Abstract
To examine the effect of TBMS1on breast cancer metastasis, and investigate the potential mechanism by which Tubeimoside-1 (TBMS1) inhibits the CXCR4 expression in breast cancer cells. The expression of CXCR4 in breast cancer cell lines was determined by immunoblotting and real-time PCR. The effect of TBMS1 on NF-κB binding activity was evaluated by EMSA assay and ChIP analysis. Cell proliferation and invasion were analyzed by MTT assay and transwell invasion assay, respectively. The effect of TBMS1 on breast cancer metastasis was further evaluated in a metastasis model of nude mice. TBMS1 suppressed the expression of CXCR4 through inhibition of NF-κB binding activity. TBMS1 inhibited CXCL12-induced invasion in breast cancer cells, while ectopic expression of CXCR4 abolished the inhibitive activity of TBMS1. TBMS1 suppressed breast cancer metastasis in the metastatic model of nude mice. TBMS1 suppressed the CXCR4-mediated metastasis of breast cancer by inhibiting NF-κB binding activity. [BMB Reports 2016; 49(9): 502-507]
Keywords: Breast cancer, CXCR4, Metastasis, Tubeimoside-1
INTRODUCTION
Worldwide, breast cancer is the leading cause of cancer mortality in females, and more than 90% of breast cancer-associated deaths are related to cancer metastasis (1). However, the mechanism of breast cancer metastasis remains poorly understood. The stromal cell-derived factor 1α [SDF-1α, also known as Cysteine X Cysteine (CXC) chemokine ligand 12 (CXCL12)] and its receptor chemokine receptor 4 (CXCR4) have been shown to play an important role in promoting breast cancer metastasis (2,3). CXCR4 is overexpressed in metastatic breast cancer cells, and CXCL12, the ligand of CXCR4, is expressed at typical metastatic sites of breast cancer, including bone marrow, lung, liver and lymph nodes (4-6). It has been demonstrated that the activation of CXCR4 by CXCL12 promoted the migration and invasion of tumor cells (5). Agents that down-regulate the expression of CXCR4 or interrupt the CXCR4-CXCL12 cell-signaling pathway have the potential to suppress tumor metastasis (7-9). Thus, developing and identifying new drugs to antagonize the CXCL12-CXCR4 signaling pathway is a promising strategy to inhibit breast cancer metastasis.
TBMS1 is a natural compound isolated from the Chinese medicinal herb Bolbostemma paniculatum (Maxim) Franquet (Cucurbitaceae) (10). It was traditionally used for the treatment of snake venoms and inflammation (10). Recent studies demonstrated that TBMS1 exerted potential anticancer effects in several types of cancer including lung cancer, ovarian cancer, colon cancer and human gliomas (11-15). However, the effects of TBMS1 on human breast cancer have not been studied, and the underlying mechanisms of its anti-tumor activity remain elusive. In this study, we investigated the anti-tumor activity of TBMS1 in breast cancer and demonstrated that TBMS1 inhibited CXCL12-induced metastasis of breast cancer by reducing the expression of CXCR4 in breast cancer cells.
RESULTS
TBMS1 down-regulated the CXCR4 expression in breast cancer cells
TBMS1 was recently identified as a potent anti-tumor compound and has been shown to induce apoptosis in some types of cancer (11,14,16). However, the underlying mechanisms of its anti-tumor activity remain elusive. To explore the potential anti-tumor mechanisms of TBMS1 in breast cancer, we performed a gene expression microarray analysis in MDA-MB-231 cells with/out TBMS1 treatment. We found that CXCR4, a chemokine receptor that is highly expressed in breast cancer and promotes tumor metastasis (17), was downregulated after TBMS1 treatment (data not shown).
