(10):1269–1275. doi: 10.1586/14737159.2015.1081057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang J. P.-W., Trzupek K. The current status of molecular diagnosis of inherited retinal dystrophies. Current Opinion in Ophthalmology. 2015;26(5):346–351. doi: 10.1097/icu.0000000000000185. [DOI] [PubMed] [Google Scholar]
- 9.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38(16, article e164) doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X., Jian X., Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Human Mutation. 2011;32(8):894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Human Mutation. 2016;37(3):235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong C., Wei P., Jian X., et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Human Molecular Genetics. 2015;24(8):2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desmet F.-O., Hamroun D., Lalande M., Collod-Bëroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Research. 2009;37(9, article e67) doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reese M. G., Eeckman F. H., Kulp D., Haussler D. Improved splice site detection in Genie. Journal of Computational Biology. 2009;4(3):311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 16.Jian X., Boerwinkle E., Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Research. 2014;42(22):13534–13544. doi: 10.1093/nar/gku1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong H. Y., Alipanahi B., Lee L. J., et al. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347(6218) doi: 10.1126/science.1254806.1254806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milenkovic V. M., Röhrl E., Weber B. H. F., Strauss O. Disease-associated missense mutations in bestrophin-1 affect cellular trafficking and anion conductance. Journal of Cell Science. 2011;124(17):2988–2996. doi: 10.1242/jcs.085878. [DOI] [PubMed] [Google Scholar]
- 19.Braun T. A., Mullins R. F., Wagner A. H., et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Human Molecular Genetics. 2013;22(25):5136–5145. doi: 10.1093/hmg/ddt367.ddt367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collin R. W., den Hollander A. I., van der Velde-Visser S. D., Bennicelli J., Bennett J., Cremers F. P. Antisense oligonucleotide (AON)-based therapy for leber congenital amaurosis caused by a frequent mutation in CEP290. Molecular Therapy Nucleic Acids. 2012;1, article e14 doi: 10.1038/mtna.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bacchi N., Casarosa S., Denti M. A. Splicing-correcting therapeutic approaches for retinal dystrophies: where endogenous gene regulation and specificity matter. Investigative Ophthalmology and Visual Science. 2014;55(5):3285–3294. doi: 10.1167/iovs.14-14544. [DOI] [PubMed] [Google Scholar]
- 22.Simonelli F., Ziviello C., Testa F., et al. Clinical and molecular genetics of Leber's congenital amaurosis: a multicenter study of Italian patients. Investigative Ophthalmology & Visual Science. 2007;48(9):4284–4290. doi: 10.1167/iovs.07-0068. [DOI] [PubMed] [Google Scholar]
- 23.Ziviello C., Simonelli F., Testa F., et al. Molecular genetics of autosomal dominant retinitis pigmentosa (ADRP): a comprehensive study of 43 Italian families. Journal of Medical Genetics. 2005;42(7):p. e47. doi: 10.1136/jmg.2005.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierrottet C. O., Zuntini M., Digiuni M., et al. Syndromic and non-syndromic forms of retinitis pigmentosa: a comprehensive Italian clinical and molecular study reveals new mutations. Genetics and Molecular Research. 2014;13(4):8815–8833. doi: 10.4238/2014.october.27.23. [DOI] [PubMed] [Google Scholar]
- 25.Krämer F., White K., Pauleikhoff D., et al. Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration. European Journal of Human Genetics. 2000;8(4):286–292. doi: 10.1038/sj.ejhg.5200447. [DOI] [PubMed] [Google Scholar]
- 26.Sheffield V. C., Stone E. M. Genomics and the eye. The New England Journal of Medicine. 2011;364(20):1932–1942. doi: 10.1056/nejmra1012354. [DOI] [PubMed] [Google Scholar]
- 27.Ellingford J. M., Barton S., Bhaskar S., et al. Molecular findings from 537 individuals with inherited retinal disease. Journal of Medical Genetics. 2016;53(11):761–767. doi: 10.1136/jmedgenet-2016-103837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang X.-F., Huang F., Wu K.-C., et al. Genotype-phenotype correlation and mutation spectrum in a large cohort of patients with inherited retinal dystrophy revealed by next-generation sequencing. Genetics in Medicine. 2015;17(4):271–278. doi: 10.1038/gim.2014.138. [DOI] [PubMed] [Google Scholar]
- 29.Ge Z., Bowles K., Goetz K., et al. NGS-based Molecular diagnosis of 105 eyeGENE(Ⓡ) probands with Retinitis Pigmentosa. Scientific Reports. 2015;5:p. 18287. doi: 10.1038/srep18287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanein S., Perrault I., Gerber S., et al. Leber congenital amaurosis: comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Human Mutation. 2004;23(4):306–317. doi: 10.1002/humu.20010. [DOI] [PubMed] [Google Scholar]
- 31.Coppieters F., Casteels I., Meire F., et al. Genetic screening of LCA in Belgium: predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Human Mutation. 2010;31(10):E1709–E1766. doi: 10.1002/humu.21336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenassi E., Vincent A., Li Z., et al. A detailed clinical and molecular survey of subjects with nonsyndromic USH2A retinopathy reveals an allelic hierarchy of disease-causing variants. European Journal of Human Genetics. 2015;23(10):1318–1327. doi: 10.1038/ejhg.2014.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liquori A., Vaché C., Baux D., et al. Whole USH2A gene sequencing identifies several new deep intronic mutations. Human Mutation. 2016;37(2):184–193. doi: 10.1002/humu.22926. [DOI] [PubMed] [Google Scholar]
- 34.den Hollander A. I., Koenekoop R. K., Yzer S., et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. The American Journal of Human Genetics. 2006;79(3):556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical information for the patients with identified pathogenic mutations is reported in Supplementary Table1. Supplementary Table2 lists the genes included in the designed NGS panel and their associated retinal disease. Primers used for PCR and Sanger sequencing are reported in Supplementary Table 3.