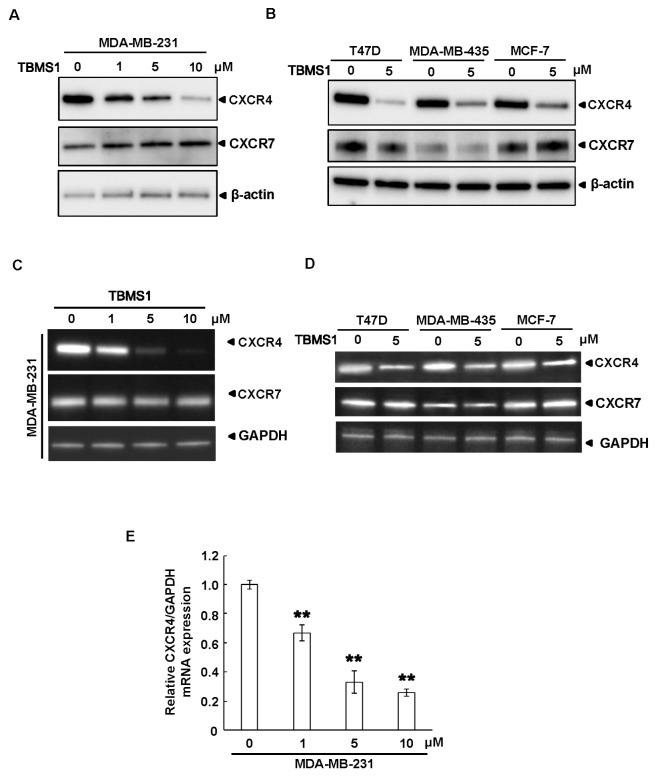
We then performed an immunoblotting assay to examine the protein expression of CXCR4 in MDA-MB-231 cells. As shown in Fig. 1A, TBMS1 inhibited the expression of CXCR4 in a dose-dependent manner. To further confirm the inhibitory effect of TBMS1 on CXCR4 expression, other breast cancer cell lines were analyzed by immunoblotting after TBMS1 treatment. We found that TBMS1 also suppressed the expression of CXCR4 in T47D, MDA-MB-435 and MCF-7 cells (Fig. 1B). Importantly, TBMS1 had no effect on CXCR7, another chemokine receptor, indicating the specific effect of TBMS1 on the regulation of CXCR4 expression (Fig. 1A and B). To determine whether TBMS1 inhibits CXCR4 expression at the level of transcription, a RT-PCR was performed. As shown in Fig. 1C and D, TBMS1 inhibited the mRNA expression of CXCR4 in a dose-dependent manner and in multiple breast cancer cell lines. Similarly, TBMS1 had no effect on the mRNA expression of CXCR7 (Fig. 1C and D). In addition, the inhibitory effect of TBMS1 on CXCR4 expression was further confirmed by a quantitative real-time PCR in MDA-MB-231 cells (Fig. 1E). Collectively, these data suggested that TBMS1 inhibited the CXCR4 expression in multiple breast cancer cells.
Fig. 1. TBMS1 downregulates the expression of CXCR4 in breast cancer cells. (A) MDA-MB-231 cells or (B) T47D, MDA-MB-435 and MCF-7 cells were treated with TBMS1 for 24 hr at the indicated concentrations, and the expression of CXCR4, CXCR7 and β-actin were analyzed by immunoblotting. (C) MDA-MB-231 cells or (D) T47D, MDA-MB-435 and MCF-7 cells were treated with TBMS1 for 24 hr. The mRNA expression of CXCR4, CXCR7 and GAPDH were either examined by reverse-transcript PCR or (E) real-time PCR, as indicated. **P < 0.01 compared with DMSO treated cells. Representative results of three independent experiments were shown.
TBMS1 suppresses the binding of NF-κB to CXCR4 promoter
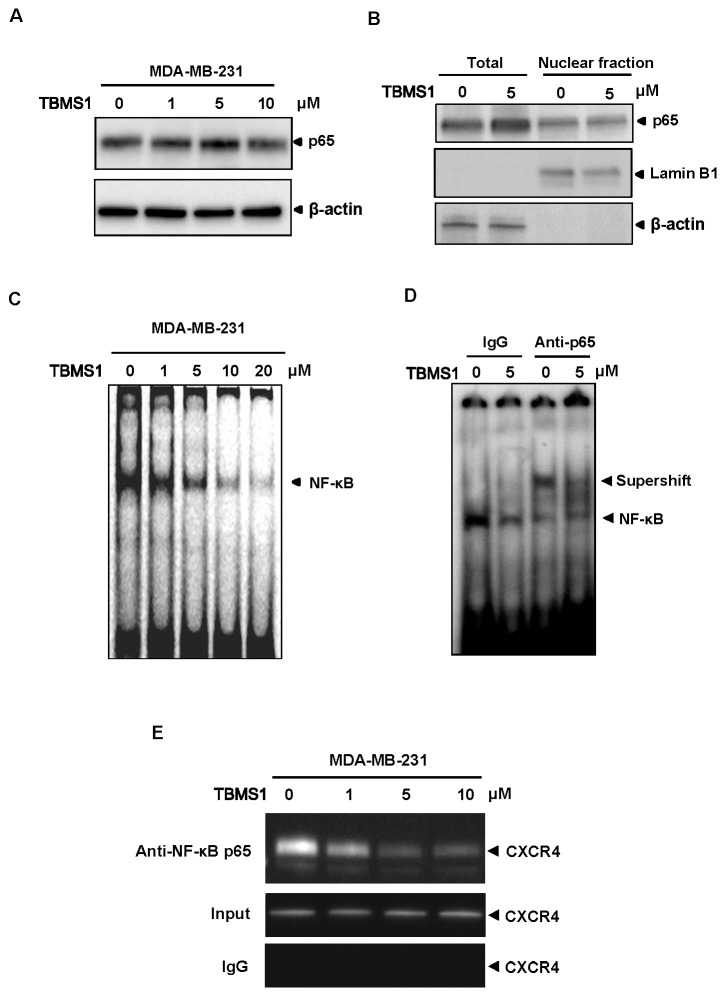
Nuclear factor-κB (NF-κB) was reported as a major transcriptional factor in regulating CXCR4 expression (18). NF-κB directly binds to the sequences within the region of CXCR4 promoter, thus activating its transcription. Since we found that TBMS1 did not influence the expression and nuclear localization of NF-κB (Fig. 2A and B), we then examined whether TBMS1 influenced the binding activity of NF-κB to CXCR4 promoter. Oligonucleotides corresponding to the NF-κB binding site in CXCR4 promoter were designed and incubated with the nuclear extracts from MDA-MB-231 cells treated with/out TBMS1, respectively. A gel EMSA assay was performed to analyze the effect of TBMS1 on NF-κB binding activity. As shown in Fig. 2C, the mobility of the labeled DNA probe was altered in the presence of nuclear protein. Remarkably, the intensity of shift band was decreased by treating the cells with TBMS1 in a dose-dependent manner (Fig. 2C). The NF-κB binding specificity for the labled DNA probe was verified by supershift, when we used anti-p65 antibody (Fig. 2D). TBMS1 further attenuated the abundance of the supershift band, suggesting a potential inhibitory effect of TBMS1 on NF-κB binding activity (Fig. 2D). We then performed a chromatin immunoprecipitation assay (ChIP) and confirmed that TBMS1 suppressed the DNA binding activity of NF-κB to CXCR4 promoter in vivo (Fig. 2E). Thus, these results suggested that TBMS1 down-regulated CXCR4 expression by attenuating the NF-κB binding activity to its promoter.
Fig. 2. TBMS1 suppresses the binding of NF-κB to CXCR4 promoter. (A) MDA-MB-231 cells were treated with TBMS1 for 24hr as indicated concentrations, and the expression of p65 and β-actin were analyzed by immunoblotting. (B) MDA-MB-231 cells were treated with 5 μM TBMS1 for 24 hr. The total cell lysate and the nuclear extracts were analyzed with the indicated antibodies by immunoblotting. The nuclear envelope protein Lamin B1 was used as a nuclear marker. (C) MDA-MB-231 cells were treated with TBMS1 for 24 hr at indicated concentrations. The nuclear extracts were used in EMSA assay with the labeled CXCR4 promoter sequence containing NF-κB binding site. (D) MDA-MB-231 cells were treated with 5 μM TBMS1 for 24 hr. The nuclear extracts were incubated with the labeled CXCR4 promoter sequence containing NF-κB binding site in the presence of 1 μg IgG control or anti-p65 antibody, as indicated. The NF-κB binding activity was analyzed by EMSA assay. Specific shift bands for NF-κB were indicated with an arrow. (E) MDA-MB-231 cells were pretreated with TBMS1 for 24 hr and the proteins were cross-linked with DNA with formaldehyde, and then subjected to ChIP assay with an anti-p65 antibody. PCR on the immunoprecipitate was performed using primers spanning -192 to +45 of the CXCR4 promoter. Reaction products were resolved by electrophoresis. Representative results of three independent experiments are shown.
TBMS1 suppresses the CXCL12-induced invasion of breast cancer cells
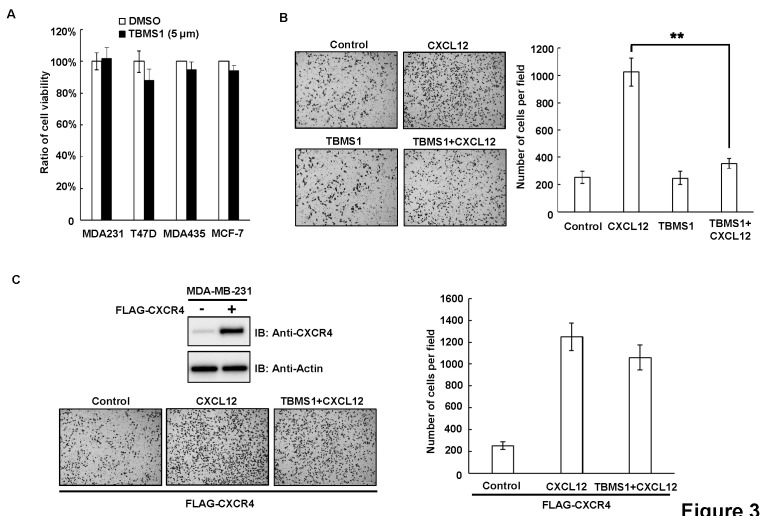
It has been reported that CXCL12-CXCR4 signaling pathway promotes breast cancer metastasis (17). As TBMS1 decreased the expression of CXCR4 in breast cancer cells, we then investigated whether TBMS1 had an influence on the CXCL12-induced invasion of breast cancer cells. MTT assay demonstrated that the treatment of breast cancer cells with 5 μM TBMS1 did not influence the cell viability and proliferation (Fig. 3A). However, the transwell migration assay indicated that TBMS1 significantly inhibited the CXCL12-induced cell invasion (Fig. 2B). Importantly, ectopic expression of CXCR4 in breast cancer cells abolished the inhibitory effect of TBMS1 on cell invasion, suggesting that TBMS1 targets CXCL12-CXCR4 signaling pathway to inhibit breast cancer cell invasion (Fig. 2C).
Fig. 3. TBMS1 suppresses the CXCL12-induced invasion of breast cancer cells. (A) MDA-MB-231, T47D, MDA-MB-435 and MCF-7 cells were treated with DMSO or TBMS1 (5 μM) for 24 hr. Relative cell viability was analyzed by MTT assay. Results shown are averages ± S.E.M. (B) MDA-MB-231 cells were treated with/out CXCL12 (300 ng/ml) combined with/out TBMS1 (5 μM) as indicated, for 24 hours. Cell invasion was analyzed by Tranwell assay. Left, representative images of Transwell assay. Right, quantitative data of migrated cells. Results shown are averages ± S.E.M. (C) MDA-MB-231 were transfected with FLAG-CXCR4 vector. Upper-left, expression levels of CXCR4 were examined by immunoblotting in empty vector and pCMV6-CXCR4 transfected MDA-MB-231 cells. Lower-left, representative images of transwell assay. Right, quantitative data of migrated cells. Results shown are averages ± S.E.M.
TBMS1 suppresses breast tumor metastasis in vivo
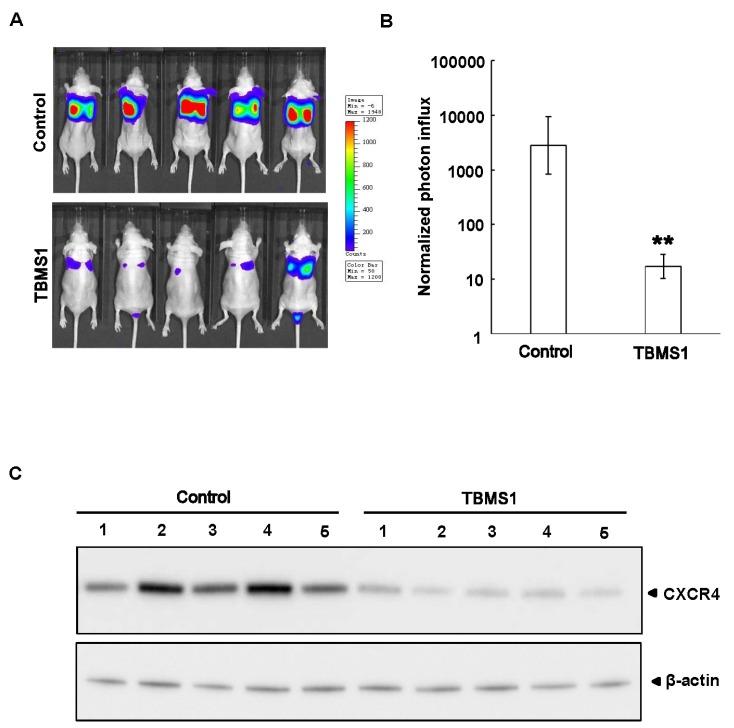
Next, in an in vivo metastasis model in nude mice, we evaluated the influence of TBMS1 on breast cancer metastasis. MDA-MB-231-Luciferase cells that can be traced by a Xenogen IVIS luminal Imager were used for tail vein injection. After the 30 days of injection, the control group had developed severe lung metastasis (5 mice each), while the TBMS1-treated group (5 mice each) showed decreased metastasis (Fig. 4A). Furthermore, the protein levels of CXCR4 in lung metastatic tumors were examined by western blotting and we found that the tumors from the TBMS1-treated group exhibited lower expression levels of CXCR4 when compared to control group (Fig. 4B). Taken together, these results demonstrated TBMS1 suppressed breast tumor metastasis to the lung.
Fig. 4. TBMS1 suppresses breast tumor metastasis in vivo. (A) MDA-MB-231-Luc cells were injected into 6-week-old female nude mice through tail veins (1 × 106 cells/mouse). These mice were intraperitoneally treated with DMSO or TBMS1 (20 mg/kg) every other day for a month, and were then photographed by Xenogen IVIS luminal Imager. The metastatic conditions correspond to the level of fluorescence, which q.v. the bar on the right side. (B) Normalized photon flux of metastasis conditions of MDA-MB-231-Luc cells treated with DMSO or TBMS1. Results shown are averages ± S.D.; **P < 0.01 vs. DMSO group. (C) Metastatic solid tumor tissues were dissected from DMSO and TBMS1 treated mice. The expression of CXCR4 and β-actin were examined by immunoblotting analysis.
DISCUSSION
CXCL12-CXCR4 signaling pathway has been shown to play a pivotal role in promoting tumor metastasis (17). Our study demonstrated that TBMS1, a potential anti-cancer compound, inhibited the CXCL12-CXCR4 signaling axis to prevent breast cancer metastasis.
As a chemokine receptor, CXCR4 has been reported to be overexpressed in a variety of cancers, including breast cancer. The patients with higher levels of CXCR4 exhibited poor clinical outcome compared to the patients with lower CXCR4 expression (19). Moreover, recent studies suggested that the activation of the CXCL12-CXCR4 pathway contributed to drug resistance in cancer chemotherapy (20). In the present study, we showed for the first time that TBMS1 attenuated CXCR4 expression in breast cancer. We observed that the mRNA level of CXCR4 was significantly down-regulated by TBMS1; thus, the decreased expression of CXCR4 was mostly not because of the proteolytic degradation. It has been reported that transcriptional factors HIF-1α, PPAR-γ and NF-κB mediated the up-regulation of CXCR4 in human cancer cells (18,21,22). TBMS1 was traditionally used as an anti-inflammatory medicine, and has been reported to inhibit NF-κB nuclear localization and activation in other types of cancer cells (16,23). Since our data demonstrated that TBMS1 did not influence the nuclear location of NF-κB in breast cancer cells, it is possible that the TBMS1-induced downregulation of CXCR4 occurs through downregulation of NF-κB activity. Although other mechanisms could contribute to the downregulation of CXCR4 by TMBS1, our data indicated that TBMS1 at least inhibited binding ability of NF-κB to CXCR4 promoter.
As a potential anti-cancer compound, the anti-cancer activity of TBMS1 was largely attributed to its anti-proliferative or pro-apoptotic effects in multiple cancer cells lines. TBMS1 has been shown to activate the caspase-dependent pathway (14), inhibit Wnt/β-catenin signaling pathway (11), and induce a p53/MDM2-dependent mechanism (13) to potentiate apoptosis and cytotoxic action. In our study, TBMS1 did not significantly influence the cellular viability and proliferation in MDA-MB-231 cells at lower concentration (5 µM). However, at this concentration, TBMS1 specifically inhibited CXCL12-induced cell invasion and metastasis. Thus, our study identified CXCR4 as a specific target for TBMS1, and revealed a new anti-cancer mechanism of TBMS1 to suppress breast cancer metastasis.
Overall, our approaches is to block the cancer dissemination but not to kill cancer cells or inhibit their growth. In the natural history of most human cancers, although patients could undergo successful surgical resection to remove primary tumors, some of tumors will ultimately metastasis to distant sites and develop later on (24). Our study suggest that TBMS1 might be used as a promising anti-metastasis agent for cancer treatment.
MATERIALS AND METHODS
Reagents
TBMS1 was purchased from the National Institute for the Control of Pharmaceutical and Biological Products (China), dissolved in phosphate buffered saline (PBS) and stored at −20℃. Antibodies to p65 and β-actin were purchased from Cell Signaling Technology (Danvers, MA, USA). Rabbit polyclonal antibody to CXCR4 and Lamin B1 was purchased from Abcam (Cambridge, MA, USA). Matrigel was purchased from Life Technology (Thermo Fisher Scientific, NY, USA).
Cell lines and cell culture
The human breast cancer cell lines MDA-MB-231, MDA-MB-435, T47D, and MCF-7 were purchased from ATCC and cultured at 37℃ with 5% CO2 in DMEM Medium (Invitrogen, USA) supplemented with 10% fetal bovine serum, 100 units/ml penicillin G, 100 μg/ml streptomycin, and 2 mM L-glutamine.
Plasmid and transfection
pCMV6-CXCR4 plasmid was purchased from Origene (Rockville, MD, USA). MDA-MB-231 cells were transfected with pCMV6-CXCR4 plasmid or empty control vector using Lipofectamin 2000 (Invitrogen, USA). After 24 hours, cells were used for experiment.
Western blotting
Cells or tumor tissues were lysed in M2 lysis buffer (150 mM NaCl, 50 mM Tris-Cl (pH 8.0), 5 mM EDTA, 1% Nonidet P-40) containing a protease inhibitor mixture (Roche Applied Science) and a phosphatase inhibitor mixture (Sigma, MO, USA). For nuclear protein extraction, cells were lysed in 500 μl of lysis buffer containing 0.5% Nonidet P-40, 10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 1 mM Na3VO4, and protease inhibitor mixture (Roche Applied Science). Intact nuclei were pelleted by centrifugation at 12,000 × g at 4℃ for 5 min, and lysed in 150 μl of nuclear lysis buffer containing 20 mM HEPES (pH 7.5), 25% glycerol, 0.42 M NaCl, 0.2 mM EDTA, 1 mM Na3VO4, and protease inhibitor mixture (Roche Applied Science). Equal amount of total protein was subjected to SDS-PAGE analysis and immunoblotting with the appropriate antibodies.
RNA analysis and reverse transcription-PCR
Total RNA was extracted using Trizol reagent according to the manufacturer’s instructions (Invitrogen Carlsbad, CA). One microgram of total RNA was converted to cDNA by PrimeScriptⓇ RT reagent Kit (TaKaRa, Tokyo, Japan). The respective forward and reverse primer sets used to analyze the relative expression of CXCR4 and CXCR7 were as follows: CXCR4, 5'-GAA GCT GTT GGC TGA AAA GG-3' and 5'-GAG TCG ATG CTG ATC CCA AT-3'; CXCR7, 5'-TCC ATC CTG CAC TAC ATC CCT TTC AC-3' and 5'-TCT ACT CAA GCA TCA AGA CCC GAA GC-3. The RT-PCR products were electrophoresed on a 2% agarose gel and then stained with ethidium bromide. Stained bands were visualized and photographed under ultraviolet light.
Quantitative real-time PCR
Total RNA was extracted from cells, and subsequently converted to cDNA, as described previously. The messenger RNA (mRNA) expression of CXCR4 was determined using quantitative real-time PCR with SYBRⓇ Premix Ex TaqTM kit (TaKaRa, Tokyo, Japan). Samples were run in triplicate to ensure amplification integrity. The primer pairs used to measure the mRNA levels of CXCR4 were the same with those used in the reverse transcription-PCR. The expression levels of genes were normalized to the expression level of GAPDH mRNA in each sample, and the threshold for positivity of real-time PCR was determined based on negative controls. The standard averages of cycle threshold (Ct) values from duplicate measurements were used to calculate the expression of the CXCR4 gene with normalization to β-actin expression.
MTT assay
Cell viability was assessed using the 3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyltetrazolium bromide (MTT) reduction assay (Sigma, St. Louis, MO). Stock solutions of MTT (5 mg/ml) were prepared in water. Cells (5 × 105 cells/ml) were seeded in 6-well plates in the absence or presence of TBMS1 at indicated concentration, for 24 hours. After this incubation period, MTT (50 μl) was added to each well and the cells were incubated for 4 h at 37℃. The medium was then removed and the converted dye was solubilized with the addition of DMSO. Absorbance, which was proportional to cell viability, was measured at 570 nm with background subtraction at 660 nm
Electrophoretic mobility shift assay
MDA-MB-231 cells (2 × 106/ml) were treated separately with different concentrations of TBMS1 at 37℃ for 24 hours. Nuclear extracts were then prepared and incubated with the oligonucleotide containing the NF-κB consensus sequence: 5'-TCCCCTGGGCTTCCCAAGCC-3' presented in the promoter of CXCR4 at −96 to −111 for 30 minutes at room temperature. The resulting DNA–protein complex was separated from the free oligonucleotides on 7.0% native polyacrylamide gel. The dried gel was directly detected by chemiluminescence. Signals were taken by LI-COR Infrared Imaged Odyssey (Gene Company).
Chromatin immunoprecipitation assay
MDA-MB-231 cells were treated with various concentrations of TBMS1 for 24 h and fixed with 1% formaldehyde. The cells were collected in lysis buffer and sonicated, after which they were immunoprecipitated with anti-p65 antibody. PCR analyses were carried out for 35 cycles with primers 5'-GGA CTC ACT ACC GAC CAC CC-3' (forward) and 5'-CGG CGT CAC TTT GCT ACC TG-3' (backward) for CXCR4, amplified DNA fragment from-192 to +45, which contains NF-κBbinding sites.
Transwell invasion assay
A transwell migration assay using 6.5-mm-diameter polycarbonate filters (8-μm pore size) was used. The filter of the Transwell plate (BD Biosciences) was coated with Matrigel. The bottom chambers were filled with 500 μl of DMEM containing 10% FBS. Cells (4 × 104) suspended in 100 μl of DMEM containing 0.5% FBS were seeded in the top chambers. Cells were allowed to migrate for 24 hours. Non-migrated cells were removed with cotton swabs, and migrated cells were fixed with cold 4% paraformaldehyde and stained with 1% crystal violet. Images were taken using an inverted microscope (10X magnification; Olympus, Tokyo, Japan), and migrated cells were quantified by manual counting.
In vivo tumor metastasis model
6-week-old female nude mice were committed to tail vein injection of MDA-MB-231-Luc cells (1 × 106 cells/mouse in 50 μl PBS). 20 mg/kg TBMS1 or DMSO were administrated intraperitoneally every other day for a month. On the 30th day, all the mice were anaesthetized by isoflurane and representative images were taken by a Xenogen IVIS luminal Imager. Animals used in the present study were maintained according to the NIH standards established in the Guidelines for the Care and Use of Experimental Animals. All of the experimental protocols were approved by the Research Ethics Committee of Hainan General Hospital, Hainan, China.
Statistical analysis
All the experiments have been done in triplicate and repeated twice. All data presented carried out the one-way ANOVA, followed by Student’s t-test. P values ≤ 0.05 were considered statistically significant.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 81300503) and the Natural Science Foundation of Hunan Province, China (Grant No. 2015SK20324).
References
- 1.Geiger TR, Peeper DS. Metastasis mechanisms. Biochim Biophys Acta. (2009);1796:293–308. doi: 10.1016/j.bbcan.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Raman D, Baugher PJ, Thu YM, Richmond A. Role of chemokines in tumor growth. Cancer Lett. (2007);256:137–165. doi: 10.1016/j.canlet.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Baruch A. Organ selectivity in metastasis: regulation by chemokines and their receptors. Clin Exp Metastasis. (2008);25:345–356. doi: 10.1007/s10585-007-9097-3. [DOI] [PubMed] [Google Scholar]
- 4.Franco R, Pirozzi G, Scala S, et al. CXCL12-binding receptors expression in non-small cell lung cancer relates to tumoral microvascular density and CXCR4 positive circulating tumoral cells in lung draining venous blood. Eur J Cardiothorac Surg. (2012);41:368–375. doi: 10.1016/j.ejcts.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Zeelenberg IS, Ruuls-Van Stalle L, Roos E. The chemokine receptor CXCR4 is required for outgrowth of colon carcinoma micrometastases. Cancer Res. (2003);63:3833–3839. [PubMed] [Google Scholar]
- 6.Taichman RS, Cooper C, Keller ET, Pienta KJ, Taichman NS, McCauley LK. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. (2002);62:1832–1837. [PubMed] [Google Scholar]
- 7.Mori T, Doi R, Koizumi M, et al. CXCR4 antagonist inhibits stromal cell-derived factor 1-induced migration and invasion of human pancreatic cancer. Mol Cancer Ther. (2004);3:29–37. [PubMed] [Google Scholar]
- 8.Richert MM, Vaidya KS, Mills CN, et al. Inhibition of CXCR4 by CTCE-9908 inhibits breast cancer metastasis to lung and bone. Oncol Rep. (2009);21:761–767. [PubMed] [Google Scholar]
- 9.Yadav VR, Sung B, Prasad S, et al. Celastrol suppresses invasion of colon and pancreatic cancer cells through the downregulation of expression of CXCR4 chemokine receptor. J Mol Med (Berl) (2010);88:1243–1253. doi: 10.1007/s00109-010-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu TX, Ma RD, Yu LJ. Structure-activity relationship of tubeimosides in anti-inflammatory, antitumor, and antitumor-promoting effects. Acta Pharmacol Sin. (2001);22:463–468. [PubMed] [Google Scholar]
- 11.Bian Q, Liu P, Gu J, Song B. Tubeimoside-1 inhibits the growth and invasion of colorectal cancer cells through the Wnt/beta-catenin signaling pathway. Int J Clin Exp Pathol. (2015);8:12517–12524. [PMC free article] [PubMed] [Google Scholar]
- 12.Gu Y, Korbel C, Scheuer C, Nenicu A, Menger MD, Laschke MW. Tubeimoside-1 suppresses tumor angiogenesis by stimulation of proteasomal VEGFR2 and Tie2 degradation in a non-small cell lung cancer xenograft model. Oncotarget. (2015);7:5258–5272. doi: 10.18632/oncotarget.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Y, Xie G, Xia J, et al. TBMS1 exerts its cytotoxicity in NCI-H460 lung cancer cells through nucleolar stress-induced p53/MDM2-dependent mechanism, a quantitative proteomics study. Biochim Biophys Acta. (2016);1864:204–210. doi: 10.1016/j.bbapap.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Jia G, Wang Q, Wang R, et al. Tubeimoside-1 induces glioma apoptosis through regulation of Bax/Bcl-2 and the ROS/Cytochrome C/Caspase-3 pathway. Onco Targets Ther. (2015);8:303–311. doi: 10.2147/OTT.S76063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen WJ, Yu C, Yang Z, et al. Tubeimoside-1 induces G2/M phase arrest and apoptosis in SKOV-3 cells through increase of intracellular Ca(2)(+) and caspase-dependent signaling pathways. Int J Oncol. (2012);40:535–543. doi: 10.3892/ijo.2011.1218. [DOI] [PubMed] [Google Scholar]
- 16.Hao W, Wang S, Zhou Z. Tubeimoside-1 (TBMS1) inhibits lung cancer cell growth and induces cells apoptosis through activation of MAPK-JNK pathway. Int J Clin Exp Pathol. (2015);8:12075–12083. [PMC free article] [PubMed] [Google Scholar]
- 17.Xu C, Zhao H, Chen H, Yao Q. CXCR4 in breast cancer: oncogenic role and therapeutic targeting. Drug Des Devel Ther. (2015);9:4953–4964. doi: 10.2147/DDDT.S84932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kukreja P, Abdel-Mageed AB, Mondal D, Liu K, Agrawal KC. Up-regulation of CXCR4 expression in PC-3 cells by stromal-derived factor-1alpha (CXCL12) increases endothelial adhesion and transendothelial migration: role of MEK/ERK signaling pathway-dependent NF-kappaB activation. Cancer Res. (2005);65:9891–9898. doi: 10.1158/0008-5472.CAN-05-1293. [DOI] [PubMed] [Google Scholar]
- 19.Nevo I, Sagi-Assif O, Meshel T, et al. The tumor microenvironment: CXCR4 is associated with distinct protein expression patterns in neuroblastoma cells. Immunol Lett. (2004);92:163–169. doi: 10.1016/j.imlet.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Duda DG, Kozin SV, Kirkpatrick ND, Xu L, Fukumura D, Jain RK. CXCL12 (SDF1alpha)-CXCR4/CXCR7 pathway inhibition: an emerging sensitizer for anticancer therapies? Clin Cancer Res. (2011);17:2074–2080. doi: 10.1158/1078-0432.CCR-10-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. (2003);425:307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 22.Richard CL, Blay J. Thiazolidinedione drugs down-regulate CXCR4 expression on human colorectal cancer cells in a peroxisome proliferator activated receptor gamma-dependent manner. Int J Oncol. (2007);30:1215–1222. [PubMed] [Google Scholar]
- 23.Huang P, Yu C, Liu XQ, Ding YB, Wang YX, He JL. Cytotoxicity of tubeimoside I in human choriocarcinoma JEG-3 cells by induction of cytochrome c release and apoptosis via the mitochondrial-related signaling pathway. Int J Mol Med. (2011);28:579–587. doi: 10.3892/ijmm.2011.727. [DOI] [PubMed] [Google Scholar]
- 24.Dalerba P, Clarke MF. Cancer stem cells and tumor metastasis: first steps into uncharted territory. Cell Stem Cell. (2007);1:241–242. doi: 10.1016/j.stem.2007.08.012. [DOI] [PubMed] [Google Scholar]